Abstract
Silencing of multiple cancer-related genes is associated with de novo methylation of linked CpG islands. Additionally, bivalent histone modification profiles characterized by the juxtaposition of active and inactive histone marks have been observed in genes that become hypermethylated in cancer. It is unknown how these ambiguous epigenetic states are maintained and how they interrelate with adjacent genomic regions with different epigenetic landscapes. Here, we present the analysis of a set of neighboring genes, including many frequently silenced in colon cancer cells, in a chromosomal region at 5q35.2 spanning 1.25 Mb. Promoter DNA methylation occurs only at genes maintained at a low transcriptional state and is characterized by the presence of bivalent histone marks, namely trimethylation of lysines 4 and 27 in histone 3. Chemically induced hyperacetylation and DNA demethylation lead to up-regulation of silenced genes in this locus yet do not resolve bivalent domains into a domain-wide active chromatin conformation. In contrast, active genes in the region become down-regulated after drug treatment, accompanied by a partial loss of chromatin domain boundaries and spreading of the inactive histone mark trimethylated lysine 27 in histone 3. Our results demonstrate that bivalent domains mark the promoters of genes that will become DNA methylated in adult tumor cells to enforce transcriptional silence. These bivalent domains not only remain upon drug induced gene reactivation, but also spread over adjacent CpG islands. These results may have important implications in understanding and managing epigenetic therapies of cancer.
Keywords: colorectal cancer, DNA methylation, epigenetic memory, transcriptional silencing
It is now clear that epigenetic events, in cooperation with genetic events, are involved in every step of tumorigenesis and play a critical role in the disruption of key cellular pathways deregulated in human cancers (1, 2).
De novo methylation of CpG islands is associated with the transcriptional silencing of many cancer-related genes (1, 2). The promoter regions of silenced genes, including those with promoter DNA methylation, contain specific histone modifications, which are a signature of transcriptional inactivation (3). Additionally, the DNA methylation mark itself can be read by specific proteins that can alter chromatin structure (4). Thus, a cross-talk exists between DNA methylation and histone modifications to orchestrate transcriptional silencing.
A growing body of evidence suggests that the two main cell memory systems implicated in the maintenance of a stem cell state, Trithorax (Trx) and Polycomb Group (PcG) proteins, may be involved in tumor-associated aberrant gene silencing and promoter DNA methylation (5, 6). In this context, the active mark, methylated lysine 4, together with the silent mark, methylated lysine 27, have been found to coexist over the promoter regions of DNA methylated genes in human cancer cells (7). This epigenetic landscape is similar to the bivalent domains characterized by the concurrence of trimethylated lysine 4 (H3K4me3) and trimethylated lysine 27 marks (H3K27me3), which have been described for a subset of key developmentally regulated genes in embryonic stem cells (ESC) (8, 9). The above scenario suggests that the aberrant de novo DNA methylation that so commonly affects cancer-related genes could be a direct consequence of the underlying chromatin environment, driven by the presence of the polycomb-mediated mark H3K27me3 (5, 6, 10–13).
Here, we report the concurrent tumor-specific DNA methylation and silencing of a new set of neighboring genes, which are embedded in a stem cell-like bivalent chromatin structure. Our results, in concordance with recent reports (7), suggest a direct involvement of the two main cell-memory systems, PcG and Trx, in the aberrant setting of promoter DNA methylation. Moreover, we show that the presence of a bivalent chromatin landscape over large distances provides a chromatin-based explanation for the concurrent promoter DNA methylation of multiple neighboring genes (14, 15).
Results
Long Range Epigenetic Silencing Affects Multiple Genes in 5q35.2 and Is a Common Event in Colorectal Carcinogenesis.
To screen for recurrent DNA methylation changes in colorectal cancer, we used the Amplification of Unmethylated Alu (AUMA) method, which tracks unmethylated SmaI sites in Alu and other repetitive elements (16). Comparing normal and tumor tissue DNA with AUMA revealed that the Complexin 2 (CPLX2) gene CpG island is frequently hypermethylated (72%) in colon cancer cells [supporting information (SI) Fig. S1].
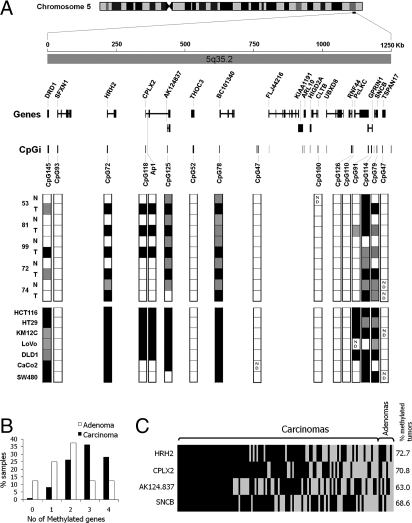
To determine whether CPLX2 hypermethylation was embedded in a larger region of epigenetic silencing, as previously described for the chromosomal region 2q14.2 (14), we performed an extended DNA methylation analysis of neighboring CpG islands in a region of 1.25 Mb in a subset of 5 normal-tumor pairs and seven colon cancer cell lines (Fig. 1A and Fig. S2). CPLX2 is located at 5q35.2, and the closest upstream CpG island, CpG72, associated with the HRH2 gene was found unmethylated in normal tissue but hypermethylated in all tumors and cell lines. Similarly, CpG145, and CpG79, associated with the DRD1 and SNCB gene promoter regions, respectively, were unmethylated in normal tissues and methylated in one or more tumors and most cell lines. Other neighboring CpG islands including CpG91, CpG114 (GPRIN1), CpG125 (AK124.837), and CpG78 (BC101340) were heavily methylated in most tumors and cell lines, although a moderate level of methylation was also observed in normal tissues. Finally, CpG islands associated with the genes SFXN1, THOC3, CLTB, RNF44 and TSPAN17 were maintained in a completely demethylated state in both normal and tumor cells (Fig. 1A).
Fig. 1.
(A) Genomic map of the 5q35.2 region covered in this study. The region spanned 1.25 Mb and included 18 genes from the most centromere-proximal DRD1 to most telomere-proximal TSPAN17. CpG islands (CpGi) across 5q35.2 are named according to the number of CpG sites they contain, except for the Ap1 fragment, which is not a CpG island itself. Bisulfite sequencing data from the five normal-tumor pairs and the seven colorectal cancer cell lines is summarized. Each box represents a CpG island, which can be found unmethylated (white), partially methylated (gray), or heavily methylated (black). ND, not determined. The complete DNA methylation dataset is found in Fig. S2. (B) Methylation data summary for the 4 analyzed genes (HRH2, CPLX2, AK124.837, and SNCB) in 118 normal-tumor (110 carcinomas, 8 adenomas) pairs. (C) Individual data for carcinomas and adenomas sorted by methylation frequency. A white box indicates unmethylated status and a black box methylated status. The methylation status was ascertained by melting curve analysis and confirmed by bisulfite sequencing in a subgroup of 50 samples.
The methylation status of the most recurrently methylated genes HRH2, CPLX2, AK124.837, and SNCB was analyzed in a large series of 121 colorectal normal-tumor pairs, including both adenomas and carcinomas. Methylation of any of the CpG islands associated with these genes occurred in 63–73% of the tumors, and virtually all tumors showed methylation of at least one of these genes (Fig. 1 B and C).
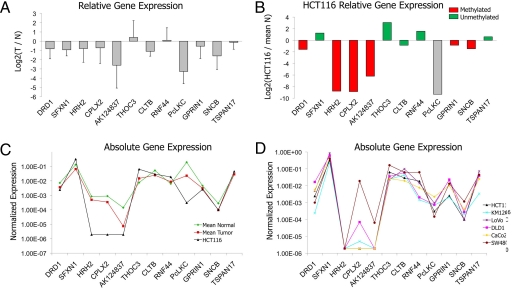
Expression profiles of 12 genes were investigated in a group of 16 microdissected tumors and their corresponding normal tissue pair. DNA-methylated genes were almost invariably down-regulated in the tumor sample (Fig. 2A and Fig. S3). Interestingly, transcriptional down-regulation also occurs in unmethylated genes (CLTB, Fig. S3), which supports the hypothesis that DNA methylation is not per se required for transcriptional down-regulation in this region. Overall, patterns of CpG island DNA methylation and gene expression were very similar between the primary tumors and the HCT116 and other colon cancer cell lines, with the exception of SW480 (Figs. 1 and 2) that globally exhibited higher expression levels for most of the genes.
Fig. 2.
Gene expression and promoter DNA methylation status of 12 genes across 5q35.2 in normal-tumor pairs and colon cancer cell lines. (A) Mean expression values (log2 of the tumor/normal ratio) of a series of 16 normal-tumor pairs. Error bars indicate standard deviation. Most genes were down-regulated in tumor samples. Individual data are illustrated in Fig. S3. (B) Relative expression of the 12 genes in HCT116 cells respectively, compared with the mean of 16 normal colonic mucosa samples. Bar color represents the methylation status of the respective promoter in HCT116 cells, except for PcLKC, that does not have a promoter CpG island. (C) Mean absolute expression values across the 12 genes analyzed for the 16 normal tissues (green), the 16 tumor tissues (red), and the HCT116 cell line (black). (D) Absolute gene expression values in six colon cancer cell lines.
Chromatin Profiles Identify Different Chromatin Domains at 5q35.2.
To gain further insights into the mechanisms that lead to transcriptional silencing in tumors along the region from DRD1 to TSPAN17 spanning 1.25 Mb, we profiled active and inactive histone modifications in the promoter region of all genes (except for GPRIN1 because of technical difficulties). The chromosomally stable HCT116 cell line was used in these studies as it is representative of the expression and methylation profiles displayed in primary tumors (Figs. 1 and 2).
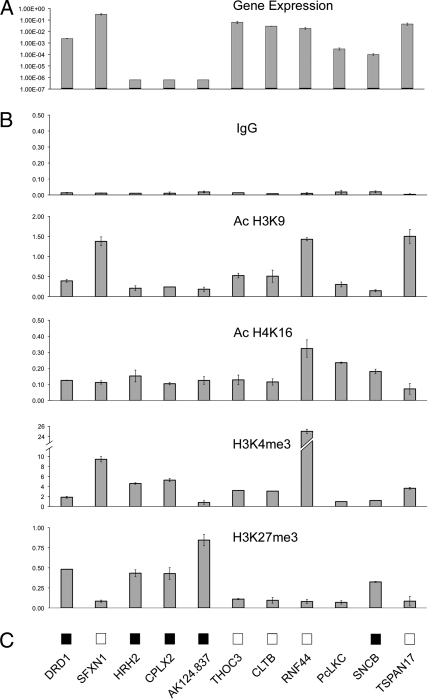
The chromatin profiling reveals the intermingled presence of active and inactive chromatin domains. Active domains, containing SFXN1, THOC3, RNF44, and TSPAN17, are characterized by many features of active chromatin: high transcription rates, unmethylated promoter CpG islands, and moderate to high levels of the active histone modifications AcH3K9 and H3K4me3 and low or undetectable levels of the repressive histone modification H3K27me3 in the promoter regions (Fig. 3 and Fig. S4). In contrast, the inactive domains are defined by low transcriptional rates, promoter DNA methylation, low levels of the active histone signature AcH3K9 and the coexistence of marks of active (H3K4me3) and inactive chromatin (H3K27me3) (Fig. 3 and Fig. S4). It is of note that the H3K4me3 mark was also present in inactive genes, in agreement with genomic analyses (17), although at lower levels than in active genes. Regardless, the enrichment in H3K4me3 was considerably higher (>50-fold) than in silenced repetitive sequences, such as Alu elements, which are devoid of this mark (data not shown). One possible explanation for the defined nature of these domains could be some insulator mechanism that functions in this region and defines coregulated genes. The CCCTC-binding factor (CTCF) is the only factor with insulator activity identified in mammals thus far and it is essential for the formation of differentially methylated imprinted domains (18). Therefore, we determined the occupancy of previously identified CTCF binding sites in this region (19) in HCT116 cells (Table S1) and found that CTCF is enriched over its previously reported binding sites compared with adjacent genomic locations (Fig. 4C). These results are consistent with a CTCF role in marking the boundaries of distinct histone methylation domains (20, 21); however, experiments using a tiling approach will be required to confirm this conclusion.
Fig. 3.
Gene expression and epigenetic profiling of genes across 5q35.2 in HCT116 cells. (A) Absolute expression levels. (B) Histone modification profiles of three active marks AcH3K9, AcH4K16, and H3K4me3 and one inactive mark H3K27me3. IgG values represent the negative control. Chromatin immunoprecipitation values are shown as the enrichment fraction over input. Error bars indicate standard deviation. (C) CpG island methylation status is noted as methylated (solid box) or unmethylated (empty box).
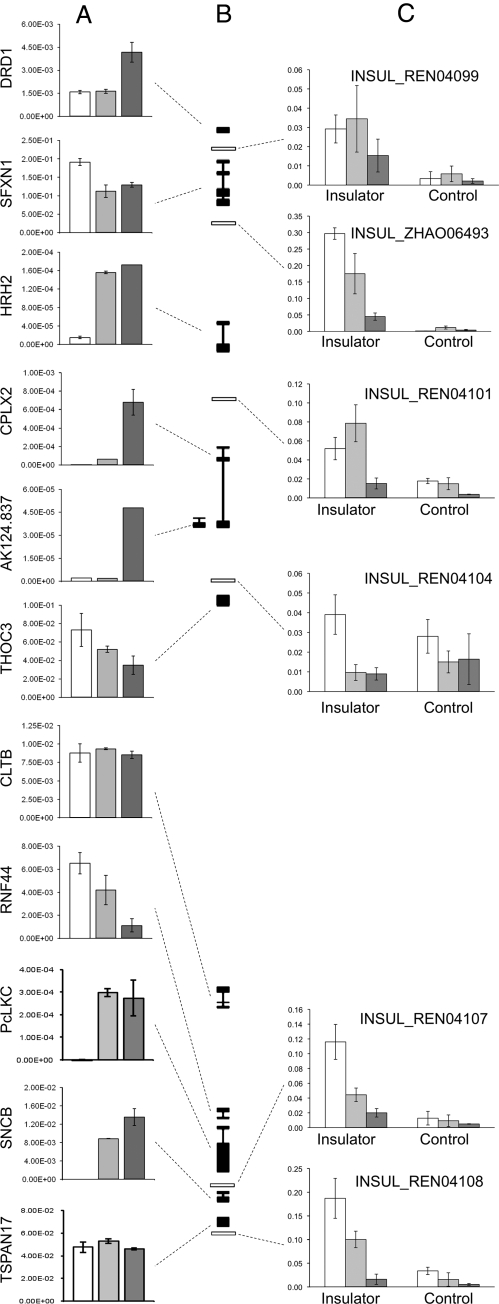
Fig. 4.
(A) Absolute expression data for genes across 5q35.2 in untreated (white bars), 5-azaC treated (light gray), and 5-AzaC/TSA (dark gray) treated HCT116 cells. (B) Genetic map of the region depicting genes (black) and CTCF-binding sites (white rectangles). (C) CTCF occupancy was determined by ChIP at the reported binding-sites (Insulator). Data were obtained from CTCFBSDB (19) and a nearby region (Control) in untreated (white bars), 5-AzaC-treated (light gray), and 5-AzaC/TSA (dark gray)-treated HCT116 cells.
Drug-Induced Gene Reactivation Results in the Disruption of Chromatin Domains and Reveals the Superseding Nature of Bivalent Signatures.
To ask how forced DNA demethylation or histone hyperacetylation would affect the observed domain structures, we treated HCT116 cells with the demethylating agent 5-AzaC and the class I and II HDAC inhibitor TSA, both alone and in combination. We have classified genes in the region according to their behavior after drug treatment and their DNA methylation and histone modification signatures. The first class of genes is that containing bivalent domains and methylated promoter CpG islands (DRD1, HRH2, CPLX2, AK124.837, and SNCB). Transcriptional up-regulation accompanied by full or partial DNA demethylation of these genes was observed after 5-azaC and 5-azaC/TSA treatments (HRH2, CPLX2, and SNCB) or only after combined treatment (DRD1 and AK124.837) (Fig. 4A and Figs. S5 and S6); TSA treatment alone was unable to up-regulate genes with methylated promoters similar to previous reports (14, 22). A second class of genes includes the silenced PcLKC gene, which has no bivalent domain in HCT116 cells and was up-regulated after both 5-azaC and 5-azaC/TSA treatments, even though this gene has no promoter CpG island. Finally, DNA unmethylated, transcriptionally active genes and enriched for active histone modifications SFXN1, THOC3, and RNF44 constitute the third class. In contrast to the response observed for the rest of the genes, these genes were down-regulated by 5-azaC and 5-azaC/TSA.
Interestingly, upon drug treatment, transcriptional changes across 5q35.2 were accompanied by the loss of CTCF at the boundaries of the different histone methylation domains (Fig. 4C), suggesting that drug treatment might also affect locus structure and organization.
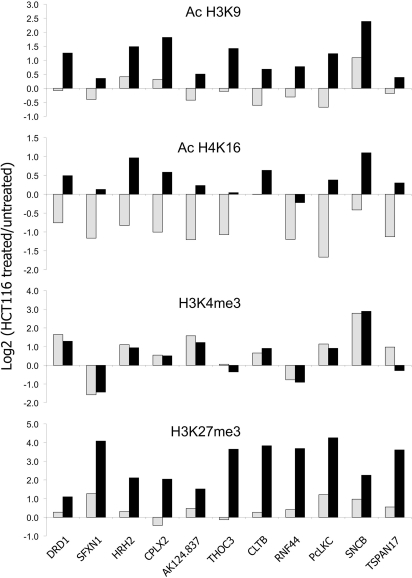
A global enrichment for the AcH3K9 mark after 5-AzaC/TSA treatment was observed at all promoters tested, but particularly those of the DNA methylated genes. Increase of the AcH3K9 mark after 5-AzaC treatment was seen only over the genes HRH2, CPLX2, and SNCB (Fig. 5) exhibiting DNA methylated promoters in cancer cells. Unexpectedly, the AcH4K16 modification was slightly depleted after treatment with 5-AzaC alone at virtually all of the regions analyzed. In contrast, 5-AzaC/TSA treatment had the opposite effect, enriching for this modification over the down-regulated genes DRD1, HRH2, CPLX2, AK124.837, CLTB, PcLKC, and SNCB, and also over the active TSPAN17 gene (Fig. 5). The highly expressed genes SFXN1, THOC3, and RNF44 showed very low or no enrichment of AcH4K16. Consistent with transcriptional down-regulation, highly expressed genes SFXN1 and RNF44 had their H3K4me3 levels reduced after the treatments, while a moderate depletion for this mark was seen only after 5-azaC/TSA treatment at the THOC3 and TSPAN17 promoters. On the other hand, silenced genes regardless of their DNA methylation status were enriched to a different degree for H3K4me3, and this was the only active mark that displayed a clear enrichment over the promoters of silenced genes after both 5-AzaC and 5-AzaC/TSA treatments.
Fig. 5.
Chromatin remodeling across 5q35.2 after drug treatment in the HCT116 cell line. The active histone modifications AcH3K9, AcH4K16, H3K4me3, and the inactive histone modification H3K27me3 in the drug-treated cells are shown as relative to the levels of the untreated cell line. Light gray bars correspond to the 5-AzaC treatment, and black bars correspond to the 5-AzaC/TSA treatment.
Finally, whereas H3K27me3 levels were moderately enriched by 5-AzaC treatment, cotreatment with TSA clearly induced a global gain of this histone modification across the entire region (Fig. 5). Enrichment for this mark affected all genes, even though it was more clearly seen at the promoters of active genes that had been described previously to have very low or undetectable levels of this mark (Fig. 3).
Components of Polycomb Repressor Complexes Are Found at the Inactive Genes.
The specific presence of H3K27me3 indicates that components of the polycomb group of proteins may be mediating, at least in part, the silencing that affects most of the genes across this region. To test this, we surveyed for the presence of the protein that catalyzes the deposition of the H3K27me3 mark, Enhancer of Zeste Homolog 2 (EZH2), identified in the context of different polycomb repressor complexes (PRC), and the protein BMI1, member of the polycomb repressor complex 1 (PRC1) that mediates the recognition of the H3K27 methylation mark (23). We show that both EZH2 and BMI1 are enriched over the promoters of the silenced genes where we had previously detected high levels of the H3K27me3 mark (Fig. S7). The highest levels of EZH2 and BMI1 were detected over the promoters of the HRH2, CPLX2 and AK124.837 genes, whereas moderate levels were detected over the SNCB and PcLKC genes. The DRD1 gene was an exception, displaying low levels of EZH2 and BMI1 despite having a bivalent promoter. DRD1 is the only gene in the region that is moderately expressed (Fig. 2) even though it displays dense promoter DNA methylation in the HCT116 cell line (Fig. 1 and Fig. S2). Additionally, specific enrichment of AcH3K16 after drug-induced reactivation over the promoter of silenced genes predicted the presence of the HDAC SIRT1 (24). Consistent with the results obtained for EZH2 and BMI1, SIRT1 was seen enriched over the promoter regions of the same genes (Fig. S7).
Both 5-AzaC and 5-AzaC/TSA treatments resulted in an increase in the EZH2 protein over the promoter regions of most genes (Fig. S7), in concordance with the enrichment of the H3K27me3 mark seen across 5q35.2. Opposite to EZH2, BMI1 was consistently depleted in all genes after drug treatment, particularly in cells treated with 5-azaC/TSA. Finally, whereas 5-azaC treatment alone increased the levels of SIRT1 at some promoters (HRH2, CPLX2, AK124.837 and CLTB), 5-AzaC/TSA treatment globally reduced the levels of this HDAC at all promoters tested, consistent with an increase of the AcH4K16 mark over the promoters of down-regulated genes after 5AzaC/TSA treatments (Fig. 5).
Discussion
In the present work we provide another example of long range epigenetic silencing (LRES) affecting genes in the genomic region 5q35.2 in colorectal cancer, characterized by concurrent DNA hypermethylation together with global down-regulation of most of the genes in the region, none of which had been previously found to be hypermethylated in cancer. A similar phenomenon has been identified in chromosomal regions 2q14.2 (14) and 3p22.2 (15) in colorectal cancer. Confirming previous studies (14, 22), DNA methylation appears to play a dominant role in the silencing mechanism, as these genes cannot be reactivated by using TSA, unless first demethylated. Moreover, the inhibitory effects of DNA methylation extend over genes without a CpG island, such as PcLKC, which can be reactivated after 5-azaC treatment, as we have previously reported for other genes in the region 2q14.2 (14).
In combination, the transcriptional, chromatin and CTCF analyses across this 1.2 Mb region provide evidence for the presence of isolated expression domains characterized by specific patterns of histone modifications and DNA methylation. The coexistence of marks of active (H3K4me3) and inactive (H3K27me3) chromatin over the promoter regions of DNA methylated genes across 5q35.2 and in many other genes in colon cancer cells (7) is noteworthy, as this signature is typical of the bivalent domains initially described in a subset of key developmentally regulated genes in ESC, which are kept at low transcriptional rates (8, 9). As it has been previously described at the genomic scale (7, 14), we find that promoter DNA methylation in the 5q35.2 region only affects genes that are transcribed at low levels, being these genes under the control of bivalent promoters. We cannot rule out allelic differences in chromatin modification to explain the apparent coexistence of both histone marks. However, consistent with our interpretation of the data, the presence of bivalent domains over essentially the same genes reported here has been described in murine (25) and human ESC (26, 27) (Table S1). Interestingly, these genes frequently undergo promoter DNA methylation in cancer cells, which prevents any further transcriptional activation (10, 11). Recently, it has been reported that bivalent chromatin is not limited to ESC: progenitor and terminal neurons also exhibit bivalent domains in specific polycomb targets (12, 13). Interestingly, when the bivalent state is resolved during cell differentiation, the active mark H3K4me2 is lost and de novo DNA methylation locks genes in a silent state (12). Here, we observe that DNA methylation is compatible with the retention of the bivalent chromatin state in cancer cells (Fig. 3), suggesting that DNA hypermethylation might override the active chromatin mark H3K4me3, becoming a primary silencing mechanism in cancer cells.
Genome-wide analysis has demonstrated specific functional and structural features, including clustering in chromosomes, of the genes methylated in cancer, leading to the postulate that EZH2 may mark these genes as preferred substrates for targeted methylation (5, 6, 28). Our findings demonstrate that the concurrent DNA methylation of neighboring genes in the 5q35.2 region is associated with the presence of bivalent promoters in these genes, supporting the hypothesis that bivalent domains may favor concurrent DNA methylation and long range epigenetic silencing of clustered genes.
Drug-induced reexpression of DNA methylated genes in 5q35.2 is accompanied by an increase in the H3K9 and H4K16 acetylation levels, together with an increase in both H3K4 and H3K27 trimethylation levels (Fig. 5). The enrichment of the inactive mark H3K27me3 over the promoters of drug-reactivated genes has been also recently noted in double-knockout HCT116 cells lacking DNA methylation (7). Although we cannot exclude a passive accumulation of these two marks because of reduced cell division induced by drugs, it can be also suggested that cellular memory systems prevent these bivalent domains from being resolved into a more active configuration during drug-induced gene reactivation by balancing the levels of active H3K4me3 and inactive H3K27me3 marks. Moreover, we provide evidence that expression domains are disrupted upon 5-azaC and 5-azaC/TSA treatments, as denoted by the loss of CTCF insulator binding (Fig. 4) and the spreading of the H3K27me3 mark over the promoter regions of active genes (Fig. 5), which also results in their transcriptional down-regulation. This occurs in the presence of an increase in the levels of the AcH3K9 mark at all promoters after drug treatment. In this context, we propose a simplified model (Fig. 6) in which domain organization is relaxed upon drug treatment and profiles of interspersed active and silent chromatin are smoothed. The relaxation of the domain boundaries does not necessarily imply changes in gene activity but would facilitate the remodeling of chromatin by altering the accessibility to modulating factors. In this new context, the reduced expression of active genes despite the rise of active marks across the promoters in 5q35.2 would be consistent with a hypothetical dominant role of H3K27me3 mark over the rest of histone modifications. On the other hand, the retention of the bivalent domains over the reexpressed genes might prevent them from being highly transcribed after drug treatment, in concordance with the low transcription rates of these types of genes in ESC (8, 13). The presence of the PRC in the promoter regions of silenced genes along 5q35.2 is consistent with the presence of trimethylated lysine 27 over the same genes and the observation that genes frequently hypermethylated in tumors tend to be occupied by proteins of the PRC in human ESC (11). Upon AZA/TSA treatment, EZH2 levels are increased, paralleling H3K27me3 changes and allowing the retention of bivalent domains in promoters originally methylated. On the other hand, partial depletion of BMI1 is consistent with a moderate decrease of the polycomb complex and transcriptional reactivation of these genes. Additionally, the partial reduction of SIRT1 might explain the increase in the levels of AcH4K16 over the reexpressed genes, favoring the formation of a more transcription-competent environment over the genes with bivalent domains. These results are of particular relevance, because SIRT1, which participates in the maintenance of a stem cell state (29) in the context of the polycomb repressor complex 4 (PRC4) (30), is over-expressed in colon cancers, and its inhibition can lead to gene reactivation in the absence of DNA demethylation (24).
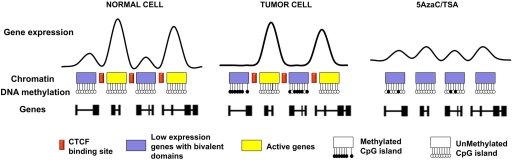
Fig. 6.
Epigenetic model for the transcriptional down-regulation and drug induced epigenetic remodeling of genes across 5q35.2. In a normal cell setting, the promoters of all genes are unmethylated (white dots) and the presence of CTCF (red box) maintains the different chromatin and expression domains isolated. During carcinogenesis, genes with bivalent domains and kept at low transcription rates become DNA-methylated (black dots), stably silenced and insensitive to differentiation or anti-proliferation cues (10, 11). This likely occurs through the recruitment of DNMT activities mediated by EZH2 (6), while active genes remain DNA methylation-free and can become up-regulated. Drug reactivation of the silenced genes not only fails to erase bivalent domains, but also induces the scattering of the silencing mark (H3K27me3) through the loss of CTCF-dependent insulation. Reactivation of silenced genes is further accompanied by an increase in the levels of active histone modifications and the partial loss of the proteins that mediate transcriptional silence.
Different studies have identified stem cell-like signatures in cancer cells and have suggested that these molecular profiles contribute to defining the biological properties of tumor cells (7, 31–33). It has been suggested that tumor-specific de novo DNA methylation could be the result of an epigenetic program that functions in embryonic stages (5). Here, we have focused on a defined genomic region, where a new set of genes with cancer-specific DNA methylation cluster. Our findings emphasize the importance of studying the relationship between DNA methylation and chromatin signatures in the context of chromatin domains rather than isolated genes, as we provide evidence that the transcriptional output for a given gene after drug treatment depends not only on its basal chromatin signatures, but also on the chromatin signatures of the neighboring genes.
Our results strengthen the concept that the cancer cell phenotype is sustained by cellular memory mechanisms directly involved in the maintenance of a bivalent chromatin state. The dominant nature of these bivalent epigenetic signatures has critical implications for understanding how cancer cells escape anti-proliferative and cell differentiation cues. The mechanisms regulating these epigenetic landscapes are likely to play a pivotal role in early stages of malignancy and endure the genotypic and phenotypic changes underlying tumor progression. Moreover, our results shed light on the complex effects of epigenetic therapies and may have important implications in the design and management of these treatments.
Materials and Methods
Full details are in the SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dirk Schübeler for comments on the manuscript and helpful suggestions and Harvey Evans for help in revising the manuscript. J.R. was supported by a Generalitat de Catalunya fellowship and a Boehringer Ingelheim travel grant. This work was supported by Spanish Ministry of Science and Innovation Grant SAF2006/351, Consolider-Ingenio Grant 2010 CSD2006/49, and National Institutes of Health Grants R01 HL65440 and R37 DK44746.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810133105/DCSupplemental.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 5.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 6.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 7.McGarvey KM, et al. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–5759. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 9.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 10.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 12.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark S. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 15.Hitchins MP, et al. Epigenetic inactivation of a cluster of genes flanking MLH1 in microsatellite-unstable colorectal cancer. Cancer Res. 2007;67:9107–9116. doi: 10.1158/0008-5472.CAN-07-0869. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez J, et al. Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells. Nucleic Acids Res. 2008;36:770–784. doi: 10.1093/nar/gkm1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Bao L, Zhou M, Cui Y. CTCFBSDB: A CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res. 2008;36:D83–87. doi: 10.1093/nar/gkm875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 23.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 24.Pruitt K, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan G, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Zhao XD, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Keshet I, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 29.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 30.Kuzmichev A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke MF, Fuller M. Stem cells and cancer: Two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.