Abstract
Purpose
Single nucleotide polymorphisms (SNPs) in the tightly linked LOC387715/ARMS2 and HTRA1 genes have been associated with age-related macular degeneration (AMD). We tested whether these SNPs are associated with AMD in Israeli populations, if they underlie variable phenotype and response to therapy in neovascular AMD (NVAMD), and if HTRA1 expression in vivo is associated with its promoter variant.
Methods
Genotyping for the rs10490924 SNP in LOC387715/ARMS2 and the rs11200638 SNP in HTRA1 was performed on 255 NVAMD patients and 119 unaffected controls from Ashkenazi and Sephardic Jewish, and from Arab origins which are the main ethnic groups composing the Israeli population. Genotyping was correlated with phenotype and response to therapy among 143 patients who underwent photodynamic therapy (PDT). HTRA1 mRNA levels in white blood cells (WBCs), measured by quantitative PCR, were correlated with genotype in 27 participants.
Results
Both SNPs were in almost complete linkage disequilibrium (D'=0.96–1). Homozygotes for the T allele of rs10490924 had an odds ratio (OR) of 8.6, with a 95% confidence interval (CI) of 3.5–20.8, and homozygotes for the A allele of rs11200638 had an OR of 10.7, with a 95% CI of 3.2–35.7, for having AMD (p<0.00001). There was no association among these SNPs and phenotype or response to PDT. HTRA1 mRNA levels in WBCs were not associated with rs11200638 genotypes.
Conclusions
The rs10490924 SNP in LOC387715/ARMS2 and the rs11200638 SNP in HTRA1 are strongly associated with NVAMD in this Israeli population. These variants do not have a major contribution to the variable phenotype and response to PDT which characterize NVAMD.
Introduction
Single nucleotide polymorphisms (SNPs) in chromosome 10q26 are associated with the risk for having age-related macular degeneration (AMD) in several populations [1-18]. Since several 10q26 SNPs are associated with disease risk, it has been difficult to determine if the T allele of rs10490924 located in the coding region of the LOC387715/ARMS2 (age-related maculopathy susceptibility 2) gene, or the A allele of rs11200638 located in the promoter region of high-temperature requirement factor A1 (HTRA1) have a role in the pathogenesis of the disease [6-9,13,19].
The HTRA1 gene encodes a heat shock serine protease which is expressed in the retina and can regulate transforming growth factor-β (TGF-β) signaling [7,20]. Evidence has conflicted with respect to the correlation between rs11200638 genotypes and HTRA1 expression levels [6-8].
The LOC387715/ARMS2 gene encodes a putative 12 kDa protein. Although it has been suggested that LOC387715/ARMS2 may not actually encode a protein, recent data has shown that it encodes a mitochondrial outer membrane protein that is also expressed in the retina [6]. The T allele of rs10490924 changes amino acid 69 from alanine to serine (A69S variant) in the putative LOC387715/ARMS2 protein [6,13]. Fritsche [21] and colleagues recently described a related variant in this gene that is associated with AMD and results in rapid mRNA turnover and undetectable expression levels in homozygous carriers.
Neovascular AMD (NVAMD) shows variable phenotype in terms of several parameters, including age of onset, neovascular lesion type and size, and response to treatment. Efforts are underway to identify genetic and other biomarkers that correlate with these various disease parameters. Of particular interest would be the identification of pharmacogenetic markers that might be able to predict response to therapy, since such information could potentially be used to guide choice, and perhaps frequency, of treatment for the individual patient. Recently, a genetic variant in the complement factor H (CFH) gene, which is strongly associated with AMD, has been reported to be associated with neovascular lesion type, size, and response to photodynamic therapy (PDT) and injections of the antivascular endothelial growth factor compound bevacizumab [22-26]. The HTRA1 polymorphism was associated with classic lesion type [26]. However, similar associations have not been consistently found in other populations [10,27,28].
To test the association of the 10q26 SNPs with NVAMD in Israeli populations and to expand our knowledge about phenotype-genotype correlation, we first evaluated HTRA1 and LOC387715/ARMS2 polymorphisms and NVAMD risk in Israel. The study included Arabs, Sephardic Jews, and Ashkenazi Jews which are the main ethnic groups composing the Israeli population. After finding a significant association, similar in magnitude to that reported in other populations, we assessed if these SNPs could account for some of the observed variation in clinical characteristics and response to PDT among the NVAMD patients. Finally, as another approach to explore whether the SNP in the 5′-upsteam region of the HTRA1 promoter influences promoter activity, we tested to see if there was a correlation between genotype and HTRA1 mRNA levels in vivo as measured in the white blood cells (WBCs) of patients and controls.
Methods
Patients and genotyping
The study included 255 NVAMD patients recruited from four retina clinics in Israel and 119 unaffected controls (from the department of Ophthalmology) who were evaluated for routine eye examination or for pathologies other than AMD, in the Department of Ophthalmology of the Hadassah – Hebrew University Medical Center in Jerusalem, Israel. Institutional Ethics Committee approval was obtained for the study, and each patient signed an informed consent form. AMD was diagnosed and graded according to the age related eye disease study (AREDS) trial classification [29]. Inclusion criteria for the control group included age over 60 years, clear cornea, lens, and vitreous which enabled ophthalmoscopy, and absence of intermediate size drusen, multiple small drusen, or retinal pigment epithelial abnormalities characteristics of AMD (AREDS category I). The data set was described in a manuscript evaluating CFH variants in the Israeli population [30]. Briefly, female:male ratio was balanced between AMD patients and controls. Mean age in the controls (70.8±8.2) was lower than that of AMD patients (78.1±7.6; p<0.05, unpaired, two-sided, t-test). Median follow-up of NVAMD patients having PDT was 16 months (range 1–156 months).
WBCs were separated from blood samples from 27 individuals included in the genotyping study (22 NVAMD patients and 5 unaffected controls). These samples were used for RNA extraction. For WBC separation, 4 CC of whole blood were drawn and shipped to the laboratory on wet ice. RNA was then extracted as described below.
The Israeli population is composed of several ethnic groups, which include Ashkenazi Jews, Sephardic Jews, and Arabs. Studies showed that these subpopulations may be genetically defined, for example, by polymorphisms and microsatellite loci on the Y chromosome [31]. Intermarriage between the groups was uncommon among parents of the elderly individuals included in the study. The control group included 10 Arabs, 40 Sephardic Jews, and 68 Ashkenazi Jews, while the study group included 11 Arabs, 75 Sephardic Jews, and 163 Ashkenazi Jews (p=0.13, χ2 test). The ethnicity of one individual from the control group and 6 NVAMD patients was unknown.
Genotyping for the rs11200638 SNP in the putative promoter region of HTRA1 and for the rs10490924 SNP in the coding region of the LOC387715/ARMS2 gene was performed. This was done by sequencing PCR products containing these SNPs (Table 1).
Table 1. Primers used for genotyping and QPCR.
| Primer | Sequence |
|---|---|
| Primers for genotyping | |
|
LOC387715/ARMS2 forward |
TACCCAGGACCGATGGTAAC |
|
LOC387715/ARMS2 reverse |
GAGGAAGGCTGAATTGCCTA |
|
HTRA1 forward |
CGGATGCACCAAAGATTCTCC |
|
HTRA1 reverse |
TTCGCGTCCTTCAAACTAATGG |
| Primers for QPCR | |
|
HTRA1 forward |
AGCTGGGACTTCGGAACTCC |
|
HTRA1 reverse |
TCCGGAATTTCCATAATTGATGA |
| GAPDH forward |
GGGGGAGCCAAAAGG GTCAT |
| GAPDH reverse | GCCCCAGCGTCAAAGGTGGA |
Sequence of primers which were used for genotyping of LOC387715/ARMS2 (rs10490924) and HTRA1 (rs11200638) SNPs and for QPCR for assessment of HTRA1 mRNA levels.
Detailed retrospective clinical information was available on a subgroup of 143 sequential NVAMD patients (of the 255 patients enrolled in the study) who were treated with PDT at the Hadassah–Hebrew University Medical Center. These patients were included in the phenotype-genotype analysis and for the purpose of analysis eyes of these patients which underwent PDT were considered as the study eye. Fluorescein angiograms were reviewed by retina specialists (I.C., E.B., I.H., and E.A.) who were masked with respect to genotype results; choroidal neovascularization was classified as classic, predominantly classic, minimally classic, or occult following the guidelines of the Macular Photocoagulation Study Group [32]. Retinal angiomatous proliferation (RAP) was classified as occult lesions. Review of the entire group of patients was also performed in a masked fashion (with respect to previous lesion type classifications and to genotypes) by one of the investigators (I.C.). There was agreement in 83.4% of cases between this investigator and the classification by the treating retina specialist (kappa measurement of agreement=0.68, p<0.0001). Classification of lesion type according to the observer who reviewed the entire group of patients was used for statistical analysis. The standard PDT protocol for NVAMD was applied [33]. Briefly, patients received 6 mg/m2 body surface area of verteporfin (Visudyne; Novartis Ophthalmics, Hettlingen, Switzerland) intravenously over 10 min. Fifteen min after commencement of the infusion exposure of 50 J/cm2 was applied at the radiant level of 600 mW/cm2 over 83 s using a diode laser (Visulas s 690, Zeiss, Switzerland), and the PDT laser lens (Volk, Mentor, OH).
The association between smoking and the risk for having NVAMD, and its interaction with the 10q26 SNPs was analyzed. Individuals were considered as smokers if they were smoking at the time of the study or if they smoked in the past. Individuals who never smoked were considered as non-smokers.
Quantitative real-time RT–PCR
HTRA1 mRNA levels were measured in WBCs from 27 participants (22 NVAMD patients and 5 unaffected controls older than 60 years of age). WBCs were separated from whole blood using hypotonic lysis buffer containing 155 mM NH4Cl (Gadot, Or Akiva, Israel), 10 mM CH2O3·NH3 (Sigma-Aldrich, St. Louis, MO), and 0.1 mM EDTA (pH 7.4; J.T. Baker, Philipsberg, NJ). Next, 8 ml of buffer was added to 4 ml of blood, which was then stored on ice for 10 min and centrifuged at 2,000x g at 4 °C for 10 min. Supernatant was discarded and the sequence was repeated an additional time. The WBC pellet was resuspended in 1 ml of TRI Reagent (Sigma). RNA was then extracted followed by treatment with DNase (DNAfree; Ambion, Austin, TX), and cDNA was synthesized from 1 ug of total RNA using the Reverse iT 1st Strand Synthesis Kit (ABgene, Epsom, UK) and oligo dT primers. Quantitative real-time RT–PCR (QPCR) was performed with HTRA1 specific primers (Table 1) using the ABI Prism 7000 SDS instrument (Applied Biosystems, Foster City, CA). Measurements of GAPDH were used for normalization of expression levels across samples.
Statistical analysis
Statistical analysis and power calculations were performed using SPSS (SPSS, Chicago, IL) and Instat software (GraphPad, San Diego, CA) as we have previously described [30]. Briefly, logistic regression and χ2 tests were applied to assess odds ratios, confidence intervals, and significance. Linkage disequilibrium was assessed by D’ calculation. Based on the number of individuals included in the analysis this study had 85% power for identification of an association between genotypes and lesion type in the magnitude described by Brantley and colleagues [22]. The study also had 94% power for identification of an association between genotypes and visual acuity following PDT in the magnitude, which was described by the same group [25].
Results
Association of 10q26 SNPs with NVAMD
Both the HTRA1 rs11200638 SNP and the LOC387715/ARMS2 rs10490924 SNP complied with Hardy–Weinberg equilibrium. Genotypes for both SNPs were in almost complete linkage disequilibrium, with D' values of 1.00 for NVAMD patients and 0.96 for controls. Distribution of the genotypes for both SNPs was significantly different between NVAMD patients and controls (p<0.0001; Table 2 and Table 3).
Table 2. Frequency of LOC387715/ARMS2 (rs10490924) alleles and genotypes in NVAMD patients and unaffected controls.
| LOC387715/ARMS2 (rs10490924) | AMD | Unaffected | p | OR (95% CI) |
|---|---|---|---|---|
| By allele | ||||
| Entire population
(G/T; %)* |
279/219
(56/44) |
190/48
(80/20) |
<0.0001 |
3.1 (2.2–4.5) |
| Ashkenazi Jews |
189/129 (59.4/40.6) |
110/26
(80.8/19.2) |
<0.0001 |
2.9 (1.8–4.7) |
| Sephardic Jews |
76/72
(51.4/48.6) |
63/17
(78.8/21.2) |
<0.0001 |
3.5 (1.9–6.6) |
| Arabs |
8/12
(40/60) |
16/4
(80/20) |
0.022 |
6 (1.5–24.7) |
| By genotype | ||||
| Entire Population |
|
|
<0.0001 |
|
| GG (%) |
91 (36.5) |
77 (64.2) |
|
|
| TG (%) |
97 (39) |
36 (30.3) |
0.0012 |
2.3 (1.4–3.7) |
| TT (%) |
61 (24) |
6 (5) |
<0.0001 |
8.6 (3.5–20.8) |
| Ashkenazi Jews |
|
|
<0.0001 |
|
| GG (%) |
67 (42.1) |
45 (66.2) |
|
|
| TG (%) |
55 (34.6) |
20 (29.4) |
0.17 |
1.6 (3.1–0.82) |
| TT (%) |
37 (23.3) |
3 (4.4) |
0.003 |
9.7 (41.6–2.2) |
| Sephardic Jews |
|
|
0.0003 |
|
| GG (%) |
20 (27) |
26 (65) |
|
|
| TG (%) |
36 (48.6) |
11 (27.5) |
0.003 |
4.3 (10.4–1.7) |
| TT (%) |
18 (24.3) |
3 (7.5) |
0.001 |
7.8 (30.3–2) |
| Arabs# |
|
|
0.036 |
|
| GG (%) |
3 (30) |
6 (60) |
|
|
| TG (%) |
2 (20) |
4 (40) |
|
|
| TT (%) | 5 (50) | 0 (0) | ||
Comparison of the frequency (%) of the LOC387715/ARMS2 (rs10490924) variant between NVAMD patients and controls in the entire Israeli population and in the Ashkenazi, Sephardic, and Arab subpopulations. Increased prevalence of the T variant was associated with the disease in the entire population as well as in Ashkenazi, Sephardic, and Arab subpopulation. CI- confidence interval, OR- odds ratio. The asterisk indicates reliable genotyping for rs10490924 was obtained from 249 of the 255 patients and for each of the 119 controls. The hash mark represents there were too few Arabs to obtain reliable ORs and CIs for analysis of genotypes in this sub population.
Table 3. Frequency of HTRA1 (rs11200638) alleles and genotypes in NVAMD patients and unaffected controls.
| HTRA1 (rs1200638) | AMD | Unaffected | p | OR (95% CI) |
|---|---|---|---|---|
| By allele | ||||
| Entire population (G/A; %) |
325/185
(63.7/36.3) |
197/41
(82.8/17.2) |
<0.0001 |
2.7 (1.9–4) |
| Ashkenazi Jews |
212/114
(65/35) |
114/22
(83.8/16.2) |
<0.0001 |
2.8 (1.7–4.6) |
| Sephardic Jews |
95/55
(63/37) |
65/15
(81.3/18.7) |
0.0065 |
2.5 (1.3–4.8) |
| Arabs |
10/12
(45.4/54.6) |
17/3
(0.85/0.15) |
0.01 |
6.8 (1.5–30.1) |
| By genotype | ||||
| Entire population |
|
|
<0.0001 |
|
| GG (%) |
116 (45.5) |
81 (68.1) |
|
|
| GA (%) |
93 (36.5) |
35 (29.4) |
0.013 |
1.8 (1.1–3.0) |
| AA (%) |
46 (18.0) |
3 (2.5) |
<0.0001 |
10.7 (3.2–35.7) |
| Ashkenazi Jews |
|
|
0.011 |
|
| GG (%) |
80 (49.1) |
48 (70.6) |
|
|
| GA (%) |
52 (31.9) |
18 (26.5) |
0.24 |
1.5 (3–0.76) |
| AA (%) |
31 (19) |
2 (2.9) |
0.009 |
7.2 (31.2–1.6) |
| Sephardic Jews |
|
|
0.02 |
|
| GG (%) |
30 (40) |
26 (65) |
|
|
| GA (%) |
35 (46.7) |
13 (32.5) |
0.044 |
2.3 (5.3–1.02) |
| AA (%) |
10 (13.3) |
1 (2.5) |
0.046 |
8.7 (71.5–1.04) |
| Arabs# |
|
|
0.05 |
|
| GG (%) |
4 (36.4) |
7 (70) |
|
|
| GA (%) |
2 (18.1) |
3 (30) |
|
|
| AA (%) | 5 (45.5) | 0 (0) | ||
Comparison of the frequency (%) of the HTRA1 (rs11200638) variant between NVAMD patients and controls in the entire Israeli population and in the Ashkenazi, Sephardic, and Arab subpopulations. Increased prevalence of the A variant was associated with the disease in the entire population as well as in Ashkenazi, Sephardic, and Arab subpopulation. CI- confidence interval, OR- odds ratio. The hash mark represents there were too few Arabs to obtain reliable ORs and CIs for analysis of genotypes in this sub population.
Homozygotes for the T allele of rs10490924 (LOC387715/ARMS2) had an odds ratio (OR) of 8.6 with a 95% confidence interval (CI) of 3.5–20.8, while heterozygotes had an OR of 2.3 (95% CI of 1.4–3.7) for having NVAMD compared with homozygotes for the wild-type allele (Table 2). Combined, individuals either homozygous or heterozygous for the T allele had an OR of 3.2 (95% CI of 2–5; p<0.0001) compared with individuals homozygous for the G allele for having AMD. Analysis according to allele distribution showed similar findings to analysis according to genotypes for each of the SNPs (Table 2).
Homozygotes for the A allele of rs11200638 had an OR of 10.7 (95% CI of 3.2–35.7), while heterozygotes had an OR of 1.8 (95% CI of 1.1–3) for having NVAMD compared with homozygotes for the wild-type allele (Table 3). Combined, individuals either homozygous or heterozygous for the A allele had an OR of 2.5 (95% CI of 1.6–4; p<0.0001) compared with individuals homozygous for the G allele for having AMD.
Subgroup analysis was performed to evaluate for association of both SNPs with NVAMD among Ashkenazi Jews, Sephardic Jews, and Arabs. The rs10490924 (LOC387715/ARMS2) SNP was associated with NVAMD among Ashkenazi Jews (p<0.0001, χ2 test), Sephardic Jews (p=0.0003, chi-square test), and Arabs (p=0.036, χ2 test). Analysis according to allele distribution showed similar findings (Table 2). The rs11200638 (HTRA1) SNP was also associated with NVAMD among Ashkenazi Jews (p=0.011, χ2 test), Sephardic Jews (p=0.044, χ2 test), and Arabs (p=0.05, χ2 test). Analysis according to allele distribution showed similar findings (Table 3).
Smoking is an established risk factor for AMD. Since this risk factor was also associated with chromosome 10q26 SNPs in other populations [11], we evaluated its effect in our population. Smoking was associated with the risk for having AMD (OR of 3; 95% CI of 1.7–5.3; p<0.001) in the entire population; however, logistic regression found no interactions between smoking and either homozygosity (p=0.359) or heterozygosity for the risk allele of rs10490924. There were also no interactions between smoking and either homozygosity (p=0.99) or heterozygosity (p=0.29) for the risk allele of rs11200638.
Association of 10q26 SNPs with phenotype of NVAMD
Following the establishment of an association among the HTRA1 rs11200638 SNP, the LOC387715/ARMS2 rs10490924 SNP, and NVAMD in the Israeli population, we explored possible correlations between the genotypes and clinical characteristics of NVAMD and response to PDT. This analysis included 143 sequential NVAMD patients who were treated in the Department of Ophthalmology of the Hadassah Medical Center and who were characterized in terms of phenotype and response to PDT. Reliable genotyping for rs11200638 was obtained from all of these participants and for rs10490924 from 139 of these patients.
There was no significant association between these SNPs and gender, history of smoking, lesion type, initial and final visual acuity, and number of PDT sessions required. The T allele of rs10490924 was associated with a positive family history for AMD while the A allele of rs11200638 showed a trend toward such an association (Table 4 and Table 5). While 23.7% of individuals carrying at least one risk allele (A) of rs11200638 had a positive family history for AMD, only 8.6% of the individuals homozygous to the wild-type allele had a positive family history (p=0.043, χ2 test). However, this p value would lose significance if any form of multiple hypothesis testing correction was applied. Similarly, while 22.8% of individuals carrying at least one risk allele (T) of rs10490924 had positive family history for AMD, only 4.4% of the individuals homozygous for the wild-type allele had a positive family history (p=0.008, chi square test). The risk alleles of both SNPs showed a trend toward an association with younger age of onset of NVAMD and with larger lesion size (Table 4 and 5).
Table 4. Correlation among demographic and phenotypic characteristics of NVAMD and genotyping for LOC387715/ARMS2 (rs10490924).
|
LOC387715/ARMS2 (rs10490924) genotype |
||||
|---|---|---|---|---|
| TT | TG | GG | p | |
| Gender (female/male) |
21/14 |
24/27 |
20/33 |
0.12 |
| Lesion type* (classic/occult) |
11/24 |
23/28 |
19/34 |
0.4 |
| Family History of AMD (yes/no) # |
9/19 |
7/35 |
2/43 |
0.006 |
| Age (mean±SD, in years) |
76.4±8 |
78.9±8.5 |
80.1±7.2 |
0.1 |
| Smoking (yes/no) # |
12/20 |
25/25 |
27/25 |
0.42 |
| Initial VA (mean±SD, logMAR) |
1.06±0.83 |
0.94±0.68 |
1.14±0.79 |
0.44 |
| Lesion size (mean±SD, in µm) |
4081±1347 |
3669±1265 |
3623±1288 |
0.34 |
| Number of PDT sessions |
2.2±2.2 |
2.6±1.9 |
2±1.6 |
0.22 |
| Final VA (mean±SD, logMAR) | 1.5±0.9 | 1.51±0.83 | 1.37±0.93 | 0.67 |
The asterisk indicates that classic lesions included predominantly classic and pure classic lesions. Occult lesions included minimally classic and occult lesions. Analysis according to four lesion types showed similar results. # Reliable information on family history was obtained from 117 out of the 143 participants included in this analysis, and smoking history was obtained from 138 of the 143 study participants.
Table 5. Correlation among demographic and phenotypic characteristics of NVAMD and genotyping for HTRA1 (rs11200638).
|
HTRA1 (rs11200638) genotype |
||||
|---|---|---|---|---|
| AA | GA | GG | p | |
| Gender (female/male) |
14/10 |
25/25 |
29/40 |
0.35 |
| Lesion type* (classic/occult) |
7/17 |
22/28 |
26/43 |
0.46 |
| Family History of AMD (yes/no) # |
5/14 |
9/31 |
5/53 |
0.08 |
| Age (mean ± SD, in years) |
75.6±7 |
79.2±8.5 |
79.5±7.8 |
0.1 |
| Smoking (yes/no) # |
11/12 |
22/26 |
33/34 |
0.93 |
| Initial VA (mean ± SD, logMAR) |
1.1±0.8 |
0.928±0.7 |
1.1±0.8 |
0.38 |
| Lesion size (mean ± SD, in µm) |
4250±1261 |
3696±1290 |
3603±1261 |
0.17 |
| Number of PDT sessions |
2.5±1.9 |
2.4±1.8 |
2.2±1.9 |
0.78 |
| Final VA (mean ± SD, logMAR) | 1.6±0.84 | 1.5±0.84 | 1.36±0.94 | 0.45 |
The asterisk indicates classic lesions included predominantly classic and pure classic lesions. Occult lesions included minimally classic and occult lesions. Analysis according to four lesion types showed similar results. # Reliable information on family history was obtained from 117 out of the 143 participants included in this analysis, and smoking history was obtained from 138 of the 143 study participants.
mRNA levels of HTRA1 in WBCs from NVAMD patients
mRNA levels of HTRA1 were measured using QPCR in WBCs from 22 NVAMD patients and 5 unaffected controls to assess the effect of the HTRA1 promoter variant on HTRA1 expression. ANOVA showed no significant differences in HTRA1 mRNA levels among individuals with different rs11200638 genotypes (Figure 1A), and between patients and controls (Figure 1B). HTRA1 expression levels were similar in homozygote of the A allele and homozygote of the G allele (p=0.2).
Figure 1.
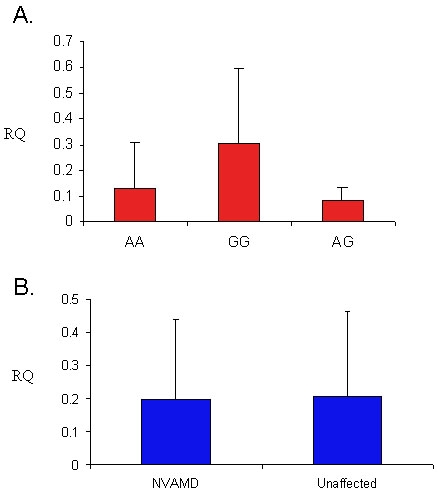
Relative expression levels of HTRA1 in NVAMD patients and unaffected controls. A: Shown are expression levels in white blood cells according to rs11200638 genotypes in individuals homozygous for the wild-type allele (GG, n=13), participants homozygous for the risk allele (AA, n=6), and in heterozygous participants (AG, n=8). Differences between the three groups were not significant (p>0.1 for each comparison). B: Shown are mRNA levels of HTRA1 in 22 NVAMD patients and 5 unaffected individuals (p=0.9). In both A and B, RQ represents relative HTRA1 mRNA levels. Error bars represent standard deviation (SD).
Discussion
This study established an association between the A allele of rs11200638, located in the putative promoter region of HTRA1, and the T allele of rs10490924 (A69S variant), located in the coding region of LOC387715/ARMS2, and NVAMD in the Israeli population. Both SNPs are strongly associated with NVAMD among Ashkenazi and Sephardic Jews. While NVAMD is relatively uncommon among Arabs in Israel [34], our data suggest that 10q26 SNPs are also associated with NVAMD in this ethnic group. Similar to findings from fair-skinned populations and Japanese populations, these SNPs are in almost complete linkage disequilibrium in the Israeli populations [1,7,8].
Increased prevalence of the same HTRA1 and LOC387715/ARMS2 variants among AMD patients was described in several fair-skinned populations as well as in cohorts from India, Japan, and China [1-8,21,35-39]. The magnitude of the association between these SNPs and NVAMD in Israel is similar to the one which was reported in other populations. For example, Rivera and colleagues [13] found an OR of 2.7 for AMD patients who were heterozygous for the LOC387715/ARMS2 A69S variant and an OR of 8.2 for patients who were homozygous for it . In the Israeli population participants who were heterozygous for this variant had an OR of 2.3 while those homozygous for it had an OR of 8.6 for having AMD. A stronger linkage of these SNPs with neovascular AMD compared with dry AMD was suggested by previous studies [2,9,40]. As our study focused on NVAMD patients, we were not able to evaluate for differences in the magnitude of the association between the HTRA1 and LOC387715/ARMS2 variants and the dry and neovascular forms of AMD.
This study also evaluated if variable phenotype and response to therapy among NVAMD patients may be attributable to the chromosome 10q26 SNPs. The LOC387715/ARMS2 A69S variant was associated with younger age of examination for AMD, but not with other phenotypic characteristics of NVAMD in a recent study [40]. In accordance with that, a trend toward younger age of onset of NVAMD was associated with both the HTRA1 and LOC387715/ARMS2 variants in our study. Another observation suggesting an aggressive phenotype in association with the LOC387715/ARMS2 variant was reported by Brantley and colleagues [10] who proposed that this variant is associated with larger neovascular lesions. A trend toward an association of both HTRA1 and LOC387715/ARMS2 variants with larger neovascular lesions was also observed in this study. While association of the HTRA1 variant with the classic lesion type was described in a French population [26], such association was not identified in another fair-skinned population which was evaluated [10] or in the Israeli population.
Brantley and coworkers did not find an association between LOC387715/ARMS2 A69S variant and response to bevacizumab injections in 86 patients. The same group did not find an association between this variant and response to PDT in 69 patients [10,25]. Yet, these authors found an association between response to bevacizumab injections and the CFH Y402H polymorphism. We confirmed this observation with respect to the LOC387715/ARMS2 variant and expanded it to include the HTRA1 variant in 143 Israeli NVAMD patients. Both variants were not associated with the visual outcome or with the number of PDT sessions required. Combined, these data do not support the existence of a major pharmacogenetic interactions among chromosome 10q26 SNPs and current therapies for NVAMD.
It is still unclear if the HTRA1 variant or the LOC387715/ARMS2 variant has a causative role in the pathogenesis of AMD. While Dewan and Young and their colleagues [7,8] suggested that the HTRA1 variant is more strongly associated with the disease, Kanda and colleagues [6] suggested that LOC387715/ARMS2 rather than the HTRA1 variants explain the association of the chromosome 10q26 region with the disease. Since the variants in these genes are in almost complete linkage disequilibrium in each of the populations reported so far, additional functional studies are required to determine which variant is involved in the pathogenesis of the disease.
In that respect, conflicting evidence has been reported regarding the functional significance of the HTRA1 promoter variant. Yang and colleagues [7] observed that this variant is associated with increased expression levels of the HTRA1 gene in 3 AMD patients homozygous to G allele of the HTRA1 promoter variant compared with 3 control patients homozygous to the A wild-type allele. Dewan and colleagues [8] described increased HTRA1 expression associated with the promoter variant. These authors speculated that altered HTRA1 expression levels might be involved in AMD. However, Kanda and colleagues [6] suggested that the same HTRA1 variant does not affect HTRA1 expression levels in cell lines or in the retina. We have also failed to identify an association between the HTRA1 variant and HTRA1 mRNA levels in WBCs. Yet, this data does not preclude an effect of the same promoter variant on retinal expression levels of HTRA1 in the context of AMD.
It is still unclear which of the chromosome 10q26 variants is involved in the pathogenesis of AMD. However, our data show that additional genetic and environmental factors which underlie variable phenotype and response to therapy in NVAMD are yet to be identified.
Acknowlegments
We thank the individuals who provided blood samples for the purpose of the study. This study was supported in part by a grant from the Israel Science Fund (624/05).
References
- 1.Yoshida T, DeWan A, Zhang H, Sakamoto R, Okamoto H, Minami M, Obazawa M, Mizota A, Tanaka M, Saito Y, Takagi I, Hoh J, Iwata T. HTRA1 promoter polymorphism predisposes Japanese to age-related macular degeneration. Mol Vis. 2007;13:545–8. [PMC free article] [PubMed] [Google Scholar]
- 2.Mori K, Horie-Inoue K, Kohda M, Kawasaki I, Gehlbach PL, Awata T, Yoneya S, Okazaki Y, Inoue S. Association of the HTRA1 gene variant with age-related macular degeneration in the Japanese population. J Hum Genet. 2007;52:636–41. doi: 10.1007/s10038-007-0162-1. [DOI] [PubMed] [Google Scholar]
- 3.Lu F, Hu J, Zhao P, Lin Y, Yang Y, Liu X, Fan Y, Chen B, Liao S, Du Q, Lei C, Cameron DJ, Zhang K, Yang Z. HTRA1 variant increases risk to neovascular age- related macular degeneration in Chinese population. Vision Res. 2007;47:3120–3. doi: 10.1016/j.visres.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Leveziel N, Souied EH, Richard F, Barbu V, Zourdani A, Morineau G, Zerbib J, Coscas G, Soubrane G, Benlian P. PLEKHA1- LOC387715/ARMS2-HTRA1 polymorphisms and exudative age-related macular degeneration in the French population. Mol Vis. 2007;13:2153–9. [PubMed] [Google Scholar]
- 5.Kondo N, Honda S, Ishibashi K, Tsukahara Y, Negi A. LOC387715/ARMS2/HTRA1 variants in polypoidal choroidal vasculopathy and age-related macular degeneration in a Japanese population. Am J Ophthalmol. 2007;144:608–12. doi: 10.1016/j.ajo.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Kanda A, Chen W, Othman M, Branham KE, Brooks M, Khanna R, He S, Lyons R, Abecasis GR, Swaroop A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci USA. 2007;104:16227–32. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, Chien J, Dewan A, Harmon J, Bernstein PS, Shridhar V, Zabriskie NA, Hoh J, Howes K, Zhang K. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–3. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 8.Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, Barnstable C, Pang CP, Hoh J. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–92. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 9.Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age- related macular degeneration. JAMA. 2007;297:1793–800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- 10.Brantley MA, Jr, Fang AM, King JM, Tewari A, Kymes SM, Shiels A. Association of Complement Factor H and LOC387715 Genotypes with Response of Exudative Age-Related Macular Degeneration to Intravitreal Bevacizumab. Ophthalmology. 2007;114:2168–73. doi: 10.1016/j.ophtha.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt S, Hauser MA, Scott WK, Postel EA, Agarwal A, Gallins P, Wong F, Chen YS, Spencer K, Schnetz-Boutaud N, Haines JL, Pericak-Vance MA. Cigarette smoking strongly modifies the association of LOC387715 and age-related macular degeneration. Am J Hum Genet. 2006;78:852–64. doi: 10.1086/503822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conley YP, Jakobsdottir J, Mah T, Weeks DE, Klein R, Kuller L, Ferrell RE, Gorin MB. CFH, ELOVL4, PLEKHA1 and LOC387715 genes and susceptibility to age-related maculopathy: AREDS and CHS cohorts and meta-analyses. Hum Mol Genet. 2006;15:3206–18. doi: 10.1093/hmg/ddl396. [DOI] [PubMed] [Google Scholar]
- 13.Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, Meitinger T, Weber BH. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–36. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 14.Tam PO, Ng TK, Liu DT, Chan WM, Chiang SW, Chen LJ, DeWan A, Hoh J, Lam DS, Pang CP. HTRA1 variants in exudative age-related macular degeneration and interactions with smoking and CFH. Invest Ophthalmol Vis Sci. 2008;49:2357–65. doi: 10.1167/iovs.07-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs D, Yang Z, Constantine R, Ma X, Camp NJ, Yang X, Chen H, Jorgenson A, Hau V, Dewan A, Zeng J, Harmon J, Buehler J, Brand JM, Hoh J, Cameron DJ, Dixit M, Tong Z, Zhang K. Further mapping of 10q26 supports strong association of HTRA1 polymorphisms with age-related macular degeneration. Vision Res. 2008;48:685–9. doi: 10.1016/j.visres.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Yang Z, Gibbs D, Yang X, Hau V, Zhao P, Ma X, Zeng J, Luo L, Pearson E, Constantine R, Kaminoh Y, Harmon J, Tong Z, Stratton CA, Cameron DJ, Tang S, Zhang K. Association of HTRA1 polymorphism and bilaterality in advanced age-related macular degeneration. Vision Res. 2008;48:690–4. doi: 10.1016/j.visres.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Hughes AE, Orr N, Patterson C, Esfandiary H, Hogg R, McConnell V, Silvestri G, Chakravarthy U. Neovascular age-related macular degeneration risk based on CFH, LOC387715/HTRA1, and smoking. PLoS Med. 2007;4:e355. doi: 10.1371/journal.pmed.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pulido JS, Peterson LM, Mutapcic L, Bryant S, Highsmith WE. LOC387715/HTRA1 and complement factor H variants in patients with age- related macular degeneration seen at the mayo clinic. Ophthalmic Genet. 2007;28:203–7. doi: 10.1080/13816810701649617. [DOI] [PubMed] [Google Scholar]
- 19.Chan CC, Shen D, Zhou M, Ross RJ, Ding X, Zhang K, Green WR, Tuo J. Human HTRA1 in the archived eyes with age- related macular degeneration. Trans Am Ophthalmol Soc. 2007;105:92–7. [PMC free article] [PubMed] [Google Scholar]
- 20.Canfield AE, Hadfield KD, Rock CF, Wylie EC, Wilkinson FL. HTRA1: a novel regulator of physiological and pathological matrix mineralization? Biochem Soc Trans. 2007;35:669–71. doi: 10.1042/BST0350669. [DOI] [PubMed] [Google Scholar]
- 21.Fritsche LG, Loenhardt T, Janssen A, Fisher SA, Rivera A, Keilhauer CN, Weber BH. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40:892–6. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 22.Brantley MA, Edelstein SL, King JM, Apte RS, Kymes SM, Shiels A. Clinical Phenotypes Associated with the Complement Factor H Y402H Variant in Age-related Macular Degeneration. Am J Ophthalmol. 2007;144:404–8. doi: 10.1016/j.ajo.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goverdhan SV, Hannan S, Newsom RB, Luff AJ, Griffiths H, Lotery AJ. An analysis of the CFH Y402H genotype in AMD patients and controls from the UK, and response to PDT treatment. Eye. 2008;22:849–54. doi: 10.1038/sj.eye.6702830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wegscheider BJ, Weger M, Renner W, Steinbrugger I, März W, Mossböck G, Temmel W, El-Shabrawi Y, Schmut O, Jahrbacher R, Haas A. Association of complement factor H Y402H gene polymorphism with different subtypes of exudative age- related macular degeneration. Ophthalmology. 2007;114:738–42. doi: 10.1016/j.ophtha.2006.07.048. [DOI] [PubMed] [Google Scholar]
- 25.Brantley MA, Jr, Edelstein SL, King JM, Plotzke MR, Apte RS, Kymes SM, Shiels A. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to photodynamic therapy. Eye. 2008 doi: 10.1038/eye.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leveziel N, Zerbib J, Richard F, Querques G, Morineau G, Fremeaux-Bacchi V, Coscas G, Soubrane G, Benlian P, Souied EH. Genotype-phenotype correlations for exudative age-related macular degeneration associated with homozygous HTRA1 and CFH genotypes. Invest Ophthalmol Vis Sci. 2008;49:3090–4. doi: 10.1167/iovs.07-1540. [DOI] [PubMed] [Google Scholar]
- 27.Seitsonen S, Jarvela I, Meri S, Tommila P, Ranta P, Immonen I. Complement factor H Y402H polymorphism and characteristics of exudative age-related macular degeneration lesions. Acta Ophthalmol. 2008;86:390–4. doi: 10.1111/j.1600-0420.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 28.Seitsonen SP, Jarvela IE, Meri S, Tommila PV, Ranta PH, Immonen IJ. The effect of complement factor H Y402H polymorphism on the outcome of photodynamic therapy in age-related macular degeneration. Eur J Ophthalmol. 2007;17:943–9. doi: 10.1177/112067210701700612. [DOI] [PubMed] [Google Scholar]
- 29.A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–36. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chowers I, Cohen Y, Goldenberg-Cohen N, Vicuna-Kojchen J, Lichtinger A, Weinstein O, Pollack A, Axer-Siegel R, Hemo I, Averbukh E, Banin E, Meir T, Lederman M. Association of complement factor H Y402H polymorphism with phenotype of neovascular age related macular degeneration in Israel. Mol Vis. 2008;14:1829–34. [PMC free article] [PubMed] [Google Scholar]
- 31.Nebel A, Filon D, Brinkmann B, Majumder PP, Faerman M, Oppenheim A. The Y chromosome pool of Jews as part of the genetic landscape of the Middle East. Am J Hum Genet. 2001;69:1095–112. doi: 10.1086/324070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subfoveal neovascular lesions in age-related macular degeneration: guidelines for evaluation and treatment in the macular photocoagulation study. Arch Ophthalmol. 1991;109:1242–57. [PubMed] [Google Scholar]
- 33.Bressler NM. Photodynamic therapy of subfoveal choroidal neovascularization in age-related macular degeneration with verteporfin: two- year results of 2 randomized clinical trials-tap report 2. Arch Ophthalmol. 2001;119:198–207. [PubMed] [Google Scholar]
- 34.Asleh SA, Chowers I. Ethnic background as a risk factor for advanced age- related macular degeneration in Israel. Isr Med Assoc J. 2007;9:656–8. [PubMed] [Google Scholar]
- 35.Cameron DJ, Yang Z, Gibbs D, Chen H, Kaminoh Y, Jorgensen A, Zeng J, Luo L, Brinton E, Brinton G, Brand JM, Bernstein PS, Zabriskie NA, Tang S, Constantine R, Tong Z, Zhang K. HTRA1 variant confers similar risks to geographic atrophy and neovascular age-related macular degeneration. Cell Cycle. 2007;6:1122–5. doi: 10.4161/cc.6.9.4157. [DOI] [PubMed] [Google Scholar]
- 36.Weger M, Renner W, Steinbrugger I, Köfer K, Wedrich A, Groselj-Strele A, El-Shabrawi Y, Schmut O, Haas A. Association of the HTRA1 - 625G>A promoter gene polymorphism with exudative age-related macular degeneration in a Central European population. Mol Vis. 2007;13:1274–9. [PubMed] [Google Scholar]
- 37.Lin JM, Wan L, Tsai YY, Lin HJ, Tsai Y, Lee CC, Tsai CH, Tsai FJ, Tseng SH. HTRA1 polymorphism in dry and wet age- related macular degeneration. Retina. 2008;28:309–13. doi: 10.1097/IAE.0b013e31814cef3a. [DOI] [PubMed] [Google Scholar]
- 38.Deangelis MM, Ji F, Adams S, Morrison MA, Harring AJ, Sweeney MO, Capone A, Jr, Miller JW, Dryja TP, Ott J, Kim IK. Alleles in the HtrA serine peptidase 1 gene alter the risk of neovascular age-related macular degeneration. Ophthalmology. 2008;115:1209–15. doi: 10.1016/j.ophtha.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaur I, Katta S, Hussain A, Hussain N, Mathai A, Narayanan R, Hussain A, Reddy RK, Majji AB, Das T, Chakrabarti S. Variants in the 10q26 Gene Cluster (LOC387715 and HTRA1) Exhibit Enhanced Risk of Age-Related Macular Degeneration along with CFH in Indian Patients. Invest Ophthalmol Vis Sci. 2008;49:1771–6. doi: 10.1167/iovs.07-0560. [DOI] [PubMed] [Google Scholar]
- 40.Shuler RK, Jr, Schmidt S, Gallins P, Hauser MA, Scott WK, Caldwell J, Agarwal A, Haines JL, Pericak-Vance MA, Postel EA. Phenotype Analysis of Patients With the Risk Variant LOC387715 (A69S) in Age-related Macular Degeneration. Am J Ophthalmol. 2008;145:303–7. doi: 10.1016/j.ajo.2007.09.027. [DOI] [PubMed] [Google Scholar]


