Abstract
Cytochrome P450 3A4 is involved in the metabolism of 50% of all swallowed drugs. The enzyme functions by means of a high-valent iron-oxo species, called Compound I (Cpd I), which is formed after entrance of the substrate to the active site. We explored the features of Cpd I using hybrid quantum mechanical/molecular mechanical calculations on various models that are either substrate-free or containing one and two molecules of diazepam as a substrate. Mössbauer parameters of Cpd I were computed. Our major finding shows that without the substrate, Cpd I tends to elongate its Fe-S bond, localize the radical on the sulfur, and form hydrogen bonds with A305 and T309, which may hypothetically lead to Cpd I consumption by H-abstraction. However, the positioning of diazepam close to Cpd I, as enforced by the effector molecule, was found to strengthen the NH---S interactions of the conserved I443 and G444 residues with the proximal cysteinate ligand. These interactions are known to stabilize the Fe-S bond, and as such, the presence of the substrate leads to a shorter Fe-S bond and it prevents the localization of the radical on the sulfur. This diazepam-Cpd I stabilization was manifested in the 1W0E conformer. The effector substrate did not influence Cpd I directly but rather by positioning the active substrate close to Cpd I, thus displacing the hydrogen bonds with A305 and T309, and thereby giving preference to substrate oxidation. It is hypothesized that these effects on Cpd I, promoted by the restrained substrate, may be behind the special metabolic behavior observed in cases of multiple substrate binding (called also cooperative binding). This restraint constitutes a mechanism whereby substrates stabilize Cpd I sufficiently long to affect monooxygenation by P450s at the expense of Cpd I destruction by the protein residues.
1. Introduction
Cytochrome P450 (P450) proteins form a family of mono-oxygenases heme containing enzymes that oxidize a variety of chemical compounds in microorganisms, plants and mammals. 1,2 P450s are versatile catalysts of chemical reactions, such as hydroxylation, epoxidation or heteroatom dealkylations. 3 The human Cytochrome P450 3A4 (CYP3A4) enzyme is responsible for the oxidation of about 50% of all drugs taken orally, 4 and is therefore of major importance in detoxification of drugs and xenobiotics. CYP3A4 oxidizes a broad range of substrates, 4 utilizing different mechanistic and metabolic paths for a given substrate, 5,6 and it is able to accommodate a few substrate molecules (usually two, but sometimes also three) in its fairly large active site, exhibiting thereby atypical kinetics. 7,8 This multiple substrate binding has been referred to also as cooperative binding (CB) and we therefore use this latter term in the paper. CB is responsible for the generally observed rate enhancement of metabolism of a substrate molecule, when an additional molecule, socalled “effector” (can be either identical to or different than the first), undergoes binding to the enzyme. This atypical steady state kinetics has been observed in the metabolism of several substrates by CYP3A4, and is typified by the sigmoidal reaction velocity vs. substrate concentration plots.7,9 Our work focuses on the impact of binding two molecules of diazepam (a substrate and an effector7) on the properties of CYP3A4 Cpd I.
Though some studies have alluded to the link between the reactivity and the ability of CYP3A4 to accommodate multiple substrates within its active site, 10 still not much is known about the effect of CB on the reactivity. More so, spectroscopic evidence,9g,11 suggest that in some cases, the effector binds to a site distant from the heme moiety, 11 while in other cases, with substrates similar to diazepam, 9g both the active and effector substrates are nestled inside the active site of CYP3A4. Here, we have chosen to focus on the effect of CB of diazepam when both the substrate and the effector are bound within the active site, with an aim of answering a key question: Does CB influence the electronic structure and geometry of the active species of the enzyme and, if so, to what extent, and what are the hypothetical reactivity consequences of that?
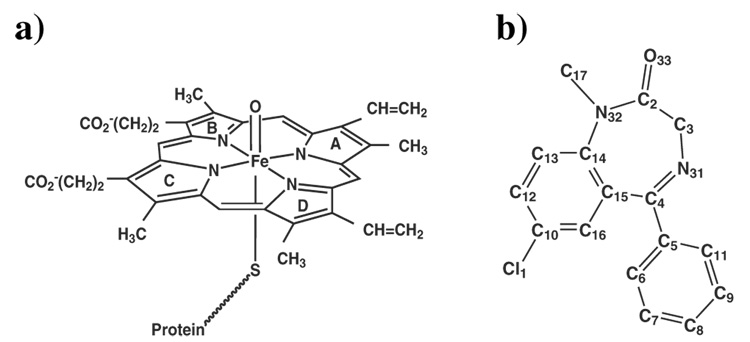
The oxidation reaction of a substrate by CYP3A4 occurs at the hydrophobic core of the protein, via a catalytic cycle, which is thought to be common to all P450s. 12–14 The active species, so-called Compound I (Cpd I) is a ferryl (FeIV)-oxo-π porphyrin cation radical (Figure 1a) with three unpaired electrons that occupy two π* orbitals and a porphyrin a2u-sulfur mixed orbital. Cpd I possesses closely lying doublet and quartet spin states, denoted as 2A2u and 4A2u respectively, 12,13 and is considered to be the species that activates chemical bonds in the substrate, e.g., by hydrogen abstraction, thereby leading to its eventual oxidation. Being one of the most reactive oxidative species in nature, Cpd I of P450s is presumably also unstable and has so far eluded detection in situ in the native cycle of the enzyme. Davydov et al.14a,b showed by ENDOR and EPR spectroscopic studies that even under cryogenic conditions, Cpd I of CYP101 (CYPcam) does not accumulate in the cycle. However, the solvent kinetic isotope effect, observed by the same group14c in the decay of the precursor ferric-hydroperoxide species, gave strong indirect evidence that Cpd I was formed in a proton assisted process. The only experimental evidence for Cpd I formation in P450 is provided from experiments whereby the species is generated using oxygen surrogate donors, e.g., in CYPcam , in CYPBM3 and in CYP119.15 More detailed information comes from structural characterization of Cpd I of the related thiolate enzyme chloroperoxidase (CPO-I) by Green et al.16 using X-ray absorption spectroscopy, and from rapid freeze quench ENDOR study of CPO-I17 which shows that the species contains a porphyrin π-cation radical species. Since CPO performs hydroxylation and epoxidation reactions similarly to P450s18 and having a similar spectroscopic profile to those of P450s,19 CPO Cpd I is suggestive of the presence of Cpd I in P450s. Thus, CYP3A4 Cpd I is going to be our target species for probing the effects of CB on reactivity. Moreover, the recent report on dramatic effects of the substrates in CYPcam on the lifetime of the last species seen in the cycle,14b,c further highlights the need to elucidate the roles of substrates in P450 isozymes, in general, and of multiple-substrate binding in CYP3A4, in particular.
Figure 1.
Structures of: a) the active oxygen species of CYP3A4, Cpd I, b) The drug diazepam (hydrogen atoms are deleted).
Among the possible tools to tackle these problems, density functional theory (DFT) is the most practical,12,13,20–22 and its hybrid method with molecular mechanics (MM), hence QM(DFT)/MM,12,23–31 is the most suitable combination for exploring electronic as well as steric effects due either to the residues of the protein in the active site or to the presence of the substrate therein. The results of QM(DFT)/MM studies and appropriately designed QM ones,31–34 show good agreement with experimental data16,17,35–37 on a variety of Cpd I species. This agreement involves the calculated23– 25,30–34 energy proximity of the doublet and quartet spin states, as is commonly observed in Cpd I species,35–37 the structural parameters,23–25 the spin density distribution23–25,33,34 and the Mössbauer spectroscopic parameters,30 which resemble the same parameters observed for the known Cpd I species of the analogous CPO enzyme.16,17 All these results show that QM(DFT)/MM, and specifically the B3LYP functional, provide credible tools for our problem. As such, our study will rely on QM(B3LYP)/MM to probe the effect of the diazepam substrate in CB and in the absence of CB on the properties of CYP3A4 Cpd I, with special focus on the sensitive parameters: the Fe-S bond length and the amount of spin density on the sulfur.17,25,33 We shall carry out also Mössbauer calculations of CYP3A4 Cpd I and compare the value to experimental and computational data.30,37
To date, there are six X-ray structures of CYP3A4 38–40 deposited in the PDB 41 that may serve as ideal starting points for QM/MM studies. Both progesterone and erythromycin, oxidized by CYP3A4, appear in their crystal structures bound to CYP3A4 in an orientation not favorable for oxidation. Thus, in order to gain information about the influence of the substrate molecule in a CB position caused by the effector molecule, two diazepam substrates were docked in silico inside the active site of CYP3A4. The enzymes unbound and bound to one and two diazepam molecules 7,42–43 were QM/MM optimized. The second diazepam, the effector molecule, was added in order to examine if the mere presence of a second substrate molecule affects Cpd I, or is it the distance of the first substrate molecule to Cpd I that makes the difference. As a way of probing the features of Cpd I, we have studied the behavior of the system along time using molecular dynamics (MD), and performed QM(B3LYP)/MM calculations for a few snapshots in different times along the equilibrium trajectory.
2. Computational methods
2.1. System setup
CYP3A4 structure was taken from the PDB 41 (codes 1TQN and 1W0E). Modeling the missing residues, addition of hydrogens, titratable groups, Cpd I and diazepam coordinates were handled as before. 44 The protein was solvated in a 49Å radius sphere of explicit water shell containing about 13700 TIP3 45 water molecules. The force field for the heme active species Cpd I was obtained from previous QM/MM calculations on Cpd I by Schöneboom et al. 25 Molecular Dynamics (MD) simulation of CYP3A4 was carried out in complex with a substrate, diazepam (Figure 1b). Diazepam was docked using the PatchDock 46 software. The docking of diazepam was based on pervious experimental work of point mutations that hindered diazepam binding to CYP3A4, 47 and on a favorable orientation for hydroxylation of the C3 carbon hydrogens (Figure 1b).
2.2. System dynamics
Two MD runs were performed here including a substrate-free and a substrate-bound enzyme. The force field used for the dynamics was from CHARMM27 48 along with the CHARMM27 protein nucleic acid topology and parameter files. The runs were performed according to Langevin dynamics with spherical boundary conditions with the CHARMM software. 49 A reaction zone including the inner 40Å water sphere was treated with the Newton equation whereas an outer 9Å water sphere was treated with the Langevin dynamics. The system energy in each run was minimized by the CHARMM software, with a decreasing harmonic force constraint on the enzyme with a tolerance gradient of 10kcal/(mol Å). The minimized system was then gradually heated to 300K and equilibrated for 0.5ns. The NVT dynamics was carried out for 0.5ns keeping the temperature at 300 K. We used a time step of 1fs. The SHAKE algorithm was applied in order to fix all bond lengths involving hydrogen atoms. We used a final non-bonded interaction cutoff radius of 12Å. Random snapshots were taken from the substrate-free MD, and the snapshots with favorable orientation for hydroxylation were selected from the substrate dynamics. The diazepam molecule was constrained with a harmonic force constant of 1kcal/(mol Å2) in order to keep the substrate in a hydroxylation position. Each snapshot was minimized for 400 steps prior to QM(B3LYP)/MM optimization.
2.3. QM/MM methodology
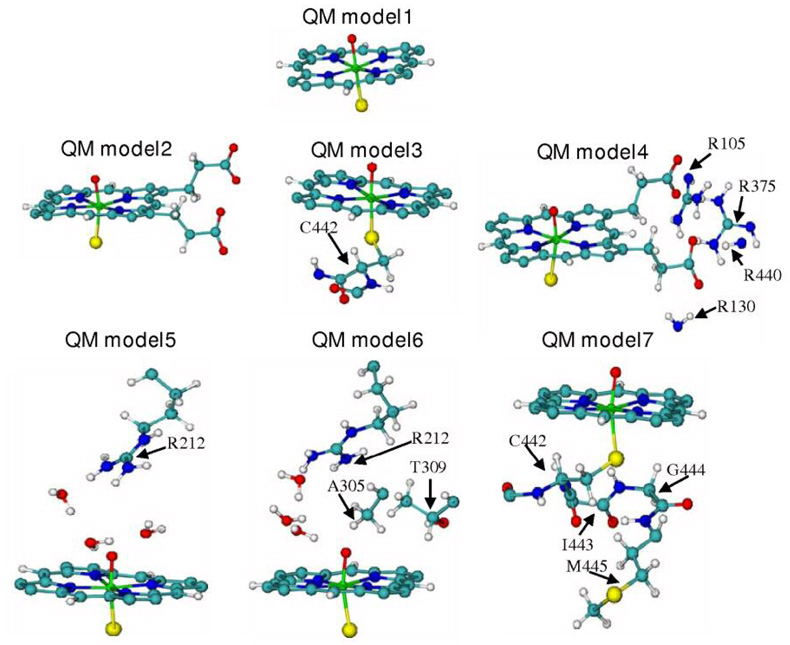
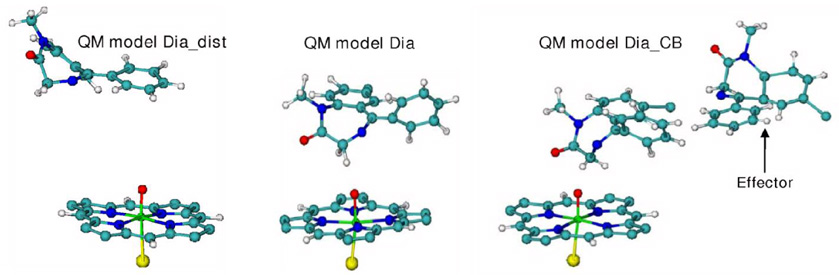
Selected snapshots from the MD trajectories were used as starting geometries for the QM(B3LYP)/MM optimization. The QM(B3LYP)/MM calculations of the substrate-free system involved seven truncated Cpd I models (depicted in Figure 2) that define these QM subsystems. In addition, as shown in Figure 3, QM(B3LYP)/MM optimizations were carried out on Cpd I with a docked diazepam molecule (Dia and Dia_dist models) inside the active site of CYP3A4 and with two diazepam molecules (Dia_CB model). The protein system was divided into two regions, one is the QM region and the other is the MM region. We used the B3LYP/CHARMM22-27 electrostatic embedding Hamiltonian, in which the fixed MM charges were included in the one-electron Hamiltonian of the QM calculation. The QM(B3LYP)/MM electrostatic interactions were evaluated from the QM(B3LYP) electrostatic potential and the MM atomic charges. Hydrogen link atoms with the charge shift model were used to treat the QM(B3LYP)/MM boundary. All QM(B3LYP)/MM calculations were carried out using the ChemShell 50 package. The program TURBOMOLE 5.7.1 51 was employed for the QM part treated with DFT by applying the B3LYP functional. The CHARMM22/27 force fields were run under the DL-POLY program and were used to compute the MM regions.
Figure 2.
The substrate-free QM models used for the QM(B3LYP)/MM optimization of Cpd I: QM model 1 includes the porphyrin ring, Fe, O and the S of the conserved C442. QM model 2 adds to model 1 the two propionate side chains. QM model 3 includes QM model 1 + the atoms of C442. QM model 4 includes QM model2 + arginines that maintain salt bridges to the propionate moieties. QM model5 includes QM model 1 + R212 and three water molecules that form a network of hydrogen bonds between R212 and the active oxygen. QM model 6 includes QM model 5 + A305 and T309. QM model 7 includes QM model 3 + I443, G444 and M445 atoms.
Figure 3.
The substrate-bound QM models used for the QM(B3LYP)/MM optimization of Cpd I. Dia_dist QM model (left) is QM model1 with a docked diazepam molecule, distal from Cpd I. Dia QM model (center) includes QM model1 and a docked diazepam molecule in the proximally restrained position (due to CB). Dia_CB model (right) has one diazepam in the in the same conformation as in the middle system and another diazepam molecule (the effector) in the MM subsystem.
A few basis sets were used in the QM(B3LYP) calculations. The basis set, denoted B0, includes an effective core potential (ECP) coupled with the LACVP valence basis set for the iron and the 6–31G basis set for all other atoms in the QM region. This basis set was used for the QM(B3LYP)/MM optimization of the QM regions. The algorithm used for the optimization was a hybrid delocalized internal coordinate optimizer (HDLC). A larger basis set, denoted B1, includes ECP and the LACVP basis set for the Iron, the 6–31G* basis set for atoms N, O and S and the 6–31G basis set for the rest of the atoms. This basis set was used in the single point calculations on the QM/MM optimized structures.
Gas phase QM(B3LYP) optimizations of Cpd I were performed with TURBOMOLE using the Relax module (QM gas phase results are shown in the SI document section 5 and section 6). Theoretical Mössbauer parameters were determined with the ORCA 52 program from single point B3LYP calculations (at the geometries obtained after QM(B3LYP)/MM optimization). The isomer shift was obtained from the electron density at the iron nucleus. 53 Iron was described by the triply polarized core properties basis set CP (PPP) 54 and the other atoms by the SV(P) basis set 55 with the inner s-functions left uncontracted. The MM point charges from the QM(B3LYP)/MM optimization were included in these calculations to probe the effect of the protein environment. The calculations were performed for the 4A2u and 2A2u states.
3. Results and discussion
Seven substrate-free QM models of Cpd I were optimized (see Figure 2). QM model 1 includes the porphyrin ring, the sulfur of C442 that is proximally ligated to the iron ion of the heme and the oxo ligand of Cpd I that is distally ligated to the iron. Four snapshots of this QM model, including the initial structure, and snapshots after 137ps, 450ps and 500ps of molecular dynamics, were taken as QM(B3LYP)/MM starting points denoted as QM model 1_0, QM model 1_137, QM model 1_450 and QM model 1_500 respectively. For the other QM models 2–7, described in Figure 2, the starting point for QM(B3LYP)/MM calculations was the minimized initial structure of PDB code 1TQN.
The influence of CB of diazepam on Cpd I was tested using the models depicted in Figure 3. In the first model, labeled Dia_dist, the substrate is quite distant from Cpd I as established previously by a 6ns MD with no harmonic constraints.44 The second model, denoted as Dia QM, includes a docked diazepam substrate, and was MD simulated in this work for 0.5ns with a harmonic force constraint acting on diazepam and restraining it close to Cpd I as it would be in CB. Five snapshots of this subsystem, including the initial structure and snapshots after 80ps, 237ps and 493ps of MD were taken for QM/MM starting points and were denoted as Dia_0 (1TQN and 1W0E conformers), Dia_80, Dia_237 and Dia_493 respectively. In the third model in Figure 3 (right hand side structure), we added to the MM subsystems of Dia_0 (1W0E conformer) and Dia_493 (1TQN conformer) an effector diazepam molecule and generated the models labeled as Dia_CB_0 and Dia_CB_493 respectively. The second and third models correspond to a restrained diazepam molecule placed close to Cpd I as found in the CB study.
3.1 Substrate-free Cpd I: QM(B3LYP)/MM optimization
3.1.1 Cpd I spin states energies
The 4A2u and 2A2u spin states of Cpd I were calculated, for each of the QM region in Figure 2. The relative energies of these states are reported in Table 1 where ΔEQM represent the QM-energy within the QM/MM calculations, while ΔEtot represent the total QM/MM energy difference, according to equation 1.
| (1) |
A negative (positive) ΔE indicates that the 4A2u (2A2u) spin state is the ground state.
Table 1.
ΔE Values for the 4A2u and 2A2u states in different QM models and snapshots
| opt B0 ΔEQMa | opt B0 ΔEtotb | sp B1 ΔEQMc | Sp B1 ΔEtotd | |
|---|---|---|---|---|
| QM model1_0 | −0.02 | 0.23 | −0.03 | 0.24 |
| QM model1_137 | −0.01 | 0.17 | −0.01 | 0.19 |
| QM model1_450 | −0.01 | 0.19 | 0.04 | 0.20 |
| QM model1_500 | −0.04 | −0.01 | −0.03 | 0.03 |
| QM model2 | 0.04 | −0.05 | 0.02 | −0.03 |
| QM model3 | 0.18 | 0.15 | 0.21 | 0.15 |
| QM model4 | 0.30 | 0.14 | 0.29 | 0.15 |
| QM model5 | 0.20 | 0.13 | 0.24 | 0.18 |
| QM model6 | 0.22 | 0.11 | 0.28 | 0.12 |
| QM model7 | 0.30 | 0.15 | 0.22 | 0.19 |
Energy difference between the QM subsystem of 4A2u and 2A2u states in kcal/mole after optimization with the B0 basis set.
Total energy difference (QM+MM energies) between 4A2u and 2A2u states in kcal/mole after optimization with the B0 basis set.
Energy difference between the QM subsystem of 4A2u and 2A2u states in kcal/mole after single point calculation with the B1 basis set.
Total energy difference (QM+MM energies) between 4A2u and 2A2u states in kcal/mole after single point calculation with the B1 basis set.
As can be seen from Table 1, in most of the QM models ΔE does not exceed 0.3 kcal/mole. This closeness of the two spin states reflects the weak interactions between the two electrons located on the Fe-O moiety and the third electron delocalized over the sulfur-porphyrin moieties. Notably, in the great majority of the QM models, the doublet state is the ground state with both basis sets (B0 and B1). This result is in accord with experimental data on the related CPO Cpd I pointing to a doublet ground state. 35,37,56–57
3.1.2 Electronic structure of Cpd I
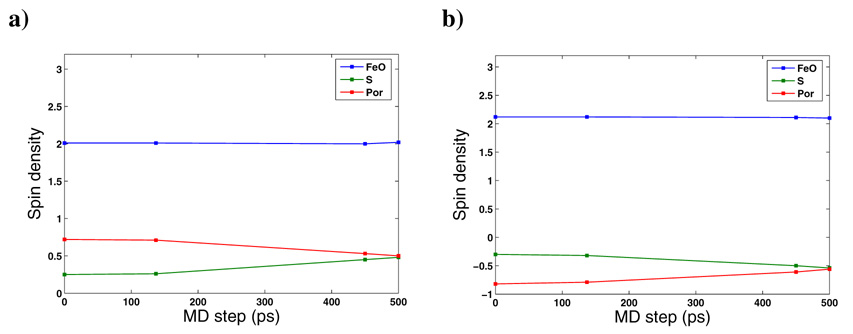
The net spin densities of Cpd I atoms for QM region 2 to 7 are listed in Table 2. Spin densities along the 0.5ns MD simulation are shown in Figure 4. In general, the spin densities on the Fe and O atoms are not affected by the size of the QM subsystem, the spin state, the basis set, or the starting conformation selected. The combined FeO spin densities are 2.01/2.11 for the 4A2u/ 2A2u spin states and it accounts for the two electrons within the two singly occupied π* orbitals. The third unpaired electron is located in QM model 1_0, QM model1_137 and QM models 2–7, mainly on the porphyrin ring with an average spin density of ~0.25 on the sulfur. However, as can be seen from Figure 4 for QM model 1, the spin density increases with time, and after 0.5ns of MD simulation the spin density on the sulfur becomes equal to the spin density on the porphyrin ring, each sharing half of the third unpaired electron. The same spin density on the sulfur was observed by Bathelt et al.29 for an MD equilibrated CYP3A4 conformer. Thus, the third unpaired electron can delocalize between the porphyrin ring a2u orbital and the ps orbital of sulfur and this electron delocalization varies over time as shown in our MD simulation.
Table 2.
Spin densities in the 4A2u (2A2u) states of Fe, S, O, and porphyrin atoms in QM models 2 to 7 after QM(B3LYP)/MM optimization.
| Fe | O | S | Porphyrin | |
|---|---|---|---|---|
| QM model2 | 1.14 (1.28) | 0.88 (0.83) | 0.25 (−0.30) | 0.72 (−0.82) |
| QM model3 | 1.13 (1.29) | 0.88 (0.83) | 0.28 (−0.34) | 0.69 (−0.78) |
| QM model4 | 1.13 (1.28) | 0.80 (0.83) | 0.28 (−0.32) | 0.69 (−0.79) |
| QM model5 | 1.17 (1.33) | 0.84 (0.79) | 0.22 (−0.27) | 0.74 (−0.86) |
| QM model6 | 1.17 (1.31) | 0.85 (0.80) | 0.19 (−0.24) | 0.76 (−0.88) |
| QM model7 | 1.14 (1.29) | 0.88 (0.82) | 0.28 (−0.33) | 0.69 (−0.78) |
Figure 4.
Spin densities of FeO, S and porphyrin atoms (Por) along the 0.5 MD of QM model 1 a) in the 4A2u spin state b) in the 2A2u spin state.
Another issue concerns the contributions of the two propionic moieties and the salt-bridging arginines to the electronic structure of Cpd I, which were investigated using QM models 2 and 4. No electron delocalization was found on the propionic moieties in accordance with prior QM/MM Cpd I optimizations in bacterial P450s25,31 and in the human CYP2C9.29 In addition, there was no influence of R212 and related hydrogen bonded water molecules or of the representation of C442 and the close residues on Cpd I electronic structure.
3.1.3 Geometry of Cpd I
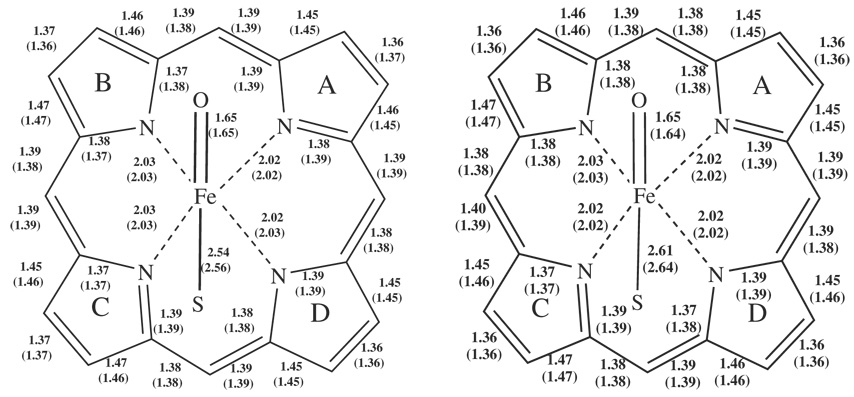
Table 3 lists key bond distances of Cpd I for QM models 2 to 7 after QM(B3LYP)/MM optimization. All the distances are in reasonable agreement with the CPO EXAFS bond lengths.16 Figure 5 depicts the bond distances of Cpd I at the starting point (QM model1_0) and at the end point (QM model1_500) of the MD simulation after QM/MM optimization. Clearly, the Fe-O bond and the Fe-N bonds to the porphyrin ring were not influenced by the QM subsystem or MD simulation. However, the Fe-S bond length was found to be more flexible and undergoes elongation from 2.52Å up to 2.64Å. The longest Fe-S bond lengths were obtained at the final snapshot of the MD simulation (QM model 1_500) and were elongated by 0.1Å.
Table 3.
Key Cpd I bond distances (in Å) of the 4A2u (2A2u) states of Cpd I atoms for QM models 2 to 7 (all are 0 snapshots) after QM(B3LYP)/MM optimization.
| Fe-S | Fe-O | Fe-Navga | |
|---|---|---|---|
| QM model2 | 2.54 (2.56) | 1.65 (1.65) | 2.02 (2.02) |
| QM model3 | 2.54 (2.56) | 1.65 (1.65) | 2.02 (2.02) |
| QM model4 | 2.55 (2.56) | 1.65 (1.65) | 2.02 (2.02) |
| QM model5 | 2.59 (2.54) | 1.65 (1.65) | 2.02 (2.02) |
| QM model6 | 2.52 (2.54) | 1.65 (1.65) | 2.02 (2.02) |
| QM model7 | 2.54 (2.56) | 1.65 (1.65) | 2.02 (2.03) |
Average distance between Fe atom and NA to ND atoms
Figure 5.
Cpd I bond distances in Å of the 4A2u (2A2u) states after QM(B3LYP)/MM optimization in: a) QM model1_0 b) QM model1_500.
3.1.4 Relationships between geometry and electronic configuration
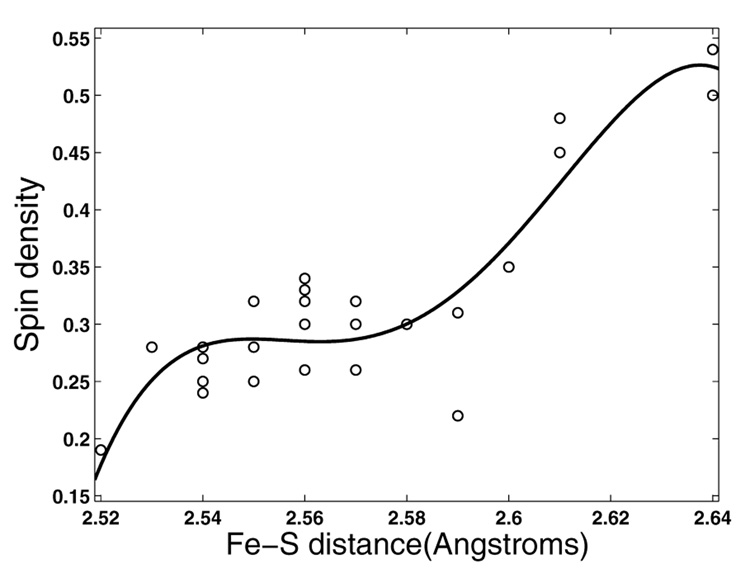
Figure 6 shows a plot of spin density vs. Fe-S bond length for all the QM(B3LYP)/MM subsystems including the snapshots taken from the dynamics with the substrate, diazepam. It seems that for a value of the Fe-S bond <2.61Å, the spin density on the sulfur is ~0.25, but above 2.61Å there is an increase in the spin density to a value of ~0.5 of spin density. The points of 28 Fe-S QM/MM optimized bond lengths, were fitted to a 4th degree polynomial with an R2 of 0.82 (using Cftool of MatLab). Even if the fitting is not statistically robust, the general tendency of the spin density of the sulfur is to be higher as the Fe-S bond length34 increases and there exists a certain length threshold from which the third unpaired electron is dispersed equally between the sulfur and the porphyrin ring.
Figure 6.
Spin densities vs. Fe-S bond lengths of all QM subsystems. The curve fitting to a 4th degree polynomial resulted in R2=0.82.
At longer bond lengths the spin will gradually be localized on the sulfur as a QM-only study showed earlier.34 The trend is reasonable, since the overlap between the a2u and ps orbitals is expected to decrease exponentially as the Fe-S bond length increases. Therefore, in some critically long bond lengths, it is reasonable that the spin density will resemble the gas phase situation and be more concentrated on the sulfur.
Table 4 and Table 5 compare the Fe-S bond lengths and sulfur spin densities, respectively, of several P450 isoforms calculated in the QM/MM study by Bathelt et al.29 with our QM model1_0 and QM model_500 subsystems. In Table 5 the QM subsystems were MD simulated whereas in the Table 4 they were close to the initial PDB structures. It seems that in the human CYP3A4 isoform, in both 1W0E and our 1TQN conformers the Fe-S bond is very flexible and can reach 2.64Å in length with a concomitant increase in the spin density on the sulfur. This flexibility of the substrate-free models may indicate a propensity of the weak Fe-S bond to cleave and perhaps destroy Cpd I in a rather short time.
Table 4.
Comparison between Fe-S distances and sulfur spin densities (ρS) in the 4A2u (2A2u) states of different P450 isoforms obtained from previous QM(B3LYP)/MM optimizations29 and the QM model1_0 snapshot. Fe-S bond length is in Å.
| Fe-S, Å | ρS | |
|---|---|---|
| 3A4 model 1_0a | 2.54 (2.56) | 0.25 (−0.30) |
| 2C9b | 2.55 (2.55) | 0.37 (−0.39) |
| 3A4 (1TQN)b | 2.56 (2.55) | 0.45 (−0.47) |
| 3A4 (1W0E)b | 2.53 (2.53) | 0.37 (−0.39) |
| 2B4 rabbitb | 2.52 (2.51) | 0.32 (−0.33) |
| CYPcamb | 2.54 (2.53) | 0.20 (−0.20) |
This study
Ref. 29
Table 5.
Comparison between Fe-S distances and sulfur spin densities (ρS) in the 4A2u (2A2u) states of different P450 isoforms obtained from previous QM(B3LYP)/MM optimizations29 and the QM model1_500 snapshot. Fe-S bond length is in Å. All isoforms are MD equilibrated.
| Fe-S, Å | ρS | |
|---|---|---|
| 3A4 model1_500a | 2.61 (2.64) | 0.48 (−0.54) |
| 3A4 (1W0E)b | 2.62 | 0.50 |
| 2B4 rabbitb | (2.62) | (−0.41) |
This study
Ref. 29.
3.2 QM(B3LYP)/MM results in the presence of diazepam
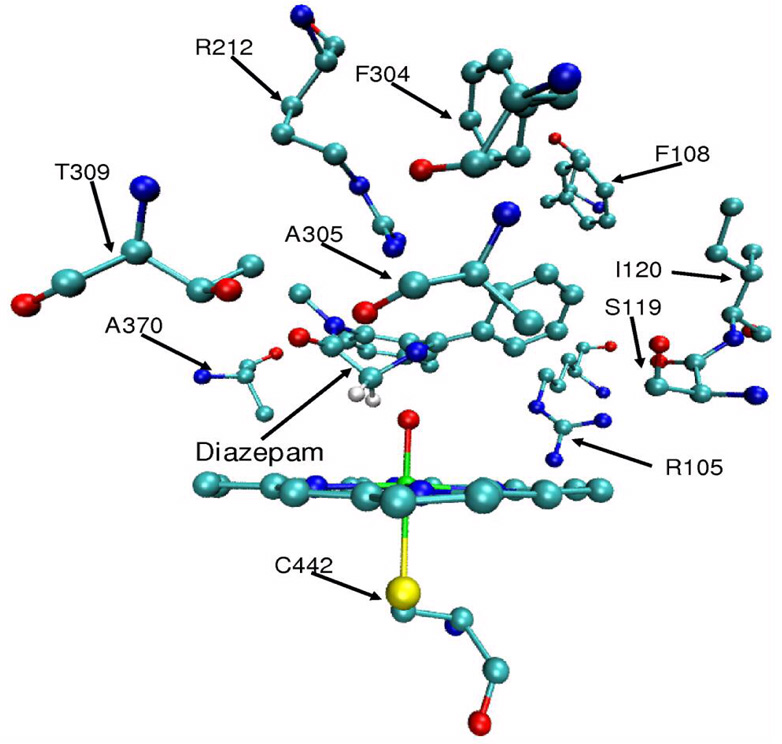
Diazepam undergoes extensive metabolism by CYP3A443 via several paths, including N-demethylation of N32 and hydroxylation of C3. Diazepam was docked inside the active site of CYP3A4 in a favorable orientation for hydroxylation with the C3 hydrogens positioned in proximity to the Cpd I active oxygen (Figure 7). The oxidized substrate interacts with the I helix (residues F304, T309), the BC loop (residues R105, F108, S119, I120) and R212 of the FF' loop (Figure 7). This docking was the starting point for the MD simulations of both unrestricted and restricted diazepam. Previous single substrate unrestricted MD simulations resulted in a long distance between the active species and the diazepam hydrogens to be abstracted. A snapshot with such a distance (Dia_dist QM model) would behave similarly to the substrate-free snapshots. On the other hand, the substrate-restricted MD (Dia QM model) simulates CB, where the effector conserves a favorable orientation of the oxidized substrate for hydroxylation. An effector added to the MM region of the Dia model (Dia_CB QM model) is therefore optimized in order to test the direct impact of the effector on Cpd I properties.
Figure 7.
Diazepam docked inside the active site of CYP3A4. Shown are close contacting residues of the protein with diazepam. Apart from diazepam the structure is without hydrogen atoms.
Table 6 describes the relative energies between 4A2u and 2A2u states (as in Table 1) of all snapshots containing diazepam (Dia_dist, Dia and Dia_CB). Table 7 presents the spin densities of Cpd I atoms in all Dia_dist, Dia and Dia_CB models. Table 8 compares the Fe-S bond lengths with the corresponding sulfur spin densities of different substrate bound QM/MM optimized snapshots.
Table 6.
ΔE between the 4A2u and 2A2u states of Cpd I obtained with the Dia_dist, Dia and Dia_CB QM models in several snapshots.
| opt B0 ΔEQMa | opt B0 ΔEtotb | ||
|---|---|---|---|
| 1 | Dia_dist1c | −0.10 | −0.06 |
| 2 | Dia_dist2c | −0.10 | 0.13 |
| 3 | Dia_0 | −0.06 | −0.03 |
| 4 | Dia_80 | −0.31 | −0.05 |
| 5 | Dia_237 | −0.03 | −0.03 |
| 6 | Dia_493 | −0.13 | −0.15 |
| 7 | Dia_0 (1W0E) | −0.02 | −0.04 |
| 8 | Dia_CB_493 | −0.24 | −0.15 |
| 9 | Dia_CB_0 (1W0E) | 0.31 | 0.18 |
Energy difference between the 4A2u and 2A2u states in kcal/mole after optimization with the B0 basis set.
Total energy difference (QM+MM energies) between 4A2u and 2A2u states in kcal/mole after optimization with the B0 basis set.
QM(B3LYP)/MM optimization with diazepam after 5.5ns (Dia_dist1) and 6ns (Dia_dist2) of the previous unrestricted MD.
Table 7.
Spin densities of atoms in Cpd I in the 4A2u (2A2u) states obtained by QM(B3LYP)/MM optimization with the Dia_dist, Dia and Dia_CB QM models and several snapshots.
| Fe | O | S | Por | ||
|---|---|---|---|---|---|
| 1 | Dia_dist1a | 1.06 (1.21) | 0.95 (0.90) | 0.35 (−0.39) | 0.64 (−0.73) |
| 2 | Dia_dist2a | 1.13 (1.30) | 0.88 (0.82) | 0.32 (−0.38) | 0.65 (−0.75) |
| 3 | Dia_0 | 1.07 (1.23) | 0.93 (0.88) | 0.28 (−0.33) | 0.69 (−0.79) |
| 4 | Dia_80 | 1.08 (1.25) | 0.93 (0.86) | 0.28 (−0.32) | 0.65 (−0.75) |
| 5 | Dia_237 | 1.09 (1.25) | 0.92 (0.87) | 0.25 (−0.29) | 0.71 (−0.83) |
| 6 | Dia_493 | 1.06 (1.22) | 0.95 (0.89) | 0.29 (−0.32) | 0.68 (−0.79) |
| 7 | Dia_0 (1W0E) | 1.04 (1.19) | 0.97 (0.91) | 0.30 (−0.34) | 0.68 (−0.77) |
| 8 | Dia_CB_493 | 1.03 (1.19) | 0.98 (0.92) | 0.29 (−0.33) | 0.67 (0.78) |
| 9 | Dia_CB_0 (1W0E) | 1.11 (1.25) | 0.90 (0.85) | 0.23 (−0.27) | 0.77 (−0.84) |
QM(B3LYP)/MM optimization with diazepam after 5.5ns (Dia_dist1) and 6ns (Dia_dist2) of the previous unrestricted MD.44
Table 8.
Comparison between Fe-S bond lengths and sulfur spin densities in the 4A2u (2A2u) states obtained from previous QM(B3LYP)/MM optimizations29 of CYP2C9 complexed with substrates and QM models Dia_dist and Dia_493.
3.2.1 QM(B3LYP)/MM results for Cpd I with a distal diazepam
Previosuly,44 diazepam was found to drift away from the CYP3A4 active site with increasing time in MD simulations, reaching a distance of ~9Å from the Cpd I atoms. Using QM/MM we optimized a snapshot after 5.5ns and the final snapshot of an unrestricted MD simulation where diazepam was placed distal to Cpd I in a non oxidizable orientation (Dia_dist1 and Dia_dist2 respectively). Along the MD trajectory it was observed that up to ~5.5ns the methyl group of T309 remains close to the oxo atom of Cpd I (2.82Å). Subsequently, there occurred a complete drift of the diazepam substrate and a movement of the flexible R212 outside of the active site that turned away the T309 methyl from the oxo atom, allowing now the hydroxyl moiety of T309 to form a close hydrogen bond (1.73Å) with the oxo atom. This hydrogen bond persisted until the end of the MD. The presence of a substrate molecule did not influence the energy difference in a major way as seen in the substrate free snapshots (Table 6, entries 1 and 2). The distal diazepam did not change the spin densities of the Fe and O atoms and the sum of their spin densities remained around 2 spin units. The spin density of the sulfur dropped slightly to ~0.4 (Table 7, entries 1 and 2) especially in the 2A2u state. The Fe-S bond length was the same as in the substrate free snapshots, 2.60–2.63Å (Table 8, entries 2 and 3). This bond flexibility was conserved (Table 8) with a slight stability in Dia_dist2 due to the T309 OH hydrogen bond to the oxo of Fe=O.
3.2.2 QM(B3LYP)/MM results for Cpd I with a proximally situated diazepam
In order to inspect the influence of a substrate on Cpd I properties, as in CB, we positioned diazepam in a productive conformation and prevented its escape. An 0.5ns MD simulation was performed with harmonic constrain on the substrate and QM(B3LYP)/MM optimized several snapshots along the MD trajectory. These Dia QM model optimizations simulate CB where the effector substrate stabilizes the close proximity of the active substrate to Cpd I. The QM/MM optimizations of diazepam resulted in a hydrogen bond elongation of 0.2Å between the diazepam hydrogen to be abstracted and the Cpd I oxo atom after ~0.5ns of MD (Dia_493) to a value of 2.2Å of distance. In the 1W0E conformer optimization, this bond showed a 0.7Å of elongation and resulted in a distance of 2.71Å between the diazepam hydrogen and the oxo atom. According to entries 3 to 7 in Table 6, there is negligible energy difference between the two spin states of Cpd I. There is no change in Fe and O spin densities. Similarly, the sulfur spin density remains around ~0.3 and does not rise to 0.5 as observed in the substrate-free MD simulation (Table 7, entries 3 to 7). The third unpaired electron was kept largely on the porphyrin ring with a spin density of ~0.7. Thus, the substrate seems to influence the distribution of the third unpaired electron in CYP3A4 Cpd I. Table 8 (entries 1 to 3) indicates that the elongation of the Fe-S bond observed in the substrate-free model after 0.5ns of MD was quenched by diazepam only when it was proximal to Cpd I. With the substrate in the vicinity of Cpd I after 0.5ns of MD the Fe-S bond length reach a maximum of 2.58Å. Table 8 (entries 4 and 5) also compares previous QM/MM results of substrate bound CYP2C9.29 When adding the substrate diclofenac to the CYP2C9 isoform, similar values were observed for the Fe-S bond length and sulfur spin density. Addition of ibuprofen resulted in the same Fe-S bond length but with elevated spin density on the sulfur. Apparently, the addition of a substrate stabilizes the Fe-S bond length and influence the delocalization of the third unpaired electron. This influence varies between substrates and between P450 isoforms due to a slight difference in the environment adjacent to the sulfur atom ligated to the heme.
3.2.3 QM(B3LYP)/MM results with active and effector diazepams
The addition of an effector to the MM region on top of the active substrate (Dia_CB QM model) revealed no change in spin state energy difference, electronic state and geometry of Cpd I in the 1TQN conformer (Table 6 and Table 7 entries 6 and 8). The sulfur spin density remained ~0.28, as in the substrate-restricted snapshots. The distance between the abstractable C–H bond and the oxo group of Cpd I bond was further elongated and resulted in 2.61Å. Thus, the effector exerted no effect on the active substrate in the 1TQN conformer.
The addition of an effector to the 1W0E conformer did not change the spin state energy difference. However, the effector pushed the substrate towards the oxo atom and the final diazepam hydrogen-oxo distance was 1.92Å that constitutes a decrease of 0.8Å in distance. This hydrogen oxygen distance shortening was coupled with a decrease in sulfur spin density in Cpd I (Table 7 entries 7 and 9) and a slight shortening of the Fe-S bond. The addition of the effector to the 1W0E conformer stabilized the QM energy by 2.14 kcal/mole and increased the energy in the 1TQN conformer in 2.70 kcal/mole. From all of the above, we observe CB in the 1W0E conformer and no CB in the 1TQN conformer as seen before in MD simulations.44 We conclude that the addition of the effector is not manifested in direct strong impact on the electronic structure of Cpd I but rather in indirect effects. Furthermore, we conclude that the positioning of diazepam proximal to Cpd I, according to our results, inhibits the elongation of the Fe-S bond and results in smaller sulfur spin density compared to the substrate free snapshots and the substrate distal snapshots (Dia_dist). When the effector substrate enters the active site, this increases the probability of finding the active substrate close to Cpd I, and thereby leading to a more geometrically robust Cpd I. The effector is important in P450 isoforms like CYP3A4, where the large binding site would have given the substrate too much dynamic freedom.
3.3 Sulfur H-bonding network and ligand influence
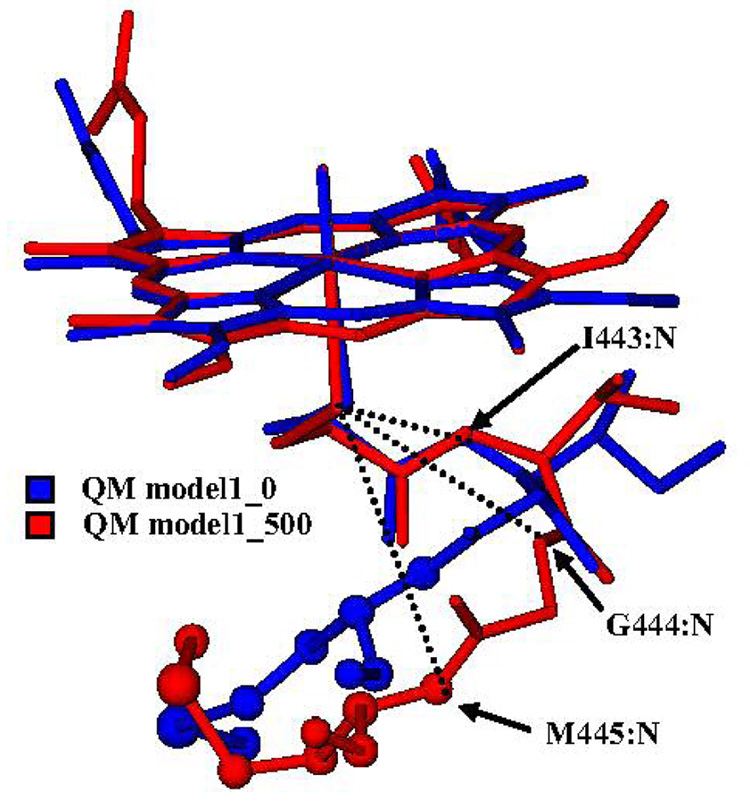
In order to find other structural determinants that influence the sulfur geometric and electronic character, we focused on the close environment of this atom. It was previously found that the proximal NH---S and C=O---S interactions that exist in CYPcam between the L358, G359 and Q360 backbone amides and the cysteine sulfur, stabilize the heme thiolate coordination, and regulate the redox potential of the heme iron.59 These residues in CYP3A4 correspond to I443, G444 and M445, respectively and all maintain NH---S interactions via their backbone amidic NH groups. Bathelt et al.29 who discussed the structural conservation of this hydrogen bonding network in different P450 isoforms, found little variance in the hydrogen bond distances. Figure 8 shows a superposition of the QM(B3LYP)/MM optimized starting and end points of the CYP3A4 substrate-free model during 0.5ns MD.
Figure 8.
Superposition of QM(B3LYP)/MM optimized Cpd I and residues C442, I443, G444 and M445 in the starting (blue) and end (red) point of the MD simulation without the substrate. In dotted lines are the conserved NH---S “hydrogen bonds” between the backbone amides of the optimized structure and the sulfur atom. The structures are without hydrogen atoms. The M445 is presented in CPK
The M445 residue exhibits flexibility and immediately after the starting point of the MDs it moves away from the heme plane by ~2Å. Addition of the substrate in a CB position did not change this movement. In Table 9 we listed NH---S distances of QM model 1 and the diazepam-containing models. Apart from the initial points in the substrate bound and substrate-free MDs (entries 1 and 7), M445 does not maintain hydrogen bonds with the cysteinate sulfur atom. In QM model 1 in snapshot 137 (Table 9, entry 2), the M445 NH---S distance is ~6.2Å and the sulfur spin density is 0.25 whereas in the QM model 1_450 (Table 9, entry 3) this distance is almost the same but the sulfur spin density is about 0.5. It seems that the fluctuations of M445 do not influence the sulfur electronic character. Yoshioka et al.59 showed by mutations, that the corresponding residue in CYPcam, Q360, increased the electron donation of the axial thiolate. Q360 maintains two hydrogen bonds with C357: one between the side chain hydrogen amide and the carbonyl oxygen of C357, and a second hydrogen bond between the backbone amide hydrogen and the C357 sulfur. Thus, Q360 exerts an electrostatic interaction on C357 in CYPcam and this can explain its influence on the sulfur electronic character. However, the hydrogen bond between the sulfur and the Q360 backbone hydrogen elongates after MD followed by QM(B3LYP)/MM optimization. 25
Table 9.
NH---S distances of the conserved hydrogen bonds between the backbone amides hydrogens of I443, G444 and M445 and the cysteinate sulfur atom. Bond lengths are in angstroms (4A2u (2A2u)).
| I443 NH---S | G444 NH---S | M445 NH---S | ||
|---|---|---|---|---|
| 1 | QM model1_0 | 3.36 (3.38) | 2.50 (2.49) | 3.30 (3.31) |
| 2 | QM model1_137 | 3.48 (3.49) | 2.69 (2.71) | 6.23 (6.22) |
| 3 | QM model1_450 | 3.12 (3.15) | 3.42 (3.42) | 6.07 (6.05) |
| 4 | QM model1_500 | 3.20 (3.22) | 3.44 (3.46) | 5.84 (5.84) |
| 5 | Dia_dist1a | 2.91 (2.92) | 2.89 (2.88) | 5.21 (5.19) |
| 6 | Dia_dist2a | 2.97 (2.98) | 2.91 (2.90) | 5.19 (5.18) |
| 7 | Dia_0 | 3.59 (3.58) | 2.65 (2.65) | 3.14 (3.13) |
| 8 | Dia_80 | 2.68 (2.72) | 3.01 (3.02) | 5.66 (5.64) |
| 9 | Dia_237 | 2.62 (2.63) | 2.96 (2.94) | 5.63 (5.64) |
| 10 | Dia_493 | 2.89 (2.91) | 2.67 (2.66) | 4.97 (4.95) |
QM(B3LYP)/MM optimization with diazepam after 5.5ns (Dia_dist1) and 6ns (Dia_dist2) of the previously unrestricted MD.44
The large flexibility of M445, in CYP3A4, manifested in all MDs is not associated with sulfur spin density variations. Coupled with the fact that in the initial structure the NH---S bond has an angle of 82°, which is not favored for a hydrogen bond we conclude that in contrast to CYPcam, the M445 residue in the CYP3A4 does not influence to a large extent the electronic structure of the thiolate ligand of Cpd I. As may be seen from Table 9 (entries 5 to 10), the addition of diazepam in proximal or distal positions relative to Cpd I resulted in shorter NH---S bonds, especially in the I443 NH---S and in the G444 NH---S hydrogen bonds. We therefore point at I443 and G444 as electronic structure modulating residues of the thiolate heme ligated atom. It appears therefore that the close contact of the substrate and Cpd I stabilizes the hydrogen bonding network surrounding the sulfur atom and thus leads to a shorter Fe-S bond of the active species, as observed in previous studies of model systems and CYPcam.24,24, 34
3.4 H-bonding network about the Fe=O Bond and the effect on the Fe-S bond
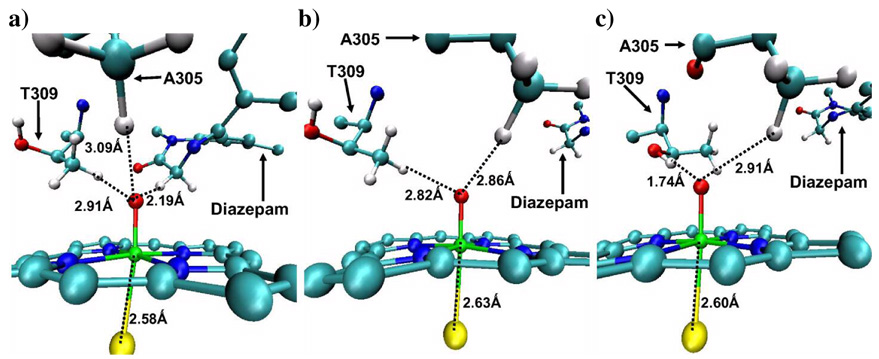
Figure 9 shows three situations: in (a) the diazepam is close to the Fe=O moiety of Cpd I, while (b) and (c), describe the situations where the substrate drifted away from Cpd I. There are a few interesting observations one can note: When diazepam is close to the oxo group, the H---O distances of T309 are longer and the Fe-S bond is shorter (Figure 9a). However, upon a drift of the diazepam away from Cpd I, T309 forms tighter hydrogen bonds to the oxo atom of Cpd I and the Fe-S bond gets longer (Figures 9b and 9c; see also the SI document). Thus, the effector places the active substrate closer to Cpd I, and for a longer period of time than in the absence of the effector, thereby indirectly stabilizing the H-bonding network around the thiolate ligand. The above-mentioned movement of T309 is facilitated by the rotation of the flexible R212 side chain, which hinders the approach of T309 toward the oxo moiety if it points downward to the heme. The likelihood of this rotation of R212 can be understood from two X-ray structures in PDB; in 1TQN, it points downward, while in 1W0E, it flips to make a salt bridge with E308. Interestingly, the methyl group of A305 was always close to the oxo moiety no matter where the substrate is in the active site, though not as close as the substrate’s C-H (Fig. 9a).
Figure 9.
Fe (green)-S (yellow) distances and distances between T309 methyl hydrogens, T309 hydroxyl hydrogens, A305 methyl hydrogens, active hydrogen atom of diazepam and the oxo atom of Cpd I in: a) Dia_493 optimized snapshot; note the close CH(diazepam)---OFe distance. b) Dia_dist1 optimized snapshot where the T309 methyl faces the oxo atom (note how far is diazepam). c) Dia_dist2 optimized snapshot where the T309 hydroxyl faces to the oxo atom (note how far is diazepam). Cpd I, T309 and diazepam are presented in CPK. The figures are for the 2A2u optimized structures.
3.5 What may the “stabilization” of Cpd I by the substrate mean?
A key point to consider is the actual meaning of the phrase “stabilization of Cpd I by the substrate and/or by CB”. In the absence of CB, the A305 and T309 residues are observed to interact with Cpd I by their methyl groups or via the tight T309-OH---O=Fe hydrogen bond (Figure 9b,c), and the Fe-S bond length increases and more radical character is found to build up on the sulfur with time (due to the floppy NH---S interactions; Fig. 8). Similarly, in the substrate-free MD the methyl groups of A305 and T309 maintain minimal distances of 2.64Å and 2.13Å between their methyl hydrogens and the oxo atom of Cpd I. As shown in the past, Cpd I can easily abstract hydrogen atoms from C-H bonds and even from O-H bonds.60 As such, the presence of potentially abstractable hydrogen atoms (C-H, O-H60), due to the proximity of the A305 and T309 to Cpd I, may result in hydrogen abstraction and conversion of Cpd I to the protonated Cpd II. In addition, the elongation of the Fe-S bond and the buildup of radical character on the sulfur makes Cpd I a better electron acceptor and could induce proton-coupled electron transfer that protonates the cysteine ligand and deactivate Cpd I. These adverse events, which are hypothetically suggested by our results, can shorten the lifetime of Cpd I and can prevent efficient monooxygenation of the substrate. By contrast, with CB when the substrate is close to Cpd I, these adverse features of Cpd I are eliminated; thus, the Fe-S bond is short, the radical is on the porphyrin, and the substrate displaces the FeO---H interactions with the close by residue such that the nearest hydrogen to the oxo group of Cpd I is that of the oxidizable C-H of the substrate. We may therefore hypothesize that the proximity of the substrate C-H to Cpd I, and the displacement of the other abstractable hydrogens, increases the rate of monooxygenation at the expense of the destructive processes. As such, CB in CYP3A4 (at least for diazepam) appears to be a mechanism that achieves optimization of substrate oxidation.
The recent results of Davydov et al14b,c may be associated with the above mechanism. Davydov et al followed the kinetics of disappearance of the ferric-hydroperoxo species of CYPcam. This species is the last observable one before the appearance of the hydroxylated camphor and its decay kinetics might be taken as the decay of the unobservable Cpd I (implicated e.g., by the presence of solvent kinetic isotope effect14c). These authors found that in the substrate-free enzyme the oxygen-active species disappeared very quickly even at cryogenic temperatures. However, in the presence of camphor,14b the lifetime of the species was at least 20- fold longer, and the enzyme performed camphor hydroxylation , while in the presence of a bulkier analog substrate,14c a 80-fold increase in lifetime was observed. Thus, in both cases the presence of a substrate molecule stabilizes the active species of the enzyme and optimizes product formation. A major difference between CYPcam and CYP3A4 is the size of the active site; CYPcam has a tight pocket while CYP3A4 has a large spacious one. As a result, what a single molecule of substrate can achieve in CYPcam must be achieved by two or more molecules (depending on their size) in CYP3A4. Therefore, the substrate effect might be a general mechanism of cycle optimization in P450s.
3.5 Mössbauer parameters
For an eventual identification of CYP3A4 Cpd I we calculated the Mössbauer parameters. The isomer shifts, quadrupole splittings and asymmetry parameters for CYP3A4 Cpd I are given in Table 10. The parameters are similar to the experimental values for CPO-I37 and different from the corresponding HRP-I parameters,56 as well as from those corresponding to Cpd II species. Furthermore, these values are different than the calculated parameters for the Cpd I species for CYPcam.27,30 The quadrupole splitting, in particular, is closer to the gas phase value for CYPcam,27,30 indicating that CYP3A4 Cpd I is less stabilized by the protein environment than the corresponding species of CYPcam. Indeed, the calculations of QM/MM stabilization energies show that CYP3A4 stabilizes its Cpd I ~86 kcal/mol relative to the gas phase optimized Cpd I (see SI document section 6), while CYPcam stabilizes its Cpd I species by ~100 kcal/mol relative to the gas phase reference structure.25,30
Table 10.
Calculated Mössbauer Parameters: Isomer shift δ (57Fe), Quadrupole Splitting ΔEQ , and Asymmetry (η) for the two Spin States of Cpd I of CYP3A4.
| 4A2u | 2A2u | |
|---|---|---|
| δ (mm/s) | 0.12 | 0.11 |
| ΔEQ (mm/s) | 1.29 | 1.36 |
| η | 0.17 | 0.17 |
| Spin densities | ||
| Fe | 1.20 | 1.29 |
| O | 0.84 | 0.79 |
| S | 0.54 | −0.52 |
| Por | 0.42 | −0.56 |
4. Conclusions
This work studied the features of Cpd I using QM/MM calculations on seven substrate-free models of different sizes, as well as three models that include substrate molecules, the drug diazepam (altogether 32 QM/MM optimized structures). Our key finding shows that without the diazepam, Cpd I undergoes elongation of its Fe-S bond and increasingly localizes the radical. In addition, A305 and T309 formed along MD simulations tight hydrogen bonds with Cpd I, a situation that may result in H-abstraction and destruction of Cpd I. However, the presence of a proximally restrained diazepam was found to strengthen the NH---S interactions of the conserved I443 and G444 residues with the thiolate ligand. Consequently, the substrate caused Cpd I to maintain a rather short Fe-S bond length, approximately 2.58Å, and prevented the localization of the radical on the sulfur.
The effector acted on the active substrate of the 1W0E conformer, stabilizing the substrate-Cpd I complex. This effect was not seen in the 1TQN conformer in accord with previous MD calculations. The effector substrate, as such, did not affect Cpd I directly but rather indirectly by positioning the active substrate close to Cpd I. In addition, and importantly, the effector substrate, at least according to our results, prevented the formation of hydrogen bonds between A305 and T309 and the oxo group of Cpd I by maintaining a rather close substrate-Cpd I distance. It is hypothesized that this stabilizing effect on Cpd I, by the restrained substrate, may be behind the special metabolic behavior observed in the cases of cooperative binding or multiple binding. By analogy to the findings with CYPcam,14b,c, and computational results of other human isozyme,29 it is further hypothesized that CB belongs to a general mechanism of Cpd I stabilization by the substrate. CYP3A4 requires CB to achieve Cpd I stabilization because its active site is large, whereas CYPcam with the tight pocket achieves the same stabilization with a single substrate molecule.
Our calculations of Mössbauer parameters of Cpd I CYP3A4 yielded isomer shift δ (57Fe) and quadrupole splitting (ΔEQ) values in agreement with experimental values of CPO but different than those of other P450 isoforms. We hope that the present calculations may contribute to a better understanding of the cooperative binding phenomena in P450 enzymes.
Supplementary Material
Complete references 48 and 50 in full, MD profiles, QM/MM optimized coordinates and QM gas phase optimized data are available in the supporting information (SI) file.
Acknowledgements
Discussions with E. Derat are acknowledged. The research at the HU (SS, HH and CH) was supported by the German Federal Ministry of Education and Research (BMBF) within the framework of the German-Israeli Project Cooperation (DIP), and by an ISF grant to SS. HH is a JSPS Postdoctoral Fellow for Research Abroad. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number N01-CO-12400. The content of this publication does not necessarily reflect the view of the policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Guengerich FP. In: cytochrome P450: Structure, Mechanism and Biochemistry. 3rd edition. Ortiz de Montellano PR, editor. New York: Plenum press; 2005. pp. 377–531. [Google Scholar]
- 2.Wrighton SA, Stevens JC. Crit. Rev. Toxicol. 1992;22:1–21. doi: 10.3109/10408449209145319. [DOI] [PubMed] [Google Scholar]
- 3.Sono M, Roach MP, Coulter ED, Dawson JH. Chem. Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 4.Wrighton SA, Schuetz EG, Thummel KE, Shen DD, Korzekwa KR, Watkins PB. Drug Metab. Rev. 2000;32:339–361. doi: 10.1081/dmr-100102338. [DOI] [PubMed] [Google Scholar]
- 5.Newcomb M, Hollenberg PF, Coon MJ. Arch. Biochem. Biophys. 2003;409:72–79. doi: 10.1016/s0003-9861(02)00445-9. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki H, Inoue K, Shaw PM, Checovich WJ, Guengerich FP, Shimada T. J. Pharmacol. Exp. Ther. 1997;283:434–442. [PubMed] [Google Scholar]
- 7.Shou M, Mei Q, Ettore MW, Jr, Dai R, Baillie TA, Rushmore TH. Biochem J. 1999;340:845–853. [PMC free article] [PubMed] [Google Scholar]
- 8.Shou M, Dai R, Cui D, Korzekwa KR, Baillie TA, Rushmore TH. J. Biol. Chem. 2001;276:2256–2262. doi: 10.1074/jbc.M008799200. [DOI] [PubMed] [Google Scholar]
- 9.(a) Harlow GR, Halpert JR. Proc. Natl. Acad. Sci. U S A. 1998;95:6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ueng YF, Kuwabara T, Chun YJ, Guengerich FP. Biochemistry. 1997;36:370–381. doi: 10.1021/bi962359z. [DOI] [PubMed] [Google Scholar]; (c) Schwab GE, Raucy JL, Johnson EF. Mol. Pharmacol. 1988;33:493–499. [PubMed] [Google Scholar]; (d) Schmider J, Greenblatt DJ, von Moltke LL, Harmatz JS, Shader RI. J. Pharmacol. Exp. Ther. 1995;275:592–597. [PubMed] [Google Scholar]; (e) Kerr BM, Thummel KE, Wurden CJ, Klein SM, Kroetz DL, Gonzalez FJ, Levy RH. Biochem. Pharmacol. 1994;47:1969–1979. doi: 10.1016/0006-2952(94)90071-x. [DOI] [PubMed] [Google Scholar]; (f) Xue L, Wang HF, Wang Q, Szklarz GD, Domanski TL, Halpert JR, Correia MA. Chem. Res. Toxicol. 2001;14:483–491. doi: 10.1021/tx000218z. [DOI] [PubMed] [Google Scholar]; (g) Cameron MD, Wen B, Allen KE, Roberts AG, Schuman JT, Campbell AP, Kunze KL, Nelson SD. Biochemistry. 2005;44:14143–14151. doi: 10.1021/bi051689t. [DOI] [PubMed] [Google Scholar]; (h) Domanski TL, He YA, Khan KK, Roussel F, Wang Q, Halpert JR. Biochemistry. 2001;40:10150–10160. doi: 10.1021/bi010758a. [DOI] [PubMed] [Google Scholar]
- 10.(a) Shou M, Grogan J, Mancewicz JA, Krausz KW, Gonzalez FJ, Gelboin HV, Korzekwa KR. Biochemistry. 1994;33:6450–6455. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]; (b) Korzekwa KR, Krishnamachary N, Shou M, Ogai A, Parise RA, Rettie AE, Gonzalez FJ, Tracy TS. Biochemistry. 1998;37:4137–4147. doi: 10.1021/bi9715627. [DOI] [PubMed] [Google Scholar]; (c) He YA, Roussel F, Halpert JR. Arch. Biochem. Biophys. 2003;409:92–101. doi: 10.1016/s0003-9861(02)00484-8. [DOI] [PubMed] [Google Scholar]
- 11.Lampe JN, Atkins WM. Biochemistry. 2006;45:12204–12215. doi: 10.1021/bi060083h. [DOI] [PubMed] [Google Scholar]
- 12.Shaik S, Kumar D, de Visser SP, Altun A, Thiel W. Chem. Rev. 2005;105:2279–2328. doi: 10.1021/cr030722j. [DOI] [PubMed] [Google Scholar]
- 13.Loew GH, Harris DL. Chem. Rev. 2000;100:407–420. doi: 10.1021/cr980389x. [DOI] [PubMed] [Google Scholar]
- 14.(a) Davydov R, Makris TM, Kofman V, Werst DE, Sligar SG, Hoffman BM. J. Am. Chem. Soc. 2001;123:1403–1415. doi: 10.1021/ja003583l. [DOI] [PubMed] [Google Scholar]; (b) Davydov R, Perera R, Yang T-C, Bryson TA, Sono M, Dawson JH, Hoffman BM. J. Am. Chem. Soc. 2005;127:1403–1413. doi: 10.1021/ja045351i. [DOI] [PubMed] [Google Scholar]; (c) Kim SH, Yang TC, Perera R, Jin S, Bryson TA, Sono M, Davydov R, Dawson JH, Hoffman BM. Dalton Trans. 2005;21:3464–3469. doi: 10.1039/b506764m. [DOI] [PubMed] [Google Scholar]
- 15.(a) Spolitak T, Dawson JH, Ballou DP. J. Biol. Chem. 2005;280:20300–20309. doi: 10.1074/jbc.M501761200. [DOI] [PubMed] [Google Scholar]; (b) Spolitak T, Dawson JH, Ballou DP. J. Inorg. Biochem. 2006;100:2034–2044. doi: 10.1016/j.jinorgbio.2006.09.026. [DOI] [PubMed] [Google Scholar]; (c) Raner GM, Thompson JI, Haddy A, Tangham V, Bynum N, Reddy GR, Ballou DP, Dawson JH. J. Inorg. Biochem. 2006;100:2045–2053. doi: 10.1016/j.jinorgbio.2006.09.025. [DOI] [PubMed] [Google Scholar]; (d) Kellner DG, Hung SC, Weiss KE, Sligar SG. J. Biol. Chem. 2002;277:9641–9644. doi: 10.1074/jbc.C100745200. [DOI] [PubMed] [Google Scholar]
- 16.Stone KL, Behan RK, Green MT. PNAS USA. 2005;102:16563–16565. doi: 10.1073/pnas.0507069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SH, Perera R, Hager LP, Dawson JH, Hoffman BM. J. Am. Chem. Soc. 2006;128:5598–5599. doi: 10.1021/ja060776l. [DOI] [PubMed] [Google Scholar]
- 18.van Rantwijk F, Sheldon RA. Curr. Opin. Biotechnol. 2000;11:554–564. doi: 10.1016/s0958-1669(00)00143-9. [DOI] [PubMed] [Google Scholar]
- 19.Dawson JH. science. 1988;240:433–439. doi: 10.1126/science.3358128. [DOI] [PubMed] [Google Scholar]
- 20.Himo F, Siegbahn PEM. Chem. Rev. 2003;103:2421–2456. doi: 10.1021/cr020436s. [DOI] [PubMed] [Google Scholar]
- 21.Harris DL. Curr. Opin. Chem. Biol. 2001;5:724–735. doi: 10.1016/s1367-5931(01)00272-1. [DOI] [PubMed] [Google Scholar]
- 22.Meunier B, de Visser SP, Shaik S. Chem. Rev. 2004;104:3947–3980. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 23.Harvey JN, Bathelt CM, Mulholland AJ. J. Comput. Chem. 2006;27:1352–1362. doi: 10.1002/jcc.20446. [DOI] [PubMed] [Google Scholar]
- 24.Derat E, Cohen S, Shaik S, Altun A, Thiel W. J. Am. Chem. Soc. 2005;127:13611–13621. doi: 10.1021/ja0534046. [DOI] [PubMed] [Google Scholar]
- 25.Schöneboom JC, Lin H, Reuter N, Thiel W, Cohen S, Ogliaro F, Shaik S. J. Am. Chem. Soc. 2002;124:8142–8151. doi: 10.1021/ja026279w. [DOI] [PubMed] [Google Scholar]
- 26.Harris D, Loew G, Waskell L. J. Inorg. Biochem. 2001;83:309–318. doi: 10.1016/s0162-0134(00)00177-x. [DOI] [PubMed] [Google Scholar]
- 27.Cohen S, Kumar D, Shaik S. J. Am. Chem. Soc. 2006;128:2649–2653. doi: 10.1021/ja056586c. [DOI] [PubMed] [Google Scholar]
- 28.Guallar V, Baik MH, Lippard SJ, Friesner RA. PNAS USA. 2003;100:6998–7002. doi: 10.1073/pnas.0732000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bathelt CM, Zurek J, Mulholland AJ, Harvey JN. J. Am. Chem. Soc. 2005;127:12900–12908. doi: 10.1021/ja0520924. [DOI] [PubMed] [Google Scholar]
- 30.Schöeneboom JC, Neese F, Thiel W. J. Am. Chem. Soc. 2005;127:5840–5853. doi: 10.1021/ja0424732. [DOI] [PubMed] [Google Scholar]
- 31.Schöneboom JC, Cohen S, Lin H, Shaik S, Thiel W. J. Am. Chem. Soc. 2004;126:4017–4034. doi: 10.1021/ja039847w. [DOI] [PubMed] [Google Scholar]
- 32.Ogliaro F, de Visser SP, Cohen S, Kaneti J, Shaik S. Chembiochem. 2001;2:848–851. doi: 10.1002/1439-7633(20011105)2:11<848::AID-CBIC848>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 33.Ogliaro F, Cohen S, de Visser SP, Shaik S. J. Am. Chem. Soc. 2000;122:12892–12893. [Google Scholar]
- 34.Ogliaro F, Cohen S, Filatov M, Harris N, Shaik S. Angew. Chem. Int. Ed. Engl. 2000;39:3851–3855. doi: 10.1002/1521-3773(20001103)39:21<3851::AID-ANIE3851>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 35.Weiss R, Mandon D, Wolter T, Trautwein AX, Müther M, Bill E, Gold A, Jayaraj K, Terner J. J. Biol. Inorg. Chem. 1996;1:377–383. doi: 10.1021/ic951058v. [DOI] [PubMed] [Google Scholar]
- 36.Hosten CM, Sullivan AM, Palaniappan V, Fitzgerald MM, Terner J. J. Biol. Chem. 1994;269:13966–13978. [PubMed] [Google Scholar]
- 37.Rutter R, Hager LP, Dhonau H, Hendrich M, Valentine M, Debrunner P. Biochemistry. 1984;23:6809–6816. doi: 10.1021/bi00321a082. [DOI] [PubMed] [Google Scholar]
- 38.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. J. Biol. Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 39.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 40.Ekroos M, Sjögren T. PNAS USA. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennardq O, Shimanouchi T, Tasumi M. Eur. J. Biochem. 1977;80:319–324. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- 42.Andrews J, Abd-Ellah MF, Randolph NL, Kenworthy KE, Carlile DJ, Friedberg T, Houston JB. Xenobiotica. 2002;32:937–947. doi: 10.1080/00498250210163289. [DOI] [PubMed] [Google Scholar]
- 43.Andersson T, Miners JO, Veronese ME, Birkett DJ. Br. J. Clin. Pharmacol. 1994;38:131–137. doi: 10.1111/j.1365-2125.1994.tb04336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fishelovitch D, Hazan C, Shaik S, Wolfson HJ, Nussinov R. J. Am. Chem. Soc. 2007;129:1602–1611. doi: 10.1021/ja066007j. [DOI] [PubMed] [Google Scholar]
- 45.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 46.Duhovny D, Nussinov R, Wolfson HJ. 2nd Workshop on Algorithms in Bioinformatics (WABI'02) Vol. 2452. Rome, Italy: Springer; 2002. pp. 185–200. Lecture Notes in Computer Science. [Google Scholar]
- 47.He YA, Roussel F, Halpert JR. Arch. Biochem. Biophys. 2003;409:92–101. doi: 10.1016/s0003-9861(02)00484-8. [DOI] [PubMed] [Google Scholar]
- 48.MacKerell , Jr, et al. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 49.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- 50.Sherwood P, et al. J. Mol. Struct. Theochem. 2003;632:1–28. [Google Scholar]
- 51.Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C. Chem. Phys. Lett. 1989;162:165–169. [Google Scholar]
- 52.Neese F. ORCA. Germany: Max-Planck-Institut für Bioanorganische Chemie: Mülheim a. d. Ruhr; 2004. version 2.4, revision 10. [Google Scholar]
- 53.Neese F. Inorg. Chim. Acta. 2002;337:181–192. [Google Scholar]
- 54.Stone KL, Hoffart LM, Behan RK, Krebs C, Green MT. J. Am. Chem. Soc. 2006;128:6147–6153. doi: 10.1021/ja057876w. [DOI] [PubMed] [Google Scholar]
- 55.Schäfer A, Horn H, Ahlrichs R. J. Chem. Phys. 1992;97:2571–2577. [Google Scholar]
- 56.Schulz CE, Rutter R, Sage JT, Debrunner PG, Hager LP. Biochemistry. 1984;23:4743–4754. doi: 10.1021/bi00315a033. [DOI] [PubMed] [Google Scholar]
- 57.Hosten CM, Sullivan AM, Palaniappan V, Fitzgerald MM, Terner J. J. Biol. Chem. 1994;269:13966–13978. [PubMed] [Google Scholar]
- 58.Antony J, Grodzicki M, Trautwein AX. J. Phys. Chem. A. 1997;101:2692–2701. [Google Scholar]
- 59.Yoshioka S, Tosha T, Takahashi S, Ishimori K, Hori H, Morishima I. J. Am. Chem. Soc. 2002;124:14571–14579. doi: 10.1021/ja0265409. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, Yang C, Wang H, Han K, Shaik S. ChemBiochem. 2007;8:277–281. doi: 10.1002/cbic.200600510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete references 48 and 50 in full, MD profiles, QM/MM optimized coordinates and QM gas phase optimized data are available in the supporting information (SI) file.