Abstract
The ability to directly visualize nanoscopic cellular structures and their spatial relationship in all three dimensions will greatly enhance our understanding of molecular processes in cells. Here, we demonstrated multicolor three-dimensional (3D) stochastic optical reconstruction microscopy (STORM) as a tool to quantitatively probe cellular structures and their interactions. To facilitate STORM imaging, we generated photoswitchable probes in several distinct colors by covalently linking a photoswitchable cyanine reporter and an activator molecule to assist bioconjugation. 3D localization was performed in conjunction with focal plane scanning and correction for refractive index mismatch to obtain whole-cell images with a spatial resolution of 20–30 nm and 60–70 nm in the lateral and axial dimensions, respectively. Using this approach, we imaged the entire mitochondrial network in fixed monkey kidney BS-C-1 cells, and studied the spatial relationship between mitochondria and microtubules. The 3D STORM images revealed mitochondrial morphologies as well as mitochondria-microtubule contacts that were obscured in conventional fluorescence images.
As a powerful imaging technique for studying cellular processes, fluorescence microscopy allows noninvasive imaging of live samples with molecular specificity. The limited resolution of fluorescence microscopy, however, leaves many biological structures too small to be observed in detail, prohibiting analyses of molecular interactions within or between these structures. The resolution of light microscopy is classically limited by diffraction to about 200–300 nm in the lateral directions and 500–800 nm in the axial direction; both dimensions substantially larger than the size of many subcellular structures. To overcome this limit, super-resolution optical microscopy approaches have been developed, attaining an order of magnitude improvement in spatial resolution. One category of approaches is based on controlling the spatial pattern of excitation, including stimulated emission depletion (STED) microscopy and its related RESOLFT technique1, and saturated structured illumination microscopy (SSIM)2. In particular, spatial resolution of about 40–45 nm in 3D has been recently reported using STED3. The other approach is based on single-molecule localization of photoswitchable fluorophores, a method independently reported under different names including stochastic optical reconstruction microscopy (STORM)4, (fluorescence) photoactivation localization microscopy ((f)PALM)5,6 and other variants7. This approach can generate 3D super-resolution images by localizing both lateral and axial positions of each photo-activated probe. A lateral resolution of 20–30 nm and an axial resolution of 50–60 nm have been reported using astigmatism imaging, allowing the morphology of nanoscopic cellular structures to be resolved8. Similar axial resolutions have been subsequently obtained using bi-focal plane imaging9.
While the ability to resolve the 3D morphology of a biological structure can be informative, much of biology is governed by interactions between structures. Colocalization analyses with multicolor imaging have been extensively used to map the likelihood of interaction between two components, but the accuracy of colocalization is inherently limited by the image resolution. Structured illumination microscopy has been recently used to image multiple cellular components with a factor of two improvement in 3D resolution compared to conventional fluorescence microscopy10. Substantially higher resolution has been realized in multicolor imaging using 2D and 3D STED3,11, whereas multicolor STORM/(f)PALM has only been achieved in 2D previously12–14. Colocalization analyses performed in 2D can, however, be ambiguous considering that biological structures are 3D in nature.
In this article, we advance STORM/(f)PALM imaging by combining both multicolor and 3D imaging capabilities. To facilitate multicolor imaging, a number of probes with distinct colors were synthesized by covalently linking the photoswitchable cyanine dyes with activator molecules to form a single, functionalized component ready for bioconjugation. To extend beyond previous 3D STORM work on relatively thin samples8, we combined focal plane scanning with a novel treatment for the spherical aberration induced by refractive index mismatch to allow for imaging of substantially thicker samples and to achieve high resolution whole cell imaging. Using these new imaging capabilities, we studied the morphology of mitochondrial network and the spatial relationship between mitochondria and microtubules in mammalian cells.
Results
Synthesis of cyanine-based photoswitchable probes in different colors
We have recently reported a family of photoswitchable probes, each composed of a reporter fluorophore that can be switched between a fluorescent and a dark state, and an activator dye that facilitates the reactivation of reporter from the dark-state12,15. A number of red or near infrared cyanine dyes can serve as the reporter. The activator has an even wider array of choices in many distinct colors. Combinatorial pairing of the activators and reporters greatly increases the number of available colors for multicolor imaging. In practice, however, labeling of target molecules with two dyes can be cumbersome. Molecules nonspecifically labeled with activators and reporters often result in heterogeneous populations with distinct switching properties due to different activator-reporter separations. Labeling molecules that contain only a limited number of attachment sites with both reporter and activator can also be difficult. It is, therefore, desirable to connect the activator and reporter into a single chemical unit.
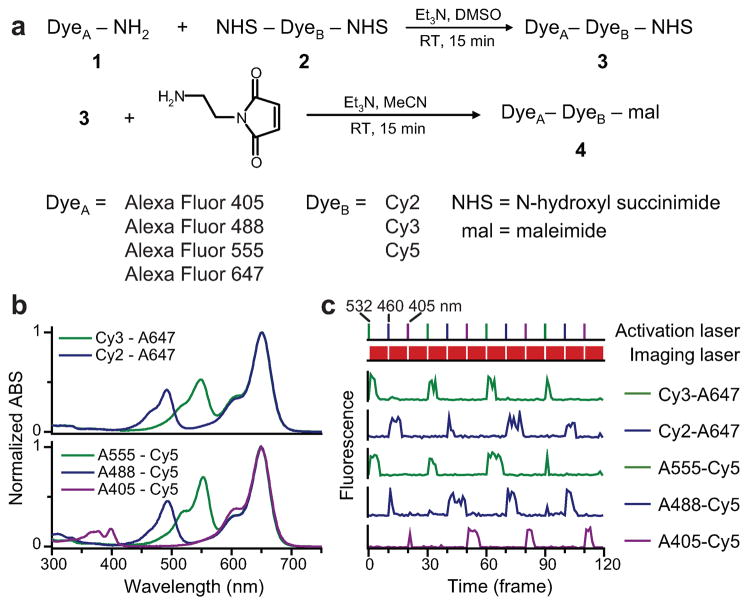
To synthesize covalently linked activator-reporter pairs, we utilized a scheme depicted in Fig. 1a16. An amine-modified dye was allowed to react with a dye that contains two amine-reactive N-hydroxyl succinimidyl (NHS) ester groups. With an excess of the bis-amine-reactive dye in the reaction, the main product generated is the linked dye-pair with a free NHS ester group, which was then purified using gravity columns or HPLC. This linked dye-pair can be used for bioconjugation via reaction with primary amines on biomolecules, such as lysine residues on proteins. To further broaden the range of labeling targets, we mixed the amine-reactive dye-pair with N-(2-aminoethyl) maleimide to create a thiol-reactive dye, which allows coupling with cysteine residues, another commonly used approach for protein labeling.
Figure 1.
Synthesis of covalently linked photoswitchable activator-reporter pairs. (a) Reaction scheme. (b) Absorption spectra of linked dye pairs in PBS at pH 7.4. (c) Photoswitching traces of linked dye pairs showing that they are specifically activated by light matching the absorption wavelength of the activators. The top panel shows the alternating sequence of green (532 nm), blue (460 nm) and violet (405 nm) activation lasers. Between activation pulses, a red (657 nm) laser is used to excite the reporters and to switch them into the dark state (second panel from top). Fluorescence traces of individual probes are shown in the five lower panels.
Both amine-modified dye and bis-amine-reactive dye have a wide range of choices (Supplementary Fig. 1 online). Most CyDyes (Cy2, Cy3, Cy5, Cy5.5 and Cy7) have bis-amine-reactive forms commercially available, among which Cy2 and Cy3 can be used as the activator, and Cy5, Cy5.5 and Cy7 can function as the photoswitchable reporter. Many amine-modified dyes, such as Alexa Fluor 405 (A405), Alexa Fluor 488 (A488) and Alexa Fluor 555 (A555), can serve as activators, and the Alexa Fluor 647 (A647), a structural analog of Cy5, can serve as the reporter.
We made five activator-reporter pairs, Cy2-A647, Cy3-A647, A555-Cy5, A488-Cy5 and A405-Cy5, using the above scheme. The absorption spectra of the purified reaction products indeed exhibit peaks at both activator and reporter absorption wavelengths (Fig. 1b). To demonstrate the specific photoswitching ability of the linked dye pairs, we attached them to antibodies and measured fluorescence emission from individual labeled antibody molecules. All of these probes were switched into a dark state upon illumination with a red (657 nm) laser, which excites the reporter dyes. Efficient reactivation of the reporters required light that specifically matches the absorption peaks of the activators (Fig. 1c). Quantitative characterization of the color specificity of activation, the brightness, and the nonspecific activation behavior of these probes, as well as the implication of these properties on super-resolution imaging, are presented in Supplementary Data and Supplementary Fig. 2 online. All of these probes can be used for STORM/(f)PALM imaging. We chose A405-Cy5, A488-Cy5 and A555-Cy5 for the subsequent imaging experiments.
Whole cell 3D imaging
To demonstrate whole cell 3D STORM, we imaged the entire mitochondrial network in mammalian cells using the covalently linked dye pairs (Fig. 2). We employed the standard indirect immunofluorescence method to stain the outer membrane of mitochondria in BS-C-1 cells. Tom20, a component of the translocase of the outer mitochondrial membrane complex and a commonly used mitochondria outer membrane marker17, was stained by primary antibodies followed by A405-Cy5-labeled secondary antibodies.
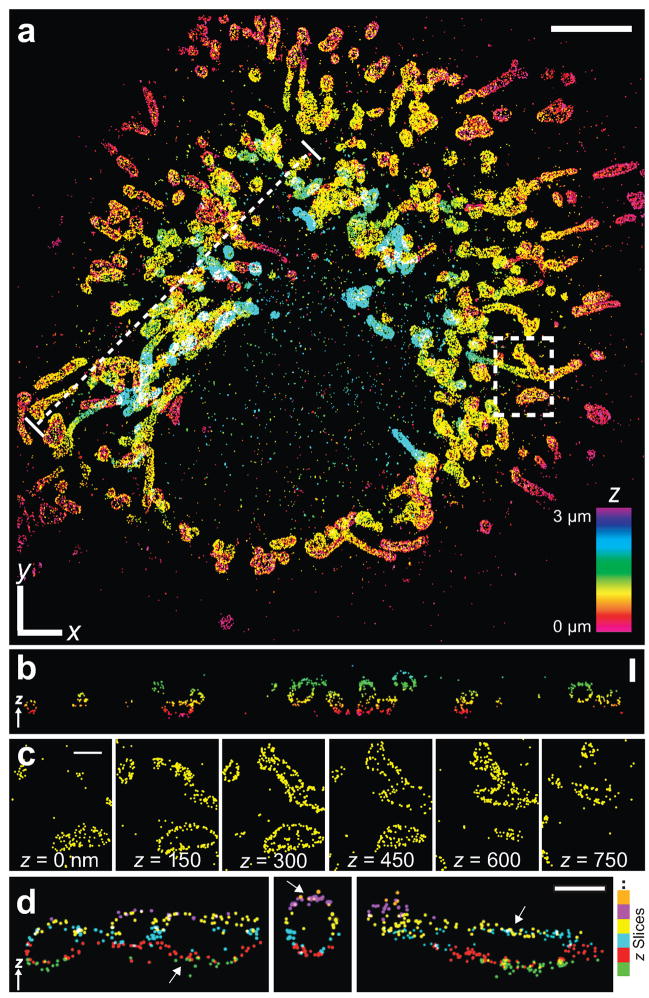
Figure 2.
Three dimensional STORM images of mitochondria in a whole mammalian cell. (a) The 3D STORM image of the mitochondrial network in a BS-C-1 cell. The image is performed in an aqueous medium with refractive index of 1.34. The z-position is color-coded according to the color scale bar. White x-y scale bar: 5 μm. See Supplementary Video 1 online for 3D views. (b) The vertical cross-section along the dotted line in a, showing the hollow shape of individual mitochondria. Scale bar: 1 μm. (c) Consecutive x-y sections of the boxed region in a. Scale bars: 1 μm. (d) Vertical cross-sections of several regions in a, color-coded by the z-slices in which they were originally recorded. Arrows indicate overlapping horizontal segments of mitochondria appearing in adjacent z-slices, where localizations from different slices are colored differently. Scale bar: 750 nm.
During STORM imaging, the A405-Cy5 probes were first switched to the dark state by the 657 nm laser. A low intensity 405 nm activation laser was then used to activate only a small subset of the probes at any given time. This condition allowed us to resolve individual activated probes and determine their positions with high precision. A super-resolution image was then constructed by plotting the measured probe positions accumulated over time. To determine the 3D position from the 2D fluorescence image of a probe, we introduced a cylindrical lens in the imaging path to create a focus offset between the two lateral directions, such that not only can the x and y coordinates of the probe be determined from the center of the image, but the z coordinate can also be determined from the ellipticity8,18. To quantify the localization precision, we analyzed the localizations from individual, isolated antibody molecules in the cell sample. The repetitive switching property of the probes allowed each molecule to be localized multiple times, and the dispersion of the localizations, 25 nm in x and y and 67 nm in z (full-width-at-half-maximum values), provides a measure of the average localization precision within a 600 nm depth near the focal plane8. The localization precision degrades when the molecule is further away from the focal plane, limiting the z imaging range if the position of the focal plane is fixed. In order to image the full depth of a mammalian cell, typically several micrometers thick, we scanned of the objective to acquire multiple image slices at focal planes deeper into the cell. These slices were then stacked to form whole cell 3D images that are ~ 50 μm wide and ~ 3 μm thick.
For 3D imaging of an aqueous sample using an oil immersion objective, the difference in refractive index between the imaging medium and the coverglass/oil/objective system must be considered. Correction for this effect is detailed in the Supplementary Methods online and recapped here briefly. This refractive index mismatch causes the light rays to bend when passing across the medium/coverglass interface, effectively increasing the apparent z position of the molecule (Supplementary Fig. 3a,b online). When the image depth is not too large, this effect can be corrected by a linear rescaling factor F as shown in the previous confocal and 3D STORM work8,19. For the imaging conditions used here (aqueous buffer containing 5% glucose with a refractive index of 1.34), the rescaling factor F is 0.72. The refractive index mismatch also causes spherical aberration because the refracted light rays no longer converge perfectly, distorting the point spread function and making it asymmetric in the axial direction. Simulations show that this effect can be quantitatively described by a second rescaling factor F2, which varies with the position of the focal plane (Supplementary Fig. 3c,d online). The value of F2 is very close to unity when the focal plane is near the interface, and therefore the effect of spherical aberration can be neglected for imaging a thin sample within several hundred nanometer of the interface8. However, when moving away from the interface, the value of F2 for molecules above the focal plane increases rapidly, whereas that below the focal plane remains close to unity. Although the average z-position can be corrected by rescaling, the localization precision also rescales with F2. Thus, to circumvent the effect of spherical aberration and to ensure the localization precision throughout a thick sample, we chose to accept only localizations below the focal plane.
Alternatively, the spherical aberration can be alleviated using an imaging medium with a higher refractive index. We have demonstrated this method using imaging media with a refractive index of 1.45, containing 80% glycerol and 5% glucose, or 60% sucrose and 5% glucose (Supplementary Fig. 4 online). Under these conditions, F2 was close to unity both below and above the focal plane (Supplementary Fig. 3c,d online), allowing localizations on both sides to be used.
Morphology of the mitochondria network
The 3D STORM images revealed the hollow shape of the mitochondrial outer membrane that is typically difficult to resolve by conventional wide-field or confocal fluorescence microscopy (Fig. 2). As shown clearly in the vertical cross-section of the STORM image (Fig. 2b), the consecutive horizontal sections (Fig. 2c), and the 3D presentations in Supplementary Video 1 online, the mitochondrial outer membrane appears as a thin envelop that encloses a hollow space, where the mitochondrial inner membrane and matrix presumably resides. Such a hollow structure was also previously resolved using 3D STED3. Some mitochondria span across multiple z-slices, allowing us to estimate the accuracy of our methods for combating spherical aberration by examining how well the same cellular structures that appeared in adjacent z-slices align with each other (Fig. 2d). By comparing the same horizontal segments of mitochondria that appeared in both slices, we measured the discrepancy between the independently determined z-positions in the adjacent slices. The discrepancy was 18 nm on average, well within our axial resolution of 60–70 nm.
The whole cell images also revealed two distinct types of mitochondrial morphology: while mitochondria appear more globular and dispersed in some cells, in other cells they emerge as a more tubular and interconnected network (Fig. 2 and Supplementary Fig. 4 online). Both types were observed using all imaging buffers mentioned above, as well as different fixation methods and cell types. The distinctive morphologies likely indicate cells in different growth stages as shown previously20,21: the interconnected tubular network of mitochondria present during interphase disintegrates during the mitotic phase22 and apoptosis23. The super-resolution images also allowed us to quantify the size of individual mitochondria. While the globular type of mitochondria were found to be relatively dispersed in size, ranging from 200 nm to 1500 nm, the diameter for the tubular structures appeared much more uniform, taking a narrow distribution of 210 ± 10 nm in B-SC-1 cells. What regulates the width of the tubular mitochondria is currently unknown.
Multicolor 3D imaging of mitochondria and microtubules
In order to maintain a dynamic morphology, mitochondria in a living cell are constantly transported and reorganized by motor proteins attached to the cytoskeleton, especially microtubules in mammalian cells24,25. To map their spatial relation, we performed two-color 3D STORM by staining Tom20 and β-tubulin. A405-Cy5 labeled secondary antibodies were used for detecting mitochondria, while the microtubules were stained by A555-Cy5 or A488-Cy5 labeled secondary antibodies. During image acquisition, an alternating sequence of 405 nm and 532nm (or 460 nm) laser pulses was used for activation of the A405-Cy5 and A555-Cy5 (or A488-Cy5) probes, respectively. The activated probes were then imaged by the 657 nm laser in between the activation pulses to determine their 3D positions. The localizations were colored according to the corresponding activation laser immediately preceding the activation events.
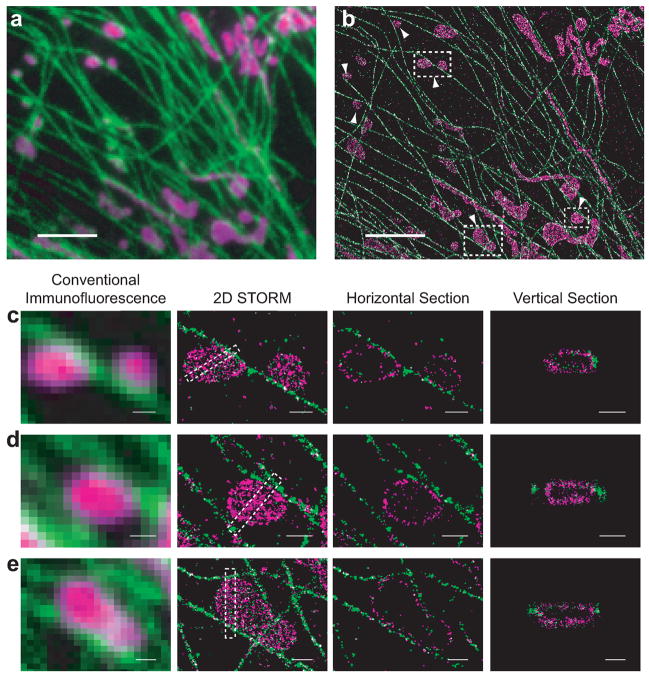
The STORM image clearly resolves mitochondria and microtubules even when they are densely packed, allowing a more precise determination of their spatial relation as compared to the conventional fluorescence image (Figs. 3a,b). The globular mitochondria (Figs. 3c,d) often contact microtubules, consistent with transport of mitochondria observed on microtubules24,25. However, many false contacts between mitochondria and microtubules were also observed in the conventional fluorescence image. For example (Fig. 3d), one mitochondrion appeared to touch two microtubules on both sides in the conventional image, whereas the STORM image clearly revealed a 150 nm separation from the left microtubule and a close contact site to the microtubule on the right. Mitochondria were often observed to be surrounded by microtubules (Fig. 3e and Supplementary Video 2 online).
Figure 3.
Two-color 3D STORM images of mitochondria and microtubules. (a) A conventional fluorescence image of mitochondria (magenta) and microtubules (green) normalized over excitation laser intensities. (b) STORM image of the same area with all localizations at different z positions stacked. The image is acquired in aqueous media and reconstructed from 500,000 localization points. It contains one 650 nm thick image slice. Scale bars: 3 μm. (c–e) Magnified views of the boxed regions in b. Panels from left to right: the conventional fluorescence image, the 2D STORM image, a horizontal cross-section, and the vertical cross-section of the boxed region in the 2D STORM image. See Supplemental Video 2 online for a 3D rendering of e. Scale bars: 500 nm.
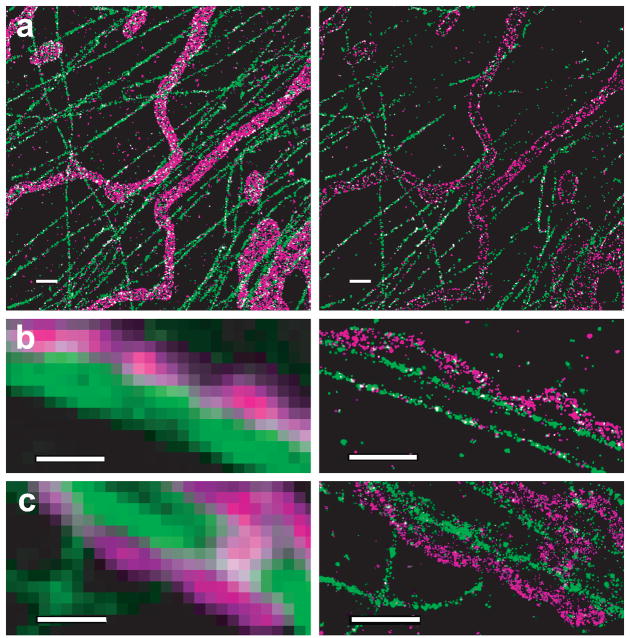
Spatial relations of the tubular mitochondria and microtubules are more complex. While some mitochondrial tubes appeared to align along the microtubules, others did not (Fig. 4 and Supplementary Fig. 5 online). Some tubular mitochondria that appeared to lie on microtubules revealed non-contiguous interaction, with contact points separated by stretches of non-contact regions in a way similar to inchworms(Figs. 4b,c) Such a spatial relationship is again obscured in the conventional fluorescent images. Though the significance of this inchworm-like contact mode is unclear, we speculate that it might make it easier to transport, as mitochondria-microtubule contacts are often mediated by motor proteins25.
Figure 4.
Two-color STORM images showing interactions between tubular mitochondria (magenta) and microtubules (green) in three BS-C-1 cells. The imaging conditions are identical to Figure 3. Images shown here are magnified view of the boxed regions of the large-field images shown in Supplementary Fig. 5 online. (a) Comparison of the z-stacked STORM image (left panel) and a 100 nm thick horizontal section displaying hollow tubes of mitochondria (right panel). (b, c) Inchworm-like interactions between tubular mitochondria and microtubules are resolved in STORM images (right panels) but not in conventional fluorescence images (left panels). Scale bars: 1 μm.
Discussion
The multicolor 3D super-resolution imaging capabilities shown here allow quantitative characterization of subcellular structures and their spatial relationship. It enables nanometer scale examination of specific molecular interactions, which is often difficult to obtain using other high-resolution imaging methods such as electron microscopy due to the low labeling efficiency of biomolecules with electron dense tags, such as immunogold. Studies on mitochondria and microtubules presented here can be extended to include motor proteins, fusion and fission related proteins, as well as other cytoskeletal structures to further elucidate how the morphology of mitochondria is regulated in cells21,24–26.
Accurate measurement of molecular structures and interactions requires a proper understanding of the factors that affect the 3D image resolution. First, the refractive index mismatch between the imaging media and the objective/coverglass system may alter the axial resolution. A high NA oil immersion objective provides high photon collection efficiency and a tight focus, both beneficial for the localization precision. However, when used to image an aqueous sample, such as cells in a physiological buffer, the observed axial position of a molecule needs to be corrected for the refractive index mismatch8,19. Moreover, the index mismatch also results in spherical aberration that can substantially reduce the z-localization precision for molecules above the imaging focal plane when it is considerably far away from the sample/coverglass interface. Interestingly, we found that spherical aberration exerted only a minimal effect on the localization precision of molecules below the imaging focal plane, providing a means to maintain high localization precision for at least several micrometers into the sample. Alternatively, index matching media may also be used to reduce the spherical aberration for oil immersion objective, although they are typically not compatible with live cell imaging. Water immersion objectives nearly eliminate spherical aberration for aqueous samples, but the lower photo collection efficiency and less tight focus due to a smaller NA are unfavorable for high localization precision.
Second, the effective resolution for resolving cellular structures not only depends on the intrinsic optical resolution but also on the label size and density. For imaging mitochondria and microtubules, we chose the commonly used immunofluorescence method, in which the label size, i.e. that of the primary and secondary antibodies, is ~10 nm. The label densities were such that the average distance between neighboring tubulin labels was ~20 nm and that between Tom20 labels was ~70 nm. In addition, a sample preparation procedure that preserves the ultrastructure of the cellular components is also crucial for maintaining a high effective resolution. For example, compared to the fixation and staining methods used in this work (as described in Supplementary Methods online), suboptimal fixation methods tend to break cytoskeletal structures and stronger permeabilization reagents tend to lower the labeling density of membrane proteins (Supplementary Fig. 6 online).
Finally, further improvement in resolution is possible considering the brightness of photoswitchable cyanine dyes. The ~5000 photons detected from these dyes per switching cycle allows, in principle, localization precision of several nanometers, assuming that the flexible linkers between the dye and the target protein allow it to freely rotate and be treated as an isotropic emitter27. The covalently linked activator-reporter pairs that we created here (Fig. 1) allow the probes to be linked directly to specific sites of target proteins. For example, the linked dye can be coupled to an enzymatic substrate, which can then be ligated to a specific peptide sequence fused to a protein of interest, allowing genetic targeting of these bright photoswitchable probes28–30. Such a direct linkage of a small photoswitchable probe to a target protein can further increase label density and reduce label size, as compared to larger labels such as antibodies and fluorescent proteins. Combination of this labeling strategy with the high localization precision afforded by these bright probes promises molecular scale resolution in future fluorescence imaging.
Methods
Synthesis of covalently linked activator-reporter pairs
Cy2, Cy3 and Cy5 bis NHS ester were purchased from GE Healthcare. A405, A488, A555 and A647 cadaverines were from Invitrogen. To synthesize an amine-reactive activator-reporter pair, the selected cadaverine-modified Alexa Fluor dye and bis NHS CyDye were dissolved in anhydrous dimethyl sulfoxide (DMSO) at a concentration of 5 mg/ml and subsequently mixed with a 1:3 molar ratio. 4× molar equivalent of triethylamine was then added to the reaction mix and the reaction was carried out for 15 min at room temperature. The product was purified on a C18 gravity column equilibrated with acetonitrile and eluted with 1:2 acetonitrile:dimethyl formamide. The product was eluted off the column in the first colored fraction. Alternatively, the product was purified by HPLC using a C18 column and eluted with an acetonitrile/water gradient in triethylamine-acetic acid buffer at pH 7.5.
To prepare thiol-reactive dyes, the amine-reactive activator-reporter pair eluted from the gravity column was mixed with 4× molar equivalent of N-aminoethyl maleimide (Sigma-Aldrich) and triethylamine. The reaction was carried out for 15 min at room temperature. The product was precipitated by adding equal volume of ethyl acetate, dissolved in DMSO and then purified using a C18 column.
STORM imaging
BS-C-1 cells (ATCC) were fixed, immunostained with rabbit anti-Tom20 (Santa Cruz Biotech) and/or mouse anti-β-tubulin (TUB2.1, Cytoskeleton) (See Supplementary Methods online for the detailed immunostaining procedure). The stained cells were imaged in PBS with the addition of 100 mM mercaptoethylamine at pH 8.5, 5% glucose (w/v) and oxygen scavenging enzymes (0.5 mg/mL glucose oxidase (Sigma-Aldrich), and 40 μg/mL catalase (Roche Applied Science)), unless otherwise mentioned. This above imaging buffer has a refractive index of 1.34. Media with higher refractive index (1.45) based on 80% (v/v) glycerol and 5% (w/v) glucose, or 60% (w/w) sucrose solution and 5% (w/w) glucose, were used in some experiments, both with the same amount of mercaptoethylamine and oxygen scavenging enzymes as described above. The slight mismatch between the medium refractive index and coverglass is needed for focus locking during imaging. Although a high concentration of mercaptoethylamine and oxygen scavenging system were used here for fixed cell imaging, the cyanine dyes also switch in buffers with lower concentrations of thiol and oxygen scavenger system compatible with live cell imaging12.
Data acquisition was performed on a fluorescence microscope as described in the Supplementary Methods online. Specifically for 3D imaging, a cylindrical lens with a focal length of 1 m was inserted into the imaging optical path for 3D localization8. To stabilize the microscope focusing during data acquisition, the reflected excitation laser from the coverglass-medium interface was directed to a quadrant photodiode. The position read out of the quadrant photodiode, which was sensitive to the distance between the coverglass and the objective focal plane, was used to provide feedback to a piezo objective positioner (Nano-F100, MadCity Labs), allowing compensation for the focus drift. The residual drift, < 40 nm (Supplementary Fig. 7 online), was corrected during data analysis. For whole cell imaging in an aqueous medium, the objective positioner was stepped in 300 nm intervals, which corresponds to a focal plane displacement of 216 nm after correcting for the refractive index mismatch at the glass-medium interface. Molecules within 270 nm below the focal plane were accepted for image reconstruction. Whole cell images were obtained from 9 partially overlapped z-slices. For imaging in media with a refractive index of 1.45, the positioner was stepped in 650 nm intervals, corresponding to an actual focal plane displacement of 580 nm. Molecules within 360 nm above and below the focal plane are accepted and whole cell images were obtained from 4 partially overlapped z-slices.
For single color imaging, the A405-Cy5 labeled sample was continuously illuminated with a 657 nm imaging laser (~30 mW). A low intensity 405 nm laser was used to activate the probes, with intensity adjusted such that only an optically resolved subset of the probes were activated at any given time. In certain cases, the 405 nm laser can be omitted because the 657 nm laser can also activate Cy5, albeit at a low rate. Emission from the fluorophores were recorded by the camera at a frame rate of 20 Hz. 3D localization of individual molecules was performed as described previously8 and described in the Supplementary Methods online. Multicolor imaging was performed by illuminating the sample repetitively with each frame of an activation laser followed by 3 frames of the 657 nm imaging laser. An alternating sequence of two activation lasers was used for two-color imaging. The 405 nm, 460 nm and 532 nm lasers were used to activated A405-Cy5, A488-Cy5, and A555-Cy5, respectively. Subtraction of crosstalk between different color channels were performed during data analysis as described in the Supplementary Methods online.
Supplementary Material
Note: Supplementary information is available on Nature Methods website.
Acknowledgments
This work is supported by in part by the National Institute of Health (to X.Z.). X.Z. is a Howard Hughes Medical Institute Investigator.
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests.
References
- 1.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 2.Gustafsson MGL. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci U S A. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt R, et al. Spherical nanosized focal spot unravels the interior of cells. Nat Methods. 2008;5:539–544. doi: 10.1038/nmeth.1214. [DOI] [PubMed] [Google Scholar]
- 4.Rust MJ, Bates M, Zhuang XW. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 6.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egner A, et al. Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophys J. 2007;93:3285–3290. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang B, Wang WQ, Bates M, Zhuang XW. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juette MF, et al. Three-dimernsional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 10.Schermelleh L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnert G, et al. Two-color far-field fluorescence nanoscopy. Biophys J. 2007;92:L67–L69. doi: 10.1529/biophysj.107.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bates M, Huang B, Dempsey GT, Zhuang XW. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bock H, et al. Two-color far-field fluorescence nanoscopy based on photoswitchable emitters. Appl Phys B-Lasers Opt. 2007;88:161–165. [Google Scholar]
- 14.Shroff H, et al. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci U S A. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bates M, Blosser TR, Zhuang XW. Short-range spectroscopic ruler based on a single-molecule optical switch. Phys Rev Lett. 2005;94:108101. doi: 10.1103/PhysRevLett.94.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conley NR, Biteen JS, Moerner WE. Cy3-Cy5 Covalent Heterodimers for Single-Molecule Photoswitching. J Phys Chem B. 2008;112:11878–11880. doi: 10.1021/jp806698p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 18.Kao HP, Verkman AS. Tracking of single fluorescent particles in 3 dimensions - use of cylindrical optics to encode particle position. Biophys J. 1994;67:1291–1300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hell S, Reiner G, Cremer C, Stelzer EHK. Aberrations in confocal fluorescence microscopy induced by mismatches in refractive-index. J Microsc. 1993;169:391–405. [Google Scholar]
- 20.Chan DC. Mitochondria: Dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Frazier AE, Kiu C, Stojanovski D, Hoogenraad NJ, Ryan MT. Mitochondrial morphology and distribution in mammalian cells. Biol Chem. 2006;387:1551–1558. doi: 10.1515/BC.2006.193. [DOI] [PubMed] [Google Scholar]
- 22.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 23.Gao W, Pu Y, Luo KQ, Chang DC. Temporal relationship between cytochrome c release and mitochondrial swelling during UV-induced apoptosis in living HeLa cells. J Cell Sci. 2001;114:2855–2862. doi: 10.1242/jcs.114.15.2855. [DOI] [PubMed] [Google Scholar]
- 24.Anesti V, Scorrano L. The relationship between mitochondrial shape and function and the cytoskeleton. Biochim Biophys Acta-Bioenerg. 2006;1757:692–699. doi: 10.1016/j.bbabio.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Boldogh IR, Pon LA. Mitochondria on the move. Trends Cell Biol. 2007;17:502–510. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Frederick RL, Shaw JM. Moving mitochondria: Establishing distribution of an essential organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enderlein J, Toprak E, Selvin PR. Polarization effect on position accuracy of fluorophore localization. Opt Express. 2006;14:8111–8120. doi: 10.1364/oe.14.008111. [DOI] [PubMed] [Google Scholar]
- 28.Chen I, Howarth M, Lin WY, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez-Suarez M, et al. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nat Biotechnol. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on Nature Methods website.