Abstract
In the brain of the fly Drosophila melanogaster, ∼150 clock-neurons are organized to synchronize and maintain behavioral rhythms, but the physiological and neurochemical bases of their interactions are largely unknown. Here we reevaluate the cellular properties of these pacemakers by application of a novel genetic reporter and several phenotypic markers. First, we describe an enhancer trap marker called R32 that specifically reveals several previously undescribed aspects of the fly's central neuronal pacemakers. We find evidence for a previously unappreciated class of neuronal pacemakers, the lateral posterior neurons (LPNs), and establish anatomical, molecular, and developmental criteria to establish a subclass within the dorsal neuron 1 (DN1) group of pacemakers. Furthermore, we show that the neuropeptide IPNamide is specifically expressed by this DN1 subclass. These observations implicate IPNamide as a second candidate circadian transmitter in the Drosophila brain. Finally, we present molecular and anatomical evidence for unrecognized phenotypic diversity within each of four established classes of clock neurons.
Indexing terms: Drosophila, circadian clock, neuropeptides, PDF receptor, IPNamide, nplp1, glass
Overt biological rhythms such as the predictable daily leaf movements of plants and the sleep/wake cycle of animals are ultimately driven by molecular oscillations. Throughout the living world such oscillations are sustained by intracellular transcriptional/translational feedback loops (reviewed by Dunlap, 1999). In Drosophila many components of the molecular clock are known in detail. Products of the clock genes period (per), timeless (tim), Clock, cycle, vrille, and PAR domain protein 1 (Pdp1) form two interconnected and self-sustained transcriptional feedback loops, the kinetics of which are regulated by clock protein interactions and attendant kinases (recently reviewed by Hardin, 2004; Schöning and Staiger, 2005; Taghert and Lin, 2005). Although the core clock genes are expressed rhythmically throughout the fly's body, only a few small groups of clock-expressing neurons in the central brain (termed pacemakers) are necessary and sufficient for the organization and maintenance of rhythmic locomotor activity (reviewed by Hall, 2005; Helfrich-Förster, 2005). When these pacemaker neurons of Drosophila are electrically silenced their molecular oscillations are lost under constant conditions, suggesting that electrical activity at the cell membrane is required for the endogenous molecular clockwork (Nitabach et al., 2002).
Approximately 150 neurons express the dynamic molecular clockwork in the adult brain and are divided into six groups based on location and size (Ewer et al., 1992; Frisch et al., 1994; Kaneko and Hall, 2000; Helfrich-Förster, 2003) (Fig. 1A). These are the large and small ventrolateral neurons (the l-LNvs and s-LNvs, respectively), the dorsolateral neurons (LNds), and three groups of dorsal neurons (DNs), the DN1s, DN2s, and DN3s. All six neuronal classes have been recognized for more than a decade (Ewer et al., 1992; Frisch et al., 1994) and no new classes of clock-neurons (i.e., neurons that support clock-gene oscillations under light:dark cycles or constant conditions) have been described in the central brain since that time (Hall, 2005). Hundreds more glial cells express clock products throughout the brain (Ewer et al., 1992), although the details of the molecular oscillations within glia and their role in directing behavioral rhythms is currently undefined.
Fig. 1.
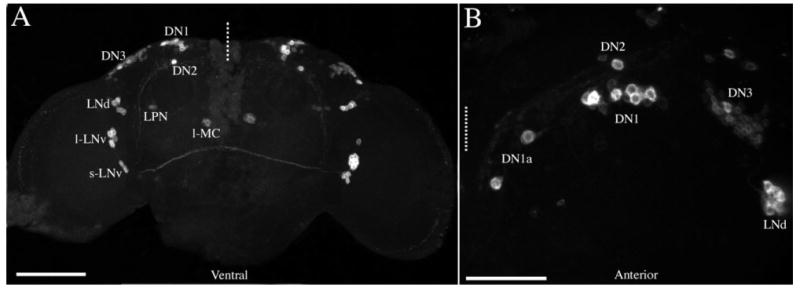
R32-mediated LacZ expression in the adult brain. A: A projected Z-series montage of an adult R32 brain labeled for LacZ and imaged through its posterior surface. Cell classes are labeled in the left hemisphere. The dashed line represents the midline and the ventral aspect is labeled. Z-series depth was 150 μm. B: A projected Z-series through a single hemisphere of the dorsal protocerebrum, imaged through the dorsal surface. Two DN1as were clearly discernable in this orientation. The dashed line indicates the midline and the anterior aspect is labeled. Z-series depth was 80 μm. Scale bars = 100 μm in A; 20 μm in B.
The extent to which locomotor rhythms require specific neuronal classes has been investigated by the targeted deletion of clock neuron classes or the specific rescue of clock function within them (reviewed by Hall, 2005; Helfrich-Förster, 2005). The LNv and LNd are necessary and sufficient for the maintenance of locomotor rhythms in the absence of environmental times cues (Frisch et al., 1994; Helfrich-Förster, 1998; Renn et al., 1999) and for normal crepuscular organization of locomotion under light:dark (LD) conditions (Renn et al., 1999). Recent mosaic analysis suggests that the morning peak of activity is controlled by the s-LNv, while the evening peak is governed by a group of neurons that include the LNd and a subset of the DN1s (Stoleru et al., 2004; Grima et al., 2004). The specific functions of the l-LNvs are not known. DNs are not required for locomotor rhythms under constant darkness (DD) and temperature, but contribute to the organization and/or entrainment of such behavior under LD conditions (Veleri et al., 2003; Klarsfeld et al., 2004).
Notwithstanding the ability to assign functions to most classes of clock neurons, the physiological and synaptic basis of these functions is poorly understood. For example, only one neurochemical output has been identified for clock neurons, a neuropeptide called pigment dispersing factor (PDF). PDF is expressed in both the large and small LNvs and is required for the maintenance of locomotor rhythms under constant conditions and the normal timing of locomotion under LD (Renn et al., 1999). The transmitter phenotypes of the remaining clock-neuron classes remain a mystery, as do the synaptic interactions that organize neuronal timekeeping.
To provide ongoing studies of circadian pacemaker networks with greater cellular resolution, we reanalyzed the identities, anatomical position, and cellular properties of the clock neurons in the fly. In the present study we employ the detection of a local enhancer by a LacZ insertion called P{wF6-84,R32} (“R32”; Schneider et al., 1993) to describe the clock-neuron classes of Drosophila's central brain. First, we show that R32 is a limited and highly accurate reporter for neuronal PER in the adult brain. Next we present evidence based on R32 expression for a previously unappreciated class of pacemakers that we have called lateral posterior neurons (LPNs), based on their earlier description as Timeless (TIM)-expressing, nonpacemakers by Kaneko and Hall (2000). We define developmental, cellular, and anatomical criteria by which identified neuronal subclasses can be recognized within the LNd, DN1, DN2, and DN3 classes. Finally, we identify a second candidate neuropeptide transmitter within the Drosophila pacemaker network, IPNamide (IPNa), a product of the gene neuropeptide-like-precursor 1 (nplp1; Baggerman et al., 2002; Verleyen et al., 2004).
Materials and Methods
Fly rearing and strains
Drosophila were reared on cornmeal/agar media supplemented with yeast and kept in either a 25°C incubator or at room temperature. For time-course experiments, flies were entrained to a 12:12 LD cycle for 4–5 days with light intensities of ∼3 × 1014 W/cm2 (measured with a LICOR LI250 photo detector) prior to dissection at 25°C. Canton S and y1w1 (hereafter referred to as y w) flies were used as wildtype controls for colabeling experiments with identical results except for lower background fluorescence in the latter line. The glass60j mutation, used here in a y w background, is described by Lindsley and Zimm (1992). Several cry-gal4 lines were used to drive GFP in clock neurons of interest. They were cry-gal4[13] (provided by P. Emery), cry-gal4[16] (provided by R. Allada), and cry-gal4[39] (provided by F. Rouyer). All three have been shown to drive expression in the LNvs, LNds, in a subset of DNs, and in other nonclock neurons (Zhao et al., 2003; Klarsfeld et al., 2004; Stoleru et al., 2004). We employed uas-cd8GFP (Flybase #5137) and uas-2XeGFP (Flybase #6874) to visualize neuronal projections. The stable reporter line y w;tim-gal4(27-1-1), uas-GFP(1010T2)/Cyo, used here to visualize the DN3s, has been reported previously (Kaneko and Hall, 2000; Helfrich-Förster, 2003).
Isolation of the enhancer trap line R32
The P element insertion line P{wF6-84 – R32} is described in this report. It is a single P insertion line generated by mobilization of the P{wF6-84} element that is inserted at cytogenic position ∼86A5. P{wF6-84} is a P{lacW} element that drives lacZ under the control of a fragment of the promoter for the neuropeptide gene dFM-RFa (Schneider et al., 1993). P{wF6-84} produced ectopic (i.e., non-dFMRFa) LacZ labeling that resembled clock neuronal populations, and also harbored a mutation of the pdf neuropeptide gene (Renn et al., 1999). It was therefore subjected to mobilization to generate the insertion (and the same ectopic labeling pattern) in a pdf+ background. Derivative lines of P{wF6-84} were either recovered as w+ (therefore named R lines) or w− (therefore named W lines); the original P insertion locus and the pdf locus were then each analyzed molecularly in derivative lines. The R32 derivative line retains the original 86A P{lacW} insertion without damage to the neighboring sequences and contains a wildtype pdf genomic sequence. The P{wF6-84} insertion site is located less than 100 bp upstream of the transcription start site for the bHLH gene stich1 (CG17100 – NM080036), which is a site shared by several other recorded P elements—e.g., P{EP(3)3470} – AQ074026.
Immunocytochemistry
Adult flies were immobilized with CO2 and pinned down under Ca2+-free fly saline (pH 7.2) in a dissecting dish by a single insect pin through the thorax. The head cuticle was disrupted using two fine forceps to allow access to the underlying brain. Fifteen to 20 flies were dissected for each timepoint or treatment on a given day and immediately placed in a well of a four-well culture plate containing 4% formaldehyde in phosphate-buffered saline (PBS, pH 7.4) with or without 7% picric acid (the latter was used when colabeling for the neuropeptides PDF and IPNa). Fixation proceeded for 1 hour at room temperature (RT) with gentle agitation, followed by a rinse in PBS. Brains were then isolated by further dissection. Brains were transferred to 2-ml round-bottomed Eppendorf tubes full of PBS-TX (PBS with 0.3% Triton X-100, pH 7.4; Sigma, St. Louis MO) and kept on ice until blocked.
Brains were blocked in 3% normal goat serum in PBS-TX for 45 minutes to 2 hours at RT with gentle agitation, rinsed in PBS-TX, and placed in primary sera for 2 nights at 4°C with gentle agitation. Brains were rinsed with five changes of PBS-TX at RT on a Lab-Quake rotator (Barnstead/Thermoline) for 15 minutes to 1 hour per rinse. Brains were placed in secondary antiserum for 1 night at 4°C with gentle agitation and rinsed as before. Care was taken to treat brains for time-course experiments identically throughout labeling and rinsing. Brains were rinsed in three changes of PBS and mounted in rows on a poly-L-lysine-coated cover slip. Typically, 8–15 brains were mounted for each treatment. Brains were dehydrated in a graded glycerol series (30%, 50%, and 70% in PBS for 5 minutes each). The coverslip was lowered onto a large drop of “Hard-Set” Vectashield fluorescent mounting medium (Vector Laboratories, Burlingame, CA) flanked by two spacer coverslips onto a microscope slide.
The primary antisera used here have all been described previously. All primary and secondary antisera were diluted in PBS-TX. Mouse anti-β-galactosidase (Promega, Madison, WI, Cat. #Z3781, Lot #149211), raised against amino acids 650–926 of E. coli β-galactosidase and tested for specificity on dot-blots by the manufacturer, was diluted 1:1,000. Rat anti-PER (provided by M. Rosbash) was raised against a fusion of the first 14 amino acids of the T7 phage coat protein and the C-terminal 1,108 amino acids of PER (Liu et al., 1992). This serum recognizes PER specifically on Western blots and fails to label tissue in per01 mutants (Liu et al., 1992). Anti-PER was used at 1:500. The rat anti-TIM serum used here (UPR41, provided by A. Seghal) was raised against His-tagged, full-length TIM and specifically recognizes a TIM band on Western blots that is missing in tim null mutants (Amita Sehgal, pers. commun.). Anti-TIM was diluted 1:500. The rabbit anti-PDP1 used here (provided by J. Blau) was raised against a GST-PDP1alpha fusion protein; it recognizes a specific PDP1 band on Western blots, and fails to label tissue in PDP1P205 mutant pupae (Cryan et al., 2003). Anti-PDP1 was diluted 1:1,000. Guinea pig anti-proPDF (PAP-59-IV) was raised against amino acids 65–79 of pre-proPDF (H-YPLILENSLGPSVPI-OH) conjugated to bovine serum albumin (BSA). This serum fails to visualize the LNvs in pdf01 mutants (Renn et al., 1999). Anti-PDF was used at 1:1,000. Mouse anti-ELAV (mAb Elav-9F8A9 supernatant, Developmental Studies Hybridoma Bank, Iowa City, IA) was raised against a fusion protein consisting of the first 260 amino acids of phage T7 gene and full-length ELAV protein (O'Neill et al., 1994). This anti-ELAV serum detects ELAV-specific bands on Western blots (e.g., Samson et al., 1995) and was used in this study at 1:10. Anti-GLASS (mAb 9B2.1 supernatant, Developmental Studies Hybridoma Bank) was raised against a GLASS-trp E fusion protein, and Glass immunosignals were lost in the photoreceptors of glass60j mutants (Moses and Rubin, 1991). Anti-GLASS was diluted 1:15. Rabbit anti-IPNa (Verleyen et al., 2004) was diluted 1:1,000. The IPNa serum used here was described by Verleyen et al. (2004), and was raised in rabbit against synthetic IPNa peptide (NVGTLARDFQLPIPNamide) coupled to thyroglobulin. Its specificity was determined by solid phase dot blot assays (Verleyen et al., 2004). Alex-aFluor 488 and 568-conjugated secondary antisera (all raised in goat; Molecular Probes, Eugene, OR) were used at 1:1,000.
Confocal microscopy
Confocal stacks were obtained by an Olympus Fluoview 500A microscope and Fluoview acquisition software using a 60X/1.20NA water objective lens. Distances were approximated by digital scale bars in Fluoview or by counting the number of 0.5-μm Z-steps between objects in different focal planes. For double-labeling experiments we used only sequential scans of the argon and krypton lasers to avoid bleed-through between channels. When imaging time-course experiments, Fluoview's “Hi-Lo” look-up table was used to set PMT voltage (typically 800 V) and laser power (0.1–4.0%) to minimize peak and trough pixel intensities (i.e., to avoid clipping) in ZT00 preparations (i.e., the most intensely labeled timepoint). Gain was maintained at 1× and offset at 0%. Typically, 5–7 scans were Kalman-averaged for each optical section. These settings were kept constant for all the timepoints of a given experiment and all brains of a given time-course were imaged in the same session with the same brain orientation so that micrographs could be directly compared for labeling intensity. The constituent optical sections of a Z-series were saved as 16-bit multi-TIFF files and exported to Image J for postacquisition analysis and Z-stack construction. Z-stack projections were adjusted for levels and contrast in Adobe Photoshop (San Jose, CA). Micrographs of PER and TIM time-course data were not adjusted in this way. For somatic size comparisons all DN1p (see below) and LNd soma of a single hemisphere were imaged by Z-series in five brains per treatment and subsequently digitally isolated from neighboring cells in Photoshop from micrographs of single optical sections.
Observation numbers and presentation of representative micrographs
Unless stated otherwise in the Results, all labeling patterns described here were unanimous within groups of 8–15 mounted brains. Many subsequent replications of each labeling experiment were then undertaken for the purpose of imaging preps in multiple orientations to attain micrographs that revealed well-separated clock neuron groups when presented in a projected Z-series. For PER and TIM time-course analysis, five brains each were imaged for each timepoint and all cell bodies of a given class within a single hemisphere were imaged per brain. The distributions of PER and TIM in these neurons were uniform among brains of a given timepoint and the micrographs presented here were selected on the basis of clarity.
Results
R32-mediated LacZ expression is a highly limited and precise reporter of neuronal PER expression
Immunocytochemical visualization of R32-mediated LacZ expression in the adult brain revealed a pattern that was strikingly similar to that of PER-expressing circadian pacemaker neurons (Fig. 1; cf. Helfrich-Förster, 2003). Clusters of cells resembling all six classes of clock neurons were clearly visible (Fig. 1). In general there was little suggestion of R32-LacZ among populations of smaller (putatively glial) cells. Three groups of R32-positive cells did not correspond to known clock-neuron classes: 1) A cluster of three to four cells in the lateral central brain were medial to the LNds and near the posterior surface of the brain (Fig. 1A). We referred to these as lateral posterior neurons (LPNs) pending confirmation as neurons (see below). 2) A single large cell was visible in the medial central brain (Fig. 1A), which we called a large medial cell (l-MC). 3) R32 brains imaged through the dorsal surface revealed two conspicuous cells separated anteriorly from the DN1s by 15–30 μm (Fig. 1B). These cells, which we called anterior DN1s (DN1as) pending the determination of their neuronal nature (see below), were not clearly distinguished in brains imaged through the anterior or posterior surfaces of the brain because they clustered with the majority of DN1s when all Z-planes were collapsed to a single micrograph as in Figure 1A (see below). To our knowledge the l-MCs and DN1as are located in brain regions not previously associated with known clock neuron classes (Kaneko and Hall, 2000; Helfrich-Förster, 2003). The LPNs were reminiscent of a cluster of TIM-expressing neurons described by Kaneko and Hall (2000) that were thought not to express PER.
Double labeling of R32 brains dissected at ZT23 in a 12:12 LD cycle (where ZT00 = lights-on) for LacZ and PER revealed that all R32-positive cells except the l-MCs expressed PER at this timepoint (Fig. 2), confirming the clock-neuron-like nature of nearly all R32 labeling. Thus, the LPNs and the DN1as were considered candidates as previously uncharacterized clock neurons. Such double labeling at ZTs 00, 06, 12, and 18 (i.e., throughout the diurnal cycle) revealed no PER labeling in the l-MCs (data not shown). The l-MCs were not considered further as clock-neuron candidates.
Fig. 2.
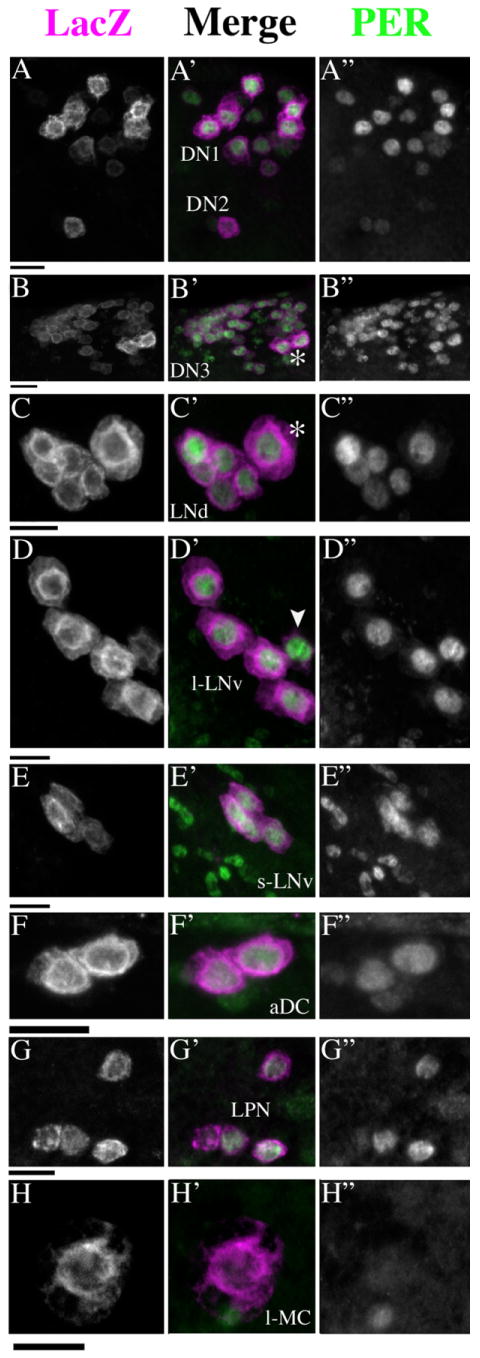
PER expression in R32-positive cells. R32 LacZ labeling is shown in the left column (A–H). The center column presents merged micrographs with R32-LacZ in magenta and PER in green (A′-H′). PER labeling is shown in the right column (A″-H″). A: The DN1 and DN2. B: The DN3, the asterisk in B′ indicates two large cells. C: Six LNd, asterisk in C′ indicates a single large cell. D: Four large LNv and fifth small LNv (indicated by the arrowhead in D′). E: Four small LNv, the smaller PER-positive cells are presumably glia. F: Two DN1as. G: Four LPNs, one cell is expressing PER very weakly. H: A single l-MC, no clear PER immunoreactivity was detected in these cells. Scale bars = 10 μm.
New class of clock neuron: the lateral posterior neurons
R32 brains double-labeled for LacZ and PER or TIM at ZTs 00, 06, 12, and 18 revealed a canonical PER/TIM oscillation in the LPNs under a 12:12 LD cycle: protein levels were high and nuclear at the end of the night and low at the end of the day (Fig. 3A). As visualized by R32, the LPNs always numbered three or four cells. These were consistently situated near the posterior surface of the brain and were frequently difficult to visualize beneath the often-high fluorescence of the brain surface. The LPNs resided in anterior/posterior planes near the posterior-most segment of the dorsal PDF projection emanating from the small LNvs (Fig. 3B–B″). These cells often straddled this projection, but were just as frequently found in positions lateral or medial to it.
Fig. 3.
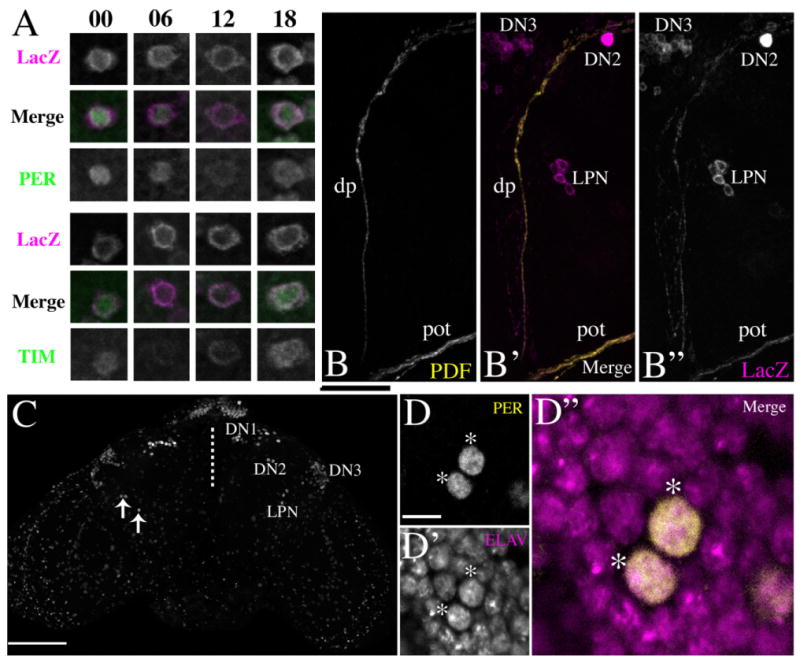
The LPNs express PER and TIM with a diurnal rhythm, are consistently situated near the posteriormost segment of the dorsal PDF projection, are present in wildtype brains, and express the neuronal marker ELAV. A: Representative micrographs of LPNs colabeled for R32 LacZ and PER (top panel) or R32 LacZ and TIM (bottom panel) at various times of day in a 12:12 LD cycle. Numbers indicate Zeitgeber Times. Merged micrographs are presented with R32 LacZ in magenta and PER and TIM in green. B: A Z-series with a depth of 5 μm through a portion of the dorsal PDF projection. B′: Merged Z-series of PDF projection (yellow) and R32lacZ (magenta). B″: Single-labeled Z-series of R32-Laz containing the LPN. The brain was imaged through the posterior surface of the brain. pot, posterior optic tract; dp, dorsal projection. C: A partial Z-series through the posterior half of a y w brain dissected at ZT23 and labeled for PER protein. Arrows in the left hemisphere indicate LPNs. The dashed line represents the midline. D: A single optical section through two LPNs labeled for PER at ZT 23. D′: The same optical section labeled for ELAV. D″: A merged micrograph with PER in yellow and ELAV in magenta. In D–D″ the colabeled neurons are marked by asterisks. Scale bars = 20 μm in B; 100 μm in C; 5 μm in D. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
To ensure that the presence of the LPNs was not an anomalous aspect specific to the R32 stock, we labeled wildtype brains (from y w and CS flies) for PER and imaged brains through the posterior surface. LPNs were clearly visible in all brains as brightly labeled nuclei at ZT23 (Fig. 3C). These nuclei were larger and much brighter than the surrounding small, weakly labeled PER cells. The latter are thought to be glia and compose the largest group of PER-expressing cells in the adult brain (Ewer et al., 1992). To determine the nature of LPNs we colabeled wildtype brains dissected at ZT23 with antisera against PER and ELAV. Every brain revealed a nuclear colocalization of PER and ELAV in all LPNs (Fig. 3D–D″). These results indicate that the LPNs comprise a previously unappreciated class of clock neurons.
DN1 class is divisible into highly reproducible anterior and posterior subclasses
R32 brains double-labeled for LacZ and PER or TIM at ZTs 00, 06, 12, and 18 revealed a canonical PER/TIM oscillation in the DN1as under a 12:12 LD cycle (Fig. 4A). As described above, the DN1as were separated anteriorly from the DN1s by 15–30 μm in R32 brains. We sought to describe the location of these cells in more detail. R32 brains were double-labeled for LacZ and PDF, the latter as an anatomical reference. The relative positions of the DN2s, DN1s, and the DN1as are most easily observed in brains imaged through the posterior aspect of the brain, but with a slight anterior bias (i.e., the brains were mounted with the posterior surface mounted against the coverslip with a slight tilt toward the dorsal surface). In brains so mounted the DN2s were always closely associated with the dorsal PDF projection, often within or ventral to the expanse of its termini, the DN1 were situated just above (i.e., dorsal) to the PDF projection, and the DN1as were further removed from the PDF projection and often closer to the dorsal brain surface than either class of DN (Fig. 4B).
Fig. 4.
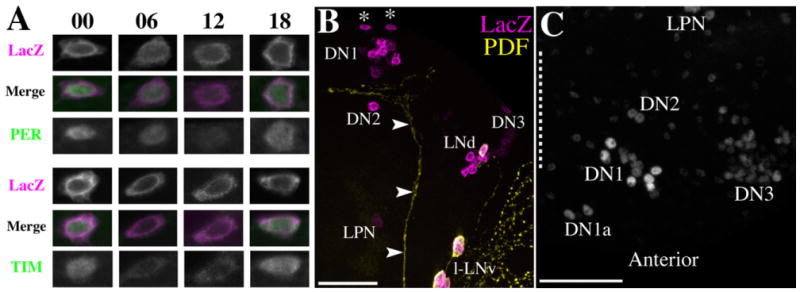
The DN1as express PER and TIM with a diurnal rhythm and are consistently situated in dorsal brain regions anterior to the DN2, DN1s, and PDF terminals. A: Representative micrographs of DN1as colabeled for R32 LacZ and PER and R32 LacZ and TIM at various times of day in a 12:12 LD cycle. Numbers indicate Zeitgeber Times. Merged micrographs are presented with R32 LacZ in magenta and PER and TIM in green. B: A Z-series with a depth of 52 μm through the dorsoposterior surface (see text) of an R32 brain colabeled for R32 LacZ (magenta) and PDF (yellow). The DN1as are indicated by asterisks and lie just beneath the dorsal surface of the brain. The dorsal PDF projection is indicated by arrowheads. C: A Z-series with a depth of 26 μm through the dorsal surface of a y w brain dissected at ZT23 and labeled for PER. The dashed line indicates the midline. The DN1as are visible as two PER-positive nuclei anterior and medial to the DN1s. Scale bars = 50 μm in B; 50 μm in C. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Using 0.5-μm Z-steps, brains mounted without an anterior bias were imaged through the posterior surface where imaged to a depth of 100 μm as a means of visualizing the relative positions of the DN1as and DN1s in the anterior/posterior axis. The DN1as were always separated by at least 15 and up to 30 μm anteriorly from the DN1s. Thus, DN1as were situated further away from the DN1s than were the DN2s. However, this was only apparent when the dorsal cells were imaged from the dorsal aspect of the brain or when relative Z-depths were accounted for in brains mounted in the anterior or posterior aspect. Clock neurons have traditionally been imaged in the anterior or posterior aspect. When imaged this way the DN1as of R32 brains usually appeared clustered with the DN1s. Therefore, when the Z-planes were collapsed the DN1as were not discernable from the DN1s (cf. Fig. 1A,B). We therefore wondered if the DN1as were truly not-previously-described clock cells or if they had been grouped with DN1s without reference to their anterior isolation (see below).
The DN1as were clearly visible in wildtype brains as brightly labeled nuclei at ZT23 in brains imaged through the dorsal surface (Fig. 4C). Thus, the presence of DN1as was not an anomalous aspect specific to the R32 stock. To determine the nature of DN1as we colabeled wildtype brains dissected at ZT23 with antisera against PER and ELAV. Every brain revealed a nuclear colocalization of PER and ELAV in all DN1as (data not shown). We hereafter refer to the larger and more posterior group of DN1s as posterior DN1s (DN1p).
DN1a are labeled by antibodies of an nplp1-derived neuropeptide
The neuropeptide IPNamide (IPNa) is widely expressed throughout the brain and ventral nerve chord of the fly (Verleyen et al., 2004). During an immunocytochemical screen of neuropeptides for expression in clock-neurons we found that antibodies directed against IPNa peptide derived from the NPLP1 precursor (Verleyen et al., 2004) specifically label the DN1a cell bodies. R32 brains double-labeled for LacZ and IPNa showed clear colocalization in the DN1a but not the DN1p and DN2s (Fig. 5A–A″). Likewise, wildtype brains dissected at ZT23 revealed cytoplasmic IPNa labeling around the PER-positive DN1a nuclei (Fig. 5B–B″). Thus, in addition to their anatomical isolation, the DN1a were differentiated from the DN1p by virtue of their IPNa expression. The DN1a represented a small minority of total IPNa labeling in the adult brain (cf. Verleyen et al., 2004) and this neuropeptide was expressed by soma that were distinct from but quite close to the DN1p (Figs. 5, 6).
Fig. 5.

The DN1a express the neuropeptide IPNa. A: A Z-series with a depth of 75 μm through posterior surface of an R32 brain labeled for R32 LacZ. A′: The same Z-series for IPNa labeling. A″: A merged micrograph of IPNa (yellow) and R32-LacZ (magenta). Dorsal is up. Asterisks indicate IPNa expression in the DN1a in A–A″. B: A Z-series with a depth of 17 μm through the dorsal surface of a y w brain dissected at ZT23 and labeled for PER. B′: The same Z-stack for IPNa labeling. B″: The merged micrographs with PER in magenta and IPNa in yellow. Anterior is up. Asterisks indicate IPNa expression in the DN1a. Scale bars = 10 μm in A; 20 μm in B. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Fig. 6.
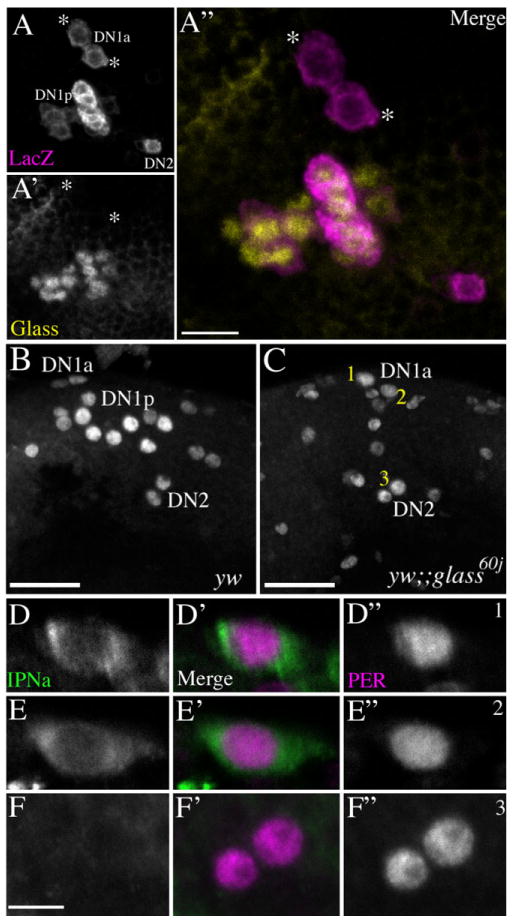
The DN1a do not express GLASS and survive the glass mutation. A: A Z-series with a depth of 23 μm through the DN1a, DN1p, and DN2 in an R32 brain, labeled for LacZ. A′: the same Z-series for GLASS labeling. A″: A merged micrograph of LacZ (magenta) and IPNa (yellow) labeling. Only the DN1p expressed glass. In A–A″ the positions of the DN1a are indicated by asterisks. This preparation was imaged through the posterior surface of the brain. Dorsal is up. B: PER expression in a y w brain dissected at ZT23. C: PER expression in a y w;;glass60j brain dissected at ZT23. Z-series in C and D are 80 μm deep and both brains were imaged through the posterior surface. Dorsal is up. Laser power was higher for the y w;;glass60j micrograph (C) to ensure the detection of all weakly PER-positive cells. For this reason PER-positive glia are visible in B but not in C. The numbering of the DN1s as 1 and 2 and the DN2 as 3 in C are for reference in the following panels. D–D″: A single optical section of the DN1 labeled 1 in panel C labeled for IPNa (D) and PER (D″) the merged image is shown in D′ with IPNa in green and PER in magenta. E–E″: A single optical section of the DN1 labeled 2 in panel C labeled for IPNa (E) and PER (E″) the merged image is shown in E′ with IPNa in green and PER in magenta. F–F″: A single optical section of the DN2s labeled 3 in C, labeled for IPNa (F) and PER (F″). The merged image is shown in F′ with IPNa in green and PER in magenta. The surviving DN1 (neurons 1 and 2 in C) display cytoplasmic IPNa (D and E), while the DN2s (labeled 3 in C) do not express this peptide (F). Scale bars = 10 μm in A; 20 μm in B,C; 5 μm in D. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DN1a are developmentally distinct from the DN1p
The glass mutation, which causes the loss of all external photoreceptors, was determined by Helfrich-Förster et al. (2001) to result in the loss of all but two DN1s. We wondered if the glass-surviving DN1s were the two DN1a. Klarsfeld et al. (2004) established that all but two DN1s express GLASS, a transcription factor necessary for photoreceptor differentiation (Moses et al., 1989). Therefore, we first asked if the DN1a express GLASS protein. R32 brains were double-labeled for LacZ and GLASS and imaged through the posterior aspect of the brain. DN1a were identified by virtue of their anterior location relative to the DN1p. In all brains the DN1p expressed GLASS and the DN1a did not (Fig. 6A–A″: cf. Klarsfeld et al., 2004). We therefore predicted that the two glass-surviving DN1s would be the DN1a. To test this supposition, y w and yw;;glass60j brains were dissected at ZT23 and colabeled for PER and IPNa, the latter as a marker for the DN1a. These brains were imaged through the posterior surface of the dorsal brain by a Z-series that commenced a few microns posterior from (i.e., superficial to) the DN2 to a depth of 80 μm, such that a Z-series always included all DN1s and DN2s of wildtype flies with tens of microns to spare. As expected, y w brains showed the normal contingent of 14–17 DN1 (Fig. 6B; Kaneko and Hall, 2000; Helfrich-Förster, 2003), while the y w;;glass brains showed only two DN1 (Fig. 6C; Helfrich-Förster et al., 2001). Single optical sections through the surviving DN1 of glass mutants revealed their expression of IPNa (Fig. 6D–E″). Identical scans through the DN2 neurons in glass mutants revealed no such expression (Fig. 6F–F″). We conclude that glass mutants lack all DN1ps and retain the DN1as.
The larval brain contains fewer clock-expressing neurons than the adult: just four to five LNvs, two DN2s, and two DN1s (Kaneko and Hall. 2000). Klarsfeld et al. (2004) hypothesized that the two DN1s that lacked GLASS expression in the adult were the same DN1s present in larvae. We therefore predicted that the larval DN1s would express IPNa. We initially conducted colabeling experiments in R32 larvae, but LacZ labeling revealed many nonclock neurons (Fig. 7A–A″), making it difficult to identify the larval DN1s unambiguously. We therefore conducted larval labeling in brains doubly heterozygous for Cry-gal4[16] and uas-cd8GFP. Only the LNvs and DN1s are clearly marked in such brains (Fig. 7B; Klarsfeld et al., 2004). IPNa was always detectable in the larval DN1 (Fig. 7B–B″). As predicted by Klarsfeld et al. (2004), these results support the hypothesis that the DN1a in the adult nervous system represent persistent larval DN1s.
Fig. 7.

The larval DN1s express IPNa. A: A Z-series with a depth of 31 μm from an R32 larval brain labeled for LacZ at ZT 23. A′: A merged micrograph of the same Z-series for LacZ (magenta) and PER (yellow). A″: The same Z-series for PER only. LacZ expression in the larval brain reveals many neurons that do not express PER. These are marked with asterisks in the left hemisphere in A′. B: A Z-series with a depth of 22 μm within a larval brain containing the targeted expression of GFP by Cry-gal4(16). Visualization of GFP reveals the larval DN1s and LNvs, but not the DN2s. B′: IPNa labeling in the same Z-series. B″: Merged micrographs of IPNa (magenta) and GFP (green). IPNa was expressed in both larval DN1s. An asterisk indicates the position of the DN1s in B–B″. Scale bars = 20 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DN1a project toward the PDF terminals and accessory medulla
Kaneko and Hall (2000) observed that the DN1s were not homogeneous in their axonal projections. Whereas most DN1s sent projections either medially or laterally across the superior protocerebrum, a smaller number of DN1s traced the dorsal PDF projection of the s-LNvs down toward the accessory medulla, while a few others projected ventrally toward the midline along the posterior surface of the brain toward the esophageal foramen (Kaneko and Hall, 2000). We wondered which of these projection patterns the DN1a followed. We visualized the projections of the larval DN1s and the adult DN1a through the directed expression of GFP or cd8GFP, driven by one of several Cry-Gal4 elements (see Materials and Methods). Larval DN1s project directly toward the PDF terminals of the LNvs (Kaneko and Hall, 2000; Fig. 8A) and form an adjacent net of termini (Fig. 8B). It is possible that the larval DN1 projection continues down the length of the PDF projection (Kaneko and Hall, 2000), but the preparations reported here did not allow for the unambiguous tracking of larval DN1 projections past the PDF termini. In the dorsal brain of Cry-Gal4[13]/uas-GFP adults, the DN1a were clearly visible, as were two DN1p and two DN3 (Fig. 8C), Cry-Gal4[13] also drove GFP in all s-LNv, l-LNv, and LNd (Klarsfeld et al., 2004; Stoleru et al., 2005; Fig. 8C). In all brains observed the DN1a projected toward the dorsal PDF termini. In most cases the DN1a projections joined this bundle of neurites and were impossible to track with confidence. Nevertheless, in several preparations at least one of the DN1a projections flanked this bundle of neurites without joining it (Fig. 8C,D). In such cases the DN1a projected ventrally toward the accessory medulla (Fig. 8C,D). We conclude that the DN1a projections maintain their association with the dorsal PDF projection after metamorphosis and that the DN1a are at least partly responsible for the innervation of the accessory medulla by the DN1 pacemaker neuron class.
Fig. 8.

The projections of larval DN1s and adult DN1as are closely associated with the dorsal PDF projection. A,B: A Z-series with a depth of 44 μm through the DN1 and LNv of a Cry-Gal4(16)/uas-cd8GFP larval brain labeled for PDF. A: GFP only. Arrowheads mark the projection of the DN1s. B: A merged micrograph of GFP (green) and PDF (magenta). Note the GFP-only terminals to the right of the PDF projection. These presumably arise from the DN1s. C: A Z-series through the DNs and LNds of a Cry-gal4(13)/uas-GFP, displaying GFP only. The DN1as are visible in this series as are two DN1p, two DN3, six LNds, and one l-LNv. The projection of a single DN1a is marked with arrowheads. D: A merged micrograph of GFP (green) and PDF (blue) from the same Z-series. This represents an atypical preparation in which a DN1a projection could be discerned from the dorsal PDF projection and followed to the accessory medulla. Scale bars = 20 μm in A (applies to B); 50 μm in C (applies to D).
R32 reveals diversity within clock-neuron classes
High-magnification imaging of R32 soma revealed unexpected diversity within previously characterized clock-neuron classes. For example, we found that the DN1ps were heterogeneous with respect to soma size and labeling intensity. Approximately half of the 14–17 DN1p somata were larger and displayed more intense R32-LacZ and PER labeling at ZT23 (Fig. 2A–A″). The DN2 pacemaker class was also reproducibly divisible by R32-LacZ labeling intensity (Fig. 2A). In most brains the more weakly labeled DN2 soma was clearly present, as visualized by PER labeling (Fig. 2A–A″). In R32 brains two large and intensely labeled DN3s stood in contrast to their typically small and weakly labeled neuronal congeners (Fig. 2B). The LNds also showed one to three cells that were clearly larger than the others (Fig. 2C). Although these results suggest diversity among the DN1p and LNd, quantification and analysis of their areas did not reveal two clear classes of cell bodies, as has been established for the large and small LNv (data not shown).
We wondered if a similar diversity of soma size was also evident in the LNd, DN1p, and DN3 of wildtype flies. For the LNd and DN1p, wildtype (CS) brains were dissected at ZT18 and double-labeled for the clock proteins PER and PDP1. At this time we observed predominantly cytoplasmic PER and nuclear PDP1 (Fig. 9B,D; cf. Cyran et al., 2003). In all brains there was a clear diversity of soma sizes within the LNd and DN1p similar to that seen in R32 brains, with cell bodies displaying a smooth range of soma sizes (Fig. 9A–D). Micrographs of individual cell bodies from the same hemisphere were isolated visually and cell body sizes were ranked qualitatively. This evaluation suggested that, unlike the LNvs, the LNd and DN1p classes could not be divided into unambiguous “large” and “small” cell body groups. Instead, these classes reproducibly displayed smoothly graded cells size differences (Fig. 9A–D). Finally, among the ∼40 neurons of the DN3 class, R32 consistently revealed two large and intensely labeled cell bodies (Figs. 2B, 9E). PER/PDP1 labeling in the DN3 was difficult to interpret because the DN3 somata are so densely arranged (data not shown). We therefore labeled brains that were doubly heterozygous for tim-gal4 and uas-GFP elements for PER and imaged the DN3s through the dorsal surface of the brain. In these preparations, four to five cells were clearly larger than the majority of the DN3s and usually expressed higher levels of GFP (Fig. 9F–F″). We propose that, similar to the LNv classes, the LNd, DN1p, DN2, and DN3 are not homogenous groups. Rather, each class may well contain identifiable neuronal subsets that will be distinguished in the future by separable molecular and anatomical phenotypes.
Fig. 9.
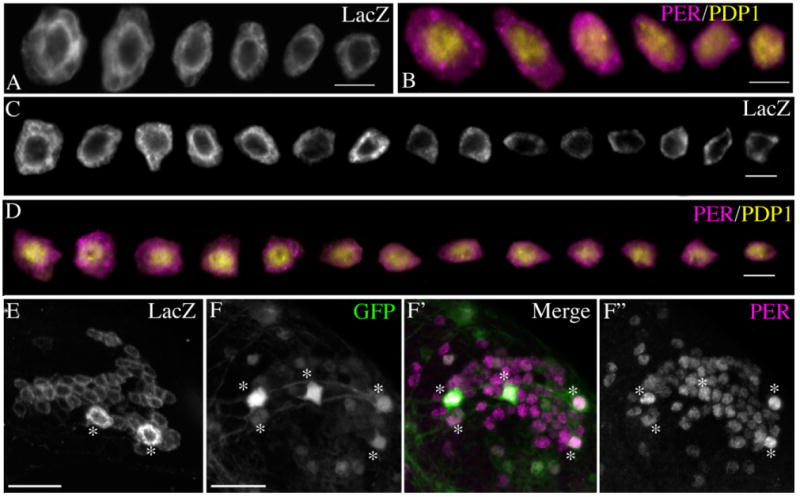
The LNd, DN1p, and DN3 each display a variety of soma sizes. A: Single optical sections through all LNds of a single R32 brain hemisphere labeled for LacZ were visually isolated from neighboring cells and ranked qualitatively from largest to smallest diameter. B: A like ordering of all LNds from a single hemisphere of a wildtype brain colabeled for PER (magenta) and PDP1 (yellow) at ZT18. C: All DN1ps from a single hemisphere of an R32 brain labeled for LacZ and presented as above. D: All DN1ps from single hemisphere of a wild-type brain co-labeled for PER (magenta) and PDP1 (yellow) at ZT18. E: A 12-μm Z-series through the DN3s from an R32 brain labeled for LacZ. DN3s were imaged through the dorsal surface of the brain. Asterisks indicate two large DN3s. F: A 13-μm Z-series through the DN3s in Tim-Gal4/uas-gfp brain dissected at ZT23 and visualized for GFP. F′: The same Z-series as a merged micrograph of GFP (green) and PER (magenta) expression. F″: The same Z-series showing PER expression only. DN3s were imaged as in E. Asterisks in F–F″ indicate five larger neurons among the DN3. Scale bars = 5 μm in A–D; Scale bar = 20 μm in E,F. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Discussion
The recognition of identified classes of neuronal pacemakers offers the possibility of analyzing behaviorally relevant neural circuits in the context of molecularly defined neuronal hierarchies. Ideally, such analysis will account for how individual neuronal pacemakers function and interact to generate and organize rhythmic outputs. To complement and extend previous work in this area, we used imaging techniques to describe the properties of circadian pacemaker neurons in Drosophila's central brain. Here we employed the novel enhancer trap line R32 to reveal several previously undescribed features of the fly's neuronal clockwork (summarized in Fig. 10). In addition to presenting new subclasses of pacemakers, this study reveals a previously uncharacterized diversity within the canonical neuronal pacemaker classes.
Fig. 10.
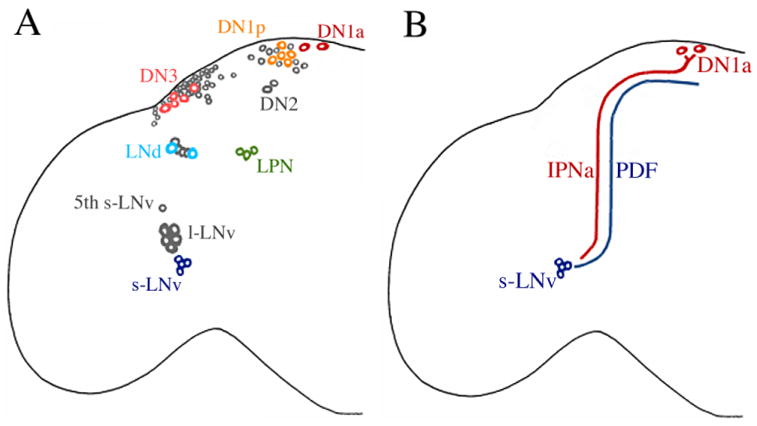
A reevaluation of the fly's neuronal clockwork. A: A map of clock neuron soma in the adult brain of Drosophila. The schematic represents only one hemisphere. Our data revealed two new classes of clock neurons, the LPNs and the DN1as. The canonical DN1s have been redesignated DN1ps. Our results also revealed a diversity of soma sizes within the LNds, DN3, and DN1ps. Larger cell bodies are presented in color. B: Peptidergic and anatomical interactions between the s-LNv and the DN1a. Our data revealed that DN1ps express IPNamide. The DN1as project toward the dorsal PDF projection and in some cases ventrally and anteriorly toward the accessory medulla. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Lateral posterior neurons
Kaneko and Hall (2000) identified a group of cells in the posterolateral brain that expressed TIM but not PER, the lack of which precluded their identification as neuronal pacemakers. Subsequently, Klarsfeld et al. (2004) alluded to PER-positive cells reminiscent of the TIM-only cells, but could not unambiguously identify them as neurons. Our data revealed a group of cells whose position and number suggest identity with this novel group. We have shown that these cells are indeed neurons (Fig. 3D–D″) and that they are diurnally rhythmic—they express both PER and TIM with a rhythm under LD conditions (Fig. 3A). We have referred to these cells, in deference to Kaneko and Hall (2000), as lateral posterior neurons (LPNs) and their recognition as pacemakers now permits investigation of their specific contributions to circadian physiology. During the preparation of this report, a study by Yoshii et al. (2005) reported that the LPN support a PER oscillation under temperature cycles and constant light, but did not detect such rhythms under LD conditions. These results, along with the detection of PER expression under LD in our study, suggest that the PER/TIM oscillations are stronger under temperature entrainment than under LD conditions, suggesting a special role for these neurons in temperature entrainment (Yoshii et al., 2005).
Anterior DN1s
Klarsfeld et al. (2004) previously described two dorsally separated DN1s and, based on GLASS labeling, hypothesized that these were the persistent larval DN1s. Furthermore, Kaneko and Hall (2000) could often identify the persistent larval DN1s as medially isolated in metamorphosing pupae. In the present study the isolated DN1a were often but not always found in brain regions medial and dorsal to the rest of the DN1s. Imaging of R32 brains through the dorsal aspect of the brain revealed an even more striking anterior separation of two DN1s that was not previously noted (Figs. 1B, 4B,C). These two neurons are situated further from the majority of DN1s (DN1ps) than were the DN2 (Figs. 1B, 4C). The DN1as, unlike the DN1ps, expressed the neuropeptide IPNa (Fig. 5), but did not express the transcription factor GLASS (Fig. 6; Klarsfeld et al., 2004). IPNa labeling in larvae suggested that the two larval DN1s persist through metamorphosis as the anteriorly separated DN1as (Fig. 7B–B″). Thus, we submit that the DN1s as previously classified (reviewed in Helfrich-Förster, 2003) actually comprise at least two distinct classes of clock neurons that are distinguishable by anatomical, immunological, genetic, and developmental criteria. Specifically, the DN1a are situated in dorsal brain regions 15–30 μm anterior of the DN1p and DN2s, express the neuropeptide IPNa, do not express GLASS, survive the glass mutation, and represent the persistent DN1s of larvae. The DN1p are located in the posterior region of the dorsal brain adjacent to the dorsal PDF projection, do not express IPNa immunosignals, are unified by GLASS expression, and are missing in adult glass mutants. The morphological and phenotypic diversity we ascribe to the two subclasses of DN1 pacemakers suggests a corresponding functional diversity within DN1z. We submit that as our knowledge of clock-cell properties grows such distinctions will permit a fuller accounting for the division of labor between and within the clock neuron classes.
Evidence for peptidergic pacemaker feedback to the accessory medulla
The mammalian suprachiasmatic nucleus, which houses the rhythmic circuits that control circadian locomotor rhythms in mammals, displays multiple transmitter phenotypes (Leak and Moore, 2000). In Drosophila multiple amidated peptides are required for normal circadian locomotor rhythms (Taghert et al., 2001), but to date only PDF has been identified as a clock-relevant transmitter (Renn et al., 1999). Here we have shown that the DN1a pacemakers express IPNa (Figs. 5, 6D–E″), thus implicating nplp1-derived peptides as additional candidate clock output factors. We have determined that the DN1a project axons toward the dorsal PDF projection and then further on to the accessory medulla (Fig. 8). Taken together, these data suggest peptidergic interactions between the accessory medulla and the dorsal brain via PDF and IPNa (Figs. 7B–B″, 8, 10B). Such interactions between clock centers could subserve the synchronization of pacemakers, the maintenance of rhythmicity, or the regulation of its amplitude (cf. Peng et al., 2003; Lin et al., 2004).
Evidence for cellular diversity within clock neuron classes
R32 revealed a replicable diversity of cell body sizes within the DN1p, DN2, DN3, and LNd classes (Figs. 2, 9). The LNvs have long been divided into large and small classes and differ in details of their molecular timekeeping (e.g., Yang et al., 2001; Shafer et al., 2002) and their genetic regulation (e.g., Park et al., 2000). Although the LNds and the DNs were not neatly divisible into large and small subclasses, the morphological diversity displayed within the DN3s suggests fundamental functional differences similar to those of the LNv. We submit that the recognition of such phenotypic distinctions will be of great practical importance as attention is turned toward the physiology of individual circadian clock neurons. Fluorescent probes of neuronal excitability are now being employed to study the physiology of single identified neurons in Drosophila (e.g., Suh et al., 2004). As such tools are brought to bear on the circadian system, the ability to recognize subclasses of pacemaker neurons will be a necessary adjunct to physiological analysis in situ.
Advantages of R32 as a clock-neuron reporter
Under the conditions reported here, R32-mediated LacZ expression is an exceptionably clear marker of clock neurons. This is due to the strong and widespread expression of LacZ in neuronal pacemakers, the apparent absence of glial labeling, the very low numbers of nonclock-neurons labeled, and the fact that most of the neuronal projections of the clock neurons are invisible or very weakly labeled. Compared to the direct immunocytochemical detection of clock proteins or the use of Gal4-driven GFP (e.g., Kaneko and Hall, 2000), R32 offers a much clearer visualization of clock neurons, even when the entire brain is reconstructed as a projected Z-series (Fig. 1A). These factors, coupled with the relative ease of immunocytochemical detection of LacZ, make R32 an effective clock neuron reporter, particularly for colabeling experiments aimed at determining the neurochemical phenotypes of clock neurons. The regulatory sequences that drive R32-mediated expression have not yet been identified. The possibility that R32 reports on a gene that normally functions within or in proximity to the molecular pacemaker has not yet been explored and cannot be excluded. The utility of the R32 P{lacW} insert may be expanded in the future if it proves convertible into a Gal4 and/or Gal80 element, as has been demonstrated for other elements (Sepp and Auld, 1999).
Acknowledgments
We thank Justin Blau, Michael Rosbash, Liliane Schoofs, and Amita Sehgal for antibodies and Ravi Allada, Patrick Emery, and François Rouyer for fly stocks.
Grant sponsors: Washington University Medical School (Keck Fellowship and NIH Vision Training Grant to O.T.S.); Grant sponsor: National Institutes of Health (NIH); Grant number: MH067122 (to P.H.T.); Grant sponsor: Deutsche Forschungs Gemeinschaft; Grant number: Fo207-9-1 (to C.H.-F.).
Abbreviations
- DN1
Dorsal neuron group 1
- DN1a
Anterior dorsal neuron group 1
- DN1p
Posterior dorsal neuron group 1
- DN2
Dorsal neuron group 2
- DN3
Dorsal neuron group 3
- dp
Dorsal projection of the s-LNv
- GFP
Green fluorescent protein
- IPNa
IPNamide
- LacZ
β-galactosidase
- l-LNv
Large ventral lateral neuron
- l-MC
Large medial cell
- LNv
Ventral lateral neuron
- LNd
Dorsal lateral neuron
- LPN
Lateral posterior neuron
Pigment dispersing factor
- PDP1
PAR domain protein 1
- PER
Period
- pot
Posterior optic tract
- TIM
Timeless
Literature Cited
- Baggerman G, Cerstiaens A, De Loof A, Schoofs L. Peptidomics of the larval Drosophila melanogaster central nervous system. J Biol Chem. 2002;277:40368–40374. doi: 10.1074/jbc.M206257200. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop MR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–790. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells' influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hall JC. Systems approaches to biological rhythms in Drosophila. Methods Enzymol. 2005;393:61–185. doi: 10.1016/S0076-6879(05)93004-8. [DOI] [PubMed] [Google Scholar]
- Hardin PE. Transcription regulation within the circadian clock: the E-box and beyond. J Biol Rhythms. 2004;19:348–360. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol A. 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc Res Techn. 2003;62:94–102. doi: 10.1002/jemt.10357. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. Techniques that revealed the network of the circadian clock of Drosophila. Methods Enzymol. 2005;393:439–451. doi: 10.1016/S0076-6879(05)93021-8. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30:249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgeneic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A, Malpel S, Michard-Vanhée C, Picot M, Chélot E, Rouyer F. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J Neurosci. 2004;24:1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J Comp Neurol. 2000;433:312–334. doi: 10.1002/cne.1142. [DOI] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. Genetic variations of Drosophila melanogaster. San Diego: Academic Press; 1992. [Google Scholar]
- Liu X, Zwiebel LJ, Hinton D, Benzer S, Hall JC, Rosbash M. The period gene encodes a predominantly nuclear protein in adult Drosophila. J Neurosci. 1992;12:2735–2744. doi: 10.1523/JNEUROSCI.12-07-02735.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses K, Rubin GM. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- Moses K, Ellis MC, Rubin RM. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- O'Neill EM, Rebay I, Tijan R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms rquire intercellular communication. Pub Lib Sci Biol. 2003;1:32–40. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SCP, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:971–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Samson ML, Lisbin MJ, White K. Two distinct temperature-sensitive alleles at the elav locus of Drosophila are suppressed nonsense mutations of the same tryptophan codon. Genetics. 1995;141:1101–1111. doi: 10.1093/genetics/141.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider LE, Roberts MS, Taghert PH. Cell type-specific transcriptional regulation of the Drosophila FMRFamide neuropeptide gene. Neuron. 1993;10:279–291. doi: 10.1016/0896-6273(93)90318-l. [DOI] [PubMed] [Google Scholar]
- Schöning JC, Staiger D. At the pulse of time: protein interactions determine the pace of circadian clocks. FEBS Lett. 2005;579:3246–3252. doi: 10.1016/j.febslet.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Auld VJ. Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics. 1999;151:1093–1101. doi: 10.1093/genetics/151.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins Period and Timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Ping Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behavior in Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Suh GS, Wong AM, Hergarden AC, Wang JW, Simon AF, Benzer S, Axel R, Anderson DJ. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature. 2004;431:854–8599. doi: 10.1038/nature02980. [DOI] [PubMed] [Google Scholar]
- Taghert PH, Lin Y. Tick-talk: the cellular and molecular biology of Drosophila circadian rhythms. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive molecular insect science. Amsterdam: Elsevier; 2005. pp. 357–395. [Google Scholar]
- Taghert PH, Hewes RS, Park JH, O'Brien MA, Han M, Peck ME. Multiple amidated neuropeptides are required for normal circadian rhythmicity in Drosophila. J Neurosci. 2001;21:6673–6686. doi: 10.1523/JNEUROSCI.21-17-06673.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleri S, Brandes C, Helfrich-Förster C, Hall JC, Stanewsky R. A self-sustaining, light-entrainable circadian oscillator in the Drosophila brain. Curr Biol. 2003;13:1758–1767. doi: 10.1016/j.cub.2003.09.030. [DOI] [PubMed] [Google Scholar]
- Verleyen P, Baggerman G, Wiehart W, Schoeters E, Van Lommel A, De Loof A, Schoofs L. Expression of a novel neuropeptide, NVGT-LARDFQLPIPNamide, in the larval and adult brain of Drosophila melanogaster. J Neurochem. 2004;88:311–319. doi: 10.1046/j.1471-4159.2003.02161.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Heshiki Y, Ibuki-Ishibashi T, Matsumoto A, Tanimura T, Tomioka K. Temperature cycles drive Drosophila circadian oscillation in constant light that otherwise induces behavioural arrhythmicity. Eur J Neurosci. 2005;22:1176–1184. doi: 10.1111/j.1460-9568.2005.04295.x. [DOI] [PubMed] [Google Scholar]
- Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, Allada R. Drosophila Clock can generate ectopic circadian clocks. Cell. 2003;13:755–766. doi: 10.1016/s0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]


