Abstract
Rat lines congenic for the rat N-acetyltransferase 2 [(RAT)Nat2] gene were constructed and characterized. F344 (homozygous Nat2 rapid) males were mated to WKY (homozygous Nat2 slow) females to produce heterozygous F1. F1 females were then backcrossed to F344 males. Heterozygous acetylator female progeny from this and each successive backcross were identified by rat Nat2 genotyping and mated with F344 rapid acetylator males. Following ten generations of backcross mating, heterozygous acetylator brother/sister progeny were mated to produce the homozgygous rapid and slow acetylator Nat2 congenic rat lines. p-Aminobenzoic acid (selective for rat NAT2) and 4-aminobiphenyl N-acetyltransferase activities were expressed in all tissues examined (liver, lung, esophagus, stomach, small intestine, colon, pancreas, kidney, skin, leukocytes, and urinary bladder in male and female rats and in breast of female and prostate of male rats). NAT2 expression in rat extrahepatic tissues was much higher than in liver. In each tissue, activities were Nat2-genotype dependent, with highest levels in homozygous rapid acetylators, intermediate levels in heterozygous acetylators, and lowest in homozygous slow acetylators. Sulfamethazine (selective for rat NAT1) N-acetyltransferase activities were observed in all tissues examined in both male and female rats except for breast (females), bladder and leukocytes. In each tissue, the activity was Nat2-genotype independent, with similar levels in homozygous rapid, heterozygous, and homozygous slow acetylators. These congenic rat lines are useful to investigate the role of NAT2 genetic polymorphism in susceptibility to cancers related to arylamine carcinogen exposures.
Introduction
N-acetyltransferase 1 (NAT1) and 2 (NAT2) catalyze the N-acetylation of aromatic amines (Hein et al., 1993). Genetic polymorphism in NAT2 segregates humans and other mammals such as rats into rapid and slow acetylators (Hein, 2002; Boukouvala and Fakis, 2005). Homozygous rapid (F344) and slow (WKY) acetylator inbred rats have been characterized as an animal model for investigations of the N-acetylation polymorphism (Hein et al., 1991a,b; Juberg et al., 1991). (RAT)Nat1 and (RAT)Nat2 genes from rapid and slow acetylator rats each contain an intronless 870 bp open reading frame (Doll and Hein, 1995). Rats also possess a third N-acetyltransferase locus (RAT)Nat3 (Walraven et al., 2006) that does not differ between F344 and WKY inbred strains (Walraven et al., 2007). Slow acetylator WKY inbred rats are homozygous for a rat Nat2 allele with four single nucleotide polymorphisms (SNPs): G361A (Val121→ Ile), G399A (synonymous), G522A (synonymous), and G796A (Val266→ Ile), as compared to the Nat2 allele in the F344 rapid acetylator inbred rat (Doll and Hein 1995). NAT2 from F344 rat has been reported to exhibit significantly higher N-acetyltransferase activities than WKY in liver, kidney, colon, prostate, and urinary bladder (Hein et. al., 1991a,b) and following recombinant expression in bacteria (Doll and Hein, 1995; Zhang et al., 2006). In contrast, Nat1 coding regions from rapid and slow acetylator rats are identical to each other and their recombinant proteins exhibit equivalent N-acetyltransferase activities (Doll and Hein, 1995).
The Nat2 genetic polymorphism in the rat model has previously been shown to modify metabolism and toxicity of 3,2′dimethyl-4-aminobiphenyl (Feng et al., 1997; Jiang et al., 1999) 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (Purewal et al., 2000 a,b), 6-hydroxydopamine (Grundmann et al., 2004) and 4,4′-methylene dianiline (Zhang et al., 2006). However, F344 and WKY inbred rat strains differ in many other characteristics, including other metabolic enzymes and DNA repair. Thus, we constructed and characterized rat lines congenic for the rat N-acetyltransferase 2 [(RAT)Nat2] gene and compared the rat NAT1 and NAT2 expression to previously reported mouse and Syrian hamster congenic lines.
Materials and Methods
Animals
F344 (rapid Nat2 acetylator) and WKY (slow Nat2 acetylator) rats were purchased from Charles River Laboratories (Wilmington, MA). All procedures were approved by the University of Louisville Institutional Animal Care and Use Committee.
Rat Nat2 genotyping
Rat genomic DNA was isolated from blood or tail clips using proteinase K followed by phenol/chloroform extraction. Approximately 0.25 to 0.5 inches of tail was removed, minced with a scalpel, and homogenized in a buffer containing 1% sodium dodecyl sulfate, 1 mM EDTA, 25 mM Tris-HCl, pH 7.4 and 0.5 mg proteinase K. Samples were incubated at 37°C for 1 hr and then extracted with equal volumes of phenol, phenol/chloroform and chloroform. The DNA in the aqueous layer was precipitated by the addition of 1/10 volume of 5 M NaCl and 3 volumes of 95% ethanol. The DNA was washed twice with 70% ethanol, dried in air, and resuspended in 10 mM Tris-HCl, 1 mM EDTA, pH 8.0. Rat Nat2 was amplified by polymerase chain reaction (PCR) and digested with the restriction enzyme RsaI. The PCR reaction contained 10 mM Tris-HCl, 50 mM KCl, 3.0 mM MgCl2, 0.2 mM of each dNTP, 0.2 μg of each primer (5′-AGGACACCAAAACTGCATAT-3′ and 5′-CAGCATCCTTGTTTACAAGT-3′) 0.6 U Taq DNA Polymerase (Applied Biosystems, Foster City CA) and 10-100 ng of rat genomic DNA. Samples were incubated at 94°C for 5 min, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min. The reaction was concluded with incubation at 72°C for 5 min. The 257 bp PCR product was digested with the restriction enzyme RsaI following manufacturer’s instruction. Digested PCR products were separated on a 2% agarose gel and visualized under UV light. Samples homozygous for the rapid Nat2 allele possessed bands of 160 and 97 bp. Samples homozygous for the slow Nat2 allele had a single band at 257 bp. Samples heterozygous for Nat2 alleles had bands of 257, 160 and 97 bp (Figure 1).
Figure 1.
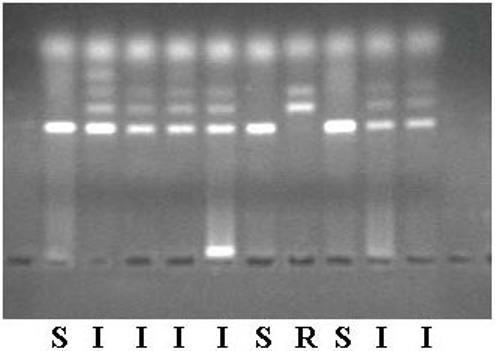
Restriction fragment length polymorphism Nat2 genotyping. The 257 bp PCR product was digested with the restriction enzyme RsaI following manufacturer’s instruction. Digested PCR products were separated on a 2% agarose gel and visualized under UV light. Samples homozygous for the rapid Nat2 allele (R) possessed bands of 160 and 97 bp. Samples homozygous for the slow Nat2 allele (S) had a single band at 257 bp. Samples heterozygous for Nat2 alleles (I) had bands of 257, 160 and 97 bp.
Construction of rapid and slow acetylator rat lines congenic at the Nat2 locus
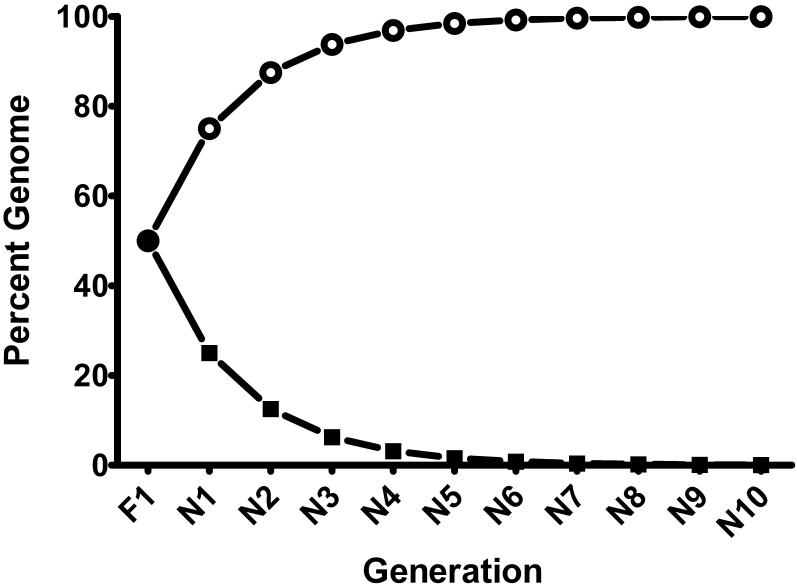
Rapid and slow acetylator congenic rat lines were constructed using methods previously used by our laboratory to construct rapid and slow acetylator congenic Syrian hamster lines (Hein et al., 1994). F344 (homozygous Nat2 rapid) males were mated to WKY (homozygous Nat2 slow) females to produce the obligate heterozygous F1 generation. F1 females were then backcrossed to F344 males. Heterozygous F1 acetylator female progeny from this and each successive backcross were identified by rat Nat2 genotyping (as described above) and mated with F344 rapid acetylator males. Following ten generations of backcross mating, heterozygous acetylator brother/sister progeny were mated to produce the homozygous rapid and slow acetylator congenic rat lines. The genome from the WKY inbred rat was the source for the slow acetylator Nat2 allele which was reduced by double dilution each backcross generation (Figure 2).
Figure 2.
Construction of F344 Nat2 congenic lines. The percentage of F344 genome (open circles) and WKY genome (solid squares) is illustrated for the F1 and each congenic (N) generation. Heterozyous Nat2 acetylator males and females from the N10 generation were bred to produce homozygous rapid and homozygous slow Nat2 acetylator F344 rats.
Preparation of Tissue Cytosols
Adult rats were sacrificed by carbon dioxide asphyxiation and the liver, lung, colon stomach, breast (females), prostate (males), small intestine, pancreas, urinary bladder, kidney, esophagus, leukocytes and skin were collected. Tissues were homogenized in 20 mM sodium phosphate (pH 7.4), 1 mM EDTA, 1 mM dithiothreitol, 100 μM phenylmethanesulfonyl fluoride, and 10 μg/ml aprotinin and 1 μM pepstatin. Homogenates were centrifuged at 100,000 x g for one hour and supernatants aliquoted and stored at -80°C until used. Protein concentrations were determined using the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA).
N-acetyltransferase Assays
Tissue cytosols were assayed for the level of NAT2 activity using the NAT2-selective substrate PABA and for NAT1 activity using the NAT1-selective substrate SMZ as described previously (Leff et al., 1999). No selective substrates for rat NAT3 have been identified and it does not catalyze the N-acetylation of PABA or SMZ (Walraven et al., 2006). Briefly, reactions containing tissue cytosol (<2 mg protein/ml), 300 μM PABA or SMZ, and 1 mM acetyl coenzyme A were incubated at 37°C for 10 minutes. Reactions were terminated by the addition of 1/10 volume 1 M acetic acid. The reaction tubes were centrifuged to precipitate protein and supernatant was injected onto a Lichrospher 100 RP-18 (125 mm × 4 mm; 5 μm) reverse phase column. Reactants and products were separated by high performance liquid chromatography (HPLC) (Beckman, Fullerton, CA). N-acetyl-PABA and N-acetyl-SMZ were quantitated by their absorbance at 280 and 260 nm, respectively. ABP N-acetyltransferase assays were carried out as previously described (Hein et al., 2006). Briefly, reactions containing suitably diluted cytosol, ABP (1 mM) and acetyl coenzyme A (1 mM) were incubated for 10 min at 37°C and terminated by the addition of 1/10 volume of 1 M acetic acid. The reaction tubes were centrifuged to precipitate protein and supernatant was injected onto a Lichrospher 100 RP-18 (125 mm × 4 mm; 5 μm) reverse phase column. Reactants and products were separated by HPLC (Beckman). N-acetyl-ABP was measured by its absorbance at 260 nm.
Results
NAT2 activities
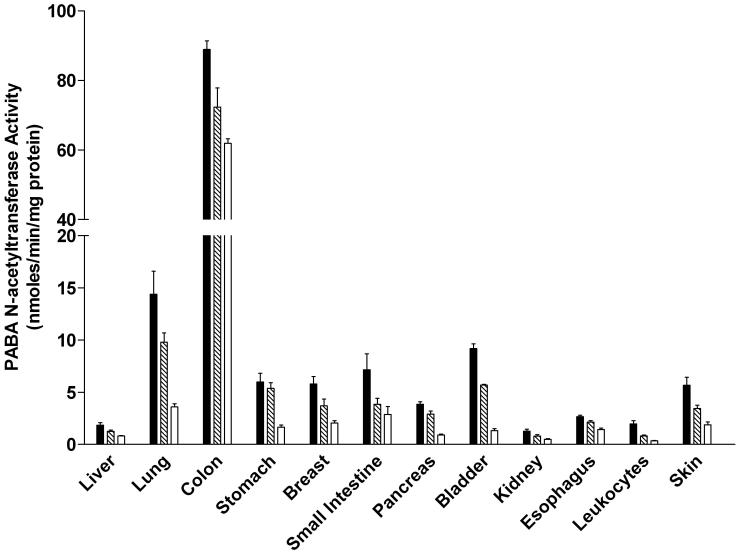
PABA (selective for rat NAT2) N-acetyltransferase activities were readily detected in all tissues examined in both male and female rats (Figure 3). In each tissue, the activity was Nat2-genotype dependent, with highest levels in homozygous rapid acetylators, intermediate levels in heterozygous acetylators, and lowest in homozygous slow acetylators. The magnitude of activity differences between rapid and slow acetylators ranged from 1.4 to 8.3-fold. NAT2 activities differed with tissue in the relative order: colon >>> lung>small intestine, urinary bladder > stomach, breast or prostate, pancreas, skin > liver, kidney, leukocytes. NAT2 activity levels did not differ significantly between males and females in any tissue examined.
Figure 3.
PABA N-acetyltransferase activities in female (top panel) and male (bottom panel) tissues as listed on the abscissa. PABA N-acetyltransferase activities differed significantly (p<0.05) with respect to Nat2 genotype in every tissue. Solid bars represent homozygous Nat2 rapid acetylator genotype, crossed bars represent heterozyous intermediate acetylator Nat2 genotype, and open bars represent homozygous Nat2 slow acetylator genotype. Each bar is the Mean ± SEM for five individual rats.
NAT1 activities
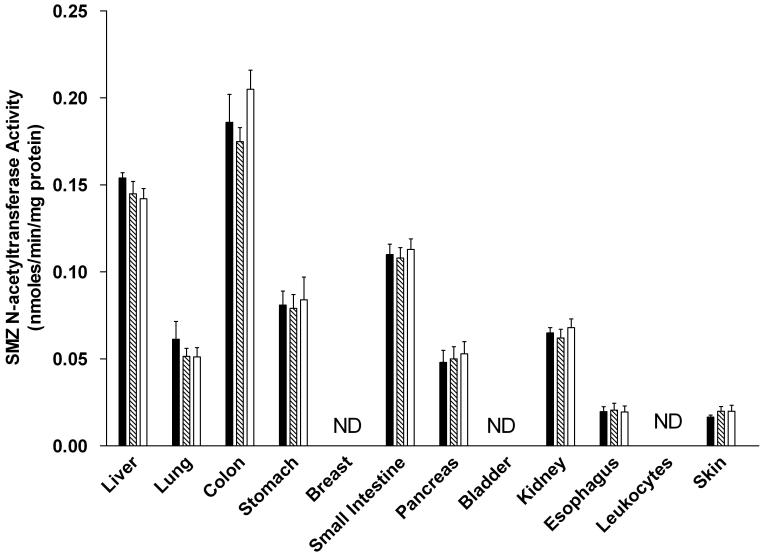
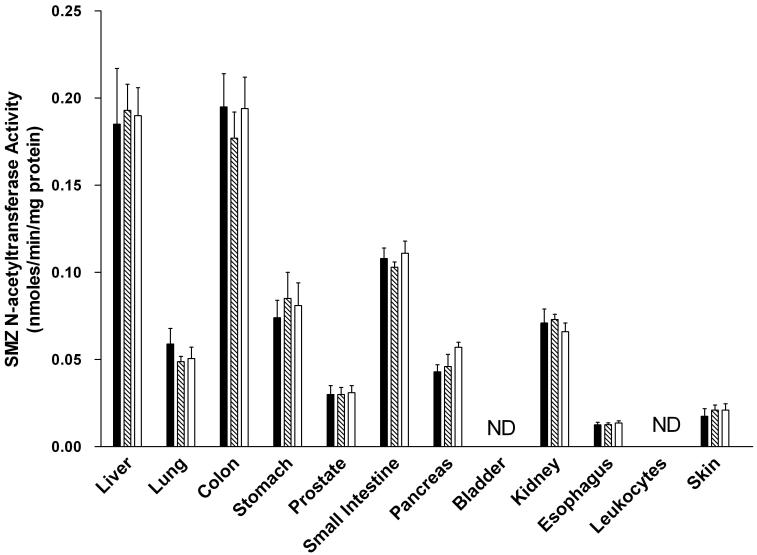
SMZ (selective for rat NAT1) N-acetyltransferase activities were observed in all tissues examined in both male and female rats except for breast (females), bladder and leukocytes (Figure 4). In each tissue, the activity was Nat2-genotype independent, with similar levels in homozygous rapid, heterozygous, and homozygous slow acetylators. NAT1 activities differed with tissue in the relative order: colon, liver >small intestine > stomach, kidney, lung, pancreas >esophagus, skin, prostate (males). NAT1 activity levels did not differ significantly between males and females in any tissue examined.
Figure 4.
SMZ N-acetyltransferase activities in female (top panel) and male (bottom panel) tissues as listed on the abscissa. SMZ N-acetyltransferase activities did not differ significantly (p>0.05) with respect to Nat2 genotype in any tissue. Solid bars represent homozygous Nat2 rapid acetylator genotype, crossed bars represent heterozyous intermediate acetylator Nat2 genotype, and open bars represent homozygous Nat2 slow acetylator genotype. Each bar is the Mean ± SEM for five individual rats. ND is non-detectable activity.
ABP N-acetyltransferase activities
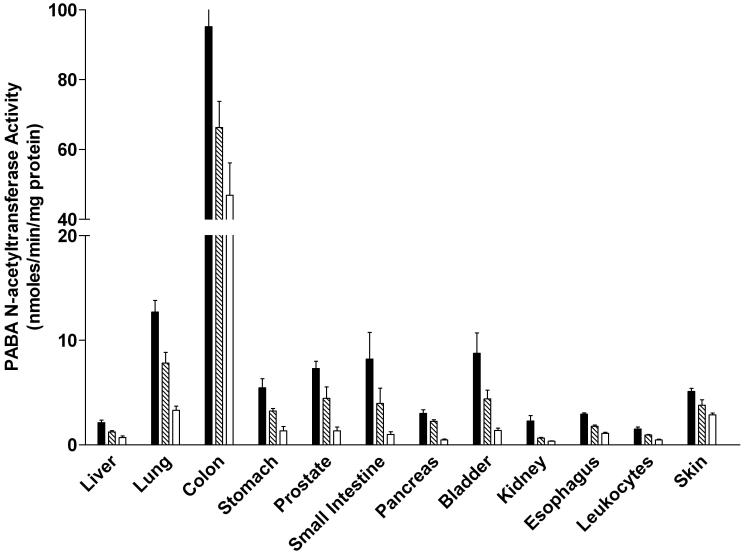
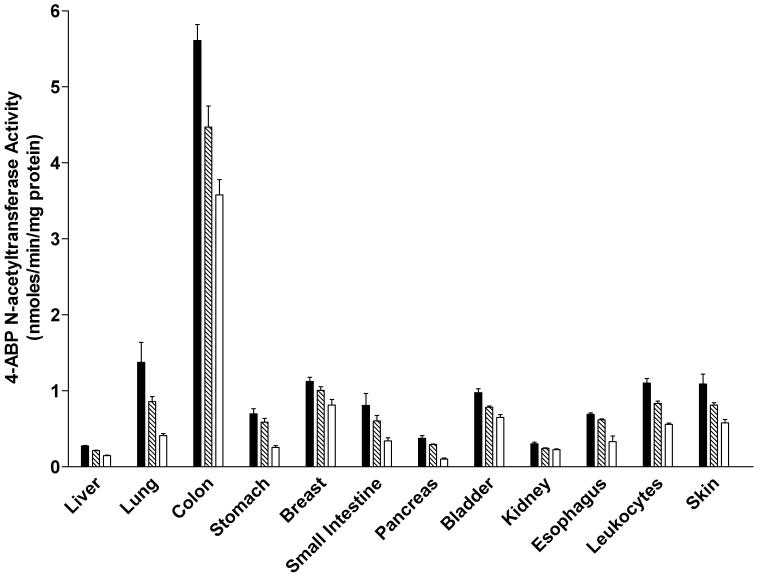
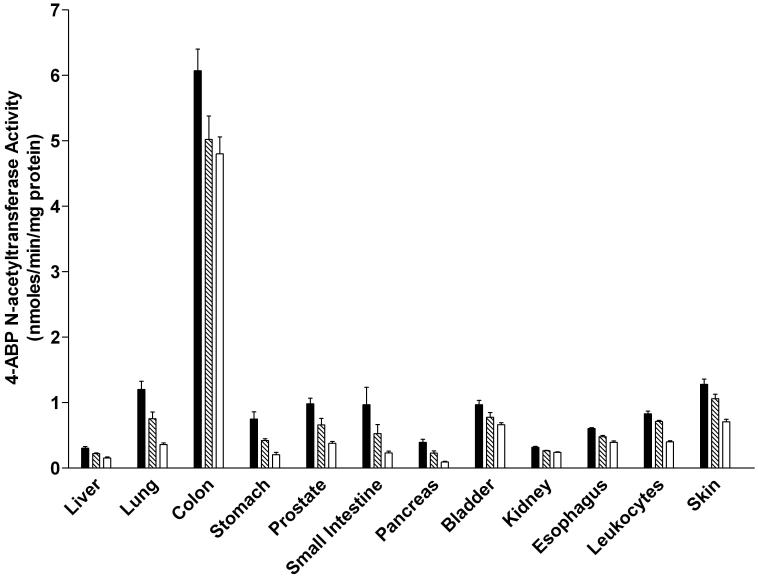
ABP N-acetyltransferase activities were readily detected in all tissues examined in both male and female rats (Figure 5). In each tissue, the activity was Nat2-genotype dependent, with highest levels in homozygous rapid acetylators, intermediate levels in heterozygous acetylators, and lowest in homozygous slow acetylators. The magnitude of activity differences between rapid and slow acetylators were more modest than observed with PABA, reflecting the fact that ABP is not a NAT2-selective substrate in the rat. ABP N-acetyltransferase activities differed with tissue in the relative order: colon >>> lung, small intestine, urinary bladder, stomach, breast (female), prostate (male), skin, leukocytes > liver, kidney, pancreas. ABP N-acetyltransferase activity levels did not differ significantly between males and females in any tissue examined.
Figure 5.
ABP N-acetyltransferase activities in female (top panel) and male (bottom panel) tissues as listed on the abscissa. ABP N-acetyltransferase activities differed significantly (p<0.05) with respect to Nat2 genotype in every tissue. Solid bars represent homozygous Nat2 rapid acetylator genotype, crossed bars represent heterozyous intermediate acetylator Nat2 genotype, and open bars represent homozygous Nat2 slow acetylator genotype. Each bar is the Mean ± SEM for five individual rats.
Discussion
Nat1 and Nat2 in animal models such as rabbit, mouse, Syrian hamster, and rat are highly homologous to both human NAT1 and NAT2 (Hein, 2002; Boukouvala and Fakis, 2005). Several different mechanisms are responsible for Nat2 polymorphisms in non-human species. The molecular basis for slow acetylator phenotype is Nat2 gene deletion in the rabbit (Blum et al., 1989), a nonsense single nucleotide polymorphism (SNP) yielding a truncated NAT2 enzyme in the Syrian hamster (Ferguson et al., 1994; Nagata et al.,1994), and non-synonomous SNP(s) in the mouse (Martell et al., 1991) and rat (Doll and Hein, 1995). The molecular basis for slow acetylator phenotype in humans likewise is due to non-synonomous SNPs in the NAT2 coding region (Hein, 2002).
ABP is a widespread environmental carcinogen present in cigarette smoke (Stabbert et al., 2003) and cooking oil fumes (Chiang et al., 1999) that induces breast tumors in the rat (Tanaka et al., 1985). ABP DNA adducts have been detected in human breast (Gorlewska-Roberts et al., 2002; Faraglia et al., 2003; Ambrosone et al., 2007) and NAT2 genotype has been shown to modify breast cancer risk in smokers (van der Hel et al., 2003; Ambrosone et al., 2008). Thus, ABP bioassays in rapid and slow acetylator rats have been proposed. PABA (selective for rat NAT2) and ABP N-acetyltransferase activities were expressed in all tissues examined (liver, lung, esophagus, stomach, small intestine, colon, pancreas, kidney, skin, leukocytes, and urinary bladder in male and female rats and in breast of female and prostate of male rats). In each tissue, activities were clearly Nat2-genotype dependent, with highest levels in homozygous rapid acetylators, intermediate levels in heterozygous acetylators, and lowest in homozygous slow acetylators. Higher levels of N-acetyltransferase activity in F344 versus WKY inbred rat strains has previously been reported in several rat tissues (Hein et al., 1991a,b; Juberg et al., 1991; Martell and Weber, 1993; Ware et al., 1995; Ware and Svensson, 1996) In contrast, SMZ N-acetyltransferase activities were Nat2-genotype independent, with similar levels in homozygous rapid, heterozygous, and homozygous slow acetylators. These results reflect the substrate specificity of PABA for rat NAT2 and SMZ for rat NAT1 (Walraven et al., 2006). While a rat NAT3-selective substrate has yet to be identified, rat Nat3 transcripts are at least 100-1000-fold lower than rat Nat1 and Nat2 transcripts in rat hepatic and extrahepatic tissues (Walraven et al., 2007; Barker et al., 2008).
In a congenic rat model in which all slow acetylators are homozygous for a single rat slow Nat2 allele and obligate heterozygotes all possess the same combination of rapid and slow rat Nat2 alleles, the Nat2 acetylation polymorphism clearly segregates into three phenotypes in hepatic and extrahepatic tissues. This trimodal distribution of rapid, intermediate and slow acetylator phenotypes also is clearly evident in the congenic Nat2 Syrian hamster model (Hein et al., 1991c; 1992; 1994). Although many human studies often exhibit bimodal distributions of rapid and slow acetylator NAT2 phenotypes, studies often yield trimodal distributions of rapid, intermediate, and slow acetylator phenotypes (Hein, 2006).
A widely held hypothesis is that human NAT2 is expressed primarily in liver and gastrointestinal tract whereas human NAT1 has widespread tissue distribution. This hypothesis derives from studies in the rabbit model where N-acetyltransferase activities reflected the Nat2 genetic polymorphism in liver and gut, but not in other tissue cytosols suggesting either absence or a much smaller contribution of rabbit NAT2 in these other tissues (Hearse and Weber, 1973). Studies have shown widespread tissue distribution of human NAT1 and NAT2 mRNA (Boukouvala and Fakis, 2005; Barker et al., 2006; Husain et al., 2007a,b). Although extrahepatic expression of N-acetyltransferase activities have been reported in rat (Hein et al., 1991a), mouse (Chung et al., 1993; Stanley et al., 1997; Sugamori et al., 2003; Wakefield et al., 2008), and Syrian hamster (Hein et al., 1991c; 1992; 1994), substrates were not selective for NAT1 or NAT2.
Recent studies with substrates selective for NAT1 versus NAT2 reported widespread distribution of both NAT1 and NAT2 catalytic activities in the rapid and slow acetylator congenic hamster (Hein et al., 2006). NAT2 catalytic activity in the Syrian hamster followed the relative order: liver >small intestine, cecum > colon, rectum, lung, pancreas, stomach, bladder > prostate, esophagus, heart. Thus the rat differs markedly from the Syrian hamster with respect to hepatic versus extrahepatic expression of NAT2 activity. NAT1 catalytic activity in the Syrian hamster followed the relative order: liver >>> small intestine, cecum, colon, rectum, pancreas, stomach, esophagus, lung > with prostate and heart (Hein et al., 2006). Thus, the relative hepatic versus extrahepatic NAT1 expression appears to be relatively similar in rat and Syrian hamster. No differences were observed between rapid and slow acetylators in NAT1 expression, consistent with our findings in the rat. ABP N-acetyltransferase activity in the Syrian hamster followed the relative order: liver, small intestine > cecum, colon, rectum, pancreas, stomach, lung, prostate, esophagus, bladder, heart (Hein et al., 2006). Thus, the contribution of hepatic versus extrahepatic NAT2 towards 4-aminobiphenyl metabolism is much less in the rat compared to the Syrian hamster. Lower and Bryan (1973) previously reported that ABP N-acetyltransferase activity was much higher in hamster than in rat liver. Previous reports clearly found that PABA N-acetyltransferase activity was much higher in rat colon (Hein et al., 1991a) than liver (Hein et al., 1991b). Subsequent reports confirmed that rat PABA N-acetyltransferase activity was much lower in rat liver than in the intestine (Ware et al., 1995) or colon (Purewal et al., 2000a).
Mouse N-acetyltransferase catalytic activity was found in all tissues examined except blood plasma and seminal vesicles (Chung et al., 1993). Lymphoid tissue, in general, was high as was skin and much of the digestive system. The NAT2 polymorphism with PABA and AF was apparent in most tissues (except brain, muscle, parotid gland, submaxillary gland and testis). Subsequently, Loehle et al (2006) reported that mouse Nat1 and Nat2 mRNA and catalytic activities were present in all mouse tissues examined (i.e., liver, gut, pancreas, bladder, and prostate). Similar results were reported by Sugamori et al (2006; 2007) with activities in the relative order: liver > kidney, colon, spleen > lung, bladder > cortex. Thus, NAT2 expression in the rat differs from both Syrian hamster and mouse. Mouse N-acetyltransferase activities were relatively constant across tissues and gender except kidney in which males were about 2-fold higher than females (Sugamori et al., 2007). Previous rat studies on gender have been inconsistent with one study reporting that female N-acetyltransferase activities were lower than male in several rat tissues (Juberg et al., 1991) while another study reported that female N-acetyltransferase activity was higher in female than male kidney (Ware et al., 1995). In contrast, we did not observe gender-related differences in any tissue for PABA, SMZ, or ABP N-acetyltransferase activity in the rat.
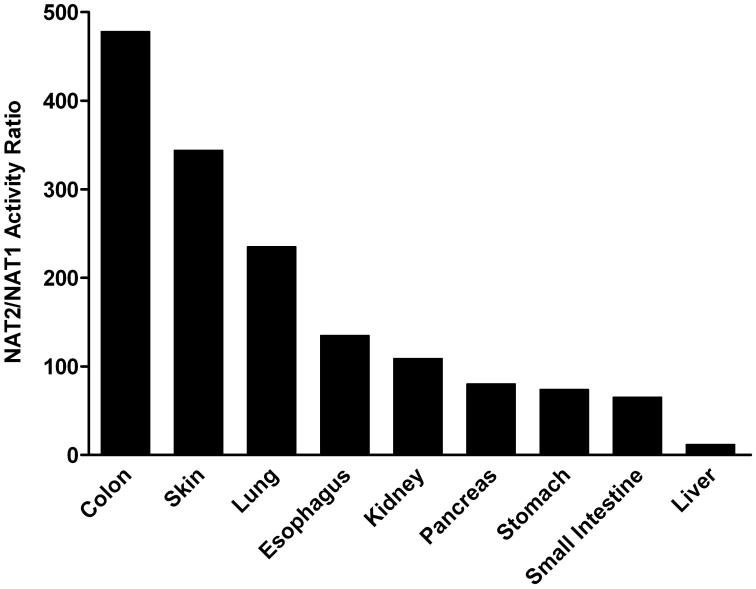
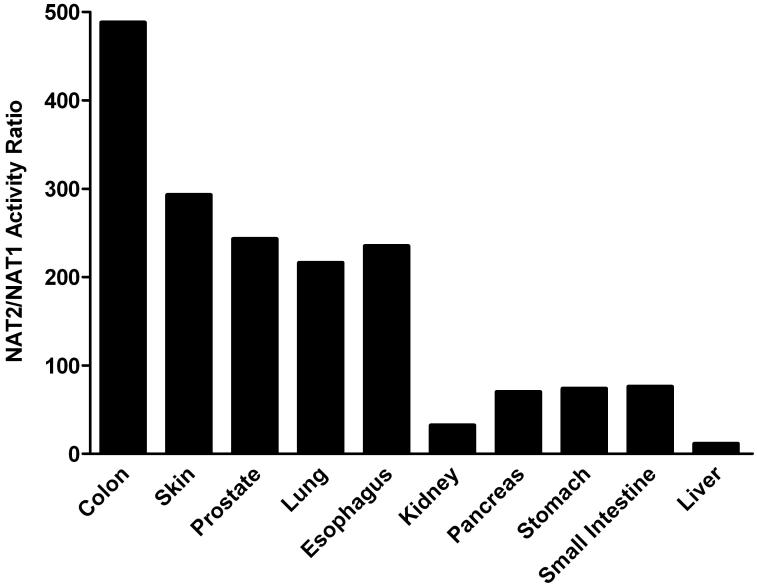
NAT2 activities were higher than NAT1 in all tissues. The NAT2/NAT1 activity ratio varied considerably from tissue to tissue but did not vary much between males and females (Figure 6). Differences between NAT2 and NAT1 activity were highest in colon (500-fold) and lowest in liver (2-fold). Thus, the tissue with the highest (colon) and lowest (liver) N-acetyltransferase activity also showed the highest and lowest difference between NAT2 and NAT1 activity.
Figure 6.
NAT2/NAT1 (PABA/SMZ) N-acetyltransferase activity ratios in female (top panel) and male (bottom panel) homozygous rapid acetylator tissues as listed on the abscissa. Ratios derived from data illustrated in Figures 3 and 4.
In conclusion, rat lines congenic for the rat N-acetyltransferase 2 [(RAT)Nat2] gene were constructed and characterized. The NAT2 expression across various tissues differed markedly from that previously reported in mouse and Syrian hamster congenic lines. Since many carcinogenic bioassay studies are conducted in the rat, particularly for arylamine carcinogens which share target organ specificity with humans, these congenic rat lines are useful to investigate the role of Nat2 genetic polymorphism in susceptibility to cancers related to arylamine carcinogen exposures. Bioassays with ABP are in progress.
Acknowledgments
This study was supported by United States Public Health Service grants R01-CA034627 and P30-ES014443.
Abbreviations
- PABA
p-aminobenzoic acid
- SMZ
sulfamethazine
- ABP
4-aminobiphenyl
- PCR
polymerase chain reaction
- AcCoA
acetyl coenzyme A
- HPLC
high performance liquid chromatography
- SNP
single nucleotide polymorphism
- Nat1 or NAT1
N-acetyltransferase 1
- Nat2 or NAT2
N-acetyltransferase 2
- PCR
polymerase chain reaction
Footnotes
A preliminary account of this work was presented at the Fourth International Workshop on the Arylamine N-acetyltransferases, Alexandroupolis, Greece, September 2007.
References
- Ambrosone CB, Abrams SM, Gorlewska-Roberts K, Kadlubar FF. Hair dye use, meat intake, and tobacco exposure and presence of carcinogen-DNA adducts in exfoliated breast ductal epithelial cells. Arch Biochem Biophys. 2007;464:169–175. doi: 10.1016/j.abb.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Ambrosone CB, Kropp S, Yang J, Yao S, Shields PG, Chang-Claude J. Cigarette smoking, N-acetyltransferase 2 genotypes, and breast cancer risk: pooled analysis and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17:15–26. doi: 10.1158/1055-9965.EPI-07-0598. [DOI] [PubMed] [Google Scholar]
- Barker DF, Husain A, Neale JR, Martini BD, Zhang X, Doll MA, et al. Functional properties of an alternative, tissue-specific promoter for human arylamine N-acetyltransferase 1. Pharmacogenet Genomics. 2006;16:515–525. doi: 10.1097/01.fpc.0000215066.29342.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DF, Walraven JM, Ristagno EH, Doll MA, States JC, Hein DW. Quantitative tissue and gene specific differences and developmental changes in Nat1, Nat2 and Nat3 mRNA expression in the rat. Drug Metab Dispos. 2008 doi: 10.1124/dmd.108.023564. Epub ahead of print September 17, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, Grant DM, Demierre A, Meyer UA. Nucleotide sequence of a full-length cDNA for arylamine N-acetyltransferase from rabbit liver. Nucleic Acids Res. 1989;17:3589. doi: 10.1093/nar/17.9.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukouvala S, Fakis G. Arylamine N-acetyltransferases: what we learn from genes and genomes. Drug Metab Rev. 2005;37:511–564. doi: 10.1080/03602530500251204. [DOI] [PubMed] [Google Scholar]
- Chiang TA, Pei-Fen W, Ying LS, Wang LF, Ko YC. Mutagenicity and aromatic amine content of fumes from heated cooking oils produced in Taiwan. Food Chem Toxicol. 1999;37:125–134. doi: 10.1016/s0278-6915(98)00081-7. [DOI] [PubMed] [Google Scholar]
- Chung JG, Levy GN, Weber WW. Distribution of 2-aminofluorene and p-aminobenzoic acid N-acetyltransferase activity in tissues of C57BL/6J rapid and B6.A-NatS slow acetylator congenic mice. Drug Metab Dispos. 1993;21:1057–1063. [PubMed] [Google Scholar]
- Doll MA, Hein DW. Cloning, sequencing and expression of NAT1 and NAT2 encoding genes from rapid and slow acetylator inbred rats. Pharmacogenetics. 1995;5:247–251. doi: 10.1097/00008571-199508000-00009. [DOI] [PubMed] [Google Scholar]
- Faraglia B, Chen SY, Gammon MD, Zhang Y, Teitelbaum SL, Neugut AI, Ahsan H, Garbowski GC, Hibshoosh H, Lin D, Kadlubar FF, Santella RM. Evaluation of 4-aminobiphenyl-DNA adducts in human breast cancer: the influence of tobacco smoke. Carcinogenesis. 2003;24:719–725. doi: 10.1093/carcin/bgg013. [DOI] [PubMed] [Google Scholar]
- Feng Y, Fretland AJ, Rustan TD, Jiang W, Becker WK, Hein DW. Higher frequency of aberrant crypt foci in rapid than slow acetylator inbred rats administered the colon carcinogen 3,2′-dimethyl-4-aminobiphenyl. Toxicol Appl Pharmacol. 1997;147:56–62. doi: 10.1006/taap.1997.8259. [DOI] [PubMed] [Google Scholar]
- Ferguson RJ, Doll MA, Baumstark BR, Hein DW. Polymorphic arylamine N-acetyltransferase encoding gene (NAT2) from homozygous rapid and slow acetylator congenic Syrian hamsters. Gene. 1994;140:247–249. doi: 10.1016/0378-1119(94)90552-5. [DOI] [PubMed] [Google Scholar]
- Gorlewska-Roberts K, Green B, Fares M, Ambrosone CB, Kadlubar FF. Carcinogen-DNA adducts in human breast epithelial cells. Environ Mol Mutagen. 2002;39:184–192. doi: 10.1002/em.10060. [DOI] [PubMed] [Google Scholar]
- Grundmann M, Earl CD, Sautter J, Henze C, Oertel WH, Bandmann O. Slow N-acetyltransferase 2 status leads to enhanced intrastriatal dopamine depletion in 6-hydroxydopamine-lesioned rats. Exp Neurol. 2004;187:199–202. doi: 10.1016/j.expneurol.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Hearse DJ, Weber WW. Multiple N-acetyltransferases and drug metabolism. Tissue distribution, characterization and significance of mammalian N-acetyltransferase. Biochem J. 1973;132:519–526. doi: 10.1042/bj1320519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506-507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- Hein DW. N-acetyltransferase 2 genetic polymorphism: effects of carcinogen and haplotype on urinary bladder cancer risk. Oncogene. 2006;25:1649–1658. doi: 10.1038/sj.onc.1209374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein DW, Doll MA, Nerland DE, Fretland AJ. Tissue distribution of N-acetyltransferase 1 and 2 catalyzing the N-acetylation of 4-aminobiphenyl and O-acetylation of N-hydroxy-4-aminobiphenyl in the congenic rapid and slow acetylator Syrian hamster. Mol Carcinog. 2006;45:230–238. doi: 10.1002/mc.20164. [DOI] [PubMed] [Google Scholar]
- Hein DW, Doll MA, Rustan TD, Gray K, Feng Y, Ferguson RJ, et al. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis. 1993;14:1633–1638. doi: 10.1093/carcin/14.8.1633. [DOI] [PubMed] [Google Scholar]
- Hein DW, Doll MA, Rustan TD, Gray K, Ferguson RJ, Feng Y. Construction of Syrian hamster lines congenic at the polymorphic acetyltransferase locus (NAT2): acetylator genotype-dependent N- and O-acetylation of arylamine carcinogens. Toxicol Appl Pharmacol. 1994;124:16–24. doi: 10.1006/taap.1994.1003. [DOI] [PubMed] [Google Scholar]
- Hein DW, Rustan TD, Bucher KD, Furman EJ, Martin WJ. Extrahepatic expression of the N-acetylation polymorphism toward arylamine carcinogens in tumor target organs of an inbred rat model. J Pharmacol Exp Ther. 1991a;258:232–236. [PubMed] [Google Scholar]
- Hein DW, Rustan TD, Bucher KD, Martin WJ, Furman EJ. Acetylator phenotype-dependent and -independent expression of arylamine N-acetyltransferase isozymes in rapid and slow acetylator inbred rat liver. Drug Metab Dispos. 1991b;19:933–937. [PubMed] [Google Scholar]
- Hein DW, Rustan TD, Bucher KD, Miller LS. Polymorphic and monomorphic expression of arylamine carcinogen N-acetyltransferase isozymes in tumor target organ cytosols of Syrian hamsters congenic at the polymorphic acetyltransferase locus. J Pharmacol Exp Ther. 1991c;259:699–704. [PubMed] [Google Scholar]
- Hein DW, Rustan TD, Martin WJ, Bucher KD, Miller LS, Furman EJ. Acetylator genotype-dependent N-acetylation of arylamines in vivo and in vitro by hepatic and extrahepatic organ cytosols of Syrian hamsters congenic at the polymorphic acetyltransferase locus. Arch Toxicol. 1992;66:112–117. doi: 10.1007/BF02342504. [DOI] [PubMed] [Google Scholar]
- Husain A, Zhang X, Doll MA, States JC, Barker DF, Hein DW. Functional analysis of the human N-acetyltransferase 1 major promoter: quantitation of tissue expression and identification of critical sequence elements. Drug Metab Dispos. 2007a;35:1649–1656. doi: 10.1124/dmd.107.016485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain A, Zhang X, Doll MA, States JC, Barker DF, Hein DW. Identification of N-acetyltransferase 2 (NAT2) transcription start sites and quantitation of NAT2-specific mRNA in human tissues. Drug Metab Dispos. 2007b;35:721–727. doi: 10.1124/dmd.106.014621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Feng Y, Hein DW. Higher DNA adduct levels in urinary bladder and prostate of slow acetylator inbred rats administered 3,2′-dimethyl-4-aminobiphenyl. Toxicol Appl Pharmacol. 1999;156:187–194. doi: 10.1006/taap.1998.8621. [DOI] [PubMed] [Google Scholar]
- Juberg DR, Bond JT, Weber WW. N-acetylation of aromatic amines: genetic polymorphism in inbred rat strains. Pharmacogenetics. 1991;1:50–57. doi: 10.1097/00008571-199110000-00008. [DOI] [PubMed] [Google Scholar]
- Leff MA, Epstein PN, Doll MA, Fretland AJ, Devanaboyina US, Rustan TD, et al. Prostate-specific human N-acetyltransferase 2 (NAT2) expression in the mouse. J Pharmacol Exp Ther. 1999;290:182–187. [PubMed] [Google Scholar]
- Loehle JA, Cornish V, Wakefield L, Doll MA, Neale JR, Zang Y, et al. N-acetyltransferase (Nat) 1 and 2 expression in Nat2 knockout mice. J Pharmacol Exp Ther. 2006;319:724–728. doi: 10.1124/jpet.106.108662. [DOI] [PubMed] [Google Scholar]
- Lower GM, Jr., Bryan GT. Enzymatic N-acetylation of carcinogenic aromatic amines by liver cytosol of species displaying different organ susceptibilities. Biochem Pharmacol. 1973;22:1581–1588. doi: 10.1016/0006-2952(73)90024-5. [DOI] [PubMed] [Google Scholar]
- Martell KJ, Vatsis KP, Weber WW. Molecular genetic basis of rapid and slow acetylation in mice. Mol Pharmacol. 1991;40:218–227. [PubMed] [Google Scholar]
- Martell KJ, Weber WW. N-acetylation polymorphism in liver and pancreas of inbred rats. Drug Metab Dispos. 1993;21:965–966. [PubMed] [Google Scholar]
- Nagata K, Ozawa S, Miyata M, Shimada M, Yamazoe Y, Kato R. Primary structure and molecular basis of polymorphic appearance of an acetyltransferase (AT-II)* in hamsters. Pharmacogenetics. 1994;4:91–100. doi: 10.1097/00008571-199404000-00006. [DOI] [PubMed] [Google Scholar]
- Purewal M, Fretland AJ, Schut HA, Hein DW, Wargovich MJ. Association between acetylator genotype and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) DNA adduct formation in colon and prostate of inbred Fischer 344 and Wistar Kyoto rats. Cancer Lett. 2000a;149:53–60. doi: 10.1016/s0304-3835(99)00346-8. [DOI] [PubMed] [Google Scholar]
- Purewal M, Velasco M, Fretland AJ, Hein DW, Wargovich MJ. 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine induces a higher number of aberrant crypt foci in Fischer 344 (rapid) than in Wistar Kyoto (slow) acetylator inbred rats. Cancer Epidemiol Biomarkers Prev. 2000b;9:529–532. [PubMed] [Google Scholar]
- Stabbert R, Schafer KH, Biefel C, Rustemeier K. Analysis of aromatic amines in cigarette smoke. Rapid Commun Mass Spectrom. 2003;17:2125–2132. doi: 10.1002/rcm.1161. [DOI] [PubMed] [Google Scholar]
- Stanley LA, Mills IG, Sim E. Localization of polymorphic N-acetyltransferase (NAT2) in tissues of inbred mice. Pharmacogenetics. 1997;7:121–130. doi: 10.1097/00008571-199704000-00005. [DOI] [PubMed] [Google Scholar]
- Sugamori KS, Brenneman D, Grant DM. In vivo and in vitro metabolism of arylamine procarcinogens in acetyltransferase-deficient mice. Drug Metab Dispos. 2006;34:1697–1702. doi: 10.1124/dmd.106.010819. [DOI] [PubMed] [Google Scholar]
- Sugamori KS, Brenneman D, Wong S, Gaedigk A, Yu V, Abramovici H, et al. Effect of arylamine acetyltransferase Nat3 gene knockout on N-acetylation in the mouse. Drug Metab Dispos. 2007;35:1064–1070. doi: 10.1124/dmd.107.015396. [DOI] [PubMed] [Google Scholar]
- Sugamori KS, Wong S, Gaedigk A, Yu V, Abramovici H, Rozmahel R, et al. Generation and functional characterization of arylamine N-acetyltransferase Nat1/Nat2 double-knockout mice. Mol Pharmacol. 2003;64:170–179. doi: 10.1124/mol.64.1.170. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Barnes WS, Williams GM, Weisburger JH. Multipotential carcinogenicity of the fried food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in rats. Jpn J Cancer Res. 1985;76:570–576. [PubMed] [Google Scholar]
- van der Hel OL, Peeters PH, Hein DW, Doll MA, Grobbee DE, Kromhout D, Bueno de Mesquita HB. NAT2 slow acetylation and GSTM1 null genotypes may increase postmenopausal breast cancer risk in long-term smoking women. Pharmacogenetics. 2003;13:399–407. doi: 10.1097/00008571-200307000-00005. [DOI] [PubMed] [Google Scholar]
- Wakefield L, Cornish V, Long H, Kawamura A, Zhang X, Hein DW, et al. Mouse arylamine N-acetyltransferase 2 (Nat2) expression during embryogenesis: a potential marker for the developing neuroendocrine system. Biomarkers. 2008;13:106–118. doi: 10.1080/13547500701673529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walraven JM, Barker DF, Doll MA, Hein DW. Tissue expression and genomic sequences of rat N-acetyltransferases rNat1, rNat2, rNat3, and functional characterization of a novel rNat3*2 genetic variant. Toxicol Sci. 2007;99:413–421. doi: 10.1093/toxsci/kfm159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walraven JM, Doll MA, Hein DW. Identification and characterization of functional rat arylamine N-acetyltransferase 3: Comparisons with rat arylamine N-acetyltransferases 1 and 2. J Pharmacol Exp Ther. 2006;319:369–375. doi: 10.1124/jpet.106.108399. [DOI] [PubMed] [Google Scholar]
- Ware JA, Svensson CK. Longitudinal distribution of arylamine N-acetyltransferases in the intestine of the hamster, mouse, and rat. Evidence for multiplicity of N-acetyltransferases in the intestine. Biochem Pharmacol. 1996;52:1613–1620. doi: 10.1016/s0006-2952(96)00567-9. [DOI] [PubMed] [Google Scholar]
- Ware JA, Divakaruni P, Svensson CK. Comparison of acetyl coenzyme A:arylamine N-acetyltransferase activity in the liver, kidney, and intestine of male and female rats from three strains. Drug Metab Dispos. 1995;23:295–297. [PubMed] [Google Scholar]
- Zhang X, Lambert JC, Doll MA, Walraven JM, Arteel GE, Hein DW. 4,4′-methylenedianiline-induced hepatotoxicity is modified by N-acetyltransferase 2 (NAT2) acetylator polymorphism in the rat. J Pharmacol Exp Ther. 2006;316:289–294. doi: 10.1124/jpet.105.093302. [DOI] [PubMed] [Google Scholar]