Abstract
Activation of the classical and noncanonical NF-κB pathways by ligation of the lymphotoxin (LT)-β receptor (LTβR) plays a crucial role in lymphoid organogenesis and in the generation of ectopic lymphoid tissue at sites of chronic inflammation. Within these microenvironments, LTβR signaling regulates the phenotype of the specialized high endothelial cells. However, the direct effects of LTβR ligation on endothelial cells remain unclear. We therefore questioned whether LTβR ligation could directly activate endothelial cells and regulate classical and noncanonical NF-κB-dependent gene expression. We demonstrate that the LTβR ligands LIGHT and LTα1β2 activate both NF-κB pathways in HUVECs and human dermal microvascular endothelial cells (HDMEC). Classical pathway activation was less robust than TNF-induced signaling; however, only LIGHT and LTα1β2 and not TNF activated the noncanonical pathway. LIGHT and LTα1β2 induced the expression of classical NF-κB-dependent genes in HUVEC, including those encoding the adhesion molecules E-selectin, ICAM-1, and VCAM-1. Consistent with this stimulation, LTβR ligation up-regulated T cell adhesion to HUVEC. Furthermore, the homeostatic chemokine CXCL12 was up-regulated by LIGHT and LTα1β2 but not TNF in both HUVEC and HDMEC. Using HUVEC retrovirally transduced with dominant negative IκB kinase α, we demonstrate that CXCL12 expression is regulated by the noncanonical pathway in endothelial cells. Our findings therefore demonstrate that LTβR ligation regulates gene expression in endothelial cells via both NF-κB pathways and we identify CXCL12 as a bona fide noncanonical NF-κB-regulated gene in these cells.
Vascular endothelial cells (EC)3 play a crucial role in normal immune and inflammatory responses and are a target of immunoregulatory and proinflammatory cytokines. Activation of EC by these factors up-regulates a wide variety of genes including chemokines, leukocyte adhesion molecules, antiapoptotic proteins, growth factors, immunomodulatory proteins, and regulators of vascular cell function. Dysregulation of this gene expression underlies the pathological role of EC in many diseases including autoimmunity, acute and chronic inflammation, and cardiovascular diseases such as atherosclerosis (1, 2). Consequently, the signaling pathways that control EC activation have been extensively studied and the transcription factor NF-κB has been identified as critical for expression of many of the genes associated with EC pathology (1–3).
NF-κB describes a family of five related proteins named p65 (RelA), RelB, c-Rel, p105/p50 (NF-κB1), and p100/p52 (NF-κB2) that homodimerize or heterodimerize to form either transcriptionally active or repressive versions of NF-κB (4). NF-κB dimers are maintained inactive in the cytoplasm by inhibitory proteins named IκB, and NF-κB activation is initiated by phosphorylation of the IκB proteins leading to their ubiquitination and degradation by the proteasome. This allows NF-κB to translocate to the nucleus where it binds to target gene promoters and regulates their expression. The kinase that phosphorylates IκB proteins is a component of the IκB kinase (IKK) complex that consists of two catalytic subunits named IKKα (IKK1) and IKKβ (IKK2) and a noncatalytic regulatory component named NEMO (NF-κB essential modulator) or IKKγ (5– 8). Intriguingly, genetic studies targeting the IKK complex have delineated two distinct pathways leading to the activation of separate NF-κB proteins that regulate discrete panels of target genes (9 –11).
The best studied of these mechanisms activated by most NF-κB-inducing stimuli including TNF and LPS is termed the “classical” NF-κB pathway (4, 9, 12). This pathway functions in the absence of IKKα but requires both NEMO and IKKβ and is hallmarked by IKKβ-mediated IκB phosphorylation and activation of NF-κB proteins including the ubiquitously expressed p50:p65 heterodimer. Classical NF-κB signaling has been extensively studied in EC and underlies the regulated expression of most proinflammatory EC genes, including E-selectin, ICAM-1, VCAM-1, IL-1κ, and TNF (2, 3, 13, 14). The more recently described “noncanonical” or alternative NF-κB pathway is activated by a limited number of stimuli, including lymphotoxin (LT)- β receptor (LTβR) ligation by LTα1β2 and LIGHT (LT-related inducible ligand that competes for glycoprotein D binding to herpesvirus entry mediator on T cells, TNFSF14) (10, 15–20). LTβR ligation also activates the classical NF-κB pathway thereby distinguishing it from proinflammatory cytokines such as TNF that only induce the classical mechanism (12). Despite the wealth of knowledge concerning the role of the classical pathway in regulating the EC phenotype, functional activation of the noncanonical pathway in EC has not yet been demonstrated.
Noncanonical NF-κB activation involves NF-κB-inducing kinase (NIK)- and IKKα-dependent phosphorylation of the p100 NF-κB2 precursor protein leading to its processing to p52 (10, 11, 15, 16, 19, 21, 22). This processing results in the nuclear translocation of p52:RelB heterodimers that target a subset of genes not activated by classical NF-κB (9, 10), including the homeostatic, proangiogenic, and proinflammatory chemokine CXCL12 and the lymphoid chemokines CXCL13, CCL19, and CCL21 required for lymph node development (9, 10). Consistent with this result, LTβR signaling is critical for the normal development of secondary lymphoid organs and for the generation of ectopic lymphoid structures that frequently arise during chronic inflammation (20, 23–30). Intriguingly, intact LTβR and IKKα are required to maintain the phenotype of the specialized high EC (HEC) within lymph nodes (29–31) that also appear in certain chronic inflammatory lesions (31), strongly suggesting that LTβR ligation on nonspecialized EC regulates their phenotype and function. Despite this evidence, very little is known about the effects of direct LTβR ligation on EC.
Early studies demonstrated that HUVECs express LTβR, although its ligation did not activate NF-κB (32) or induce gene expression in these cells (33). As LTβR ligation activated NF-κB in non-EC cell lines, this led Mackay and colleagues (32, 33) to conclude that LTβR-induced NF-κB activation in LTβR-positive cells is cell type-specific and does not occur in EC. More recently LTα1β2 was shown to induce gene expression in HUVEC (34), although the signaling mechanisms underlying this remain unknown. Furthermore, as noncanonical NF-κB signaling has not been directly investigated in EC, it is not known whether this pathway or noncanonical NF-κB-dependent genes are regulated in response to LTβR ligation in these cells. To address these unknowns, we sought to determine whether the classical and noncanonical NF-κB pathways are activated in EC by the LTβR ligands LIGHT and LTα1β2 and whether these mechanisms regulate gene expression in vascular EC.
We report in this study that LIGHT and LTα1β2 activate both the classical and noncanonical NF-κB pathways in EC, and up-regulate T cell adhesion to human EC. Furthermore, LTβR ligation induces the expression of classical NF-κB-dependent genes in EC including E-selectin and CXCL2, although the levels of expression are less than that induced by TNF. We also show that LIGHT and LTα1β2 but not TNF induces CXCL12 expression in HUVEC and human dermal microvascular EC (HDMEC) thereby establishing CXCL12 as a cytokine-regulated gene in EC. Using HUVEC retrovirally transduced with dominant negative IKKα and IKKβ, we demonstrate that LTβR-induced CXCL12 expression requires IKKα and hence the noncanonical NF-κB pathway. Our study therefore establishes LTβR ligation as a mechanism that directly activates EC via both the classical and noncanonical NF-κB pathways and we provide direct evidence of noncanonical NF-κB-dependent gene expression in vascular EC.
Materials and Methods
Materials
Recombinant human TNF, LIGHT, and LTα1β2 were purchased from R&D Systems. Rabbit Anti-p100/p52 was from Upstate Biotechnology, rabbit anti-IκBα (C-21) was from Santa Cruz Biotechnology, rabbit anti-histone-3, rabbit anti-c-jun, and phospho-c-jun were from Cell Signaling Technology. Abs for EMSA supershift: p65 (C20) was from Santa Cruz Biotechnology, p52 was from Upstate Biotechnology. Isotype control IgG, anti-ICAM-1 (11C81), and VCAM-1 (1E10) were purchased from R&D Systems and FITC-conjugated goat anti-mouse IgG was from Chemicon International. Mouse anti-TNFR1 and LTβR were also from R&D Systems. Mouse anti-E-selectin (H4/18) was a gift from Dr. J. Pober (Yale University, New Haven, CT). NF-κB consensus oligonucleotide probes and PCR primers were purchased from Integrated DNA Technologies. Collagenase was from Worthington Biochemical. Real-time reagents: Taqman Fast Universal Master Mix, Power SYBR, and Taqman primer probe sets were purchased from Applied Biosystems.
Cell culture
HUVECs were isolated from discarded tissue in accordance with a protocol approved by the University of Pennsylvania Institutional Review Board. Following collagenase digestion (1 mg/ml in PBS) from the canulated umbilical vein, EC were serially cultured on gelatin-coated (J.T. Baker) tissue culture plastic (Falcon) in Medium 199 (M199) supplemented with 20% FCS, 200 μM L-glutamine (all Invitrogen Life Technologies), 50 μg/ml EC growth factor (Collaborative Biomedical Products), 100 μg/ml porcine heparin (Sigma-Aldrich), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen Life Technologies). Cells were passaged using trypsin/EDTA (0.05%; Invitrogen Life Technologies) and all experiments were performed using HUVEC at passage 2–3. HDMEC were purchased from Lonza Walkersville and grown in EGM2-MV medium (Lonza Walkersville).
T cell adhesion assays
For the fluorescence adhesion assay, ex vivo T cells were prepared from blood obtained from volunteer donors using the RosetteSep T cell isolation kit (StemCell Technologies) according to the manufacturer’s instructions. The use of these samples was approved by the Children’s Hospital of Philadelphia Institutional Review Board for the Protection of Human Subjects. Isolated T cells were washed twice with PBS and adjusted to a density of 5 × 106 cells/ml in RPMI 1640 before incubation with calcein AM (5 μM) for 30 min. Following incubation, cells were washed twice with M199 and resuspended to a density of 5 × 106 cells/ml before addition of 5 × 105 cells/well to the pretreated EC that were grown to confluence in 24-well trays. EC were incubated with T cells for 1 h before gentle washing of wells with M199 plus HEPES to detach nonadherent T cells. Cells were maintained in M199/HEPES and EC/T cell adhesion was determined by measuring the fluorescence (λEx = 494 nm, λEm = 517) using a Varioskan Microplate Reader (Thermo Scientific). For the cell count assay, adherent T cells in six randomly selected fields of view were counted microscopically (at a magnification ×100) by a blinded investigator. T cells were easily distinguished as small round-phase light cells compared with the EC monolayer. Statistical analysis of the stimulated vs control samples for both assays was performed using the TTEST function of Microsoft Excel.
Retroviral transduction
Phoenix cells were transiently transfected with retroviral constructs LZRS-EGFP, LZRS-IKKβK44M, or LZRS-IKKαSSAA using Fugene reagent (Roche) and selected for gene expression 24 h following transfection using puromycin (1 μg/ml). Puromycin-resistant cells were used to derive conditioned medium to provide a retroviral stock for HUVEC transduction. For transduction of primary HUVEC, M199 containing EC growth factor was removed, cells were washed with HBSS and incubated 5– 8 h with retroviral conditioned medium containing polybrene (8 μg/ml; Sigma-Aldrich). After incubation, retrovirus was removed and replaced with normal growth medium overnight. The transduction process was repeated a further 3–5 times with intermittent cell passage as required. Using this protocol, the percentage of HUVEC expressing the transgene is routinely >90%.
Immunoblotting
For all immunoblots, each well of a 12- or 6-well plate containing a confluent HUVEC monolayer was washed twice in ice-cold PBS then lysed by the addition of 50 –100 μl of TNT lysis buffer (50 mM Tris · Cl (pH 6.8), 150 mM NaCl, 1% Triton X-100) containing Complete Protease Inhibitor Cocktail (Roche), 2 mM NaF, and 2 mM β-glycerophosphate. After 20 min on ice, lysates were harvested by scraping. Protein content was determined using Coomassie Plus Reagent (Pierce), and for each sample, an equal amount of protein (10–20 μg) was fractionated by SDS-PAGE then transferred electrophoretically to polyvinylidene difluoride membrane (Immobilon-P; Millipore) and immunoblotted with the appropriate primary and species-specific HRP-conjugated secondary Abs (Jackson ImmunoResearch Laboratories). Detection of the bound Ab by chemiluminescence was performed using Luminol reagent according to the manufacturer’s instructions (Santa Cruz Biotechnology). Densitometry was performed on each resulting immunoblot using a Gel-Doc EQ image analysis system and the QuantityOne software package (Bio-Rad). Data are presented as representative blots and densitometry histograms derived from one of at least three replicate experiments performed using separate EC cultures.
Preparation of nuclear and cytosolic extracts and EMSA
EC were harvested after treatment by scraping into PBS at 4°C. Samples (1.5 × 106/sample) were centrifuged (1500 × g/5 min), then pellets were resuspended in 100 μl of NAR A buffer (10 mM HEPES (pH 7.9), 10 mM KCl, 0.1 mM EDTA (pH 8)) supplemented with sodium vanadate, sodium fluoride, and Complete Protease Inhibitor Cocktail (Roche) and left to swell on ice for 10 min. After incubation, Nonidet P-40 was added to generate a final concentration of 0.1% and samples were vortexed for 30 s before centrifugation at 4000 × g for 3 min. Supernatants (cytosolic extracts) were removed, and the nuclear pellet was resuspended in 50 μl of NAR C buffer (20 mM HEPES, 0.4 M NaCl, 1 mM EDTA) supplemented with protease and phosphatase inhibitors as described for NAR A. Samples were agitated for 15 min at 4°C then centrifuged at 16,000 × g for 20 min to generate the nuclear samples. All samples were either used immediately or snap frozen and stored at −80°C.
For gel-shift analysis, single-stranded complimentary oligonucleotides encompassing a consensus NF-κB site (upper strand: 5′-AGTTGAGGG GACTTTCCCAGGC-35′) were annealed and end-labeled with γ-32P[ATP] using T4 polynucleotide kinase (New England Biolabs). Labeled probe was purified using MiniQuick Spin Columns (Roche) according to the manufacturer’s instructions. For EMSA, 2–5 μg of nuclear extracts supplemented with 1 μg of polydeoxyinosinic-polydeoxycytidylic acid were incubated with an equal volume of 2X binding buffer (40 mM Tris · Cl (pH 7.9), 100 mM NaCl, 10 mM MgCl2, 2 mM EDTA, 20% Glycerol, 0.2% Nonidet P-40, 2 mM DTT, 1 mg/ml BSA) on ice for 10 min. After incubation, 1 μl of labeled probe was added and the sample was mixed and incubated at room temperature for 20 min before protein-DNA samples were separated in nondenaturing gels (89 mM Tris, 89 mM boric acid, 10 mM EDTA, 5% acrylamide) that were then dried and exposed to film. Supershift analysis was performed as described with an additional preincubation of samples with 1 μl of Ab at room temperature for 2 h before the addition of polydeoxyinosinic-polydeoxycytidylic acid and 32P γ-ATP. All EMSA and Supershift experiments were performed at least three times using separate EC cultures and the data shown are from one representative experiment.
Indirect immunofluorescence and FACS analysis
Indirect immunofluorescence was used to quantify the surface amount of TNFR1 and the LTβR. EC were harvested by collagenase digestion and washed in PBS containing 1% FCS before incubation for 30 min at 4°C with saturating amounts of each Ab or nonbinding isotype control IgG. Cells were washed three times before the addition of FITC-conjugated anti-mouse secondary Ab and incubation for a further 30 min at 4°C. Labeled cells were washed a further three times, fixed in paraformaldehyde (2%) and analyzed by FACS (FACSort; BD Biosciences). For indirect analysis of adhesion molecule expression EC were cytokine treated for times up to 24 h before harvesting cells with trypsin and proceeding as described incubating for 30 min with saturating amounts of H4/18 for E-selectin, 11C81 for ICAM-1 or 1E10 for VCAM-1. All FACS analyses were performed at least three times using separate EC cultures and the data shown in each experiment are from one representative experiment.
mRNA isolation of RNA and analysis
EC were treated as described before medium was removed, and RNA was extracted using RNeasy according to the manufacturer’s instructions (Qiagen). Samples were subjected to on column DNase digestion. First strand cDNA was derived from 500 ng of RNA from each treatment group using SuperScript II and oligo(dT) according to the manufacturer’s instructions (Invitrogen Life Technologies). Semiquantitative amplification of the target cDNA was performed using Taq polymerase (Invitrogen Life Technologies) using primers pairs as follows: E-selectin, 5′-GCTTCTGGCAGTTTCCGTT ATGG-3′ and 5′-ACAGCGAGCAAGGGAGAGTTAGAA-3′; CXCL2, 5′-GCCAGTGCTTGCAGACC-3′ and 5′-ATGGGAGAGTGTGCAAGTAG-3′; CXCL12, 5′-GAA CGC CAA GGT CGT GGT CG-3′ and 5′-CTT TAG CTT CGG GTC AAT G-3′; and β-actin, 5′-TCAGCAAGCAGGAGTATGA CGAG-3′ and 5′-ATTGTGAACTTTGGGGGATGC-3′.
Targets were amplified using the following PCR parameters: 94°C for 2 min, then 25–35 cycles of 94°C for 45 s, 58°C for 30s followed by 70°C for 30 s. Replicates of samples were removed at each cycle between 25 and 35 cycles to ensure product amplification was in the linear range and this was established for each reaction. No reverse transcriptase controls were included in each experiment (data not shown) to ensure no genomic DNA contamination.
For quantitative real-time PCR analysis of E-selectin, and β-actin primers were used as described using Power SYBR (Applied Biosystems), according to the manufacturer’s instructions. Using the ABI 7500 Real-Time PCR system, PCR products were generated in triplicate or quadruplicate and normalized to β-actin also generated in triplicate or quadruplicate. Relative quantification (RQ) was derived from the difference in cycle threshold (Ct) between the gene of interest and β-actin using the equation RQ = 2−Δ ΔCt and analyzed using SDS v1.3 Software. Data in each experiment are displayed as the average RQ plus RQmax calculated on 95% confidence intervals and are representative of at least three independent experiments. PCR product specificity was confirmed by performing a dissociation curve at the end of each experiment. For the quantitative analysis of CXCL2, CXCL12, and β-actin, TaqMan primer probe sets were used (Applied Biosystems). PCR product generation and analysis was performed as described.
Results
LTβR ligation activates NF-κB in HUVEC
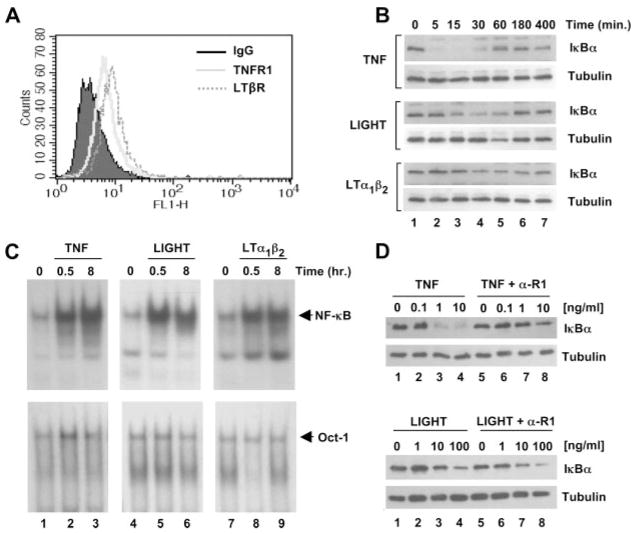
We performed FACS analysis to verify that our HUVEC cultures expressed the LTβR and confirmed that surface expression was similar to that observed for TNFR1 (Fig. 1A). To compare the downstream effects of LTβR ligation with TNF-induced NF-κB activation, we incubated HUVEC for a range of times up to 400 min with TNF, LIGHT, or LTα1β2. Surprisingly in light of previous studies (32), we found that both LIGHT and LTα1β2 incubation led to the degradation and reappearance of IκBα, which is central to the classical NF-κB activation pathway (Fig. 1B). Although we consistently observed stronger IκBα degradation in response to LIGHT than LTα1β2, maximal effects for both cytokines occurred after 30 min (Fig. 1B, lane 4) and IκBα levels returned to basal after 180 – 400 min (Fig. 1B, lanes 6 and 7). The response to LIGHT and LTα1β2 was not as robust or rapid as TNF-induced IκBα degradation that was complete after 5 min (Fig. 1B, upper panel, lane 2) and returned to basal levels after 60 min (Fig. 1B, upper panel, lane 5). Despite this difference, EMSA revealed that all three cytokines induce the nuclear accumulation of DNA-binding NF-κB complexes in HUVEC (Fig. 1C). NF-κB was detected by EMSA 30 min after incubation with each cytokine (Fig. 1C, lanes 2, 5, and 8) and remained detectable after 8 h (Fig. 1C, lanes 3, 6, and 9), demonstrating that similar to TNF, LIGHT, and LTα1β2 induce rapid and sustained NF-κB activity in HUVEC. To ensure that the effects of LIGHT and LTβR were not due to nonspecific activation of TNFR1 or by release of TNF from EC leading to feedback activation, we incubated HUVEC with a blocking anti-TNFR1 Ab and analyzed IκBα degradation. Anti-TNFR1 blocked TNF-induced IκBα degradation (Fig. 1D, top panels), whereas degradation in response to LIGHT (Fig. 1D, bottom panels) or LTα1β2 (data not shown) remained intact. We therefore conclude that LTβR ligation directly activates HUVEC leading to IκBα degradation and NF-κB activation.
FIGURE 1.
LIGHT and LTα1β2 activate NF-κB in HUVEC. A, HUVEC were harvested by collagenase digestion, stained by indirect immunofluorescence and analyzed by FACS for the expression of TNFR1 (gray solid histogram) or LTβR (gray dotted histogram) compared with staining using an isotype-matched control IgG (filled histogram). B, HUVEC treated for the times indicated with TNF (10 ng/ml), LIGHT (100 ng/ml), or LTα1β2 (100 ng/ml) were lysed and immunoblotted using either anti-IκBαor anti-tubulin as a loading control. C, HUVEC were either untreated or incubated with TNF (10 ng/ml), LIGHT (100 ng/ml), or LTα1β2 (100 ng/ml) for 0.5 or 8 h then nuclear extracts were analyzed by EMSA using a γ-32P[ATP]-labeled NF-κB oligonucleotide probe (top row). The same samples were also analyzed using an OCT-1 oligonucleotide probe to ensure equal loading of nuclear lysates (bottom row). D, HUVEC were either untreated or incubated for 30 min with a range of concentrations of either TNF (top panels) or LIGHT (bottom panels) in the absence or presence of anti-TNFR1 (α-R1) as indicated. Lysates were immunoblotted using either anti-IκBαor anti-tubulin as a loading control.
LTβR ligation up-regulates adhesion molecule expression and T cell adhesion to HUVEC
A major proinflammatory response elicited by TNF and IL-1 in EC is the induction or up-regulation of the leukocyte adhesion molecules E-selectin, ICAM-1, and VCAM-1 (2, 13); however, Mackay and colleagues (32, 33) suggested that these are not targets of LTβR signaling in HUVEC. As these adhesion molecules are well-characterized targets of the classical NF-κB pathway (3, 13), we compared their expression levels in HUVEC stimulated with TNF, LIGHT, and LTα1β2. The percentage of HUVEC positive for E-selectin, ICAM-1, and VCAM-1 after cytokine treatment was determined by FACS analysis and as expected TNF strongly induced surface expression of all three molecules on EC (Fig. 2A). Consistent with their effects on IκBα degradation (Fig. 1B), LIGHT and LTα1β2 also increased surface expression of all three proteins, although this expression was substantially lower for each molecule than that induced by TNF. To determine whether this increased expression could lead to changes in the adhesion of lymphocytes to LTβR-stimulated EC, we performed adhesion assays using calcein-labeled human T cells. As shown in Fig. 2B, incubation of HUVEC for 24 h with TNF induced a 4.5 ± 0.4-fold increase in T cell adhesion compared with adhesion to unstimulated EC. Consistent with the effects on adhesion molecule expression (Fig. 2A), both LIGHT and LTα1β2 increased T cell adhesion albeit to a lower level than maximal adhesion induced by TNF (2.5 ± 0.3-fold and 2.5 ± 0.5-fold, respectively). To ensure that the effects on T cell adhesion were not influenced by the calcein-labeling procedure, we microscopically counted unlabeled-adhered lymphocytes. Consistent with the fluorometric analysis, TNF induced a 3.9 ± 0.9-fold increase in adhered T cells per field of view, and LIGHT and LTα1β2 up-regulated adhesion by 2.2 ± 0.4-fold and 2.2 ± 0.2-fold, respectively, over control untreated HUVEC (Fig. 2C). Hence, in contrast to previous reports (32, 33), Figs. 1 and 2 clearly demonstrate that LTβR ligation activates EC by inducing the classical NF-κB pathway and by up-regulating their adhesive phenotype.
FIGURE 2.
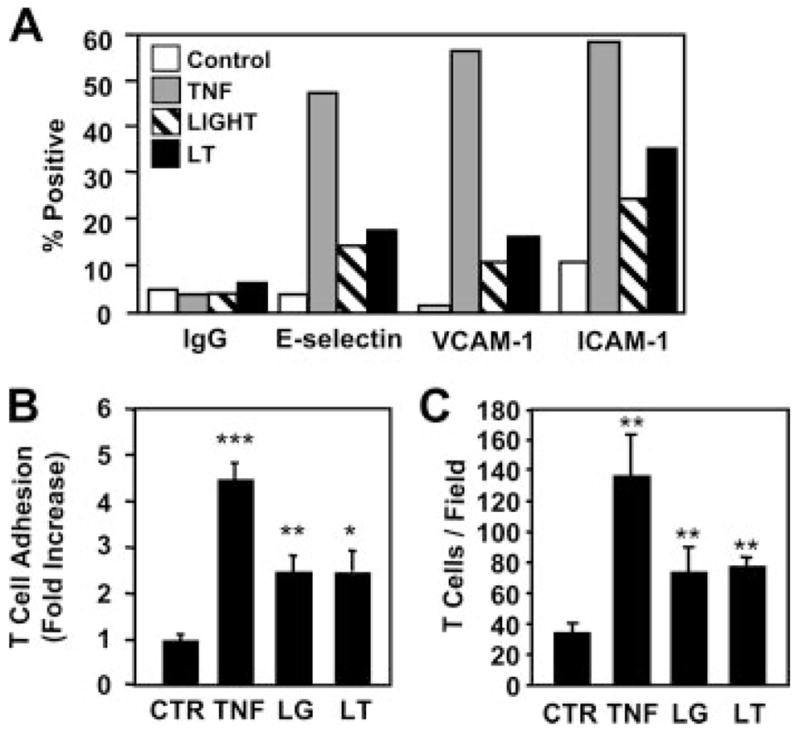
LTβR ligation induces adhesion molecule expression and up-regulates T cell adhesion to HUVEC. A, HUVEC were treated for 4 h with TNF (10 ng/ml), LIGHT (100 ng/ml), or LTα1β2 (100 ng/ml) (LT) then harvested and stained using control IgG, anti-E-selectin, anti-VCAM-1, or anti-ICAM-1 and analyzed by FACS. Population data were analyzed and the percentage of each population that was positive for adhesion molecule expression compared with control IgG was determined. Data shown are from one experiment representative of four performed. B, HUVEC were either untreated (CTR) or treated for 24 h with TNF (10 ng/ml), LIGHT (100 ng/ml) (LG), or LTα1β2 (100 ng/ml) (LT) then incubated with calcein-labeled T cells for 1 h. Fold increase in adhesion relative to unstimulated HUVEC was determined and the results are expressed as mean fold increase ± SEM from three independent experiments. ***, p < 0.001; **, p < 0.01; *, p < 0.05. C, HUVEC were treated as described for B then incubated with unlabeled T cells for 1 h. After washing, adherent lymphocytes were counted in six randomly selected fields of view for each treatment. Data from three separate experiments are presented as mean ± SEM. **, p < 0.01.
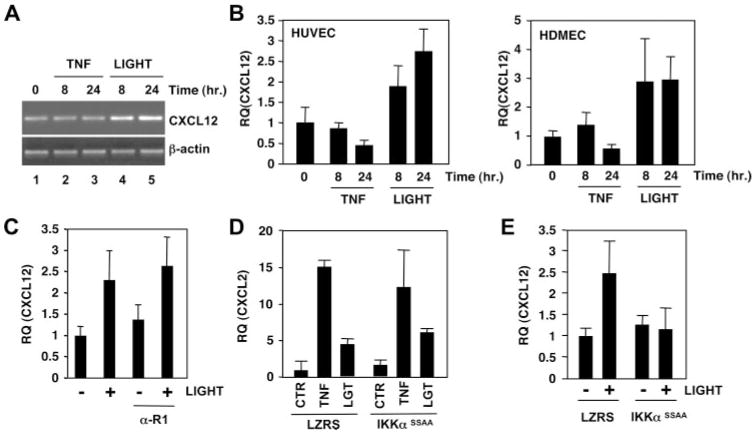
LIGHT and LTα1β2, but not TNF, induce p100 processing to p52 in HUVEC
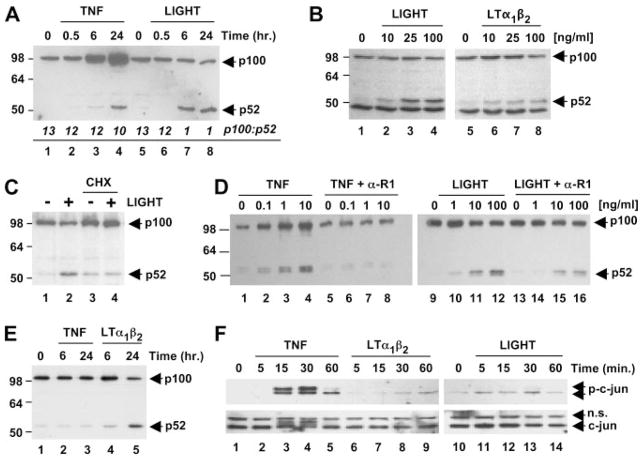
LTβR ligation activates both classical and noncanonical NF-κB signaling in murine embryonic fibroblasts with maximal noncanonical activation occurring later (i.e., after 6 h) than IκBαdegradation (10, 17, 19). To determine whether LTβR ligation could activate the noncanonical pathway in EC, we incubated HUVEC for times up to 24 h with LIGHT or LTα1β2 and examined non-canonical NF-κB activation by immunoblotting using anti-p100. This Ab recognizes full-length p100 and the processed p52 fragment and as shown in Fig. 3A, LIGHT incubation led to an increase in p52 levels after 6 h that remained elevated after 24 h (Fig. 3A, lanes 7 and 8). LTα1β2 induced similar effects with the same kinetics (data not shown), although the levels of p52 were consistently less than those stimulated by LIGHT (Fig. 3B). The response to both cytokines was dose-dependent with maximal effects occurring with 100 ng/ml each (Fig. 3B; lanes 4 and 8). A slight decrease in p100 levels concomitant with increased p52 was routinely observed after 24 h incubation with either LIGHT or LTα1β2. As a previously demonstrated hallmark of noncanonical signaling in murine embryonic fibroblasts (18), cycloheximide treatment inhibited LIGHT-induced p52 accumulation (Fig. 3C, compare lane 2 with lane 4) confirming that protein synthesis is required for signal-induced p100 processing. Taken together, these findings therefore demonstrate that LIGHT and LTα1β2 induce protein synthesis-dependent p100 processing to p52 and hence activate the noncanonical NF-κB pathway in HUVEC.
FIGURE 3.
LIGHT and LTα1β2 activate the noncanonical NF-κB pathway in HUVEC and HDMEC. A, HUVEC were incubated with TNF (10 ng/ml) or LIGHT (100 ng/ml) for the times indicated then lysates were immunoblotted using anti-p100/p52. The ratio of p100 to p52 was calculated by densitometry and is shown for each treatment in italics. B, Cells incubated for 24 h with the range of concentrations of LIGHT or LTα1β2 indicated were lysed and immunoblotted using anti-p100/p52. C, HUVEC were treated for 8 h with LIGHT (100 ng/ml) in the presence or absence of cycloheximide (2.5 μg/ml) (CHX). Samples were then analyzed for p100/p52 processing by immunoblotting. D, HUVEC were either untreated or incubated for 24 h with a range of concentrations of either TNF (lanes 1– 8) or LIGHT (lanes 9 –16) in the absence or presence of anti-TNFR1 (α-R1) as indicated then lysates were immunoblotted using anti-p100/p52. E, HDMEC were treated with TNF or LTα1β2 for 6 or 24 h and lysates were analyzed for the expression of p100/p52. F, HUVEC were incubated for times up to 60 min with either TNF (10 ng/ml), LTα1β2 (100 ng/ml), or LIGHT (100 ng/ml) and lysates were immunoblotted using anti-c-jun (bottom panels) and anti-phospho-c-jun (top panels).
In contrast to LTβR ligation, TNF only activates the classical NF-κB pathway in fibroblasts (10, 17, 19) and consistent with this result, we did not observe TNF-induced p100 processing in HUVEC. We routinely observed increased p52 levels after incubation with TNF for 24 h (Fig. 3A, lane 4). However, unlike LTβR signaling (Fig. 3A, lanes 5– 8), this increase was accompanied by substantial up-regulation of p100 levels in these samples (Fig. 3A, lanes 3 and 4). It has been established that nfkb2 (the gene encoding p100) is a classical NF-κB-dependent gene and that elevated p100 expression in response to TNF occurs via activation of this pathway (10, 17–19). In contrast, although LIGHT and LTα1β2 also up-regulate p100 via the classical pathway, they induce its signal-dependent processing resulting in either a stabilization or reduction in p100 levels (Fig. 3A, compare p100 in lane 4 with lane 8). Densitometric analysis confirmed signal-induced p100 processing in LIGHT-treated HUVEC as the ratio of p100 to p52 decreased from 13:1 in untreated cells (Fig. 3A, lanes 1 and 5) to 1:1 in response to LIGHT (Fig. 3A, lanes 7 and 8). In contrast, the ratio of p100 to p52 following TNF treatment was not significantly different from untreated cells (Fig. 3A, lane 4 compared with lanes 1 and 5). Hence, consistent with previous reports (10, 17–19), the increased p52 levels in TNF-stimulated HUVEC most likely result from basal processing of up-regulated p100 and not from signal-induced processing as observed with LIGHT and LTα1β2. To confirm that the effects of LTβR ligation on p100 processing were direct and not due to LTβR-induced release of TNF, we incubated HUVEC with anti-TNFR1 then determined the levels of p100 and p52 following TNF and LIGHT stimulation. As shown in Fig. 3D, lanes 1– 8, anti-TNFR1 completely blocked TNF-induced up-regulation of p100 and the concomitant increase in p52. In contrast, anti-TNFR1 did not block the generation of p52 in response to LIGHT (Fig. 3D, lanes 9 –16), confirming that the effects of TNF and LTβR-ligation on p100 processing are entirely distinct.
To determine whether LTβR ligation could induce p100 processing in separate types of EC, we compared the effects of LTβR ligands with TNF on signaling in HDMEC (Fig. 3E). In these cells, TNF failed to up-regulate p100 expression and did not induce its processing to p52 (Fig. 3E, lanes 1–3), whereas processing was readily detected in response to LTα1β2 (Fig. 3E, lanes 1, 4, and 5) and LIGHT (data not shown).
In addition to the classical and noncanonical NF-κB pathways, LTβR ligation also activates the JNK signaling pathway and induces the transcription factor AP-1 in some cell types (35). As this pathway has not been examined in EC following LTβR ligation, we explored JNK signaling in HUVEC by immunoblotting for c-jun and phosphorylated c-jun. As shown in Fig. 3F, lanes 3 and 4, TNF induced a rapid and transient phosphorylation of c-jun that was detected by anti-phospho-c-jun and up-shift of the c-jun band. Phosphorylation of c-jun also occurred in response to LTα1β2 and LIGHT (Fig. 3F, top panels, lanes 6 –14), although this was substantially weaker than activation by TNF.
Together with the data in Fig. 1, these results demonstrate that LTβR ligation directly activates the classical and noncanonical NF-κB pathways and weakly induces JNK signaling in EC. In contrast, TNF robustly activates classical NF-κB and JNK signaling but does not induce signal-dependent p100 processing to p52.
TNF and LTβR ligation induce the nuclear localization of distinct NF-κB proteins in HUVEC
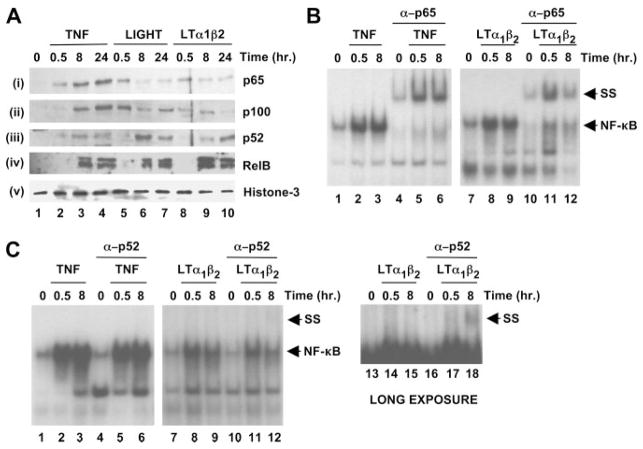
To determine whether LTβR ligation induced nuclear accumulation of typical classical (i.e., p65) and noncanonical (i.e., p52 and RelB) NF-κB proteins in HUVEC, we immunoblotted nuclear lysates from TNF-, LIGHT-, or LTα1β2-stimulated cells. As shown in Fig. 4A, nuclear localization of p65 was observed after 30 min of incubation with all three cytokines (Fig. 4A, panel i, lanes 2, 5, and 8). Following LIGHT and LTα1β2 treatment, p65 accumulation was transient and diminished after longer incubation times (8 –24 h) (Fig. 4A, panel i, lanes 6, 7, 9, and 10). In contrast, nuclear p65 levels increased over time in TNF-stimulated HUVEC (Fig. 4A, panel i, lanes 3 and 4). Similar to p65, rapid nuclear accumulation of p100 was observed following LIGHT and LTα1β2 stimulation and these levels diminished after 8 –24 h (Fig. 4A, panel ii, lanes 5–10) accompanied by increased accumulation of p52 (Fig. 4A, panel iii, lanes 5–10). Contrasting with these effects but consistent with the TNF-induced up-regulation of p100 shown in Fig. 3A, nuclear p100 accumulated with slower kinetics (Fig. 4A, panel ii, lanes 2–4) and was associated with only a modest increase in nuclear p52 levels following TNF treatment (Fig. 4A, panel iii, lanes 2–4). RelB accumulated in the nucleus following treatment for 8–24 h with all three cytokines (Fig. 4A, panel iv).
FIGURE 4.
LIGHT and LTα1β2 induce nuclear translocation of classical and noncanonical NF-κB proteins in EC. A, Nuclear extracts prepared from HUVEC incubated with either TNF (10 ng/ml), LIGHT (100 ng/ml), or LTα1β2 (100 ng/ml) for the times indicated were immunoblotted using anti-p65, anti-p100/p52, anti-RelB, or anti-histone-3 (loading control) as indicated. B, Nuclear extracts of HUVEC treated with either TNF (10 ng/ml; left) or LTα1β2 (100 ng/ml; right) were subjected to EMSA using a γ-32P[ATP]-labeled NF-κB consensus oligonucleotide probe. Samples in lanes 4 – 6 and lanes 10–12 were incubated with anti-p65 (2 μg/sample) in the EMSA reaction and the supershifted (SS) complex in these lanes is indicated. Samples in lanes 1–3 and lanes 7–9 were incubated with an isotype-matched nonspecific Ig (2 μg/sample). C, Supershift analysis was performed as described in B using anti-p52. The panel at the far right (lanes 13–18) is a longer exposure of the LTα1β2 experiment in the middle panel (lanes 7–12).
To further investigate the NF-κB proteins activated by LTβR ligation in HUVEC, we performed EMSA and Supershift analysis. For supershifting, we used anti-p65 to identify typical classical NF-κB heterodimers (e.g., p65:p50) and anti-p52 to detect potential noncanonical NF-κB complexes (i.e., p52:RelB). Following incubation with TNF for 0.5 and 8 h, anti-p65 completely shifted the retained probe (Fig. 4B, lanes 5 and 6), indicating that p65 was present in the complexes formed at both time points. In contrast, anti-p65 incompletely shifted the DNA-bound complex formed after LTα1β2 treatment (Fig. 4B, lanes 11 and 12). Consistent with the activation of a nuclear NF-κB complex that does not contain p65 after longer incubation with LTα1β2, anti-p52 shifted a complex in samples generated following 8 h of LTα1β2 treatment (Fig. 4C, lanes 12 and 18). This shift was accompanied with a decrease in the unshifted NF-κB band (Fig. 4C, compare lane 9 with lane 12). No effect was observed on the TNF-induced complexes at either time point following incubation with anti-p52 (Fig. 4C, lanes 4–6).
The data in Fig. 4 confirm that the major NF-κB complexes that migrate to the nucleus and bind to DNA in response to TNF in HUVEC are classical complexes containing p65. In contrast, although LTβR ligation rapidly induces p65 migration, analysis of longer time points reveals the nuclear translocation and DNA-binding of p52-containing complexes. Like LTα1β2, TNF also induced the migration of RelB (Fig. 4A); however, this RelB may be part of a transcriptionally inactive nuclear complex with p100 that is not processed to p52. These findings therefore demonstrate that the components of both classical and noncanonical NF-κB complexes are differentially induced to migrate to the nucleus in HUVEC following treatment with either TNF or ligation of the LTβR.
Dominant negative IKKαand IKKβselectively inhibit noncanonical and classical NF-κB signaling, respectively, in HUVEC
Activation of the classical NF-κB pathway requires IKKβwhereas noncanonical NF-κB signaling is IKKβ-independent but requires intact IKKα (12). We therefore generated retroviral LZRS constructs encoding dominant negative versions of each kinase (IKKβK44M or IKKαSSAA) (8, 36) to selectively inhibit these pathways in EC. Virus harvested from Phoenix cells stably transfected with these constructs infected HUVEC with >90% transduction efficiency as observed by expression of enhanced GFP (data not shown). Stimulation of LZRS transduced HUVEC with a range of concentrations of TNF (0.1, 1, and 10 ng/ml) for 30 min dose-dependently induced IκBαdegradation (Fig. 5A, lanes 1–4) and this was blocked in cells transduced with LZRS-IKKβK44M (Fig. 5A, lanes 9–12). Unlike LZRS-IKKβK44M, transduction with LZRS-IKKαSSAA did not prevent TNF-induced IκBαdegradation (Fig. 5A, lanes 5–8). Consistent with the effects of the dominant negative kinases on TNF stimulation, LZRS-IKKβK44M but not LZRS-IKKαSSAA inhibited LIGHT-induced IκBαdegradation (Fig. 5B, lanes 9–12 compared with lanes 5–8) thereby confirming that LTβR ligation activates the classical NF-κB pathway via IKKβ.
FIGURE 5.
Selective inhibition of the classical and noncanonical NF-κB pathways in HUVEC. A and B, HUVEC transduced with LZRS, LZRS-IKKαSSAA, or LZRS-IKKβK44M were either untreated or incubated with a range of concentrations of TNF (0.1, 1, and 10 ng/ml) (A) or LIGHT (1, 10, and 100 ng/ml) (B) for 30 min then lysates were analyzed by immunoblotting using anti-IκBα (upper), anti-IKKα, anti-IKKβ (two middle), or anti-tubulin (lower). C, HUVEC transduced with LZRS or LZRS-IKKβK44M were either untreated or incubated for 24 h with a range of concentrations of LIGHT (1, 10, and 100 ng/ml). D, Samples were analyzed for p100/p52 or tubulin by immunoblotting and densitometry was performed on the resulting immunoblot. E and F, HUVEC transduced with LZRS or LZRS-IKKαSSAA were treated and analyzed as described for C and D. Data shown are from a single experiment representative of at least three performed using separate transduced EC cultures.
To determine the effects of IKKβK44M and IKKαSSAA on the noncanonical NF-κB pathway in EC, transduced HUVEC were incubated for 24 h with a range of concentrations of LIGHT (1, 10, and 100 ng/ml) and p100 processing was determined by immunoblotting using anti p100/p52. As shown in Fig. 5C, LZRS-IKKβK44M did not block LIGHT-induced p100 processing and the generation of p52. Densitometric analysis of immunoblots (Fig. 5D) revealed that generation of p52 occurred in LZRS- IKKβK44M transduced cells with a concomitant decrease in p100 levels reflecting classical pathway inhibition and a subsequent lack of induced p100 expression. In contrast, transduction with LZRS-IKKαSSAA blocked LIGHT-induced p100 processing that was observed as both a lack of p52 generation and a concomitant sustained expression of p100 (Fig. 5E, lanes 1–4, compared with lanes 5–8 and Fig. 5F).
LTβR ligation induces classical NF-κB-dependent gene expression by HUVEC
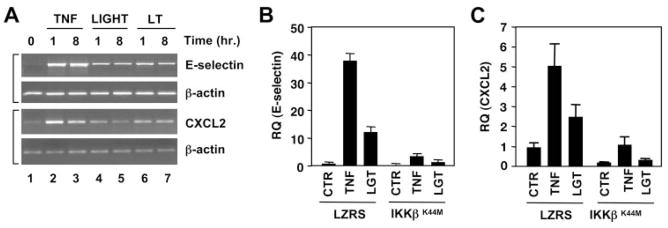
Data presented in Fig. 2A demonstrate that LTβR ligation up-regulates adhesion molecule expression by HUVEC. To further analyze LTβR-induced classical NF-κB-dependent gene expression in EC, we performed RT-PCR analysis for E-selectin (Fig. 6A, top panels) and the chemokine CXCL2 (Fig. 6A, bottom panels). Consistent with the FACS analysis (Fig. 2A), LIGHT and LTα1β2 induced E-selectin mRNA (Fig. 6A, lanes 4 –7, top panels) albeit to a lower level than the maximal induction observed in response to TNF (Fig. 6A, lanes 2 and 3). Similar results were observed with CXCL2 (Fig. 6A, bottom panels) and other well-characterized classical NF-κB-dependent genes including those encoding ICAM-1, VCAM-1, p100, IκBα, and CX3CL1 (data not shown). These results indicate that LIGHT and LTα1β2 can induce classical gene expression in HUVEC, although the response to these cytokines is less robust than the response to TNF. To definitively establish that LTβR-induced expression of these genes in EC results from classical pathway activation, we examined the effect of LZRS-IKKβK44M transduction on LIGHT and TNF-induced expression of E-selectin and CXCL2. As shown in Fig. 6, B and C, quantitative real-time PCR analysis demonstrated that induction of E-selectin (Fig. 6B) and CXCL2 (Fig. 6C), in response to both cytokines, was blocked by IKKβK44M. Identical effects of IKKβK44M were seen on LIGHT- and TNF-induced CX3CL1 expression (data not shown). In contrast, LZRS-IKKαSSAA (which only blocks the noncanonical pathway) (Fig. 5) did not inhibit TNF- or LIGHT-induced expression of any of these classical NF-κB dependent genes in HUVEC (see Fig. 7D, and data not shown).
FIGURE 6.
LTβR ligation induces classical NF-κB-dependent gene expression in EC. A, Messenger RNA was isolated from HUVEC following incubation with TNF (10 ng/ml), LIGHT (100 ng/ml), or LTα1β2 (100 ng/ml) (LT) for the times indicated. Semiquantitative RT-PCR analysis was performed using primers specific for E-selectin, CXCL2, or β-actin as indicated. B and C, LZRS or LZRS-IKKβK44M transduced HUVEC were stimulated with either TNF (10 ng/ml) or LIGHT (100 ng/ml) (LGT) for 8 h and real-time quantitative PCR was performed to determine the expression levels of E-selectin (B) and CXCL2 (C). Data shown are from a single experiment representative of at least three performed using separate transduced EC cultures.
FIGURE 7.
LTβR ligation induces noncanonical NF-κB-dependent expression of CXCL12 in EC. A, HUVEC were either untreated or incubated for 8 or 24 h with TNF (10 ng/ml) or LIGHT (100 ng/ml) then mRNA was isolated and used in semiquantitative RT-PCR using primer pairs specific for CXCL12 (35 cycles) or β-actin (25 cycles). B, HUVEC (left) or HDMEC (right) were stimulated for 8 or 24 h with TNF (10 ng/ml) or LIGHT (100 ng/ml) then mRNA was isolated and used for quantitative real-time PCR analysis of CXCL12. C, HUVEC were either untreated (−) or incubated (+) for 8 h with LIGHT (100 ng/ml) in the absence or presence of anti-TNFR1 (α-R1) as indicated then mRNA was isolated and used for quantitative real-time PCR analysis of CXCL12. D, LZRS- or LZRS-IKKαSSAA-transduced HUVEC were stimulated for 8 h with TNF (10 ng/ml) or LIGHT (100 ng/ml) (LGT) then quantitative real-time PCR was performed to determine the expression levels of CXCL2. E, LZRS- or LZRS-IKKαSSAA-transduced HUVEC were incubated with (+) or without (−) LIGHT (100 ng/ml) for 24 h then quantitative real-time PCR analysis was performed to determine the expression levels of CXCL12. Data shown are from a single experiment representative of at least three performed using separate transduced EC cultures.
LTβR ligation induces noncanonical NF-κB-dependent CXCL12 expression in EC
LTβR-induced noncanonical NF-κB activation up-regulates CXCL12 expression in murine splenocytes (10) and previous studies have suggested that CXCL12 plays a role in lymphocyte trans-endothelial migration (37–39). However, it remains unclear whether CXCL12 is intrinsically expressed by EC and whether this expression is regulated following cell activation. To address this question, we investigated the effects of LTβR ligation on CXCL12 expression by HUVEC using semiquantitative PCR and found that LIGHT but not TNF up-regulated its expression after 8 and 24 h incubation (Fig. 7A). Using quantitative real-time PCR, we further established that LIGHT (and LTα1β2, data not shown) but not TNF induced an increase in CXCL12 mRNA in HUVEC (Fig. 7B, left) and HDMEC (Fig. 7B, right) comparable in magnitude to the increased expression observed in splenocytes activated in vivo by an agonistic LTβR Ab (10). In four separate identical experiments, incubation of HUVEC for 24 h with LIGHT induced a mean 2.96 ± 0.23-fold increase in CXCL12 expression over unstimulated control cells ( p < 0.01), whereas TNF had no significant effect (0.88 ± 0.21-fold compared with control; p = 0.61). Consistent with the failure of anti-TNFR1 to block LTβR-induced p100 processing (Fig. 3D), incubation of HUVEC with anti-TNFR1 had no effect on LIGHT-induced CXCL12 expression (Fig. 7C) thereby confirming that the downstream effects of LTβR ligation are completely distinct from TNF-induced increases in p100 and p52 levels. Furthermore, anti-TNFR1 did not block LIGHT-induced CXCL2 expression, whereas it completely inhibited CXCL2 induction by TNF (data not shown). To determine whether the increase in CXCL12 expression in LIGHT-stimulated EC was dependent upon the noncanonical NF-κB pathway, we retrovirally transduced HUVEC with LZRS-IKKαSSA. Consistent with its previously described redundant role in TNF-induced expression of classical NF-κB-dependent genes, IKKαSSAA did not affect either TNF- or LIGHT-induced CXCL2 (Fig. 7D). In contrast, IKKαSSAA completely blocked LIGHT-induced expression of CXCL12 in HUVEC (Fig. 7E).
Discussion
We initiated this study to investigate the effects of LTβR ligation on noncanonical NF-κB signaling and gene expression in vascular EC. However, in marked contrast to a previous report (32), we found that both LIGHT and LTα1β2 activate the classical NF-κB pathway in EC. It is not clear why our findings differ from those of Mackay et al. (32) who did not detect active NF-κB in LTα1β2-stimulated HUVEC, as the treatment conditions were similar between our studies. Their lack of observed effects led Mackay et al. (32) to propose that LTβR-induced NF-κB activation in LTβR-positive cells is cell-type specific and does not occur in EC. However, our findings clearly conflict with this conclusion. Instead, our data demonstrate that LTβR signaling in EC is similar to that in stromal cells in which it rapidly activates the classical pathway (10, 17–19). In fibroblasts, LTβR-induced classical NF-κB activation requires IKKβand NEMO in a manner analogous to TNF and IL-1R signaling (10, 19). We confirmed that the classical pathway in HUVEC requires IKKβthereby strongly supporting a model in which the classical signaling “cassette” of NEMO-dependent IKKβ-mediated IκBαphosphorylation and liberation of p50:p65 NF-κB heterodimers (4, 12) is the mechanism activated by LTβR ligation in EC. Our accumulated findings therefore lead us to conclude that LIGHT and LTα1β2 are novel inducers of classical NF-κB activity in vascular EC.
We also found that LTκR ligation induces or increases the expression of classical NF-κB-dependent proinflammatory genes in EC, including those encoding E-selectin, VCAM-1, ICAM-1, and CXCL2. This observation again conflicts with an earlier report suggesting that LTβR ligation does not induce gene expression in LTβR-positive EC (33). However our data are consistent with a more recent study showing that LTβR ligation on HUVEC regulates gene expression (34). In addressing the mechanism responsible for this expression, we found that LTβR-induced E-selectin, and CXCL2 expression is dependent upon the classical NF-κB pathway because it was inhibited in HUVEC by dominant negative IKKβbut not dominant negative IKKα. We also found that LTβR ligation up-regulates T cell adhesion to HUVEC thereby confirming that in addition to activating NF-κB and inducing gene expression, LTβR signaling results in functionally relevant phenotypic changes to EC.
We consistently observed that the levels of classical NF-κB-dependent genes induced by LIGHT and LTα1β2 were lower than levels induced by TNF. Furthermore, we found that activation of the classical NF-κB-driven luciferase reporter constructs pBIIX-luc, E-selectin, and E-selectinΔCRE was lower in response to LIGHT and LTα1β2 than TNF (data not shown). This lower level of classical pathway-dependent gene expression may reflect the less robust IκBαdegradation and p65 nuclear translocation that we observed in response to LIGHT and LTα1β2 compared with TNF. It is also possible that expression of these genes requires additional transcriptional mechanisms that are elicited by TNF but not by LTβR ligation. Hence activation of the classical pathway might not in itself be sufficient for maximal induction of the classical NF-κB-dependent genes we explored and these may require other signals for full expression. Further studies will be required to determine the precise profile of classical gene expression in LTβR-stimulated EC and whether this consistently occurs at a lower level than the response to TNF. Nevertheless, our study definitively establishes that LTβR ligation activates the classical NF-κB pathway and regulates classical NF-κB-dependent proinflammatory gene expression in EC.
The primary goal of our study was to investigate the effects of LTβR ligation on the noncanonical NF-κB pathway in EC and we demonstrate that this is indeed a major signaling mechanism activated by this receptor in HUVEC. Importantly, we observed identical effects in HDMEC, confirming that this is not unique to large vessel-derived EC and occurs in cells from distinct vascular beds. To our knowledge, this is the first demonstration of direct activation of this pathway in human EC because the majority of studies of noncanonical signaling in response to LTβR ligation have used murine embryonic fibroblasts or splenocytes (10, 17, 19). Consistent with these earlier studies, however, the noncanonical pathway was only activated by LIGHT and LTα1β2 and was not induced by TNF demonstrating the specificity of signaling via the LTβR in EC. Noncanonical NF-κB activation in EC occurred with slower kinetics than classical signaling and was completely blocked by cycloheximide confirming the previously reported requirement for protein synthesis in the noncanonical pathway (18). In this regard, we have found by RT-PCR that classical NF-κB-dependent synthesis of p100 occurs before LTβR-induced processing to p52 via the noncanonical pathway (data not shown). We also determined by immunoblotting and by PCR (data not shown) that up-regulation of p100 in response to TNF occurs via the classical pathway in HUVEC. However, unlike LTβR ligation, TNF did not induce processing to p52. The increased p52 levels that appeared concomitantly with increased p100 in response to TNF do not contribute to noncanonical NF-κB-dependent gene expression, as TNF did not up-regulate CXCL12 levels. Furthermore, incubation of HUVEC with anti-TNFR1 blocked the up-regulation of both p100 and p52 levels in response to TNF but not LTβR ligation, and anti-TNFR1 did not block LIGHT-induced CXCL12 expression. These findings therefore separate the downstream functional effects of LTβR-induced noncanonical NF-κB activation and the enhanced p52 levels that occur via classical signaling induced by TNF.
Signal-induced p100 processing requires TNFR-associated factor (TRAF)3 degradation that in turn stabilizes the cytoplasmic levels of newly synthesized NIK (40, 41). It has been further suggested that noncanonical signaling is regulated by TRAF2, although it is not yet clear whether TRAF2 functions as a negative or positive regulator or whether its role following LTβR ligation is cell type-dependent (42, 43). Although HUVEC express both TRAF2 and TRAF3 (44 – 46), we do not yet know whether these factors function in LTβR-induced p100 processing in EC. However, the blockade with cycloheximide that we observed suggests that similar regulatory mechanisms do occur in these cells. Furthermore, although we have demonstrated that noncanonical NF-κB activation in HUVEC is blocked by dominant negative IKKα, it remains to be determined whether NIK also plays a role in these cells. However, in light of the strong genetic evidence implicating interplay between NIK and IKKαin the noncanonical pathway in other cell types (11, 21), it is likely that NIK functions in this capacity in EC.
While addressing the hypothesis that LTβR ligation regulates noncanonical NF-κB-dependent gene expression in EC, we found that LIGHT and LTα1β2 but not TNF up-regulated CXCL12 mRNA levels in both HUVEC and HDMEC. The levels of induction in both EC types were entirely consistent with the magnitude of induced changes in CXCL12 expression reported previously in splenocytes (10). More importantly LTβR-induced CXCL12 expression was completely blocked in EC that were retrovirally transduced with dominant negative IKKα. These results therefore identify CXCL12 as the first bona fide noncanonical NF-κB gene directly regulated by LTβR ligation in vascular EC.
Induction of mRNA for CXCL12 by LTβR ligation in HUVEC and HDMEC is intriguing as previous studies failed to detect increased expression in cytokine-stimulated EC in vitro (47). However, in their study, Calderon et al. (47) treated HUVEC with IL-1 and TNF that did not activate the noncanonical NF-κB pathway thereby supporting our findings that only stimuli that activate the noncanonical pathway can regulate CXCL12 expression in EC. It is well-established that EC express only a limited number of chemokine genes and that surface expression of many chemokines occurs via transcytosis from separate cellular sources followed by binding to specific cell surface receptors (37, 48 –52). This model of transcytosed surface expression has been described for the non-canonical NF-κB-dependent chemokines CCL19 and CXCL13 on HEC within the lymphoid microenvironment (48, 49) and consistent with this, we did not detect any LTβR-induced up-regulation of CCL19 or CXCL13 mRNA in either HUVEC or HDMEC (data not shown). We did routinely observe increased CCL21 in LTβR-stimulated EC by semiquantitative RT-PCR supporting previous in vivo studies of mRNA expression in high EC (50). However, we were unable to definitively confirm enhanced expression by quantitative real-time PCR in either HUVEC or HDMEC (data not shown). Hence our findings suggest that unlike lymphoid stromal cells in which LTβR induces or up-regulates the full array of mR-NAs encoding noncanonical NF-κB-dependent chemokines (10, 53, 54), EC respond to LTβR ligation by regulating only a subset of noncanonical genes that include CXCL12.
CXCL12 expression by EC has been described in both lymphoid tissue and inflammatory lesions where it regulates lymphocyte transendothelial migration (37, 39, 52, 55–57). Similar to CCL19 and CXCL13, transcytosis has been proposed as the mechanism underlying CXCL12 surface expression on high EC in mouse lymph nodes (37) and this mechnism has also been suggested for CXCL12 expression on synovial EC in patients with rheumatoid arthritis (52). In each of these scenarios, mRNA for CXCL12 was not detected in situ in EC, suggesting that it is not transcriptionally regulated in these cells. However mRNA encoding CXCL12 has been described in situ in large and small vessel EC in human skin (56), in HEC derived from rat lymph nodes (38) and in human microvascular EC derived from brain tumors (57). Furthermore, enhanced levels of CXCL12 mRNA have been demonstrated in microvascular EC derived from early (edematous) but not late lesions in systemic sclerosis patients, suggesting that CXCL12 levels are differentially regulated in EC during ongoing inflammatory responses (55). Intriguingly the cellular influx in early systemic sclerosis consists predominantly of T cells (55) possibly facilitating interaction between LTα1β2 and LIGHT expressed on the T cell surface with LTβR on EC. Further in vivo investigation of the timing of CXCL12 gene expression in EC during distinct inflammatory responses and lymphoid organogenesis is clearly required. However, these accumulated studies combined with our data demonstrate that CXCL12 can be transcriptionally regulated in both large vessel and microvascular EC.
In conclusion, our study identifies LIGHT and LTα1β2 as direct regulators of the EC phenotype. We show that LTβR ligation directly activates both the classical and noncanonical NF-κB pathways in HUVEC and HDMEC and that activation of these pathways results in altered patterns of gene expression in EC (modeled in Fig. 8). We have also demonstrated that LTβR ligation up-regulates T cell adhesion to EC thereby confirming that signaling from this receptor alters the function of EC. Finally, our study definitively establishes CXCL12 as a bona fide noncanonical NF-κB-dependent gene expressed in EC in response to LTβR ligation. These findings therefore identify the LTβR-induced classical and noncanonical NF-κB pathways as potential targets for novel therapeutic strategies aimed at blocking the altered EC phenotype at sites of chronic inflammation.
FIGURE 8.

Model of TNFR1 and LTβR ligation-induced signaling in vascular EC. This model depicts the relative strengths of activation of classical and noncanonical NF-κB signaling and the JNK pathway following TNF and LTβR ligation in vascular EC. Strong activation is indicated by the thick arrow, weaker induction by the thin arrow, and minor activation by the dotted arrow, respectively.
Acknowledgments
We gratefully acknowledge the staff of the Labor and Delivery Department at the Hospital of the University of Pennsylvania for collecting umbilical cords.
Footnotes
This work was supported by Grant R01 HL080612 from the National Institutes of Health and by a McCabe Foundation Award (to L.A.M.).
Abbreviations used in this paper: EC, endothelial cell; HDMEC, human dermal microvascular EC; IKK, IκB kinase; LT, lymphotoxin; LTβR, LT-β receptor; NIK, NF-κB-inducing kinase; TRAF, TNFR-associated factor HEC, high EC.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Choi J, Enis DR, Koh KP, Shiao SL, Pober JS. T lymphocyte-endothelial cell interactions. Annu Rev Immunol. 2004;22:683–709. doi: 10.1146/annurev.immunol.22.012703.104639. [DOI] [PubMed] [Google Scholar]
- 2.De Martin R, Hoeth M, Hofer-Warbinek R, Schmid JA. The transcription factor NF-κB and the regulation of vascular cell function. Arterioscler Thromb Vasc Biol. 2000;20:E83–E88. doi: 10.1161/01.atv.20.11.e83. [DOI] [PubMed] [Google Scholar]
- 3.Collins T, Read MA, Neish AS, Whitley MZ, Thanos D, Maniatis T. Transcriptional regulation of endothelial cell adhesion molecules: NF-κB and cytokine-inducible enhancers. FASEB J. 1995;9:899 –909. [PubMed] [Google Scholar]
- 4.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 5.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548 –554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 6.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860 – 866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 7.Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-γ is an essential regulatory subunit of the IκB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 8.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKαand IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 9.Bonizzi G, Bebien M, Otero DC, Johnson-Vroom KE, Cao Y, Vu D, Jegga AG, Aronow BJ, Ghosh G, Rickert RC, Karin M. Activation of IKKα target genes depends on recognition of specific κB binding sites by RelB:p52 dimers. EMBO J. 2004;23:4202– 4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dejardin E, Droin NM, Delhase M, Haas E, Cao Y, Makris C, Li ZW, Karin M, Ware CF, Green DR. The lymphotoxin-β receptor induces different patterns of gene expression via two NF-κB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 11.Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M. Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 12.Bonizzi G, Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280 –288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Denk A, Goebeler M, Schmid S, Berberich I, Ritz O, Lindemann D, Ludwig S, Wirth T. Activation of NF-κB via the IκB kinase complex is both essential and sufficient for proinflammatory gene expression in primary endothelial cells. J Biol Chem. 2001;276:28451–28458. doi: 10.1074/jbc.M102698200. [DOI] [PubMed] [Google Scholar]
- 14.Kempe S, Kestler H, Lasar A, Wirth T. NF-κB controls the global pro-inflammatory response in endothelial cells: evidence for the regulation of a pro-atherogenic program. Nucleic Acids Res. 2005;33:5308 –5319. doi: 10.1093/nar/gki836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-κB2 in maturing B cells. Nat Immunol. 2002;3:958 –965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 16.Coope HJ, Atkinson PG, Huhse B, Belich M, Janzen J, Holman MJ, Klaus GG, Johnston LH, Ley SC. CD40 regulates the processing of NF-κB2 p100 to p52. EMBO J. 2002;21:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derudder E, Dejardin E, Pritchard LL, Green DR, Korner M, Baud V. RelB/p50 dimers are differentially regulated by tumor necrosis factor-α and lymphotoxin-β receptor activation: critical roles for p100. J Biol Chem. 2003;278:23278 –23284. doi: 10.1074/jbc.M300106200. [DOI] [PubMed] [Google Scholar]
- 18.Mordmüller B, Krappmann D, Esen M, Wegener E, Scheidereit C. Lymphotoxin and lipopolysaccharide induce NF-κB-p52 generation by a co-translational mechanism. EMBO Rep. 2003;4:82– 87. doi: 10.1038/sj.embor.embor710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller JR, Siebenlist U. Lymphotoxin β receptor induces sequential activation of distinct NF-κB factors via separate signaling pathways. J Biol Chem. 2003;278:12006 –12012. doi: 10.1074/jbc.M210768200. [DOI] [PubMed] [Google Scholar]
- 20.Yilmaz ZB, Weih DS, Sivakumar V, Weih F. RelB is required for Peyer’s patch development: differential regulation of p52-RelB by lymphotoxin and TNF. EMBO J. 2003;22:121–130. doi: 10.1093/emboj/cdg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao G, Cvijic ME, Fong A, Harhaj EW, Uhlik MT, Waterfield M, Sun SC. Retroviral oncoprotein Tax induces processing of NF-κB2/p100 in T cells: evidence for the involvement of IKKα. EMBO J. 2001;20:6805– 6815. doi: 10.1093/emboj/20.23.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao G, Fong A, Sun SC. Induction of p100 processing by NF-κB-inducing kinase involves docking IκB kinase α (IKKα) to p100 and IKKα-mediated phosphorylation. J Biol Chem. 2004;279:30099 –30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- 23.De Togni P, Goellner J, Ruddle NH, Streeter PR, Fick A, Mariathasan S, Smith SC, Carlson R, Shornick LP, Strauss-Schoenberger J, et al. Abnormal development of peripheral lymphoid organs in mice deficient in lymphotoxin. Science. 1994;264:703–707. doi: 10.1126/science.8171322. [DOI] [PubMed] [Google Scholar]
- 24.Koni PA, Sacca R, Lawton P, Browning JL, Ruddle NH, Flavell RA. Distinct roles in lymphoid organogenesis for lymphotoxins α and β revealed in lymphotoxin β-deficient mice. Immunity. 1997;6:491–500. doi: 10.1016/s1074-7613(00)80292-7. [DOI] [PubMed] [Google Scholar]
- 25.Paxian S, Merkle H, Riemann M, Wilda M, Adler G, Hameister H, Liptay S, Pfeffer K, Schmid RM. Abnormal organogenesis of Peyer’s patches in mice deficient for NF-κB1, NF-κB2, and Bcl-3. Gastroenterology. 2002;122:1853–1868. doi: 10.1053/gast.2002.33651. [DOI] [PubMed] [Google Scholar]
- 26.Shinkura R, Kitada K, Matsuda F, Tashiro K, Ikuta K, Suzuki M, Kogishi K, Serikawa T, Honjo T. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-κb-inducing kinase. Nat Genet. 1999;22:74 –77. doi: 10.1038/8780. [DOI] [PubMed] [Google Scholar]
- 27.Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, Ryseck RP, Lira SA, Bravo R. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-κB/Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 28.Yin L, Wu L, Wesche H, Arthur CD, White JM, Goeddel DV, Schreiber RD. Defective lymphotoxin-β receptor-induced NF-κB transcriptional activity in NIK-deficient mice. Science. 2001;291:2162–2165. doi: 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- 29.Browning JL, Allaire N, Ngam-Ek A, Notidis E, Hunt J, Perrin S, Fava RA. Lymphotoxin-β receptor signaling is required for the homeostatic control of HEV differentiation and function. Immunity. 2005;23:539 –550. doi: 10.1016/j.immuni.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Drayton DL, Ying X, Lee J, Lesslauer W, Ruddle NH. Ectopic LT α β directs lymphoid organ neogenesis with concomitant expression of peripheral node addressin and a HEV-restricted sulfotransferase. J Exp Med. 2003;197:1153–1163. doi: 10.1084/jem.20021761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drayton DL, Liao S, Mounzer RH, Ruddle NH. Lymphoid organ development: from ontogeny to neogenesis. Nat Immunol. 2006;7:344 –353. doi: 10.1038/ni1330. [DOI] [PubMed] [Google Scholar]
- 32.Mackay F, Majeau GR, Hochman PS, Browning JL. Lymphotoxin β receptor triggering induces activation of the nuclear factor κB transcription factor in some cell types. J Biol Chem. 1996;271:24934 –24938. doi: 10.1074/jbc.271.40.24934. [DOI] [PubMed] [Google Scholar]
- 33.Hochman PS, Majeau GR, Mackay F, Browning JL. Proin-flammatory responses are efficiently induced by homotrimeric but not heterotrimeric lymphotoxin ligands. J Inflamm. 1995;46:220 –234. [PubMed] [Google Scholar]
- 34.Pablos JL, Santiago B, Tsay D, Singer MS, Palao G, Galindo M, Rosen SD. A HEV-restricted sulfotransferase is expressed in rheumatoid arthritis synovium and is induced by lymphotoxin-α/ β and TNF-α in cultured endothelial cells. BMC Immunol. 2005;6:6. doi: 10.1186/1471-2172-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen MC, Hwang MJ, Chou YC, Chen WH, Cheng G, Nakano H, Luh TY, Mai SC, Hsieh SL. The role of apoptosis signal-regulating kinase 1 in lymphotoxin-β receptor-mediated cell death. J Biol Chem. 2003;278:16073–16081. doi: 10.1074/jbc.M208661200. [DOI] [PubMed] [Google Scholar]
- 36.Delhase M, Hayakawa M, Chen Y, Karin M. Positive and negative regulation of IkappaB kinase activity through IKKβ subunit phosphorylation. Science. 1999;284:309 –313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 37.Okada T, V, Ngo N, Ekland EH, Forster R, Lipp M, Littman DR, Cyster JG. Chemokine requirements for B cell entry to lymph nodes and Peyer’s patches. J Exp Med. 2002;196:65–75. doi: 10.1084/jem.20020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips R, Ager A. Activation of pertussis toxin-sensitive CXCL12 (SDF-1) receptors mediates transendothelial migration of T lymphocytes across lymph node high endothelial cells. Eur J Immunol. 2002;32:837– 847. doi: 10.1002/1521-4141(200203)32:3<837::AID-IMMU837>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 39.Scimone ML, Felbinger TW, Mazo IB, Stein JV, Von Andrian UH, Weninger W. CXCL12 mediates CCR7-independent homing of central memory cells, but not naive T cells, in peripheral lymph nodes. J Exp Med. 2004;199:1113–1120. doi: 10.1084/jem.20031645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hauer J, Puschner S, Ramakrishnan P, Simon U, Bongers M, Federle C, Engelmann H. TNF receptor (TNFR)-associated factor (TRAF) 3 serves as an inhibitor of TRAF2/5-mediated activation of the noncanonical NF-κB pathway by TRAF-binding TNFRs. Proc Natl Acad Sci USA. 2005;102:2874 –2879. doi: 10.1073/pnas.0500187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF-κB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 42.Grech AP, Amesbury M, Chan T, Gardam S, Basten A, Brink R. TRAF2 differentially regulates the canonical and noncanonical pathways of NF-κB activation in mature B cells. Immunity. 2004;21:629 – 642. doi: 10.1016/j.immuni.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 43.Kim YS, Nedospasov SA, Liu ZG. TRAF2 plays a key, non-redundant role in LIGHT-lymphotoxin β receptor signaling. Mol Cell Biol. 2005;25:2130 –2137. doi: 10.1128/MCB.25.6.2130-2137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng X, Gaeta ML, Madge LA, Yang JH, Bradley JR, Pober JS. Caveolin-1 associates with TRAF2 to form a complex that is recruited to tumor necrosis factor receptors. J Biol Chem. 2001;276:8341– 8349. doi: 10.1074/jbc.M007116200. [DOI] [PubMed] [Google Scholar]
- 45.Madge LA, Sierra-Honigmann MR, Pober JS. Apoptosis-inducing agents cause rapid shedding of tumor necrosis factor receptor 1 (TNFR1): a nonpharmacological explanation for inhibition of TNF-mediated activation. J Biol Chem. 1999;274:13643–13649. doi: 10.1074/jbc.274.19.13643. [DOI] [PubMed] [Google Scholar]
- 46.Urbich C, Mallat Z, Tedgui A, Clauss M, Zeiher AM, Dimmeler S. Upregulation of TRAF-3 by shear stress blocks CD40-mediated endothelial activation. J Clin Invest. 2001;108:1451–1458. doi: 10.1172/JCI13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calderon TM, Eugenin EA, Lopez L, Kumar SS, Hesselgesser J, Raine CS, Berman JW. A role for CXCL12 (SDF-1α) in the pathogenesis of multiple sclerosis: regulation of CXCL12 expression in astrocytes by soluble myelin basic protein. J Neuroimmunol. 2006;177:27–39. doi: 10.1016/j.jneuroim.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 48.Baekkevold ES, Yamanaka T, Palframan RT, Carlsen HS, Reinholt FP, von Andrian UH, Brandtzaeg P, Haraldsen G. The CCR7 ligand elc (CCL19) is transcytosed in high endothelial venules and mediates T cell recruitment. J Exp Med. 2001;193:1105–1112. doi: 10.1084/jem.193.9.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebisuno Y, Tanaka T, Kanemitsu N, Kanda H, Yamaguchi K, Kaisho T, Akira S, Miyasaka M. Cutting edge: the B cell chemokine CXC chemokine ligand 13/B lymphocyte chemoattractant is expressed in the high endothelial venules of lymph nodes and Peyer’s patches and affects B cell trafficking across high endothelial venules. J Immunol. 2003;171:1642–1646. doi: 10.4049/jimmunol.171.4.1642. [DOI] [PubMed] [Google Scholar]
- 50.Gunn MD, Tangemann K, Tam C, Cyster JG, Rosen SD, Williams LT. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc Natl Acad Sci USA. 1998;95:258 –263. doi: 10.1073/pnas.95.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyasaka M, Tanaka T. Lymphocyte trafficking across high endothelial venules: dogmas and enigmas. Nat Rev Immunol. 2004;4:360 –370. doi: 10.1038/nri1354. [DOI] [PubMed] [Google Scholar]
- 52.Santiago B, Baleux F, Palao G, Gutierrez-Canas I, Ramirez JC, Arenzana-Seisdedos F, Pablos JL. CXCL12 is displayed by rheumatoid endothelial cells through its basic amino-terminal motif on heparan sulfate proteoglycans. Arthritis Res Ther. 2006;8:R43. doi: 10.1186/ar1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carragher D, Johal R, Button A, White A, Eliopoulos A, Jenkinson E, Anderson G, Caamano J. A stroma-derived defect in NF-κB2 −/− mice causes impaired lymph node development and lymphocyte recruitment. J Immunol. 2004;173:2271–2279. doi: 10.4049/jimmunol.173.4.2271. [DOI] [PubMed] [Google Scholar]
- 54.Drayton DL, Bonizzi G, Ying X, Liao S, Karin M, Ruddle NH. IκB kinase complex α kinase activity controls chemokine and high endothelial venule gene expression in lymph nodes and nasal-associated lymphoid tissue. J Immunol. 2004;173:6161– 6168. doi: 10.4049/jimmunol.173.10.6161. [DOI] [PubMed] [Google Scholar]
- 55.Cipriani P, Franca Milia A, Liakouli V, Pacini A, Manetti M, Marrelli A, Toscano A, Pingiotti E, Fulminis A, Guiducci S, et al. Differential expression of stromal cell-derived factor 1 and its receptor CXCR4 in the skin and endothelial cells of systemic sclerosis patients: pathogenetic implications. Arthritis Rheum. 2006;54:3022–3033. doi: 10.1002/art.22047. [DOI] [PubMed] [Google Scholar]
- 56.Pablos JL, Amara A, Bouloc A, Santiago B, Caruz A, Galindo M, Delaunay T, Virelizier JL, Arenzana-Seisdedos F. Stromal-cell derived factor is expressed by dendritic cells and endothelium in human skin. Am J Pathol. 1999;155:1577–1586. doi: 10.1016/S0002-9440(10)65474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salmaggi A, Gelati M, Pollo B, Frigerio S, Eoli M, Silvani A, Broggi G, Ciusani E, Croci D, Boiardi A, De Rossi M. CXCL12 in malignant glial tumors: a possible role in angiogenesis and cross-talk between endothelial and tumoral cells. J Neurooncol. 2004;67:305–317. doi: 10.1023/b:neon.0000024241.05346.24. [DOI] [PubMed] [Google Scholar]