Abstract
Ultraviolet resonance Raman spectroscopy reveals that, when heated at pH 3, a substantial fraction (30 %) of cytochrome c converts to a β-sheet structure, at the expense of turns and helices. β-sheet formation is rapid, exhibiting a 2 µs rise time, following a temperature jump. It is proposed that a short β-sheet segment, comprised of residues 37–39 and 58–61 extends itself into the large 37–61 loop, when the latter is destabilized by protonation of H27, which forms an anchoring H-bond to loop residue P44. This conformation change ruptures the Met80-Fe bond, as revealed by changes in ligation-sensitive heme-resonant Raman bands. It also induces peroxidase activity with the same temperature profile. This process is suggested to model the apoptotic peroxidation of cardiolipin by cytochrome c.
We report resonance Raman (RR) spectroscopic evidence for a hitherto unrecognized conformational transition to β-sheet structure in cytochrome c (cyt c), which may have important functional consequences.
In addition to its electron-transfer activity in mitochondria, cyt c plays a key role in apoptosis,1 and partial unfolding seems to be a critical element in the mechanism. Jemmerson et al. have observed that, in association with lipid vesicles, cyt c binds an antibody that recognizes an unfolded region around residue Pro44, and that the same response is seen in apoptotic cells.2 Belikova et al. report that binding to cardiolipin induces peroxidase activity in cyt c, producing cardiolipin hydroperoxides that are required for release of pro-apoptotic factors.3 It seems likely that this activity is triggered by changes in heme ligation when cyt c interacts with lipids.4 In addition to cardiolipin,5 oleic acid6 has been observed to destabilize the cyt c fold; the oleic acid effect can be partially reversed by ATP, a component of the apoptosome.6
We find that heating cyt c under destabilizing conditions (pH 3) not only unfolds a significant fraction of the protein, but converts it to a β-sheet structure. This conversion is reversible unless the concentration exceeds 50 µM, when cyt c precipitates upon heating. Moreover β-sheet formation is remarkably fast, occurring on the microsecond time scale. This structural change is associated with rupture of the Fe-S bond (heme-Met80), and induction of peroxidase activity.
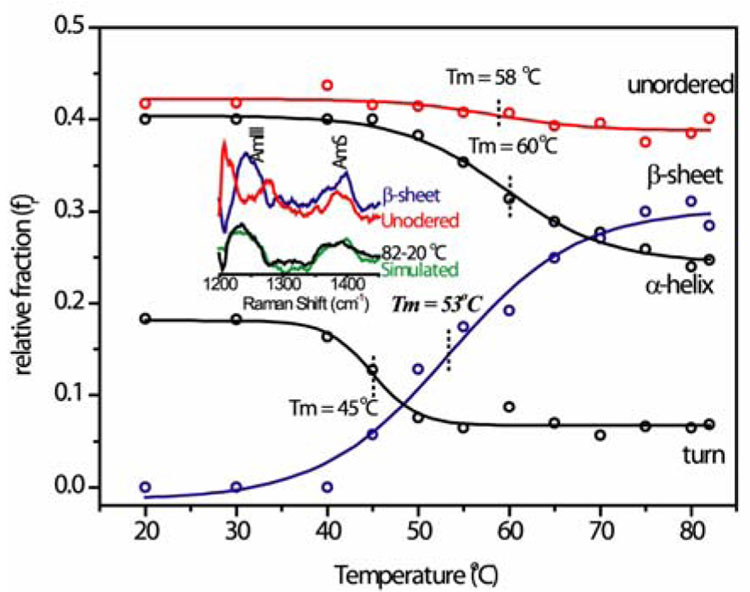
The evidence for β-sheet formation comes from UVRR spectroscopy. Excitation at 197 nm produces optimal enhancement of amide vibrational modes, whose frequencies and intensities are diagnostic of secondary structure.7a UVRR signatures have been extracted from a suite of structurally characterized proteins, allowing quantitation of secondary structure.7b Figure 1 shows the result of applying this procedure (supporting info, S1) to UVRR spectra of equine FeIII cyt c at pH 3. At 20°C, the secondary structure is 18 % β-turn, 40 % α-helix, and 42 % unordered structure. This composition is consistent with the crystal structure of native cyt c8. As the temperature rises, all three of these elements diminish, while the β-sheet content rises from near-zero to 30 %. The far-UV circular dichroism temperature profile (supporting info, S2), shows loss of α-helix at about the same temperature as the UVRR analysis; however CD does not readily distinguish unordered structure from β-sheet. In contrast, the UVRR signatures are distinctive, especially in the amide III and S region, which is highly sensitive to conformation, as can be seen in the inset to Figure 1.
Figure 1.
FeIII cyt c secondary structure contributions at pH 3 (50 µM) extracted from 197 nm-excited UVRR spectra. The inset shows the distinctive spectral profiles7b for β-sheet and unordered structure in the amide III and S region, and the quality of data fitting via simulation of the 82-20° C difference spectrum.
Cyt c is known to form β-sheet aggregates, e.g. when heated in the presence of denaturant,9 and intermolecular association has been detected in the course of cyt c folding studies.10 However, reversible β-sheet formation has not previously been reported. To investigate the rate of this process, we applied a laser temperature jump, using a recently described 1 kHz OPO solid state laser, timed to our UVRR probe laser,11a and obtained a 2.2 µs time constant for β-sheet formation (supporting info, S3). Thus the structural conversion is rapid, on the same time scale as, for example, the melting of helices in apomyoglobin.11b Aggregation to intermolecular β-sheets is a much slower process.11c
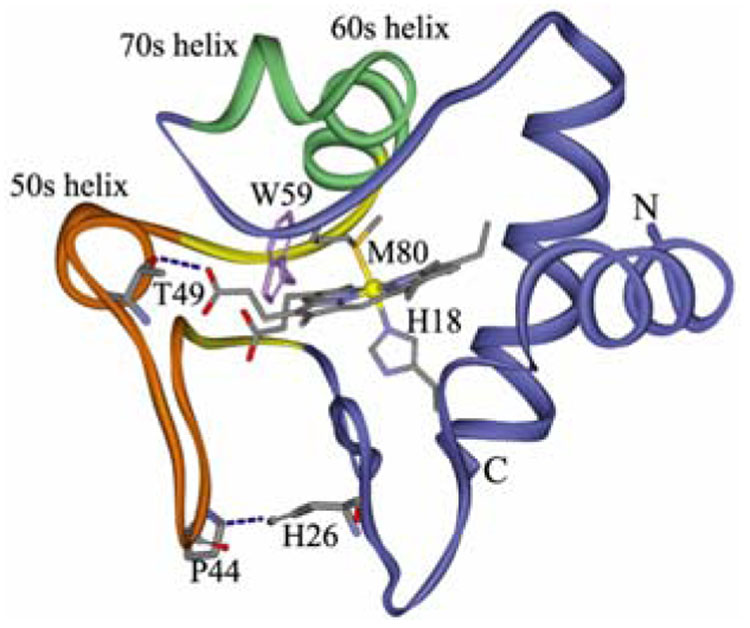
What is the nature of intramolecular β-sheet formation in cyt c? Figure 2 shows the protein fold. The left-most segment is a large Ω loop12 (40s Ω loop), containing residues 40–57 (orange), which has been identified by Englander and coworkers13 as the least stable of the cyt c ‘foldons’, structural elements found via NMR to unfold as cooperative units. At the ends of the 40s Ω loop is a ‘neck’ of residues, 37–39 and 58–61 (colored yellow in Figure 2), which are arranged in a short anti-parallel β-sheet. The 40s Ω loop is labilized at low pH, probably because anchoring H-bonds, His26⋯Pro4414 and Thr49⋯heme propionate, are disrupted by protonation.13
Figure 2.
Ribbon diagram of cyt c, highlighting the 40s Ω loop (orange) and the 37–61 foldon with its 37–39/58–61 β-sheet neck (yellow). This β-sheet segment is suggested to extend into the 40s Ω loop when the latter is destabilized by disruption of the H26…P44 H-bond via protonation of H26. The conformational switch is proposed to displace the heme via the propionate H-bonds, inducing Met80-Fe bond rupture.
Our hypothesis is that upon heating at pH 3, the 37–39/58–61 β-sheet neck extends itself into the 40s Ω loop, forming additional β-sheet at the expense of the three β-turns and the short 50s helix contained in the loop. The pre-existence of the β-sheet neck can explain the fast rate of the process. To account for the more extensive helix loss at higher temperature (Figure 1), we suggest that the sheet structure continues to grow at the expense of the adjacent 60s and 70s helices (Figure 2). These helices, the β-neck, and the 40s Ω loop together account for 36 % of the residues, consistent with the extent of β-sheet formation. The proposed sequence of events is supported by the sharper transition and lower temperature for the loss of turns than of helix (45° vs 60° C – Figure 1). The β-sheet profile is broad, and covers both the turn and helix transition temperatures.
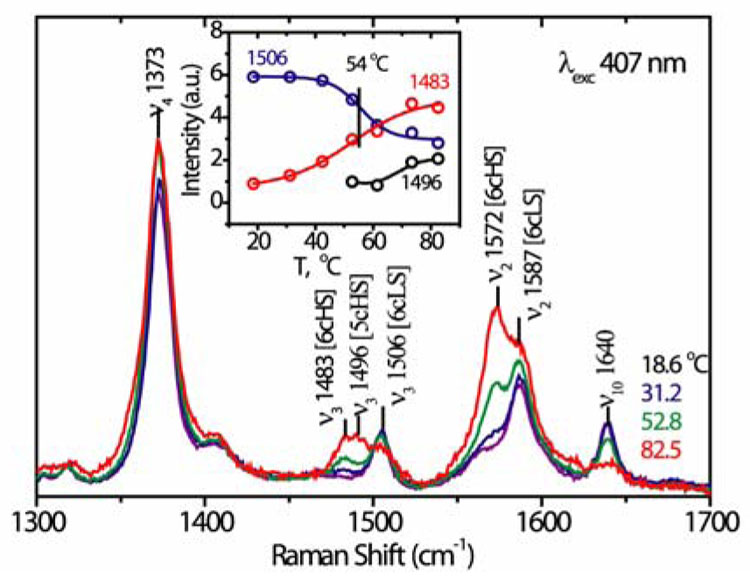
A connection to function can be seen in the visible RR spectra (Figure 3), which contain markers of the heme ligation state.15 At low temperature, the bands are characteristic of 6-coordinate low-spin (6cLS) heme, but 6-coordinate high-spin (6cHS) heme bands grow in as the temperature rises, as does a small contribution from 5-coordinate high-spin (5cHS) heme. We infer that the Met80 ligand, which is known to be labile at low pH (we find that the 695 nm absorption band, which is diagnostic of Met ligation, has half the native protein intensity at pH 3 at 20° C), is replaced by a water ligand16 producing a temperature dependent 6cLS/6cHS mixture; 17 the water is partially lost at high temperature (5cHS). The temperature dependence of the deconvoluted bands (Figure 3 inset) reveals the same Tm, 54° C, for the LS/HS conversion as for β-sheet formation (Figure 1). The heme propionate substituents form H-bonds with the backbone CO of Thr49 and the indole sidechain of Trp59; both residues are on the 37–61 foldon (Figure 2). We suggest that the switch to β-sheet displaces the heme, via these H-bonds, augmenting the Met80-Fe bond rupture.
Figure 3.
The replacement of low-spin by high-spin heme in cyt c (50 µM) at pH 3 is signaled by 407 nm-excited porphyrin RR bands.15 The deconvoluted ν3 band heights reveal the same transition temperature as for β-sheet formation; at higher temperature, a small fraction of 5-coordinate heme appears (1496 cm−1).
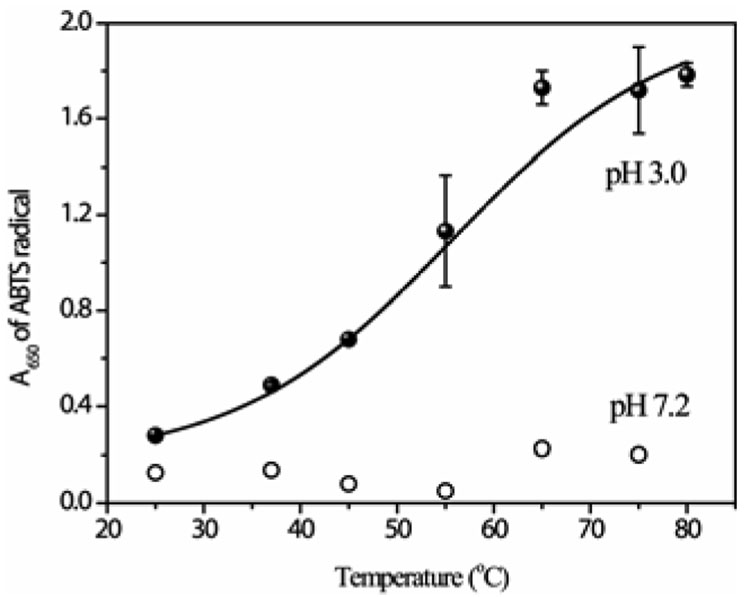
Loss of the Met80 ligand provides access to exogenous ligands, including hydrogen peroxide, which can easily displace the weakly bound water molecule. We found that peroxidase activity is indeed induced, with a Tm near 54° C, when cyt c is heated at pH 3 (Figure 4). Peroxidase activity is negligible at pH 7, regardless of temperature. Thus peroxidase activity and β-sheet formation are directly correlated.
Figure 4.
Peroxidase activity (measured spectrophotometrically with ABTS as substrate – see supporting info, S4) of cyt c (0.5 µM) is induced at pH 3 (but not at pH 7), with the same transition temperature as β-sheet formation.
It is possible that cardiolipin binding to cyt c induces a similar conformational switch to the β-sheet structure,18 resulting in heme exposure and cardiolipin peroxidation, as part of the apoptotic mechanism. The trigger for this switch could be H-bonding of the anionic cardiolipin head group to His26, whose protonation is likely responsible for the pH 3 response at elevated temperature.
Supplementary Material
UVRR spectral analysis, CD heating profile, T-jump kinetic trace, and details of peroxidase activity monitoring. This material is available free of charge via the internet at http://acs.pub.org.
Acknowledgment
This work was supported by NIH grant GM 25158. (to T.G.S.) and GM 036298 (to J. T. G.).
REFERENCES
- 1.Green DR, Reed JC. Science. 1998;281:1309. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 2.Jemmerson R, Liu J, Hausauer D, Lam KP, Mondino A, Nelson RD. Biochemistry. 1999;38:3599. doi: 10.1021/bi9809268. [DOI] [PubMed] [Google Scholar]
- 3.Belikova NA, Vladimirov YA, Osipov AN, Kapralov AA, Tyurin VA, Potapovich MV, Basova LV, Peterson J, Kurnikov IV, Kagan VE. Biochemistry. 2006;45:4998. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Nantes IL, Zucchi MR, Nascimento OR, Faljoni-Alario A. J. Biol. Chem. 2001;276:153. doi: 10.1074/jbc.M006338200. [DOI] [PubMed] [Google Scholar]; (b) Oellerich S, Lecomte S, Paternostre M, Heimburg T, Hildebrandt P. J. Phys. Chem. B. 2004;108:3871. [Google Scholar]
- 5.(a) Spooner PJR, Watts A. Biochemistry. 1991;30:3871. doi: 10.1021/bi00230a010. [DOI] [PubMed] [Google Scholar]; (b) Spooner PJR, Watts A. Biochemistry. 1991;30:3880. doi: 10.1021/bi00230a011. [DOI] [PubMed] [Google Scholar]
- 6.Sinibaldi F, Mei G, Polticelli F, Piro MC, Howes BD, Smulevich G, Santucci R, Ascoli F, Fiorucci L. Protein Sci. 2005;14:1049. doi: 10.1110/ps.041069405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Balakrishnan G, Hu Y, Nielsen SB, Spiro TG. Appl. Spectrosc. 2005;59:776. doi: 10.1366/0003702054280702. [DOI] [PubMed] [Google Scholar]; (b) Huang CY, Balakrishnan G, Spiro TG. J. Raman Spectrosc. 2006;37:277. [Google Scholar]
- 8.Bushnell GW, Louie GV, Brayer GD. J. Mol. Biol. 1990;214:585. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 9.Dong AC, Randolph TW, Carpenter JF. J. Biol. Chem. 2000;275:27689. doi: 10.1074/jbc.M005374200. [DOI] [PubMed] [Google Scholar]
- 10.(a) Nawrocki JP, Chu RA, Pannell LK, Bai YW. J. Mol. Biol. 1999;293:991. doi: 10.1006/jmbi.1999.3226. [DOI] [PubMed] [Google Scholar]; (b) Segel DJ, Eliezer D, Uversky V, Fink AL, Hodgson KO, Doniach S. Biochemistry. 1999;38:15352. doi: 10.1021/bi991337k. [DOI] [PubMed] [Google Scholar]
- 11.(a) Balakrishnan G, Hu Y, Spiro TG. Appl. Spectrosc. 2006;60:347. doi: 10.1366/000370206776593799. [DOI] [PubMed] [Google Scholar]; (b) Huang CY, Balakrishnan G, Spiro TG. Biochemistry. 2005;44:15734. doi: 10.1021/bi051578u. [DOI] [PubMed] [Google Scholar]; (c) JiJi RD, Balakrishnan G, Hu Y, Spiro TG. Biochemistry. 2006;45:34. doi: 10.1021/bi051507v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leszczynski JF, Rose GD. Science. 1986;234:849. doi: 10.1126/science.3775366. [DOI] [PubMed] [Google Scholar]
- 13.(a) Krishna MMG, Lin Y, Rumbley JN, Englander SW. J. Mol. Biol. 2003;331:29. doi: 10.1016/s0022-2836(03)00697-1. [DOI] [PubMed] [Google Scholar]; (b) Krishna MMG, Maity H, Rumbley JN, Lin Y, Englander SW. J. Mol. Biol. 2006;359:1410. doi: 10.1016/j.jmb.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 14.Sinibaldi F, Howes BD, Piro MC, Caroppi P, Mei G, Ascoli F, Smulevich G, Santucci R. J. Biol. Inorg. Chem. 2006;11:52. doi: 10.1007/s00775-005-0057-6. [DOI] [PubMed] [Google Scholar]
- 15.Spiro TG, Li XY. Resonance Raman Spectroscopy of Metalloporphyrins. In: Spiro TG, editor. Biological Applications of Raman Spectroscopy. Vol. 3. New York: Wiley & Sons, Inc.; 1988. pp. 1–37. [Google Scholar]
- 16.Takahashi S, Yeh SR, Das TK, Chan CK, Gottfried DS, Rousseau DL. Nat. Str. Biol. 1997;4:44. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 17.(a) Jordan T, Eads JC, Spiro TG. Protein Sci. 1995;4:716. doi: 10.1002/pro.5560040411. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Oellerich S, Wackerbarth H, Hildebrandt P. J. Phys. Chem. B. 2002;106:6566. [Google Scholar]
- 18.Mixing cyt c with anionic lipids induces only small changes in the amide I’ IR band, but large temperature-dependent enhancement of amide H/D exchange rates. Muga A, Mantsch HH, Surewicz WK. Biochemistry. 1991;30:7219. doi: 10.1021/bi00243a025. Heimburg T, Marsh D. Biophys. J. 1993;65:2408. doi: 10.1016/S0006-3495(93)81299-2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
UVRR spectral analysis, CD heating profile, T-jump kinetic trace, and details of peroxidase activity monitoring. This material is available free of charge via the internet at http://acs.pub.org.