Abstract
G-protein coupled chemoattractant receptors signal transiently upon ligand binding to effect cell orientation and motility, but then are rapidly desensitized. The importance of desensitization has been unclear, since mutated nondesensitizable receptors mediate efficient chemotaxis. We hypothesized that homologous receptor desensitization is required for cellular navigation in fields of competing attractants. Modeling of receptor-mediated orientation shows that desensitization allows integration of attractant signals: Cells expressing normal receptors are predicted to 1) orient preferentially to distant gradients; 2) seek an intermediate position between balanced agonist sources; 3) and can be repositioned between chemoattractant-defined microenvironmental domains by modest changes in receptor number. In contrast, in the absence of desensitization, orientation is dominated by local agonist sources, precluding continued navigation. Furthermore, cell orientation in competing ligand gradients depends on the relative kinetic rates of receptor desensitization and recycling. We propose that homologous receptor desensitization is critical for cellular navigation in complex chemoattractant fields.
Keywords: cell trafficking, chemotaxis
Introduction
Extracellular clues that direct the migration of cells are involved in many biological processes including the immune response (1) and cancer metastasis (2). In addition to the complex nature of the migration process itself (3–8), another layer of complexity stems from the spatial and temporal presentation of the guiding extracellular chemoattractant gradients. It is now clear that most if not all leukocytes and neoplastic cells have the capacity to respond to multiple different chemokines and other attractants (9, 10) and that they must be able to integrate or interpret multiple chemoattractant gradients to direct their navigation in vivo (11, 12). Defining the mechanisms by which cells effectively navigate through such complex gradient fields would advance our understanding of cell migration and chemotaxis at the system level.
Previous studies using under agarose assays showed that neutrophils preferentially migrate toward a distant chemoattractant source in competing gradients of two chemoattractants (11, 13–15). Similar observations were reported in a study employing a microfluidic device that can produce stable concentration gradients with well-defined shapes (16). In these studies, Interleukin 8 (IL-8) and Leukotriene B4 (LTB4) were selected to generate competing gradients. The preferred chemotactic migration toward the distant gradient in competing gradients has been proposed to be a mechanism allowing leukocyte and possibly other eukaryotic cells to find their ultimate target in complex gradient microenvironment through multiple steps (i.e. multi-step chemotactic navigation) (11, 14).
Many of the most potent cellular chemotactic signals are processed by ligand-specific receptors of the G-protein coupled receptor (GPCR) family. All G-protein coupled chemoattractant receptors studied to date undergo ligand-dependent receptor desensitization (17), in which ligand-bound receptors are turned off after a transient period of signaling. The universality of homologous receptor desensitization suggests that it is a critically important property of chemoattractant receptors, and yet several studies have shown that mutated chemokine receptors that are unable to be desensitized by ligand nonetheless mediate efficient chemotactic responses, comparable to those directed by wild type receptors (18, 19). These assays focused on chemotaxis in a single gradient, however; whereas complex gradient fields are more likely the norm in physiologic settings. The potential significance of homologous receptor desensitization for chemotactic navigation in complex gradient environments has not been tested, either experimentally or through existing models for cell gradient sensing and chemotaxis (20–22). Notably, although several studies have attempted to approach multi-step chemotaxis using stochastic models, the cellular mechanisms involved have not been considered (23, 24). Here, we consider the hypothesis that ligand-induced homologous receptor desensitization may be essential for chemotactic navigation in competing attractant fields, providing the key to the ability of cells to integrate signals effectively, to preferentially orient to distant or newly arising agonist sources (11, 14), and to undergo regulated repositioning between chemoattractant-defined microenvironmental domains. We describe and apply a model of cellular chemotactic orientation to examine the role of receptor desensitization and recycling. The results suggest that homologous receptor desensitization mechanisms are essential for chemotactic navigation.
Results
Establishing a Model for Receptor-Mediated Orientation
We adapted models from previous studies to describe the dynamics of ligand-induced receptor modifications (25). We first consider ligand-induced homologous receptor desensitization and recycling of a model cellular unit in a uniform ligand field. Briefly, the cell is modeled as a geometry-free receptor-expressing unit. At the beginning of the process, receptors for the ligand are expressed on the cell surface. Upon ligand binding, receptors form receptor-ligand complexes and are activated to signal. The activated receptor-ligand complexes are then rapidly desensitized, a process associated with conversion of complexes from the low affinity state to the high affinity state. Desensitization is followed by a relatively slow process of receptor internalization and recovery. The internalized ligand is assumed to be degraded in a process that is irrelevant to orientation: in the model, dissociation of the internalized receptor-ligand complex is thus assumed to be instantaneous. On the other hand, since previous studies suggest that ligand binds almost irreversibly to desensitized receptors, dissociation of ligand from desensitized receptors on the cell surface is assumed to be negligible and is not considered in the current model (25). This process is illustrated in the diagram below.
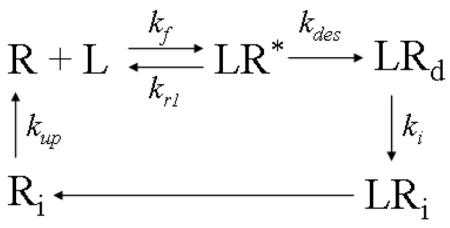
L: ligand concentration;
R: free surface receptors;
Ri: intracellular free receptors;
LR*: active (signaling) receptor-ligand complex;
LRd: desensitized receptor-ligand complex;
LRi: internalized receptor-ligand complex;
kf: ligand receptor association rate;
kr1: low affinity ligand receptor dissociation rate;
kdes: desensitization rate;
ki: internalization rate;
kup: up-regulation rate;
The kinetics of this process is described by a standard series of ordinary differential equations.
| [1] |
| [2] |
| [3] |
The total number of receptors Rtot, and the ligand concentration L are considered constants, providing the boundary conditions for the system.
| [4] |
| [5] |
The initial conditions for the system are configured as follows:
| [6] |
| [7] |
| [8] |
| [9] |
Thus, the values of the variables R, Ri, LR*, and LRd (here only three of these variables are independent variables due to total receptor conservation in Eq.[4]) at different time points can be determined by solving the differential equations as described above.
Using this simple model as a building block, we construct a model for cell gradient sensing in 1-D ligand gradients. The cell is now modeled as two receptor-expressing units representing the front and the back of the cell. The two units are connected by an isolating string representing the length of the cell, l = 10μm. Initially, the front unit and the back unit express equal number of surface receptors (i.e. 25,000 receptors for each unit). The ligand concentration for each unit is determined by the location of the cell and the profile of the ligand gradient. The ligand-induced receptor modifications at the front and the back unit are considered local events as described previously. Thus, internalized receptors from the front or the back unit are only allowed to be recycled back to the same side of the cell.
In a 1-D single gradient of ligand L, actively signaling ligand-receptor complexes LR* for the front and the back units of the cell in response to L are calculated. Then the difference of LR* between the two units of the cell is obtained to determine the orientation of the cell:
| [10] |
As the model is one dimensional, cell orientation is considered binary. Issues of threshold levels of signaling (magnitude of ΔLR*) required for effective orientation are considered below.
Previous studies of the chemotactic orientation of neutrophils to the chemotactic peptide N-formylnorleucylleucylphenylalanine (FNLLP) suggest that the cell can orient and chemotax in response to a ~1% ligand concentration difference across the cell length at the optimal concentration that is close to the dissociation constant kd, and that the receptor occupancy difference across the cell length required for robust chemotactic orientation can be as low as 10 receptors (26). Moreover, it has been suggested that the fluctuation of ligand concentration sensed by the cell around the kd due to the stochastic receptor-ligand binding is of about the same order (27). Based on these considerations, we assume that a differential signaling receptor occupancy across the cell length of ~10 would be required for orientation; and we define a threshold of |ΔLR*|≥ 10 for chemotactic orientation in our model. If ΔLR* is less than 10, the cell is considered to orient randomly.
In 1-D competing gradients of two different ligands, L1 and L2, the model cell is allowed to express two sets of ligand-specific receptors, R1 and R2, each equally represented in the front and the back unit. The two sets of receptors bind to their ligands and undergo receptor modifications using the formalism as described above. The total signaling ligand-receptor complexes are calculated for the front and the back units of the cell in response to the local concentrations of L1 and L2. Here we assume the front unit is always the right edge of the cell.
| [11] |
| [12] |
Then the total difference of signaling LR* (ΔLR*total) between the two units of the cell is obtained:
| [13] |
This definition is equivalent to the scalar sum of signaling ΔLR* to the gradients of L1 and L2 (ΔLR*total=ΔLR1*+ΔLR2*; ΔLR1*=LR1*front−LR1*back; ΔLR2*=LR2*front−LR2*back). While different GPCRs can potentially trigger distinct downstream signaling events, here we assume that pathways triggering orientation are shared. Thus in our model, ΔLR*total determines the chemotactic orientation of the cell in response to the combined gradients of L1 and L2.
For the configurations of competing ligand gradients considered in the model, the cell will orient up the gradient of L1 if ΔLR*total is positive or up the gradient of L2 if ΔLR*total is negative.
In our model, we consider fixed 1-D gradients, and the general gradient profile is approximated using a power function:
| (14) |
where Lmax is the highest ligand concentration at one border of the region; L0 is the lowest (basal) concentration and is set to zero at the opposite border for simplicity; x is the position in the gradient region; W is the width of the functional gradient region and is set to 1mm; n is the power representing the nonlinearity of the gradient (n=3 in our examples below). A nonlinear gradient is chosen because it is likely to be encountered by cells in vivo and in many in-vitro chemotaxis assays. In the model, the gradient of L1 is set to increase from the left to the right over the 1mm gradient region while the gradient of L2 is configured in the opposite direction with the same characteristics (i.e. same Lmax and n).
Modeling Cell Orientation in a Single Gradient
Having the model established, we wanted to examine the role of ligand-induced homologous receptor desensitization in cell gradient sensing in single and competing ligand gradients. The values of the parameters used in the model are listed below and the kinetic rates are adapted from the literature in experimental studies of formyl peptide receptors on neutrophils (25, 28, 29).
Lmax: 17.6nM;
kf: 8.4×107 M−1s−1;
kr1: 0.37 s−1;
kdes: 0.065 s−1 for desensitizable receptor, 0 for nondesensitizable receptor;
ki: 0.0033 s−1;
kup: 0.004 s−1;
The low affinity dissociation constant kd of 4.4nM is determined from the ratio of kr to kf. Lmax is chosen so that the average ligand concentration in the modeled region is equivalent to the kd.
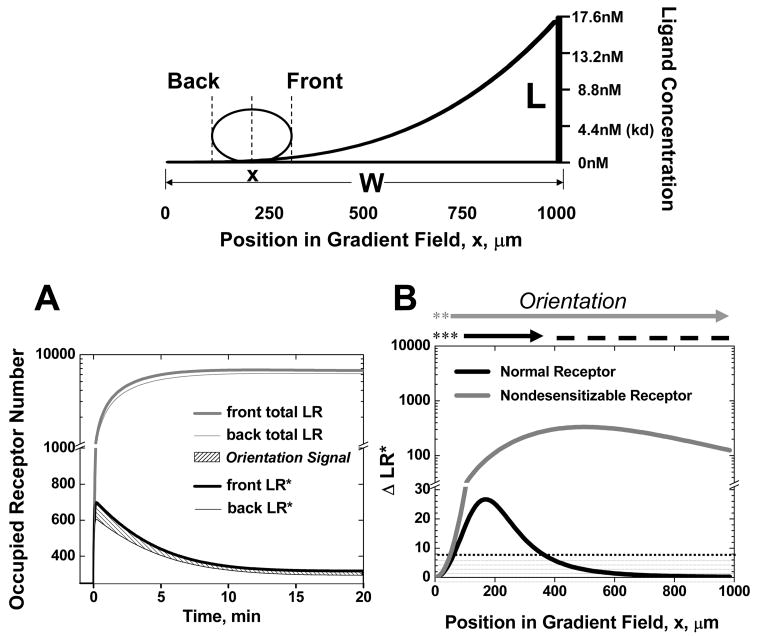
We first tested the model in a gradient of a single ligand. The orientation of the cell is determined by the difference in the number of active receptor-ligand complexes across the cell length (Fig. 1A). Compared with the total number or receptor-ligand complexes (i.e. total LR = LR* + LRd) in the front and the back of the cell, the number of active receptor-ligand complexes LR* is dramatically reduced because of rapid desensitization but relatively slow recycling and up-regulation of the surface receptors (Fig. 1A, x=0.2W in this example).
Figure 1. Chemotactic Orientation in a Gradients of a Single Ligand: (Top panel).
Illustration of a model cell in a single ligand gradient. (A) Total ligand-occupied receptor numbers (i.e. total LR = LR* + LRd) and active (signaling) receptor-ligand complexes (i.e. active LR*) in the front and back of a cell at the positions indicated as a function of time after ligand stimulation. Orientation is determined by the difference in active LR* across the cell length (shaded region). (B) Comparison of orientation of a cell expressing normal or nondesensitizable receptors in a single ligand gradient at the equilibrium state. The threshold of ΔLR* for orientation is set to 10 (shaded region). Dashed lines indicate the predicted stable regions of random migration (where ΔLR* is below the threshold and the cell will be drawn back if it migrates into the surrounding orienting regions). Asterisks indicate an unstable region of poor orientation (ΔLR* is below the threshold but the cell will be directed away from these zones if it migrates into the orienting region).
We calculated the ΔLR* (the orientation signal) of cells at equilibrium (d ΔLR*/dt=0) as a function of position in the single ligand field (Fig. 1B). In the context of our model, both normal desensitizable receptors and nondesensitizable receptors effectively orient the model cell toward the agonist source: The model cell can sense the gradient in a large area of the left side of the gradient region (ΔLR*≥10). In the right side of the gradient region, where the cell experiences high agonist concentrations, nondesensitizable receptors, but not the normal receptors, can mediate cell orientation up the gradient. Moreover, the orientation signal ΔLR* to the gradient is in fact substantially higher for the cell expressing nondesensitizable receptors. These results are consistent with experimental studies showing comparable or even higher levels of chemotaxis of cells mediated by nondesensitizable receptor mutants (18, 19).
Modeling the Role of Desensitization in Cell Orientation and Positioning in Competing Gradients
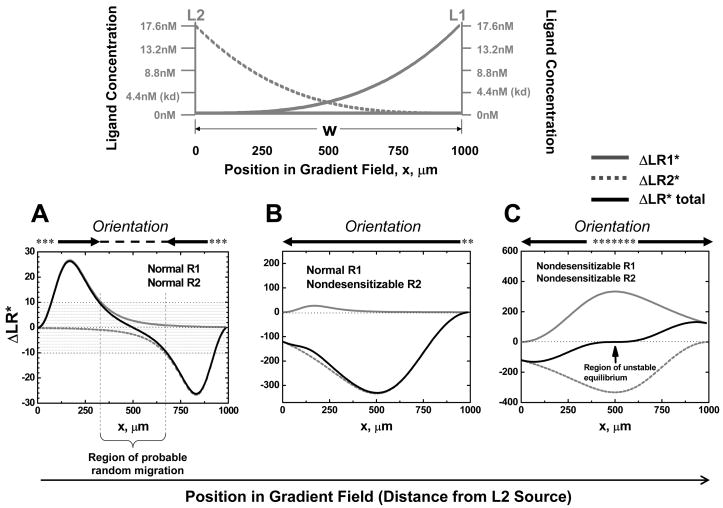
The consequences of the nondesensitizable mutant phenotypes of the receptor, however, are dramatically different in the setting of competing gradients of two ligands. The gradient configuration modeled is illustrated in Figure 2. At the equilibrium state, we compare the orientation of the model cell expressing desensitizable R1 receptors to L1, and desensitizable or nondesensitizable R2 receptors to L2 in competing gradients of L1 and L2. Our results show that a model cell in the left side of the gradient region can orient towards the distant gradient of L1 (ΔLR*total>10 for cells at ~0–0.3W) and (symmetrically) the cell in the right side of the gradient region can sense the distant gradient of L2 (ΔLR*total < −10 for cells at ~0.7–1W) if the cell expresses wild type (desensitizable) receptors for both ligands (Fig. 2A). In the area close to the mid-point of the gradient region, ΔLR*total is below the threshold of ~10 receptors for orientation, indicating that the cell cannot establish a preferred orientation to either gradient as the signals from the two sources are balanced (we consider this a target zone or a region for probable random migration; see Fig. 2A). In contrast, if the cell expresses desensitizable receptors for L1 but nondesensitizable receptors for L2, then the cell can no longer effectively sense the gradient of L1 (Fig. 2B); ΔLR*total is negative regardless of the cell’s position in the field, indicating orientation to the nondesensitizing agonist. Additionally, if the cell expresses nondesensitizable receptors for both L1 and L2, the model predicts preferred orientation of the cell toward the closest agonist source: a model cell in the left side of the gradient region orients toward the local gradient of L2 (~0–0.5W) and (symmetrically) the cell in the right side of the gradient region orients to the local gradient of L1 (~0.5–1W) (Fig. 2C).
Figure 2. Chemotactic Orientation in Competing Ligand Gradients Predicted by the Model: Top panel.
Illustration of competing ligand gradients. Lower panels: Equilibrium orientation signals for cells expressing the indicated receptor types (normal/desensitizable or nondesensitizable) in competing gradients of L1 and L2. In each case, ΔLR1* to L1 gradient, ΔLR2* to L2 gradient, and ΔLR*total at equilibrium are compared as a function of position. The shaded area indicates ΔLR*total levels below the threshold for orientation, set at ±10. Above each plot is a summary of predicted cell orientation and positioning at equilibrium: Arrows indicate the orienting zones (where ΔLR*total is above the threshold). Dashed lines indicate predicted target zones, stable regions of random migration (where ΔLR*total is below the threshold and the cell will be drawn back if it migrates into the surrounding orienting regions). Asterisks indicate an unstable region of poor orientation (ΔLR*total is below the threshold but the cell will be directed away from these zones if it migrates into the orienting region). (A) Equilibrium orientation of a cell expressing normal receptors to L1 and L2 (normal R1 and R2). ΔLR*total is below the threshold for effective orientation within the parenthesis. (B) Equilibrium orientation of a cell expressing normal R1 and nondesensitizable R2 receptors. (C) Equilibrium orientation of a cell expressing nondesensitizable receptors to both L1 and L2 (nondesensitizable R1 and R2). The arrow indicates the unstable central position at which cells expressing nondesensitizable receptors experience no net orientation signal.
When the cell is located equidistant to both sources (i.e. x=0.5W), and the gradients are again of equal magnitude, the cell expressing desensitizable receptors integrates the two signals, resulting in no net orientation (ΔLR*total =0 at 0.5W at equilibrium; Fig. 2A). From this example it can be seen that cells expressing normal desensitizable receptors for two competing gradients will seek an intermediate position between the agonist sources; in the case of real cells, they would be expected to undergo random migration within a central area between balanced attractant fields, being drawn back to the center each time they stray. Such a stable region of random migration can be considered a target zone. In contrast, for the cell expressing normal R1, but nondesensitizable R2, at the mid-point of the gradient region, a net orientation toward the nondesensitizing gradient of L2 is predicted (Fig. 2B). If both receptors for L1 and L2 are nondesensitizable, an unstable equilibrium can form at the mid-point of the gradient region (Fig. 2C): in contrast to the cell expressing normal receptors, however, these cells will migrate away from the equilibrium position each time they stray from center (which cells would normally do due to chemokinesis). Thus wild type and nondesensitizable receptors have diametrically opposing effects on cellular orientation responses in competing gradients.
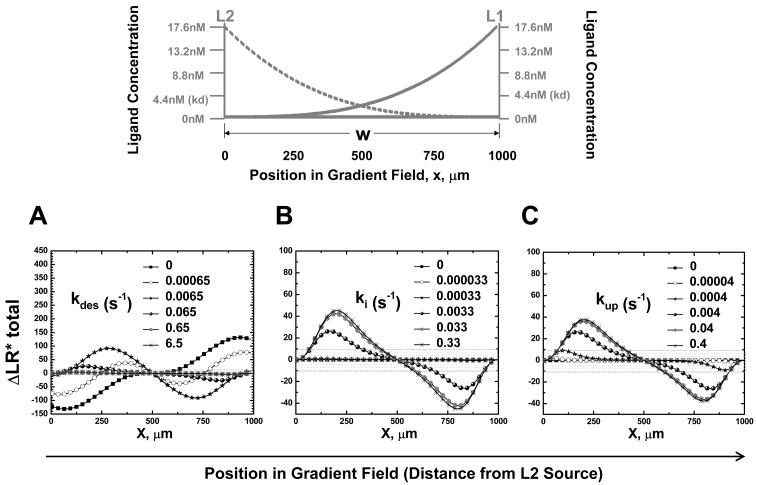
To test the effect of relative kinetic rates of receptor desensitization and recycling on cell gradient sensing in competing gradients, we compare the equilibrium orientation signal for a range of kinetic rates of receptor desensitization (kdes), internalization (ki) and up-regulation (kup). When ki and kup are fixed at 0.0033s−1 and 0.004s−1 respectively and kdes is varied for both R1 and R2, the cell orients toward the local gradient at kdes=0, and can sense the distant gradient as kdes increases (i.e. kdes=0.00065s−1, 0.0065s−1, 0.065s−1)(Fig. 3A). When kdes continues to increase (i.e. kdes=0.65s−1, 6.5s−1), the orientation signal even at optimum is below the threshold and the cell is predicted to orient randomly (Fig. 3A). These results suggest that receptor desensitization within an appropriate range is required for cell orientation to the distant gradient in competing gradients. Similarly, when kdes is fixed at 0.065s−1 and either ki or kup is varied for both R1 and R2, the orientation signal for the cell is below the threshold at low ki or kup, but cells will orient to the distant gradient in competing gradients at high ki or kup (Fig. 3B and 3C). However, the cell can not sense the distant gradient if ki and kup are both so high in relation to kdes that receptors are recycled almost as soon as they are desensitized (data not shown). These results indicate that receptor recycling at an appropriate rate relative to receptor desensitization is important for cell orientation to the distant gradient in competing gradients as well.
Figure 3. Effect of Relative Kinetic Rates of Desensitization and Recycling on Predicted Chemotactic Orientation in Competing Ligand Gradients: Top panel.
Illustration of competing ligand gradients. Lower panels: Equilibrium orientation signals (ΔLR*total) for cells expressing receptors to ligand L1 and L2 with different kinetic rates for desensitization (kdes)(A), internalization (ki)(B) and up-regulation (kup)(C) in competing gradients of L1 and L2 (except when varied as indicated, kdes is fixed at 0.065s−1, ki at 0.0033s−1 and kup at 0.004s−1.). The shaded area indicates ΔLR*total levels below the threshold for orientation, set at ±10.
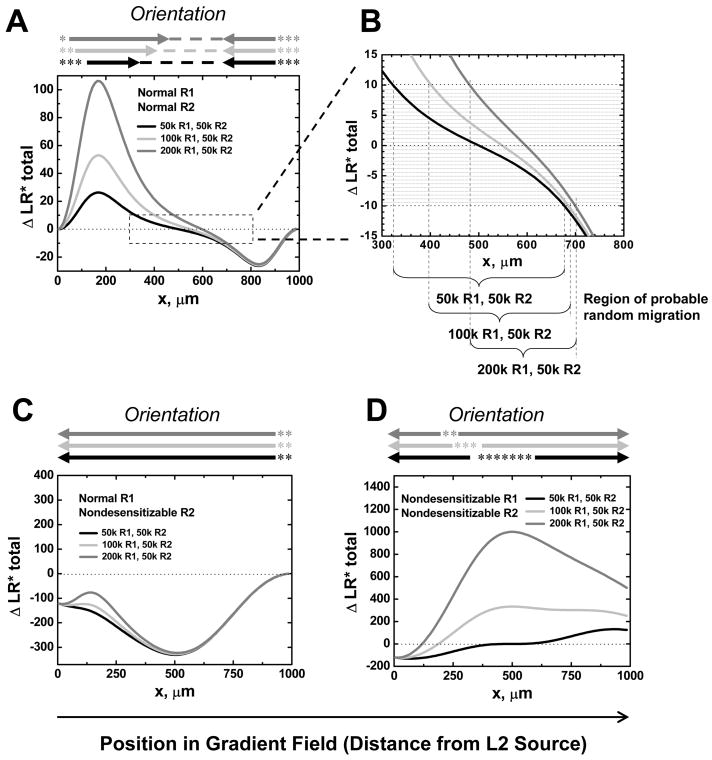
Previous study has shown that modest changes in receptor expression levels can have dramatic effects on cell positioning within chemoattractant-defined environments. For example, a 2–4 fold change in expression of certain chemokine receptors during activation of B lymphocytes can target them from the B zone follicles to the junctional zone between T and B cell areas in secondary lymphoid tissues (12). We were therefore interested in modeling the effects of different receptor numbers on the equilibrium positioning of a model cell in balanced competing ligand gradients (Fig. 4). As above, the central positions at which the net orientation signal is below the threshold (|ΔLR*total| <10) are indicated; these regions are assumed to represent a target zone within the static gradient fields modeled. The cell is allowed to express 2 or 4 time more total receptors for L1 than L2 (i.e. R1=2R2 or R1=4R2. R2 is fixed at 50,000). In this situation, we calculated the shift of the equilibrium position in the gradient fields. Our results show a significant shift of the equilibrium position (ΔLR*total =0) toward L1, for which the cell expresses more receptors (~50μm shift from 0.5W for R1=2R2 and ~100μm shift from 0.5W for R1=4R2) (Fig. 4A). Furthermore, the target zones, to which a migrating cell would be drawn, shifts toward L1 when R1 level increases (Fig. 4B); the shift occurs primarily on one side of the ‘target zone”, in essence narrowing the zone by enhancing the orientation response to L1. Thus, the equilibrium position of a migrating cell expressing normal receptors is predicted to depend on the relative number of receptors expressed specific for the two agonists. In contrast, the ability of regulated receptor numbers to modulate cell positioning by balancing two gradients is severely impaired when one receptor (R2) is nondesensitizable: ΔLR*total remains negative for all positions in the gradient region and thus an equilibrium position could not be reached (ΔLR*total <0 for R1=R2, or R1=2R2, or R1=4R2. R2 is fixed at 50,000)(Fig. 4C). If the cell expresses nondesensitizable receptors for both L1 and L2, the unstable equilibrium position dramatically shifts from the mid-point toward L2 when R1 level increases: in all cases, however, the cell will ultimately target either one source or the other, since the equilibrium positions in this setting are unstable. Parallel results were obtained for all three situations if R1 is held constant at 50,000 and R2 is reduced (data not shown). Thus, homologous receptor desensitization allows cellular positioning within attractant fields to be regulated as a function of receptor expression levels.
Figure 4. Effect of Receptor Numbers on Cell Positioning in Competing Gradients.
Equilibrium orientation signals for cells expressing the indicated receptor types and numbers in competing gradients of L1 and L2 configured as in Figure 2. ΔLR*total at equilibrium are compared as a function of position. The shaded area in B highlights ΔLR*total levels below the threshold for orientation, set at ±10. (A) The cell is allowed to express 2 or 4 times more desensitizable surface receptors R1 to L1 than desensitizable receptors to L2 (normal R1 and R2; R1=2R2 or R1=4R2. R2 is fixed at 50,000). In competing gradients, the equilibrium position shifts significantly toward the L1 gradient, for which the cell expresses more receptors. (B) Enlarged view comprising the target zone where the signals from the two agonists are balanced, resulting in no net orientation (ΔLR*total < threshold). The target zone (i.e. the region of probable random migration, indicated by parentheses) shifts toward L1 when R1 level increases. (C) If R2 is nondesensitizable, changes in R1 levels have no effect on orientation. (D) If both receptors are nondesensitizable, receptors numbers dramatically shift the position of unstable equilibrium of signals, but cells are predicted to migrate to one agonist source or the other. Same as in Figure 2, orientation (arrows), target zones (dashed lines), and zones of unstable random migration (asterisks) are summarized above the plot, and highlight the differences in predicted cell behaviors induced by wild type versus nondesensitizable receptors.
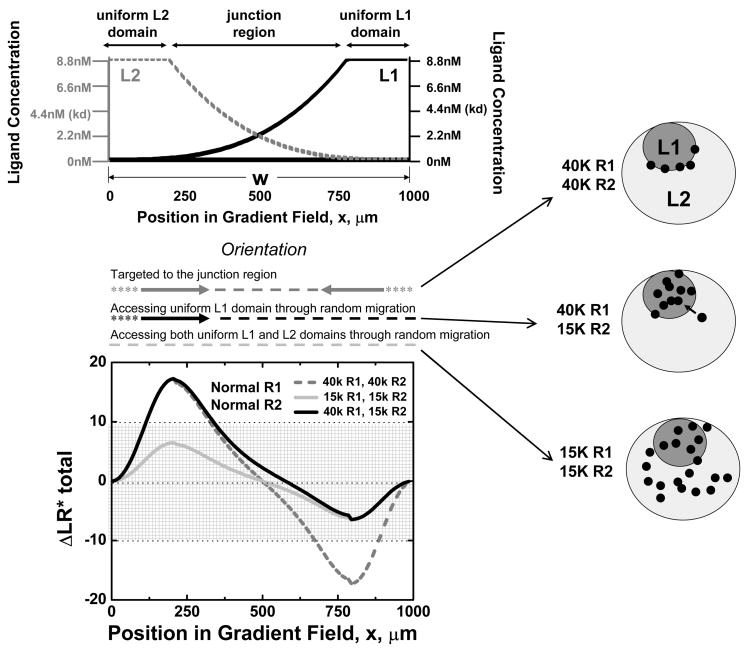
In the example cited above of lymphocyte subset positioning in lymphoid tissues, and likely in other physiologic settings, cells normally occupy particular domains within tissues (e.g. in lymph nodes resting B cells occupy follicles, whereas T cells occupy paracortical zones), while being excluded from adjacent domains to which other cells home. We asked if regulation of receptor numbers could control access of cells to adjacent domains of uniform chemoattractant concentration, separated by a junctional gradient zone similar to that modeled above. We consider two domains of uniform L1 or L2 concentration as illustrated in Figure 5, with L1 and L2 gradients (n=3) in a junctional region between the domains. The concentration of ligands L1 and L2 in the uniform fields is set at 2kd. As above, we evaluated ΔLR*total at equilibrium as a function of position for a cell expressing desensitizable receptors R1 and R2. When R1=R2=40,000, ΔLR*total exceeds the threshold for orientation in the left and right sides of the junctional zone: thus randomly migrating cells in either domain would ultimately be targeted to the junctional region. If R1=R2=15,000, however, ΔLR*total remains below the threshold at all positions, implying that cells would have access through random motility to the entire field. On the other hand, if R1=40,000 or more and R2=15,000, cells will be “captured” by the uniform L1 domain, accessing the uniform L1 domain and the junctional region but being excluded from the uniform L2 domain. Whereas desensitizing receptors thus permit regulated targeting and positioning between chemoattractant-defined microenvironments as a function of modest changes in receptor levels, nondesensitizable receptors would force cells to migrate to one domain or the other (as implied by Figure 2B, 2C, 4C and 4D); repositioning in this instance would require a nearly complete down-regulation of the restricting receptor for the initially targeted agonist.
Figure 5. Effect of Receptor Numbers on Cell Access to Different Ligand Domains: (Top panel).
Illustration of adjacent uniform chemoattractant fields (domains) separated by a junctional region of overlapping gradients. (Bottom panel; and see schematics) When the cell expresses 40,000 wild type receptors of both R1 and R2, orientation (ΔLR*total) signals are predicted to target it to the junctional region. If R1 and R2 are reduced to 15,000, the orientation of the cell is below the threshold at all positions and thus the cell will have random access to both uniform L1 domain and uniform L2 domain and the junctional region. If only one receptor (R2 illustrated here) is reduced to 15,000 and R1 remains at 40,000, then a cell wandering into the junctional region will be preferentially directed towards uniform L1 domain; at long time points therefore cells will be restricted to the L1 domain. As for earlier figures, orientation (arrows), target zones (dashed lines), and zones of unstable random migration (asterisks) are indicated above the plot, and summarize the ability of modest changes in receptor numbers to control cell positioning and access to attractant-field defined microenvironmental domains. The schematics illustrate the predicted differential targeting in the context of two adjacent chemoattractant-defined domains as a function of receptor expression levels.
Together, these results support a critical role for mechanisms of homologous receptor desensitization in cell orientation and positioning in the context of competing gradients and attractant-defined microenvironments.
Discussion
In this paper, we modeled the role of ligand-induced homologous receptor desensitization in cell gradient sensing in conflicting gradients. The modeling results suggest that ligand-induced homologous receptor desensitization is important for cells to integrate signals from different chemoattractants: desensitization allows cells to preferentially respond to distant or newly emerging agonist sources in the presence of competing ligand gradients, and to undergo repositioning in complex attractant fields in response to modest changes in receptor numbers. The results derived from these studies provide a molecular explanation for previous studies of the experimental behaviors of neutrophils in competing chemoattractant gradients, and support the multi-step model of chemotactic navigation (11, 13–16). Moreover, they provide a potential explanation for the evolutionary conservation and universal expression of mechanisms of homologous receptor desensitization by G-protein coupled chemoattractant receptors.
In our model, nondesensitizable receptors mediate effective cell chemotaxis to single ligand gradients, a prediction that is consistent with experimental chemotaxis studies that have examined the effect of several nondesensitizable chemokine receptor mutants (18, 19). In addition, not unexpectedly, our modeling predicts that nondesensitizing ligands dominate cell orientation when in direct competition with a desensitizing attractant. However, the results emphasize further that cells expressing nondesensitizable receptors are unable to integrate attractant signals, fail to navigate effectively in complex gradient environments, and will become ‘trapped’ by local agonist sources.
Numerous settings in which multiple chemoattractants contribute in a complex manner to cell migration have been reported. For example, the neutrophil chemoattractants IL-8 and LTB4 both contribute to neutrophil recruitment to the airways in chronic obstructive pulmonary disease (COPD) (30); chemokines CXCL12, CXCL13, CCL19 and CCL21 all contribute to regulate B cell homing to lymph nodes and Peyer’s patches, including recruitment from the blood following by chemotaxis into the follicular zones (31, 32); CCL17 and CCL27 participate in memory T cell homing to the skin, and CCL27 is then thought to recruit subsets of T cells to the epidermis (33–35). The chemokine receptors involved in these events are universally sensitive to homologous ligand desensitization, a fact that our study suggests may be essential to the successful step-by-step migration of cells in these complex situations.
A particularly well characterized example is the finding that the balance of receptor expression for B cell zone chemokines (CXCR5 for the follicular chemokine CXCL13) and T cell zone chemokines (CCR7 for T zone chemokines CCL19 and CCL21) controls the positioning of antigen-engaged B cells at the T cell zone border (12). As shown in our model, such balanced integration of conflicting signals requires mechanisms of homologous desensitization: desensitization allows cells to migrate down a local attractant gradient in response to a distant one. This in turn allows cells ultimately to seek an intermediate position between two chemokine sources, rather than becoming ‘trapped’ by a local high chemokine concentration. The levels of receptors expressed then will determine whether the cell moves more towards one environment or the other; and, as we have shown, modest changes in relative receptor levels can target cells to one or another adjacent chemoattractant-defined microenvironment domain. In the context of B cell positioning, resting B cells normally express both CXCR5 and CCR7, yet they selectively occupy “B cell follicles”, sites of stromal cell based CXCL13 production. Antigen activation induces a 2–3 fold increase in CCR7 expression, associated with a shift of the equilibrium position of B cells from a dispersed distribution within the follicle to the junctional region between the B cell zone and T cell zone (B/T junction); this is presumed to enhance interactions between B cells and T cells required for the immune response (12, 36). In a parallel but opposing manner, although many memory T cells (e.g. most ‘central’ memory cells in human tonsils) like naïve B cells co-express CXCR5 and CCR7 (37), most resting CXCR5+ T cells are found in the T cell zone; in this case, activation causes down-regulation of CCR7 on a subset of T cells, which then home to the B cell follicles where they support the B cell germinal center response (36, 38). Our model raises the additional possibility that CXCR5+CCR7+ resting memory T cells can in fact access both T and B zones, migrating randomly between them until receiving a specific signal to alter expression levels of one or the other receptor. We imagine that in the physiologic setting of lymphoid tissue microenvironments, the follicular chemokine CXCL13 and the T cell zone chemokine CCL19, which are expressed by dispersed stromal cells in these microenvironments, are likely to be present as qualitatively uniform fields in the follicles and T cell zone respectively; and that a steep competing gradient of the two ligands is present at the B/T junction. Our modeling of cells expressing normal receptors in competing gradients separating uniform attractant fields show that modest changes in receptor expression levels have the potential to determine whether a cell occupies one or another domain, or instead is targeted to the junctional region, just as in the examples of lymphocyte targeting in vivo. Key features of the model of cellular orientation required for this behavior include not only homologous receptor desensitization, but also the existence of a threshold level of differential receptor occupancy (between the front and back of the cell) below which the cell migrates randomly.
Last, we discuss the basis and limitations of our model. In our model, parameter values such as the total receptor number (50,000 in most of our analyses), the dissociation constant (4.4nM) and other kinetic rates are adapted from experimental studies of formyl peptide receptors on neutrophils (25, 28, 29). The values of some key parameters are similar in other chemoattractant receptor systems as well (e.g. 75,000 total receptors and kd=4nM for chemokine IL-8 on human neutrophils). In our model, changing the total receptor number or kd alone by up to 10-fold does not alter the fundamental importance of homologous receptor desensitization for cell positioning in competing gradients as shown in our examples. Recycling of desensitized receptors, however, is required to maintain sufficient signaling receptor-ligand complexes at equilibrium for orientation in our model, and the relative rate of receptor internalization and recycling compared with receptor desensitization will affect cell orientation as shown in Figure 3B and 3C: if insufficient unoccupied receptors are recycled to the surface, the cells lose the ability to orient at equilibrium. Similarly, the rate of receptor desensitization relative to receptor internalization and recycling is also important for robust orientation to the distant gradient in competing gradients. The threshold of signaling receptor occupancy difference required for chemotactic orientation is set to 10 in our model based on 1) the minimal receptor occupancy difference (~10) calculated to be required for orientation by neutrophils, which can chemotax directionally in response to a ~1% ligand concentration difference across the cell length (26); and on 2) considerations mentioned above regarding predicted stochastic fluctuations in receptor-ligand binding near the kd (27). The physiologic threshold for effective orientation is likely to be influenced by a number of factors, including the kd of the receptor, the local ligand concentration (determining the number of receptors remaining on the cell surface, as well as the level of stochastic variation in receptor-ligand complexes), and the dimensions of the cell. Moreover, a physiologic “threshold” is not expected to be absolute as modeled here; the efficiency of orientation is not a step function, but rather would improve as ΔLR* increases until an optimum is achieved.
The ligand gradient in our model is generated by a power function and a nonlinear profile is chosen with n=3, so that agonist concentration changes with the cube of the distance. This profile is an approximation of the nonlinear gradients generated in some in-vitro migration assays (e.g. the under agarose assay (11, 14), or the micropipette-based assay (39)). Lmax for simple gradient modeling is set arbitrarily at 17.6 nM so that the mean concentration in the gradient region modeled equals the kd (4.4 nM). Additionally, we consider that nonlinear gradients are likely to be the rule in vivo. In the simplest in-vivo scenario, a single cell produces a finite amount of freely diffusing ligand at a constant rate into an environment, resulting in concentration decreasing nonlinearly as a function of the distance from the cell. If as is likely the ligand is degraded everywhere with first order kinetics, nonlinear gradients with n>2 will be expected. In addition, most chemokines bind with low affinity to glycosaminoglycan (GAG) on cells and in tissue stroma. Reversible interactions with stromal elements are expected to have the effect of steepening gradients even further. For these reasons, we chose to focus on a gradient with a nonlinear profile as presented rather than modeling the gradient based on free diffusion as such a gradient will become uniform or linear at equilibrium. Quantitative aspects of cell orientation in competing gradients may also be sensitive to gradient profiles, but this issue is not addressed here.
Previous studies have shown that there is an intracellular signaling hierarchy in which end-target chemoattractants such as C5a and fMLP dominate host-derived chemoattractants such as IL-8 and LTB4 in attracting neutrophils in competing gradients, an effect mediated by heterologous receptor desensitization and/or, as shown in recent studies, by alterations in intracellular orientation signals used by host chemokines (11, 13–15). Since heterologous receptor desensitization in essence eliminates the contribution of the competing distant host ligand, orientation in this context is equivalent to orientation to a single local ligand gradient. Thus we have limited our modeling here to host-derived non-cross-desensitizing chemoattractants, which trigger common downstream signaling pathways in cells through their binding receptors.
In addition, our model is limited to chemotactic orientation and does not describe cell migration. Numerous studies indicate that, regardless of gradient configuration, and even in uniform ligand fields, cells are motile when in the presence of ligand. Thus, orientation as modeled here would determine the direction of this continuous migration, and regions in which there is insufficient ΔLR* to support orientation are considered zones of random migration. Furthermore, we consider only fixed 1-D ligand gradients and point cellular units, avoiding complications such as time-dependent gradient shape and ligand concentration, and diffusion of receptors on the cell surface. Ligand-induced receptor modifications at the front and the back unit are considered local events, as well. While this is likely a simplification of the reality of receptor cycling, during which signaling events may influence preferential receptor cycling to the front of the cell (40), we consider that there is not adequate data available on this point for modeling. Moreover, the effects of preferential recycling to the front would be to enhance orientation at equilibrium and thus would not alter our conclusions qualitatively.
In summary, the current model provides a theoretical framework that has allowed us to consider the role of ligand-induced homologous receptor desensitization in cell gradient sensing in competing ligand gradients. Our results suggest a critical role for ligand-dependent receptor desensitization in the orientation and positioning of cells in complex physiologic settings.
Acknowledgments
Grant Support: F. L. is supported by the postdoctoral training grant (5T32AI07290-20) from the NIH-NIAID, Immunology Program, Stanford University, and a grant from the Stanford Bio-X Interdisciplinary Initiative Program. This study is supported in part by grants from the NIH and an award from the Department of Veterans Affairs to E.C.B..
We thank Dr. Martin Meier-Schellersheim and Dr. Eric Kunkel for helpful discussion. We thank Dr. Eric Kunkel, Dr. Tau-Mu Yi and Dr. Fabio Baldessari for valuable comments on the manuscript.
Nonstandard Abbreviation
- B/T junction
the junctional region between the B cell zone and T cell zone
- GPCR(s)
G-protein coupled receptor(s)
Footnotes
Disclosures: The authors have no financial conflict of interest.
This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.
References
- 1.Kubes P. Introduction: The complexities of leukocyte recruitment. Seminars in Immunology. 2002;14:65–72. doi: 10.1006/smim.2001.0343. [DOI] [PubMed] [Google Scholar]
- 2.Condeelis J, Jones J, Segall J. Chemotaxis of metastatic tumor cells: clues to mechanisms from the Dictyostelium paradigm. Cancer Metastasis Rev. 1992;11:55–68. doi: 10.1007/BF00047603. [DOI] [PubMed] [Google Scholar]
- 3.Chung CY, Funamoto S, Firtel RA. Signaling pathways controlling cell polarity and chemotaxis. Trends in Biochemical Sciences. 2001;26:557–566. doi: 10.1016/s0968-0004(01)01934-x. [DOI] [PubMed] [Google Scholar]
- 4.Dekker LV, Segal AW. SIGNAL TRANSDUCTION:Signals to Move Cells. Science. 2000;287:982–985. doi: 10.1126/science.287.5455.982. [DOI] [PubMed] [Google Scholar]
- 5.Harms BD, Bassi GM, Horwitz AR, Lauffenburger DA. Directional Persistence of EGF-Induced Cell Migration Is Associated with Stabilization of Lamellipodial Protrusions. Biophys J. 2005;88:1479–1488. doi: 10.1529/biophysj.104.047365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liddington RC, Bankston LA. The Structural Basis of Dynamic Cell Adhesion: Heads, Tails, and Allostery. Experimental Cell Research. 2000;261:37–43. doi: 10.1006/excr.2000.5058. [DOI] [PubMed] [Google Scholar]
- 7.Zicha Allen, Brickell Kinnon, Dunn Jones, Thrasher Chemotaxis of macrophages is abolished in the Wiskott-Aldrich syndrome. British Journal of Haematology. 1998;101:659–665. doi: 10.1046/j.1365-2141.1998.00767.x. [DOI] [PubMed] [Google Scholar]
- 8.Zigmond SH. How WASP Regulates Actin Polymerization. J Cell Biol. 2000;150:117F–120. doi: 10.1083/jcb.150.6.f117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mantovani A. The chemokine system: redundancy for robust outputs. Immunology Today. 1999;20:254–257. doi: 10.1016/s0167-5699(99)01469-3. [DOI] [PubMed] [Google Scholar]
- 10.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 11.Foxman EF, Kunkel EJ, Butcher EC. Integrating Conflicting Chemotactic Signals: The Role of Memory in Leukocyte Navigation. J Cell Biol. 1999;147:577–588. doi: 10.1083/jcb.147.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reif K, Ekland EH, Ohl L, Nakano H, Lipp M, Forster R, Cyster JG. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature. 2002;416:94–99. doi: 10.1038/416094a. [DOI] [PubMed] [Google Scholar]
- 13.Heit B, Tavener S, Raharjo E, Kubes P. An intracellular signaling hierarchy determines direction of migration in opposing chemotactic gradients. J Cell Biol. 2002;159:91–102. doi: 10.1083/jcb.200202114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foxman EF, Campbell JJ, Butcher EC. Multistep Navigation and the Combinatorial Control of Leukocyte Chemotaxis. J Cell Biol. 1997;139:1349–1360. doi: 10.1083/jcb.139.5.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heit B, Colarusso P, Kubes P. Fundamentally different roles for LFA-1, Mac-1 and Alpha-4-integrin in neutrophil chemotaxis. Journal of Cell Science. 2005;118:5205–5220. doi: 10.1242/jcs.02632. [DOI] [PubMed] [Google Scholar]
- 16.Lin F, Nguyen C, Wang S, Saadi W, Gross S, Jeon N. Neutrophil migration in opposing chemoattractant gradients using microfluidic chemotaxis devices. Ann Biomed Eng. 2005;33:475–482. doi: 10.1007/s10439-005-2503-6. [DOI] [PubMed] [Google Scholar]
- 17.Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. Desensitization of G protein-coupled receptors and neuronal functions. Annu Rev Neurosci. 2004;27:107–144. doi: 10.1146/annurev.neuro.27.070203.144206. [DOI] [PubMed] [Google Scholar]
- 18.Arai H, Monteclaro FS, Tsou CL, Franci C, Charo IF. Dissociation of Chemotaxis from Agonist-induced Receptor Internalization in a Lymphocyte Cell Line Transfected with CCR2B. Evidence that Directed Migration Does not Require Rapid Modulation of Signaling at the Receptor Level. J Biol Chem. 1997;272:25037–25042. doi: 10.1074/jbc.272.40.25037. [DOI] [PubMed] [Google Scholar]
- 19.Mueller SG, White JR, Schraw WP, Lam V, Richmond A. Ligand-induced Desensitization of the Human CXC Chemokine Receptor-2 Is Modulated by Multiple Serine Residues in the Carboxyl-terminal Domain of the Receptor. J Biol Chem. 1997;272:8207–8214. doi: 10.1074/jbc.272.13.8207. [DOI] [PubMed] [Google Scholar]
- 20.Iglesias PA, Levchenko A. Modeling the cell’s guidance system. Sci STKE. 2002;2002:RE12. doi: 10.1126/stke.2002.148.re12. [DOI] [PubMed] [Google Scholar]
- 21.Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J Biol Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 22.Krishnan J, Iglesias PA. Uncovering directional sensing: where are we headed? Systems Biology, IEE Proceedings. 2004;1:54–61. doi: 10.1049/sb:20045001. [DOI] [PubMed] [Google Scholar]
- 23.Oelz D, Schmeiser C, Soreff A. Multistep navigation of leukocytes: a stochastic model with memory effects. Mathematical Medicine and Biology. 2005;22:291–303. doi: 10.1093/imammb/dqi009. [DOI] [PubMed] [Google Scholar]
- 24.Painter KJ, Maini PK, Othmer HG. Development and applications of a model for cellular response to multiple chemotactic cues. J Math Biol. 2000;41:285–314. doi: 10.1007/s002850000035. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman JF, Linderman JJ, Omann GM. Receptor Up-regulation, Internalization, and Interconverting Receptor States. Critical Components of a Quantitative Description of N-Formyl Peptide-Receptor Dynamics in the Neutrophil. J Biol Chem. 1996;271:18394–18404. doi: 10.1074/jbc.271.31.18394. [DOI] [PubMed] [Google Scholar]
- 26.Zigmond SH. Consequences of chemotactic peptide receptor modulation for leukocyte orientation. J Cell Biol. 1981;88:644–647. doi: 10.1083/jcb.88.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tranquillo RT, Lauffenburger DA, Zigmond SH. A stochastic model for leukocyte random motility and chemotaxis based on receptor binding fluctuations. J Cell Biol. 1988;106:303–309. doi: 10.1083/jcb.106.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norgauer J, Eberle M, Fay SP, Lemke HD, Sklar LA. Kinetics of N-formyl peptide receptor up-regulation during stimulation in human neutrophils. J Immunol. 1991;146:975–980. [PubMed] [Google Scholar]
- 29.Sklar LA, Bokoch GM, Button D, Smolen JE. Regulation of ligand-receptor dynamics by guanine nucleotides. Real- time analysis of interconverting states for the neutrophil formyl peptide receptor. J Biol Chem. 1987;262:135–139. [PubMed] [Google Scholar]
- 30.Beeh KM, Kornmann O, Buhl R, Culpitt SV, Giembycz MA, Barnes PJ. Neutrophil Chemotactic Activity of Sputum From Patients With COPD: Role of Interleukin 8 and Leukotriene B4. Chest. 2003;123:1240–1247. doi: 10.1378/chest.123.4.1240. [DOI] [PubMed] [Google Scholar]
- 31.Okada T, V, Ngo N, Ekland EH, Forster R, Lipp M, Littman DR, Cyster JG. Chemokine Requirements for B Cell Entry to Lymph Nodes and Peyer’s Patches. J Exp Med. 2002;196:65–75. doi: 10.1084/jem.20020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hargreaves DC, Hyman PL, Lu TT, Ngo VN, Bidgol A, Suzuki G, Zou YR, Littman DR, Cyster JG. A Coordinated Change in Chemokine Responsiveness Guides Plasma Cell Movements. J Exp Med. 2001;194:45–56. doi: 10.1084/jem.194.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiss Y, Proudfoot AE, Power CA, Campbell JJ, Butcher EC. CC Chemokine Receptor (CCR)4 and the CCR10 Ligand Cutaneous T Cell-attracting Chemokine (CTACK) in Lymphocyte Trafficking to Inflamed Skin. J Exp Med. 2001;194:1541–1547. doi: 10.1084/jem.194.10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morales J, Homey B, Vicari AP, Hudak S, Oldham E, Hedrick J, Orozco R, Copeland NG, Jenkins NA, McEvoy LM, Zlotnik A. CTACK, a skin-associated chemokine that preferentially attracts skin-homing memory T cells. PNAS. 1999;96:14470–14475. doi: 10.1073/pnas.96.25.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sigmundsdottir H, Pan J, Debes GF, Alt C, Habtezion A, Soler D, Butcher EC. DCs metabolize sunlight-induced vitamin D3 to ‘program’ T cell attraction to the epidermal chemokine CCL27. Nat Immunol. 2007;8:285–293. doi: 10.1038/ni1433. [DOI] [PubMed] [Google Scholar]
- 36.Arnold Carrie N, Campbell Daniel J, Lipp M, Butcher Eugene C. The germinal center response is impaired in the absence of T cell-expressed CXCR5. European Journal of Immunology. 2007;37:100–109. doi: 10.1002/eji.200636486. [DOI] [PubMed] [Google Scholar]
- 37.Campbell DJ, Kim CH, Butcher EC. Separable effector T cell populations specialized for B cell help or tissue inflammation. Nat Immunol. 2001;2:876–881. doi: 10.1038/ni0901-876. [DOI] [PubMed] [Google Scholar]
- 38.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T Cells: B Helper Activity Is Focused in a Germinal Center-localized Subset of CXCR5+ T Cells. J Exp Med. 2001;193:1373–1382. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohof AM, Quillan M, Dan Y, Poo MM. Asymmetric modulation of cytosolic cAMP activity induces growth cone turning. J Neurosci. 1992;12:1253–1261. doi: 10.1523/JNEUROSCI.12-04-01253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallin J, Seligmann B. Mobilization and adaptation of human neutrophil chemoattractant fMet-Leu-Phe receptors. Fed Proc. 1984;43:2732–2736. [PubMed] [Google Scholar]