Abstract
The interactions between herbivorous insects and their symbiotic micro-organisms can be influenced by the plant species on which the insects are reared, but the underlying mechanisms are not understood. Here, we identify plant nutrients, specifically amino acids, as a candidate factor affecting the impact of symbiotic bacteria on the performance of the phloem-feeding aphid Aphis fabae. Aphis fabae grew more slowly on the labiate plant Lamium purpureum than on an alternative host plant Vicia faba, and the negative effect of L. purpureum on aphid growth was consistently exacerbated by the bacterial secondary symbionts Regiella insecticola and Hamiltonella defensa, which attained high densities in L. purpureum-reared aphids. The amino acid content of the phloem sap of L. purpureum was very low; and A. fabae on chemically defined diets of low amino acid content also grew slowly and had elevated secondary symbiont densities. It is suggested that the phloem nutrient profile of L. purpureum promotes deleterious traits in the secondary symbionts and disturbs insect controls over bacterial abundance.
Keywords: aphid, Buchnera aphidicola, plant usage, secondary symbiont, symbiosis
1. Introduction
Many herbivorous insects are chronically infected by symbiotic micro-organisms, some of which supplement the nutritionally poor plant diet with vitamins and essential amino acids (Buchner 1965; Douglas 1998; Moran & Telang 1998). These symbiotic micro-organisms are generally beneficial for the insect host but, as with some other symbioses (e.g. Redman et al. 2001; Backhed et al. 2005), their impact on the host is predicted to vary with host and symbiont genotype and with environmental conditions.
The plant usage traits of some insects are influenced by their complement of symbiotic micro-organisms (Chen et al. 2000; Hosokawa et al. 2007). This study explores the pattern and underlying mechanisms of microbial impacts on plant usage by the phloem-feeding aphid Aphis fabae. The microbiota of most aphids is dominated by the γ-proteobacterium Buchnera aphidicola, which provides the aphid with essential amino acids and is required by the insect: aphids experimentally deprived of B. aphidicola by antibiotic treatment (‘aposymbiotic’ aphids) grow very slowly and are reproductively sterile (Douglas 1998). Some aphids also bear bacterial ‘secondary symbionts’, e.g. Serratia symbiotica, Hamiltonella defensa and Regiella insecticola (Moran et al. 2005), which can, variously, promote resistance to parasitoids (Oliver et al. 2005) and entomopathogenic fungi (Scarborough et al. 2005), aphid usage of certain plants (Tsuchida et al. 2004) and thermal tolerance (Russell & Moran 2006). An interaction between secondary symbionts and the nitrogen nutrition of aphids is suggested by the demonstration that the density of S. symbiotica is elevated in aphids reared on low-nitrogen diets (Wilkinson et al. 2007), although other experimental data suggest that secondary symbionts do not contribute to the amino acid nutrition of aphids (Douglas et al. 2006a).
The basis for this study is the finding that untreated and aposymbiotic A. fabae grew at similar, low rates on the labiate plant Lamium purpureum (Adams & Douglas 1997), raising the possibility that symbiosis dysfunction induced by L. purpureum contributed to the poor aphid performance on this plant. Consistent with this interpretation, bacterial production of essential amino acids, especially threonine, was depressed in A. fabae on L. purpureum, even though the bacterial density was elevated (Wilkinson et al. 2001). These effects of L. purpureum cannot be attributed to low growth rates because A. fabae reared on other plant species that support poor growth rates did not have elevated bacterial densities or modified amino acid nutrition (Wilkinson et al. 2001).
Our approach to investigate how the symbiotic bacteria and plant nutrients affect A. fabae performance on L. purpureum involved experimental manipulation of the microbial complement of the aphids and analysis of aphid nitrogen nutrition. Following the evidence that aphid genotype can influence secondary symbiont impacts on aphid traits (e.g. Russell & Moran 2006), the experiments used multiple clones of A. fabae. The choice of secondary symbiont taxa for these experiments was decided by an investigation of the prevalence of secondary symbiont taxa in natural populations of A. fabae colonizing L. purpureum and other host plants of A. fabae; and these data are presented first.
2. Material and methods
(a) Field aphid populations
Vicia faba cv. The Sutton, L. purpureum, Chenopodium album and Papaver dubium were raised from seed in a common garden experiment, following the protocol of Darby et al. (2003). Twice-weekly between 10 June and 30 September 2003, eight pre-flowering plants of each species were examined for colonizing A. fabae and all aphids on the plants were removed. The bacterial complement of all adults which had colonized (as defined by proximity to young larvae, presumably deposited by the adult) was determined by diagnostic PCR.
(b) Experimental aphids and plants
Sixteen clones of Aphis fabae fabae (clones 1–8 lacking secondary symbionts, clones 9–12 with the secondary symbiont H. defensa and clones 13–16 with R. insecticola) were reared at 18°C with 18 hours L : 6 hours D on the test plants: pre-flowering V. faba cv. The Sutton, on which all A. f. fabae perform well (Stroyan 1984), and L. purpureum. All clones were reared on Rumex obtusifolius for at least 10 generations prior to this study, to ensure that their response to the test plants was not influenced by prior experience of different host plants.
To investigate aphid performance on plants, newborn aphids were caged individually in mesh-covered clip-on cages (2.5 cm internal diameter) on the test plants for 10 days, when they were final instar larvae. Aphid relative growth rate (RGR) was assessed by the formula ln(day-10 weight/day-0 weight)/10, with each of either eight aphids (for experiments with natural symbioses) or nine aphids (for manipulated symbioses) weighed to the nearest microgram on an MT5 microbalance (Mettler). For diet experiments, 10 replicate aphids were reared individually from birth for eight days on chemically defined diets containing 0.5 M sucrose and 25, 50, 100 or 150 mM amino acids (Wilkinson & Douglas 2003), and then weighed individually.
(c) Manipulations of secondary symbiont complement in aphids
The secondary symbiont complement of A. fabae clones was manipulated in aphids reared on chemically defined diets. Hamiltonella defensa was eliminated from clones 10 and 11, and R. insecticola from clones 13 and 15 by feeding on diets supplemented with 50 μg ampicillin, cefotaxime and gentamicin ml−1 (Douglas et al. 2006a). Aphids were infected orally with H. defensa (clones 5 and 6) and R. insecticola (clones 7 and 8) added to the diet (Darby & Douglas 2003), using symbionts obtained from pea aphids Acyrthosiphon pisum because A. fabae consistently performed poorly when fed on diets containing A. fabae extracts. The aphids with manipulated secondary symbiont complements were maintained for at least 10 generations on R. obtusifolius prior to analysis, and their status was confirmed by the complementary methods of diagnostic PCR assay and fluorescence in situ hybridization. Without exception, congruent results were obtained by the two methods, and the localization of bacteria in aphids infected naturally and experimentally with secondary symbionts was indistinguishable (electronic supplementary material, figure 1).
(d) Identification and quantification of symbiotic bacteria in aphids
Buchnera aphidicola, S. symbiotica, H. defensa and R. insecticola were detected by diagnostic 16S rRNA gene endpoint PCR, following the protocol of Douglas et al. (2006a). The bacteria in clone 13 were quantified by the direct counting method of Wilkinson et al. (2001) and in clones 5–8, 11, 12, 15 and 16 by TaqMan quantitative PCR (Q-PCR; Douglas et al. 2006b) using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, UK). The primers and probes designed for the dnaK gene were as follows: B. aphidicola forward TGCGGCACTTGCATATGGT, reverse CGAAAGTCCCCCCACCTAAG, probe AGGCCAAGGGAACCGGACTATAGCT GT; H. defensa forward CAAGCGGATT ATTAATGAACCCA, reverse TGGTGCT ATTCCCTTTTCCCT, probe CGCGGCCATTGCCTACGGTTT; and R. insecticola forward CCGTAAAGACGTCAATCCCG, reverse GCACCCCTCCTTGAATAGCA, probe CGAAGCGGTTGCGGTCGGTG. For the probe, FAM (6-carboxy fluorescein) was the 5′-terminal reporter dye and TAMRA (tetramethyl carboxyrhodamine) was the 3′-terminal quencher dye. Gene copy number was estimated from the standard curve generated with serial dilutions of linearized cloned dnaK gene fragments (the standards) and normalized to aphid weight. The standards and the DNA extracted from each of the eight replicate aphids per treatment were run in duplicate with template-free controls.
(e) Collection and quantification of free amino acids
Phloem sap from L. purpureum and V. faba was collected from the severed stylets of A. fabae (one sample per plant) obtained by high-frequency microcautery following the protocol of Pescod et al. (2007). Known weights of honeydew were collected over 24 hours from aphids confined to a leaf in clip-on cages lined with pre-weighed aluminium foil, and washed off the foil in a known volume of 80% methanol. Amino acids were separated by reverse-phase HPLC, following derivatization with o-phthaldialdehyde (Jones et al. 1981), using a Hewlett-Packard HP1100 Series autosampling LC system with C18 ZORBAX Eclipse XDB-C8 column (Agilent, UK) and fluorescence detection. AA-S-18 amino acid standards (Sigma, UK) supplemented with asparagine, glutamine and tryptophan were used for calibration.
(f) Statistical analysis
The frequency of aphids colonizing different plants and with different bacterial complements was analysed by Χ2-test. All other datasets were normally distributed with homogeneous variances, by the Ryan–Joiner one-sample test and Bartlett's test, respectively. The impact of two fixed factors, rearing plant (‘plant’) and either presence/absence of secondary symbionts (‘symbiosis’) or the identity of secondary symbionts (‘secondary symbiont’), on the performance and bacterial abundance of aphids was investigated by ANOVA, with clone as a random factor nested within either symbiosis or secondary symbiont. For experiments using aphids with manipulated symbioses, the term ‘treatment’ was included as a third fixed main factor in the ANOVA. The error term for each source in the ANOVA was chosen from the pattern of components in expectations of mean squares obtained using standard rules (Steele & Torrie 1960). Full outputs from the statistical tests are provided in the electronic supplementary material, table 1.
3. Results
(a) Prevalence of symbiotic bacteria in field populations of A. fabae
The field plot was colonized by 318 wild A. fabae. Buchnera aphidicola was detected in every aphid tested and secondary symbionts were detected in 179 (56%) aphids. No individual aphid bore more than one secondary symbiont. The most prevalent secondary symbiont was R. insecticola, and S. symbiotica was present at low frequencies in aphids from most plant species (table 1). The frequency of aphids with different bacterial complements varied significantly with plant species (Χ92=26.66, 0.01>p>0.001), and this could be attributed principally to the relatively high prevalence of S. symbiotica in aphids on V. faba.
Table 1.
Prevalence of symbiotic bacteria in field populations of the aphid A. fabae colonizing four plant species between June and September 2003.
| plant species | number of aphidsa | ||||
|---|---|---|---|---|---|
| total | with Ba only | with Ba+Ss | with Ba+Hd | with Ba+Ri | |
| C. album | 75 | 43 | 4 | 7 | 21 |
| L. purpureum | 80 | 24 | 7 | 17 | 32 |
| P. dubium | 71 | 38 | 1 | 9 | 23 |
| V. faba | 92 | 34 | 15 | 13 | 30 |
Ba, B. aphidicola; Ss, S. symbiotica; Hd, H. defensa; Ri, R. insecticola.
(b) Impact of rearing plant and bacterial symbiont complement on aphid performance
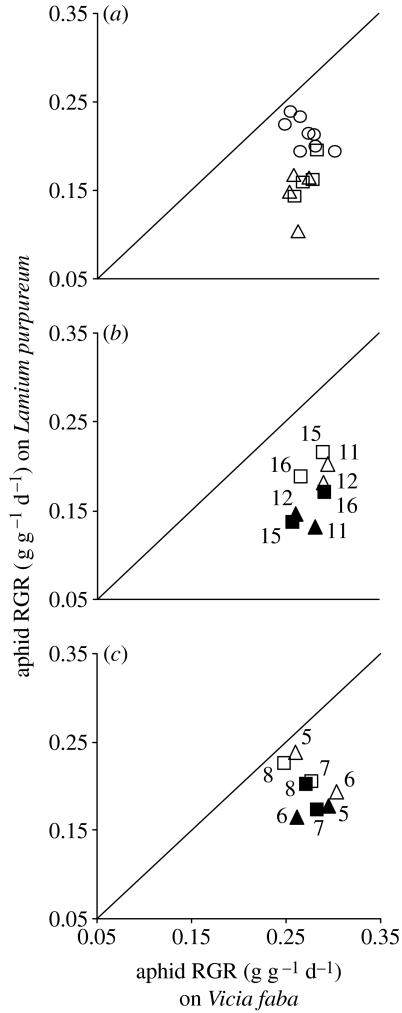
The RGR of the 16 A. fabae clones was significantly depressed in aphids on L. purpureum relative to V. faba (F1,14=225.22, p<0.001), but to a greater extent in clones with secondary symbionts than in secondary symbiont-free clones (F1,14=23.83, p<0.001). Underlying these effects was significant variation among clones (F14,224=2.38, 0.01>p>0.001), which was greater on L. purpureum than on V. faba (F14,224=2.88, p<0.001; figure 1a; with full statistical analysis in the electronic supplementary material, table 1a). The performance of aphids bearing H. defensa and R. insecticola did not differ significantly (p>0.05).
Figure 1.
Aphid performance on V. faba and L. purpureum. (a) Clones with ‘natural’ bacterial complement (n=8; open circles, secondary symbiont-free aphids; open triangles, aphids with H. defensa; open squares, aphids with R. insecticola); (b) clones cured of their natural secondary symbiont infections (n=9; filled triangles, aphids with H. defensa; filled squares, aphids with R. insecticola; open triangles, aphids cured of H. defensa; open squares, aphids cured of R. insecticola); (c) clones experimentally infected with secondary symbionts (n=9; filled triangles, aphids experimentally infected with H. defensa; filled squares, aphids experimentally infected with R. insecticola; open triangles, aphids lacking H. defensa; open squares, aphids lacking R. insecticola). Error bars are not displayed for clarity. Numbers in (b) and (c) indicate aphid clone numbers.
The negative impact of secondary symbionts on aphid performance on L. purpureum is open to two alternative explanations: that the secondary symbionts exacerbate the negative effect of L. purpureum on aphid performance; and that certain aphid genotypes are particularly prone to infection with secondary symbionts and, independently, are deleteriously affected by L. purpureum. To discriminate between these explanations, the performance of aphids with manipulated complements of secondary symbionts was investigated. Aphid performance on L. purpureum is predicted to be changed by the manipulations only if the secondary symbionts, and not aphid genotype, are responsible for the exacerbated negative effect of this plant on performance. Elimination of the secondary symbionts by antibiotic treatment improved the mean performance of all four clones tested on L. purpureum (figure 1b) to a greater extent than for aphids on V. faba-reared aphids (plant×treatment: F1,128=13.33, p<0.001). In the reverse experiment, infection of clones naturally lacking these bacteria had a more negative effect on the performance of aphids reared on L. purpureum than on V. faba (figure 1c: plant×treatment: F1,128=72.41, p<0.001). Aphid performance also varied between the clones, as influenced by curing (electronic supplementary material, table 1b), plant species and infection (electronic supplementary material, table 1c).
(c) Abundance of bacteria in aphids
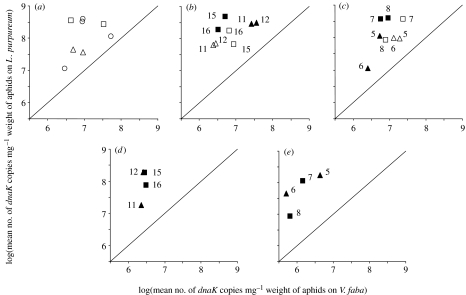
We hypothesized that aphid control over the populations of symbiotic bacteria may be disturbed in aphids reared on L. purpureum. The copy number of the B. aphidicola dnaK gene per unit aphid weight was significantly elevated in all aphids on L. purpureum; both clones with natural secondary symbiont complements (figure 2a: F1,7=54.59, p<0.001) and experimentally manipulated symbioses (figure 2b: F1,112=168.53, p<0.001; figure 2c: F1,112=103.89, p<0.001). The ANOVAs for all datasets revealed significant interclonal variation in bacterial abundance that, for the manipulated symbioses, was also influenced by treatment (electronic supplementary material, table 1d–f).
Figure 2.
Impact of rearing plant on bacterial populations, quantified as copy number of the gene dnaK. (a) Buchnera aphidicola in clones with ‘natural’ bacterial complement (open circles, secondary symbiont-free aphids; open triangles, aphids with H. defensa; open squares, aphids with R. insecticola); (b) B. aphidicola in clones cured of their natural secondary symbiont infections (filled triangles, aphids with H. defensa; filled squares, aphids with R. insecticola; open triangles, aphids cured of H. defensa; open squares, aphids cured of R. insecticola); (c) B. aphidicola in clones experimentally infected with secondary symbionts (filled triangles, aphids infected with H. defensa; filled squares, aphids infected with R. insecticola; open triangles, aphids lacking H. defensa; open squares, aphids lacking R. insecticola); (d) secondary symbionts in clones with ‘natural’ bacterial complement (filled triangles, aphids with H. defensa; filled squares, aphids with R. insecticola); (e) secondary symbionts in clones experimentally infected with secondary symbionts (filled triangles, aphids infected with H. defensa; filled squares, aphids infected with R. insecticola). For all assays, n=8. Error bars are not included for clarity. Numbers in (b) to (e) indicate aphid clone numbers.
The copy number of the dnaK gene in the secondary symbionts was also significantly elevated in aphids reared on L. purpureum relative to those on V. faba for both aphid clones bearing their natural complement of bacteria (figure 2d: F1,56=75.32, p<0.001) and aphids experimentally infected with secondary symbionts (figure 2e: F1,56=166.15, p<0.001). A significant clonal effect was obtained for the latter experiment. Further analysis revealed no relationship between the performance of the various clones and the density of secondary symbionts on either plant species (data not shown).
(d) Determinants of the aphid response to L. purpureum
The honeydew (egesta) of A. fabae feeding from L. purpureum contained 5±1.6 nmol amino acids mg−1 (n=9), 40-fold lower than 200±60 nmol amino acids mg−1 honeydew produced by A. fabae on V. faba (n=9). These data demonstrate that the amino acid content of L. purpureum phloem sap is very low. Lamium purpureum is one of a number of plants ill-suited to direct quantification of phloem sap because sap flow ceases very rapidly on stylet severance. Even so, sufficient sap was obtained from three plants of L. purpureum to establish: first, that L. purpureum phloem sap is dominated by the sugar sucrose and free amino acids; and, second, that the mean amino acid content was 77±30.1 mM (mean±s.e., n=3), a value at ‘the lower end’ of published values (50–800 mM; Douglas 2003) and just 20% of the mean amino acid content of V. faba phloem sap, at 389±51 mM (n=4).
To investigate the significance of the low amino acid content of L. purpureum for the A. fabae symbiosis, the impact of dietary amino acid concentration on the aphids was investigated using chemically defined diets with 25–150 mM amino acids (table 2). All aphids of clone 13 (which bears R. insecticola) survived the 8-day experimental period and weight gain was strongly dependent on dietary amino acid concentration. As previously observed for aphids on L. purpureum, the density of the secondary symbiont was elevated two- to threefold on the 25 mM amino acid diet relative to the other diets with higher amino acid concentrations. The density of B. aphidicola in the aphids did not vary significantly with diet.
Table 2.
Response of symbiosis in A. fabae clone 13 to dietary amino acid concentration over 8 days of larval development. (Values of mean±s.e. (10 replicates) are shown; values in one column with the same superscript letter are not significantly different by Tukey's post hoc test (p>0.05).)
| dietary amino acids (mM) | aphid weight (mg) | density of bacteria (107×number of bacteria mg−1 fresh weight) | |
|---|---|---|---|
| B. aphidicola | R. insecticola | ||
| 150 | 0.56±0.016a | 2.28±0.096 | 0.10±0.007a |
| 100 | 0.33±0.025b | 2.49±0.136 | 0.13±0.008a |
| 50 | 0.19±0.015c | 2.27±0.146 | 0.14±0.010a |
| 25 | 0.13±0.004d | 2.53±0.072 | 0.29±0.023b |
| ANOVA: F3,36 | 126.9, p<0.001 | 1.4, p>0.05 | 36.6, p<0.001 |
4. Discussion
A key result of this study is that, in A. fabae reared on L. purpureum, secondary symbionts exacerbate the deleterious impact of the plant on aphid performance (figure 1). This effect is accompanied by an elevated density of secondary symbionts in the aphids (figure 2d,e). Secondary symbiont densities are also high in aphids reared on low nitrogen diets (table 2), suggesting that the low nitrogen content of L. purpureum phloem sap (results of this study) might contribute to the high secondary symbiont density in the aphids. The amino acid composition of L. purpureum phloem sap and diets differ; and the high secondary symbiont densities on L. purpureum with 77 mM amino acids but on diets with 25 mM amino acids raise the possibility that secondary symbiont density is responsive to specific amino acids and not to the total amino acid concentration. Factors contributing to the greater responsiveness of the secondary symbionts than B. aphidicola to the diet may include differences between the bacteria in metabolic capabilities or access to aphid nutrients.
Elevated secondary symbiont densities and poor aphid performance have been identified previously for Ac. pisum reared on low nitrogen diets (Wilkinson et al. 2007) or infected with more than one secondary symbiont (Oliver et al. 2006), but poor performance is not always accompanied by high secondary symbiont numbers. For example, the poor performance of A. fabae on white clover Trifolium repens and Ac. pisum on high sucrose diets are not accompanied by elevated secondary symbiont densities (D. Adams and A. E. Douglas 1995, unpublished results; E. Jones and A. E. Douglas 2005, unpublished results). The relationship between secondary symbiont density and aphid performance may be causal, i.e. the high bacterial populations consume nutrients and cause other physiological disturbances that collectively depress aphid growth. Alternatively, the depressed growth and high bacterial populations may not be related causally, except in the sense that they are independent consequences of a dysfunction that remains to be identified.
Significantly, no consistent difference in the impact of the two secondary symbiont taxa, H. defensa and R. insecticola, on aphid usage of L. purpureum was evident, indicating that the effect is not dictated by genes specific to one taxon of secondary symbiont. Other effects of secondary symbionts on ecologically important traits of aphids are more taxon specific. For example, secondary symbiont-mediated resistance to parasitoids in the pea aphid is displayed by H. defensa and not by R. insecticola (Ferrari et al. 2004; Oliver et al. 2005). The considerable interclonal variation in the impact of secondary symbionts on aphid traits, as identified here and by various previous studies (e.g. Chen et al. 2000; Scarborough et al. 2005; Russell & Moran 2006), illustrates how the genotype of aphid and symbiont and their interactions can influence microbial impacts on insect–plant interactions.
The present study illustrates the importance of environmental context (here, rearing plant) on the impact of these micro-organisms on their hosts. Further research is required to establish whether the negative effects of secondary symbionts are an important factor contributing to the variable incidence of these bacteria in field populations of A. fabae and other aphids. The secondary symbionts, unlike B. aphidicola, are horizontally transmissible (Sandström et al. 2001; Darby & Douglas 2003; Moran & Dunbar 2006). The fitness of secondary symbionts may be enhanced by their high population density, even though their aphid host performs poorly, because the high population densities might enhance the opportunity for horizontal transmission away from the relatively unfit aphid host on L. purpureum.
In conclusion, this study offers the first identification of a candidate plant factor, low phloem amino acid concentration, contributing to the impact of secondary symbionts on aphid performance on plants. The interaction between the A. fabae symbiosis and L. purpureum is not a ‘laboratory phenomenon’. Lamium purpureum in the field is colonized by A. fabae from natural populations, including individuals bearing secondary symbionts (table 1), and the aphid numbers increase slowly on L. purpureum to form small but persistent colonies (Raymond et al. 2000). Host plant-dependent impacts of symbiotic micro-organisms on the fitness of herbivorous insects may be a widespread and currently unrecognized dimension in insect–plant interactions.
Acknowledgments
We thank Dr Neil Boonham for advice on QPCR analysis, Dr Roger Sturmey for assistance with HPLC analysis, Dr T. Fukatsu and colleagues for their advice and support with FISH analysis and Dr Terry Crawford for statistical support. BBSRC and the James Burgess Foundation provided financial support.
Supplementary Material
Statistical analyses
Fluorescence in situ hybridisation (FISH) analysis of symbiotic bacteria in Aphis fabae
Key for FISH micrographs and methods for FISH analysis
References
- Adams D, Douglas A.E. How symbiotic bacteria influence plant utilisation by the polyphagous aphid, Aphis fabae. Oecologica. 1997;110:528–532. doi: 10.1007/s004420050190. doi:10.1007/s004420050190 [DOI] [PubMed] [Google Scholar]
- Backhed F, Ley R.E, Sonnenburg J.L, Peterson D.A, Gordon J.I. Host–bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. doi:10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- Buchner P.Endosymbioses of insects with plant micro-organisms1965John Wiley & Sons; Chichester, UK [Google Scholar]
- Chen D.Q, Montllor C.B, Purcell A.H. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol. Exp. Appl. 2000;95:315–323. doi:10.1046/j.1570-7458.2000.00670.x [Google Scholar]
- Darby A.C, Douglas A.E. Elucidation of the transmission patterns of an insect-borne bacterium. Appl. Environ. Microbiol. 2003;69:4403–4407. doi: 10.1128/AEM.69.8.4403-4407.2003. doi:10.1128/AEM.69.8.4403-4407.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby A.C, Tosh C.R, Walters K.F.A, Douglas A.E. The significance of a facultative bacterium to natural populations of the pea aphid Acyrthosiphon pisum. Ecol. Entomol. 2003;28:145–150. doi:10.1046/j.1365-2311.2003.00492.x [Google Scholar]
- Douglas A.E. Nutritional interactions in insect–microbial symbioses: aphids and their symbiotic bacteria B. aphidicola. Annu. Rev. Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. doi:10.1146/annurev.ento.43.1.17 [DOI] [PubMed] [Google Scholar]
- Douglas A.E. Nutritional physiology of aphids. Adv. Insect Physiol. 2003;31:73–140. doi:10.1016/S0065-2806(03)31002-1 [Google Scholar]
- Douglas A.E, François C.L.M.J, Minto L.B. Facultative ‘secondary’ bacterial symbionts and the nutrition of the pea aphid, Acyrthosiphon pisum. Physiol. Entomol. 2006a;31:262–269. doi:10.1111/j.1365-3032.2006.00516.x [Google Scholar]
- Douglas A.E, Price D.R.G, Minto L.B, Jones E, Pescod K.V, François C.L.M.J, Pritchard J, Boonham N. Sweet problems: insect traits defining the limits to dietary sugar utilisation by the pea aphid, Acyrthosiphon pisum. J. Exp. Biol. 2006b;209:1395–1403. doi: 10.1242/jeb.02148. doi:10.1242/jeb.02148 [DOI] [PubMed] [Google Scholar]
- Ferrari J, Darby A.C, Daniell T.J, Godfray H.C.J, Douglas A.E. Linking the bacterial community in pea aphids with host–plant use and natural enemy resistance. Ecol. Entomol. 2004;29:60–65. doi:10.1111/j.1365-2311.2004.00574.x [Google Scholar]
- Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T. Obligate symbiont involved in pest status of host insect. Proc. R. Soc. B. 2007;274:1979–1984. doi: 10.1098/rspb.2007.0620. doi:10.1098/rspb.2007.0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.N, Pääbo S, Stein S. Amino acid analysis and enzymatic sequence determination of peptides by an improved o-phthaldialdehyde precolumn labelling procedure. J. Liquid Chromatogr. 1981;4:565–586. doi:10.1080/01483918108059956 [Google Scholar]
- Moran N.A, Dunbar H.E. Sexual acquisition of beneficial symbionts in aphids. Proc. Natl Acad. Sci. USA. 2006;103:12 803–12 806. doi: 10.1073/pnas.0605772103. doi:10.1073/pnas.0605772103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N.A, Telang A. Bacteriocyte associated symbionts of insects. BioScience. 1998;48:295–304. doi:10.2307/1313356 [Google Scholar]
- Moran N.A, Russell J.A, Koga R, Fukatsu T. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 2005;71:3302–3310. doi: 10.1128/AEM.71.6.3302-3310.2005. doi:10.1128/AEM.71.6.3302-3310.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver K.M, Moran N.A, Hunter M.S. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl Acad. Sci. USA. 2005;102:12 795–12 800. doi: 10.1073/pnas.0506131102. doi:10.1073/pnas.0506131102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver K.M, Moran N.A, Hunter M.S. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. R. Soc. B. 2006;273:1273–1280. doi: 10.1098/rspb.2005.3436. doi:10.1098/rspb.2005.3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescod K.V, Quick W.P, Douglas A.E. Aphid responses to plants with genetically manipulated phloem nutrient levels. Physiol. Entomol. 2007;32:253–258. doi:10.1111/j.1365-3032.2007.00577.x [Google Scholar]
- Raymond B, Darby A.C, Douglas A.E. Intraguild predation and the spatial distribution of a parasitoid. Oecologia. 2000;124:367–372. doi: 10.1007/s004420000396. doi:10.1007/s004420000396 [DOI] [PubMed] [Google Scholar]
- Redman R.S, Duvigan E.D, Rodriguez R.J. Fungal symbiosis from mutualism to parasitism: who controls the outcome, host or invader? New Phytol. 2001;151:705–716. doi: 10.1046/j.0028-646x.2001.00210.x. doi:10.1046/j.0028-646x.2001.00210.x [DOI] [PubMed] [Google Scholar]
- Russell J.A, Moran N.A. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc. R. Soc. B. 2006;273:603–610. doi: 10.1098/rspb.2005.3348. doi:10.1098/rspb.2005.3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström J.P, Russell J.A, White J.P, Moran N.A. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol. Ecol. 2001;10:217–228. doi: 10.1046/j.1365-294x.2001.01189.x. doi:10.1046/j.1365-294X.2001.01189.x [DOI] [PubMed] [Google Scholar]
- Scarborough C.L, Ferrari J, Godfray H.C. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781. doi: 10.1126/science.1120180. doi:10.1126/science.1120180 [DOI] [PubMed] [Google Scholar]
- Steele R.G.D, Torrie J.H. McGraw Hill; London, UK: 1960. Principles and procedures of statistics: a biometric approach. [Google Scholar]
- Stroyan H.L.G. Royal Entomological Society of London; London, UK: 1984. Aphids—Pterocommatinae and Aphidinae (Aphidini) [Google Scholar]
- Tsuchida T, Koga R, Fukatsu T. Host plant specialization governed by facultative symbiont. Science. 2004;303:1989. doi: 10.1126/science.1094611. doi:10.1126/science.1094611 [DOI] [PubMed] [Google Scholar]
- Wilkinson T.L, Douglas A.E. Phloem amino acids and the host plant range of the polyphagous aphid, Aphis fabae. Entomol. Exp. Appl. 2003;106:1–11. doi:10.1046/j.1570-7458.2003.00014.x [Google Scholar]
- Wilkinson T.L, Adams D, Minto L.B, Douglas A.E. The impact of host plant on the abundance and function of symbiotic bacteria in an aphid. J. Exp. Biol. 2001;204:3027–3038. doi: 10.1242/jeb.204.17.3027. [DOI] [PubMed] [Google Scholar]
- Wilkinson T.L, Koga R, Fukatsu T. Role of host nutrition in symbiont regulation: impact of dietary nitrogen on proliferation of obligate and facultative bacterial endosymbionts of the pea aphid, Acyrthosiphon pisum. Appl. Environ. Microbiol. 2007;73:1362–1366. doi: 10.1128/AEM.01211-06. doi:10.1128/AEM.01211-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical analyses
Fluorescence in situ hybridisation (FISH) analysis of symbiotic bacteria in Aphis fabae
Key for FISH micrographs and methods for FISH analysis