Abstract
Purpose
Genetic variation in Cytochrome P450 2D6 (CYP2D6) and the gene expression ratio of the homeobox 13 (HOXB13) to interleukin-17B receptor (IL-17BR) are associated with tamoxifen resistance. We sought to determine the combined effect of inherited (CYP2D6) and somatic (HOXB13/IL17BR) gene variation in tamoxifen treated breast cancer.
Experimental Design
Retrospective analysis of women with node negative breast cancer randomized to receive 5 years of tamoxifen (NCCTG 89-30-52). CYP2D6 metabolism (extensive or decreased) was based on CYP2D6*4 genotype and presence/absence of a CYP2D6 inhibitor. RT-PCR profiles for HOXB13 and IL-17BR and the cut-point separating patients into high and low risk categories according to disease-free survival (DFS) were utilized. A risk factor (CYP2D6:HOXB13/IL17BR) representing the four categories of combining CYP2D6 metabolism (extensive or decreased) and HOXB13/IL17BR (low or high) was created. The association between CYP2D6:HOXB13/IL17BR and DFS and overall survival (OS) was assessed using the log-rank test and proportional hazards modeling.
Results
CYP2D6 metabolism and HOXB13/IL17BR gene ratio was available in 110/160 (69%) patients. The combined CYP2D6:HOXB13/IL17BR risk factor was significantly associated with DFS (log rank p=0.004) and OS (p=0.009). Relative to women with extensive CYP2D6 metabolism and low HOXB13/IL17BR, those with either decreased metabolism or a high HOXB13/IL17BR ratio had significantly worse OS [adjusted hazard ratio (HR) =2.41, 95% confidence interval:1.08-5.37; p=0.031) whereas women with both decreased metabolism and high HOXB13/IL17BR had the shortest survival (adjusted HR=3.15, 95% CI:1.17-8.52, p=0.024).
Conclusions
An index comprised of inherited (CYP2D6) and tumor (HOXB13/IL17BR) gene variation identifies patients with varying degrees of resistance to tamoxifen.
Keywords: Tamoxifen, Cytochrome P450 2D6, HOXB13/IL17BR, Breast cancer
Introduction
Genetic markers are increasingly utilized to identify patients at increased risk for relapse (prognosis) and to predict the likelihood of a therapeutic response (predictive markers). Given that both tamoxifen and aromatase inhibitors (AIs) are effective therapies for the treatment of estrogen receptor (ER) positive breast cancer,1, 2 there is great interest in identifying biomarkers that may allow the individualization of therapy.
Cytochrome P450 (CYP) 2D6 is a polymorphic enzyme responsible for the metabolic activation of tamoxifen to metabolites (endoxifen and 4-OH tamoxifen) with a significantly greater affinity for the ER and greater ability to inhibit tumor cell proliferation compared with the parent drug (tamoxifen) or its primary metabolite, N-desmethyltamoxifen.3, 4 Both genetic (inherited traits) and environmental (drug-induced) factors which alter CYP2D6 enzyme activity affect the concentrations of the active tamoxifen metabolites.5, 6 We and others have demonstrated the importance of CYP2D6 as a predictor of tamoxifen benefit in the prevention,7 metastatic,8 and adjuvant settings.9-11 Based on these data, an FDA Clinical Pharmacology Subcommittee recently recommended that the tamoxifen label should be updated to reflect the increased risk of breast cancer recurrence for women who are CYP2D6 poor metabolizers, resulting from genetic variation or drug-induced inhibition of CYP2D6.
In a genome-wide microarray analysis of tumors from women with ER positive breast cancer treated with adjuvant tamoxifen, Ma et al discovered the HOXB13/IL-17BR gene ratio as an independent predictor of treatment outcome and demonstrated that ectopic expression of HOXB13 in a nontransformed human mammary epithelial cell confers increased cell migration and invasion.12 Follow-up studies have demonstrated that both HOXB13 and IL17BR are regulated by estradiol in an ER-dependent manner and this regulation is abrogated by tamoxifen.13 In hormone sensitive cell lines, HOXB13 expression rendered cells less sensitive to tamoxifen-induced apoptosis.14 Further clinical analysis of nearly 2700 patients from four independent clinical databases15-18 has repeatedly validated the H/I ratio as a prognostic marker18 and a biomarker predictive of tamoxifen benefit,15-17 with the effect most pronounced in node-negative, ER positive breast cancer.15, 16, 18
In a cohort of postmenopausal women with resected ER positive breast treated with adjuvant tamoxifen only, we reported that women with decreased CYP2D6 metabolism (defined as the presence of one or two CYP2D6 *4 alleles or the documented co-prescription of a CYP2D6 inhibitor) had significantly shorter disease free survival compared to women with extensive CYP2D6 metabolism (defined as women who did not carry a CYP2D6 *4 allele and were not co-prescribed a CYP2D6 inhibitor).9 In this same cohort, we demonstrated that a high HOXB13/IL17BR ratio was associated with significantly worse DFS and OS, but only in lymph node negative patients.15
We hypothesized that because CYP2D6 and HOXB13/IL17BR represent biologically independent markers of tamoxifen resistance, a combined index may provide a better indicator of tamoxifen treatment benefit. Therefore, using the same study population from which we derived our original findings regarding CYP2D69, 10 and HOXB13/IL17BR, 15 we evaluated the combined effect of CYP2D6 and HOXB13/IL17BR on the outcomes of DFS and OS in women with lymph node negative breast cancer treated with adjuvant tamoxifen monotherapy.
Methods
Patients
The North Central Cancer Treatment Group (NCCTG) conducted a randomized phase III clinical trial in postmenopausal women with resected ER positive breast cancer to assess the value of administering the androgen fluoxymesterone for one year during the standard 5 years of tamoxifen adjuvant therapy (NCCTG 89-30-52). A description of the clinical trial and its outcome has been published.19 The protocol was amended to assess genetic and environmental factors which may be associated with increase risk of breast cancer relapse. The trial and its amendments were approved by the institutional review board of Mayo Clinic Rochester and the individual NCCTG sites that enrolled patients onto the clinical trial.
The means by which CYP2D6 genotyping10 and HOXB13/IL17BR15 were measured and quantified has been previously published. Additionally, we published a comprehensive analysis of both CYP2D6 genotype and CYP2D6 enzyme inhibition on tamoxifen treatment outcome.9 Briefly, a woman who carried either one or two CYP2D6 *4 alleles or had any CYP2D6 genotype but co-prescribed a CYP2D6 inhibitor was considered to have decreased CYP2D6 metabolism. A woman without a CYP2D6 *4 allele and who was not co-prescribed a CYP2D6 inhibitor was considered to have extensive metabolism. A women was said to have a low HOXB13/IL17BR gene ratio if her tumor H/I gene ratio fell below <−1.339.15
Study Design and End Points
The primary objective of this study was to assess the association of effect of CYP2D6 metabolism and the HOXB13/IL17BR gene ratio on disease-free survival (DFS) and overall survival (OS) in women with lymph node negative breast cancer receiving adjuvant tamoxifen only. DFS was defined as the time from randomization to documentation of the first of the following events: any recurrence (local, regional or distant) of breast cancer, a contralateral breast cancer, a second primary cancer, or death due to any cause. Patients who were alive without any of these events were censored at the date of their last disease evaluation. OS was estimated as the time from randomization to death due to any cause.
The overall distributions of DFS and OS were estimated using the Kaplan-Meier method. Log rank tests and univariate proportional hazard models were used to assess whether the endpoint differed with respect to any one of the following factors: age greater than 65 (yes vs. no), ER status at time of entry into the trial (10-49 fmols vs.≥50 fmols vs. positive by immunohistochemistry), tumor size 3 cm or greater (yes vs. no), Nottingham grade (3 vs. 1 or 2), HER2 expression (3+ vs. 0, 1+, or 2+). For each clinical outcome, multivariate proportional hazard modeling was performed to obtain a subset of the potential prognostic factors which provided an adequate fit to the data. Residual plots were examined. A risk factor (CYP2D6:HOXB13/IL17BR) representing the 4 categories of combining CYP2D6 metabolism (extensive vs. decreased) and HOXB13/IL17BR (low vs. high) was created. Representing CYP2D6:HOXB13/IL17BR as 3 indicator variables, multivariate proportional hazard modeling was performed to assess whether any one of these factors made a significant contribution to the previous established model for that clinical endpoint.
Results
Our study population is derived from women with node-negative breast cancer (NNBC) (n=160) who were randomized from December 31, 1990 to April 6, 1995 to the tamoxifen only arm of NCCTG 89-30-52. In 110/160 patients, both HOXB13/IL17BR gene expression data and a comprehensive assessment of CYP2D6 metabolism (consisting of CYP2D6 *4 genotype and medication history) were known. The characteristics of these 110 patients are listed in Table 1 and the combined CYP2D6 and HOXB13/IL17BR phenotype is listed in Table 2.
Table 1.
Baseline Characteristics
| Characteristic | H/I and CYP2D6 phenotype available (N=110) | H/I and CYP2D6 phenotype NOT available (n=50) |
|---|---|---|
| Median Age (Range): | 65 (42-84) | 63 (47-82) |
| Race | ||
| Caucasian | 105 (95%) | 42 (84%) |
| African-American | 1 (1%) | 1 (2%) |
| Unknown | 4 (4%) | 7 (14%) |
| Extent of Surgery | ||
| Mastectomy | 91 (83%) | 38 (76%) |
| Breast Sparing | 19 (17%) | 12 (24%) |
| ER status | ||
| 10-49 fmols | 27 (25%) | 7 (14%) |
| >= 50 fmols | 65 (59%) | 34 (68%) |
| Positive | 18 (16%) | 9 (18%) |
| Tumor Size ≥ 3cm | 22 (20%) | 7 (14%) |
| Tumor Grade | ||
| 1-2 | 86 (78%) | 22 (44%) |
| 3 | 17 (15%) | 5 (10%) |
| Unknown | 7 (6%) | 23 (46%) |
| IHC Her 2 expression | ||
| 0 | 13 (12%) | 2 (4%) |
| 1+ | 34 (31%) | 10 (20%) |
| 2+ | 35 (32%) | 9 (18%) |
| 3+ | 21 (19%) | 6 (12%) |
| Unknown | 7 (6%) | 23 (46%) |
Table 2.
Risk groups as defined by CYP2D6 Metabolism and HOXB13/IL17BR gene ratio. The Kaplan Meier estimates of 5-year disease-free survival (DFS) rates and corresponding confidence intervals (CI) for each group are given. CYP2D6 inhibitors co-administered during tamoxifen in 9 patients were as follows: cimetidine (n=4), fluoxetine (n=2), paroxetine (n=2), sertraline (n=1), and haloperidol (n=1)*
| Risk Group | HOXB13/IL17BR gene ratio | CYP2D6 Metabolism Phenotype | N=110 | estimated 5 yr DFS rate (95% CI) |
|---|---|---|---|---|
| (CYP2D6*4 Genotype: CYP2D6 inhibitor use) | ||||
| 1 | < −1.339 | Extensive | 46 | 93.5% (86.6-99.9%) |
| (Wt/Wt: no) | 46 | |||
| 2 | < −1.339 | Decreased | 18 | 83.8% (67.8-99.9%) |
| (Unknown: yes | 2 | |||
| Wt/Wt:yes | 5 | |||
| Wt/*4:yes | 1 | |||
| Wt/*4:no | 9 | |||
| *4/*4:no | 1 | |||
| 3 | ≥ −1.339 | Extensive | 32 | 75.0% (61.4-91.6%) |
| (Wt/Wt: no) | 32 | |||
| 4 | ≥ −1.339 | Decreased | 14 | 57.1% (36.3-89.9%) |
| (Wt/Wt:yes | 1 | |||
| Wt/*4:no | 11 | |||
| *4/*4:no) | 2 |
One patient was co-prescribed two CYP2D6 inhibiting drugs
As of October 25 2006, 12 women were alive with disease progression, a second primary, or contralateral breast disease and 37 women had died. The median length of follow-up for those women still alive is 12.5 years (range: 5.7-15.5 years).
Univariate analyses of Clinical Outcomes
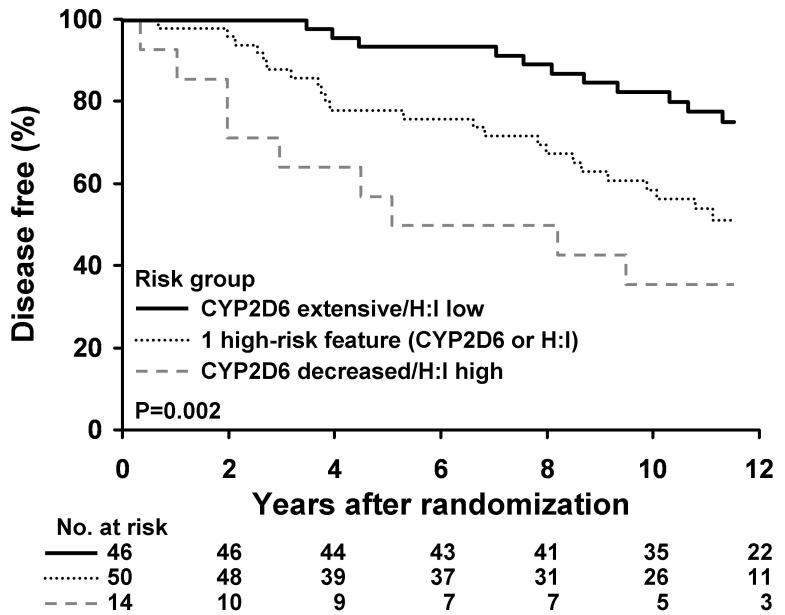
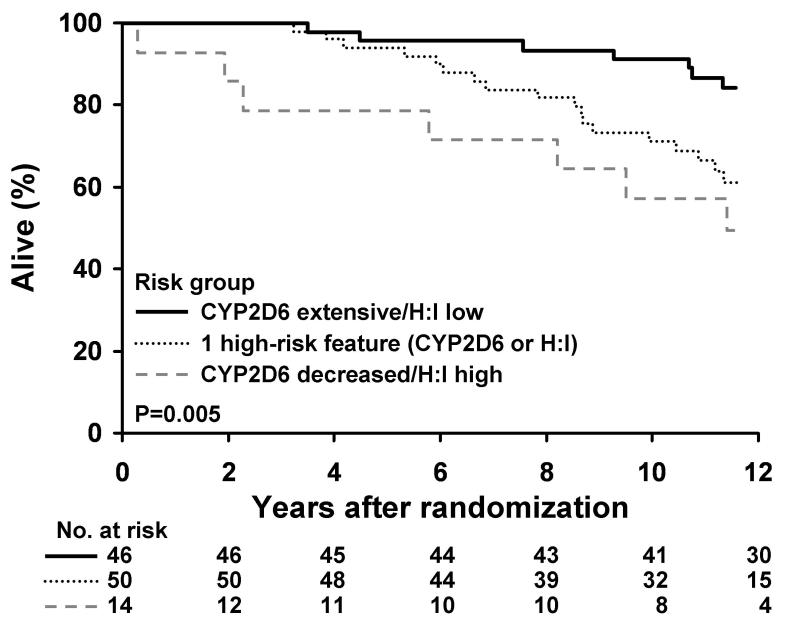
Four risk groups were formed by combining CYP2D6 metabolism and HOXB13/IL17BR (Table 2). DFS and OS were found to differ significantly with respect to these four patient categories (p=0.004 and p=0.009, respectively). Table 2 presents the Kaplan Meier estimates of 5 year DFS rate and its corresponding confidence interval for each risk group. In order to evaluate the cumulative effect of carrying 1 or more risk factors, we analyzed DFS and OS based on the absence of a risk factor (extensive CYP2D6 and HOXB13/IL17BR low), only 1 high risk feature (either CYP2D6 metabolism decreased or HOXB13/IL17BR high) and finally 2 high risk features (both CYP2D6 decreased and HOXB13/IL17BR high). In this analysis, both DFS (p=0.001) and OS (p=0.005) differed significantly according to risk groups (Figures 1 and 2)
Figure 1.
Kaplan-Meier Estimates of Disease-Free Survival in lymph node negative patients according to the presence of no high risk features (extensive CYP2D6 metabolism and HOXB13/IL17BR < −1.339), only 1 high risk feature (either decreased CYP2D6 metabolism or HOXB13/IL17BR ≥ −1.339), and 2 high risk features (both decreased CYP2D6 metabolism and HOXB13/IL17BR ≥ −1.339)
Figure 2.
Kaplan-Meier Estimates of Overall Survival in lymph node negative patients according to the presence of no high risk features (extensive CYP2D6 metabolism and HOXB13/IL17BR < −1.339), only 1 high risk feature (either decreased CYP2D6 metabolism or HOXB13/IL17BR ≥ −1.339), and 2 high risk features (both decreased CYP2D6 metabolism and HOXB13/IL17BR ≥ −1.339)
Multivariate analyses of clinical outcomes
For each endpoint, proportional hazard modeling was performed utilizing age, extent of surgery, ER status, tumor size, tumor grade and Her2 expression. Once tumor size was accounted for, none of the other traditional factors made a significant contribution to explaining the variability in these clinical outcomes. Further multivariate proportional hazard modeling demonstrated that after adjusting for tumor size, DFS and OS were significantly different among patients with 0, 1 or 2 risk factors as determined by CYP2D6 and HOXB13/IL17BR. gene ratio. Relative to women with no risk factors (extensive CYP2D6 and HOXB13/IL17BR low), women with at least one risk factor had significantly worse DFS (adjusted HR=2.03, 95% confidence interval:1.04-3.94; p=0.037) and OS (adjusted HR=2.41, 95% confidence interval:1.08-5.37, p=0.031). An even greater effect was observed in women with 2 risk factors (decreased CYP2D6 metabolism and HOXB13/IL17BR high). These women had the shortest DFS (adjusted HR=3.10, 95% CI:1.34-7.17; p=0.008), and OS (adjusted HR=3.15, 95% CI:1.17-8.52, p=0.024) (Table 3)
Table 3. Results of multivariate modeling of DFS and OS.
Multivariate modeling based on tumor size, and the three risk group model comparing presence of 1 high risk feature (defined as the presence of either decreased CYP2D6 metabolism or a HOXB13/IL17BR gene ratio ≥ −1.339) or 2 high risk features (defined as the presence of both decreased CYP2D6 metabolism and a HOXB13/IL17BR gene ratio ≥ −1.339) relative to CYP2D6 extensive metabolizers with a HOXB13/IL17BR <−1.339 Hazard Ratios (HR) and corresponding 95% confidence intervals
| Clinical Outcome | ||
|---|---|---|
| Factor | DFS | OS |
| Tumor size ≥ 3 cm | 2.64 (1.42-4.92) | 2.52 (1.23-5.14) |
| Either HOXB13:IL17BR high or decreased CYP2D6 metabolism | ||
| 2.03 (1.04-3.94) | 2.41 (1.08-5.37) | |
| Both HOXB13:IL17BR high and decreased CYP2D6 metabolism | ||
| 3.10 (1.34-7.17) | 3.15 (1.17-8.52) | |
Discussion
Our analysis of this cohort of estrogen receptor positive, lymph node negative patients treated with adjuvant tamoxifen strongly suggests that a combination of inherited (CYP2D6) and tumor (HOXB13/IL17BR) genetic variation influences the risk of breast cancer recurrence and death, independent of standard prognostic factors. Furthermore, we demonstrated a stepwise increased risk of recurrence and death, with the greatest risk in those patients who carry both high risk features (e.g. both decreased CYP2D6 metabolism and a high HOXB13/IL17BR gene ratio). In contrast to prior studies in lymph node negative breast cancer that have simply focused on tumor genetic variation,20 the paradigm of accounting for host genetic variation affecting drug metabolism, as well as tumor somatic variation, offers the potential for a more accurate estimate of drug benefit and the identification of groups of patients for whom alternative or additional therapies could be considered.
The potential synergy between HOXB13/IL17BR and CYP2D6 as biomarkers for tamoxifen resistance may be related to the differential effects of tamoxifen and its metabolites in the setting of tumor cells with dysfunctional estrogen signaling. Endoxifen exhibits a significantly greater affinity for the ER than tamoxifen3 and multiple studies suggest that only two tamoxifen metabolites, endoxifen and 4-OH tamoxifen, have significant anti-proliferative activity.3, 22 Tamoxifen and other metabolites exhibit only weak antagonist/agonist properties.23 The importance of these pharmacologic differences may be magnified in women with a high HOXB13/IL17BR gene ratio, as in vitro, HOXB13 and IL17BR are regulated by estradiol, which suppresses HOXB13 and augments IL17BR expression.13 In tumors with a high HOXB13/IL17BR ratio, the loss of both suppression of HOXB13 expression and induction of IL17BR expression may not only be a marker for impaired ER signaling, but also for increased growth factor signaling. Based on studies demonstrating that ER-positive tumors with a high HOXB13/IL17BR ratio are more likely to overexpress HER2,13 in vivo ER binding of tamoxifen and other non-potent metabolites (as opposed to endoxifen) may activate growth factor signaling and thus tumor growth18. This hypothesis is supported by preliminary data wherein nontransformed human mammary epithelial cells expressing HOXB13 demonstrated a synergistic increase in cell motility and invasion in the presence of epidermal growth factor.12
Most of the published breast cancer gene signatures appear to provide similar information in the setting of ER positive breast cancer24 and proliferation-related genes appear to be an essential component accounting for the concordance between these expression signatures.12, 20, 25-28 While “proliferation” signatures appear to identify those patients who derive the greatest benefit from adjuvant chemotherapy,29 the role of HOXB13 and IL17BR as a tool for clinical decision making has been unclear. However, our data suggest that postmenopausal patients with decreased CYP2D6 metabolism (especially CYP2D6 poor metabolizers) may preferentially benefit from alternative hormonal therapy (e.g. aromatase inhibitor) in the setting of tumors with a high HOXB13/IL17BR ratio. Further retrospective studies of large adjuvant trials comparing tamoxifen to aromatase inhibitors are necessary to validate this hypothesis.
In summary, our data suggest that an index comprised of CYP2D6 and HOXB13/IL17BR may provide a robust indicator of tamoxifen resistance. A prospective approach incorporating both factors may lead to the improved individualization of endocrine therapy.
Supplementary Material
Acknowledgment
The authors would like to thank the women who participated in this clinical trial, as well as the NCCTG investigators and clinical research associates at each site who made this translational research study possible
Dr. Matthew Goetz had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis
Funding/Support: Supported in part by CA 90628: Paul Calabresi Program in Clinical-Translational Research at Mayo Clinic (MPG); Mayo Clinic Breast Cancer Specialized Program of Research Excellence CA116201 (JNI, FJC, MPG, VJS, CR), CA 15083: Mayo Comprehensive Cancer Center Grant (MMA). CA87898 (FJC), Breast Cancer Research Foundation (JMR EAP), U-01 GM61388 from the National Institute of General Medical Sciences, Bethesda, MD (RMW, MPG, and JNI) CA-25224 (North Central Cancer Treatment Group)
Role of the Sponsor: All study funding was from public grants for scientific research. The funding organizations had no role in the design and conduct of the study; the collection, analysis, and interpretation of the data; or the preparation, review, or approval of the manuscript.
Footnotes
Financial Disclosures: X-J.M., M.G.E., and D.C.S. are named inventors on a patent to use the HoxB13:IL17BR expression ratio to ascertain breast cancer prognosis. XJM and MGE are employees of AviaraDx, San Diego, CA MPG, JNI, MMA, and FJC are named inventors on a pending patent application regarding the combined index of CYP2D6 and HOXB13:IL17BR as a predictor for tamoxifen resistance. Drs Goetz and Ingle have been consultants for Roche (2007). Drs. Goetz, Ingle, and Couch received research funding from Arcturus in 2005
Previous Presentation: This work was presented in part at the San Antonio 2006 Breast Cancer Symposium
References
- 1.Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomized trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Howell A, Cuzick J, Baum M, Buzdar A, Dowsett M, Forbes JF, Hoctin-Boes G, Houghton J, Locker GY, Tobias JS. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years' adjuvant treatment for breast cancer. Lancet. 2005;365:60–2. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 3.Johnson MD, Zuo H, Lee KH, Trebley JP, Rae JM, Weatherman RV, Desta Z, Flockhart DA, Skaar TC. Pharmacological characterization of 4-hydroxy-N-desmethy tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–9. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 4.Lim YC, Desta Z, Flockhart DA, Skaar TC. Endoxifen (4-hydroxy-N-desmethyltamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother Pharmacol. 2005;55:471–8. doi: 10.1007/s00280-004-0926-7. [DOI] [PubMed] [Google Scholar]
- 5.Gjerde J, Hauglid M, Breilid H, Lundgren S, Varhaug JE, Kisanga ER, Mellgren G, Steen VM, Lien EA. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol. 2008;19:56–61. doi: 10.1093/annonc/mdm434. [DOI] [PubMed] [Google Scholar]
- 6.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–9. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 7.Bonanni B, Macis D, Maisonneuve P, Johansson HA, Gucciardo G, Oliviero P, Travaglini R, Muraca MG, Rotmensz N, Veronesi U, Decensi AU. Polymorphism in the CYP2D6 tamoxifen-metabolizing gene influences clinical effect but not hot flashes: data from the Italian Tamoxifen Trial. J Clin Oncol. 2006;24:3708–9. doi: 10.1200/JCO.2006.06.8072. author reply 9. [DOI] [PubMed] [Google Scholar]
- 8.Lim HS, Ju Lee H, Seok Lee K, Sook Lee E, Jang IJ, Ro J. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol. 2007;25:3837–45. doi: 10.1200/JCO.2007.11.4850. [DOI] [PubMed] [Google Scholar]
- 9.Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, Visscher DW, Rynolds C, Cuch FJ, Lingle WL, Weinshilboum RM, Fritcher EG, Nibbe AM, Desta Z, Nguyen A, Flockhart DA, Perez EA, Ingle JN. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–21. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 10.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Flockhart DA, Desta Z, Perez EA, Ingle JN. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–8. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 11.Schroth W, Antoniadou L, Fritz P, Schwab M, Muerdter T, Zanger UM, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–93. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 12.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–16. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Dahiya S, Provencher H, Muir B, Carney E, Coser K, et al. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin Cancer Res. 2007;13:6327–34. doi: 10.1158/1078-0432.CCR-07-0310. [DOI] [PubMed] [Google Scholar]
- 14.Miao J, Wang Z, Provencher H, Muir B, Dahiya S, Carney E, et al. HOXB13 promotes ovarian cancer progression. Proc Natl Acad Sci U S A. 2007;104:17093–8. doi: 10.1073/pnas.0707938104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goetz MP, Suman VJ, Ingle JN, Nibbe AM, Visscher DW, Reynolds CA, et al. A two-gene expression ratio of homeobox 13 and interleukin-17B receptor for prediction of recurrence and survival in women receiving adjuvant tamoxifen. Clin Cancer Res. 2006;12:2080–7. doi: 10.1158/1078-0432.CCR-05-1263. [DOI] [PubMed] [Google Scholar]
- 16.Jansen MP, Sieuwerts AM, Look MP, Ritstier K, Meijer-van Gelder ME, van Staveren IL, et al. HOXB13-to-IL17BR expression ratio is related with tumor aggressiveness and response to tamoxifen of recurrent breast cancer: a retrospective study. J Clin Oncol. 2007;25:662–8. doi: 10.1200/JCO.2006.07.3676. [DOI] [PubMed] [Google Scholar]
- 17.Jerevall PL, Brommesson S, Strand C, Gruvberger-Saal S, Malmstrom P, Nordenskjold B, et al. Exploring the two-gene ratio in breast cancer-independent roles for HOXB13 and IL17BR in prediction of clinical outcome. Breast Cancer Res Treat. 2008;107:225–34. doi: 10.1007/s10549-007-9541-8. [DOI] [PubMed] [Google Scholar]
- 18.Ma XJ, Hilsenbeck SG, Wang W, Ding L, Sgroi DC, Bender RA, et al. The HOXB13:IL17BR expression index is a prognostic factor in early-stage breast cancer. J Clin Oncol. 2006;24:4611–9. doi: 10.1200/JCO.2006.06.6944. [DOI] [PubMed] [Google Scholar]
- 19.Ingle JN, Suman VJ, Mailliard JA, Kugler JW, Krook JE, Michalak JC, et al. Randomized trial of tamoxifen alone or combined with fluoxymesterone as adjuvant therapy in postmenopausal women with resected estrogen receptor positive breast cancer. North Central Cancer Treatment Group Trial 89-30-52. Breast Cancer Res Treat. 2006;98:217–22. doi: 10.1007/s10549-005-9152-1. [DOI] [PubMed] [Google Scholar]
- 20.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 21.Goetz MP, Reid J, Safgren S, Kuffel M, Adjei A, Goldberg R, et al. ASCO Gastrointestinal Cancers Symposium 2007. UGT1A1*28 genotype determines the maximally tolerated dose and pharmacokinetics of irinotecan-based chemotherapy: a phase I dose-escalation trial. [Google Scholar]
- 22.Buck MB, Coller JK, Murdter TE, Eichelbaum M, Knabbe C. TGFbeta2 and TbetaRII are valid molecular biomarkers for the antiproliferative effects of tamoxifen and tamoxifen metabolites in breast cancer cells. Breast Cancer Res Treat. 2008;107:15–24. doi: 10.1007/s10549-007-9526-7. [DOI] [PubMed] [Google Scholar]
- 23.Johnson MD, Westley BR, May FE. Oestrogenic activity of tamoxifen and its metabolites on gene regulation and cell proliferation in MCF-7 breast cancer cells. Br J Cancer. 1989;59:727–38. doi: 10.1038/bjc.1989.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, et al. Concordance among Gene-Expression-Based Predictors for Breast Cancer. N Engl J Med. 2006;355:560–9. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 25.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–46. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 26.Oh DS, Troester MA, Usary J, Hu Z, He X, Fan C, et al. Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol. 2006;24:1656–64. doi: 10.1200/JCO.2005.03.2755. [DOI] [PubMed] [Google Scholar]
- 27.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 29.Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–34. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.