Abstract
The long-term study of animal populations facilitates detailed analysis of processes otherwise difficult to measure, and whose significance may appear only when a large sample size from many years is available for analysis. For example, inbreeding is a rare event in most natural populations, and therefore many years of data are needed to estimate its effect on fitness. A key behaviour hypothesized to play an important role in avoiding inbreeding is natal dispersal. However, the functional significance of natal dispersal with respect to inbreeding has been much debated but subject to very few empirical tests. We analysed 44 years of data from a wild great tit Parus major population involving over 5000 natal dispersal events within Wytham Woods, UK. Individuals breeding with a relative dispersed over several-fold shorter distances than those outbreeding; within the class of inbreeding birds, increased inbreeding was associated with reduced dispersal distance, for both males and females. This led to a 3.4-fold increase (2.3–5, 95% CI) in the likelihood of close (f=0.25) inbreeding relative to the population average when individuals dispersed less than 200 m. In the light of our results, and published evidence showing little support for active inbreeding avoidance in vertebrates, we suggest that dispersal should be considered as a mechanism of prime importance for inbreeding avoidance in wild populations.
Keywords: dispersal, inbreeding, inbreeding avoidance, wild population, Parus major, great tit
1. Introduction
Natal dispersal, defined as the distance moved by an individual from its birth place to the site of its first reproduction (Howard 1960; Greenwood 1980; Clobert et al. 2001), is a key life-history event with relevance for many aspects of evolutionary biology, ecology and conservation biology: it alters gene flow; changes allele frequencies within and among populations (Clobert et al. 2001) and influences the distribution and abundance of organisms (Johnson & Gaines 1990). Three factors are most often proposed to be important contributors towards the evolution of dispersal (Gandon & Michalakis 2001). Dispersal may have evolved in order to reduce competition between relatives (Hamilton & May 1977), owing to the temporal variability of the environment (Olivieri et al. 1995; Gandon & Michalakis 1999) or it may function as a means of inbreeding avoidance. The consequence of inbreeding, defined as the mating of individuals sharing ancestors (Wright 1922), is increased genome-wide homozygosity. This, in turn, causes inbreeding depression, which is mediated either through overdominance (where heterozygous individuals have superior fitness relative to homozygous equivalents) or most often due to the expression of deleterious recessive alleles (Lynch & Walsh 1998; Charlesworth & Charlesworth 1999; Keller & Waller 2002). Thus, in the absence of other mechanisms of inbreeding avoidance, the act of dispersing from its natal site may substantially reduce the likelihood of choosing a related individual for mate.
Many studies have investigated the relationship between inbreeding and dispersal from a theoretical standpoint (Bulmer 1973; Bengtsson 1978; Waser et al. 1986; Motro 1991; Gandon 1999; Perrin & Mazalov 1999, 2000; Roze & Rousset 2005; Guillaume & Perrin 2006). The outcome of these theoretical studies varies greatly depending on their initial assumptions. Ultimately, this leads to a trade-off between simple models with reduced realism, or complex models where estimating the relative importance of each parameter and the size of interactions between them becomes increasingly difficult (Gandon & Michalakis 2001). Using a game-theoretical approach, Perrin & Mazalov (1999) emphasized that inbreeding by itself is unlikely to account for the evolution of dispersal on its own. Undoubtedly, there is much more to dispersal than just inbreeding avoidance; the question is not whether inbreeding affects dispersal or not, but in what way and by how much (Perrin & Goudet 2001). It is clear that there is a need for empirical studies that explore the interplay between inbreeding and dispersal; such tests should ideally be carried out in a natural setting, where dispersal is under natural selection.
The number of theoretical studies on inbreeding and dispersal contrasts markedly with the very few empirical studies where the costs of inbreeding, and the importance of dispersal as a mean of inbreeding avoidance, have been estimated (Greenwood et al. 1978; Schiegg et al. 2006). This paucity of empirical studies is probably partly due to the fact that in order to test the relative importance of inbreeding on the evolution of dispersal, very large numbers of dispersal events, together with a continuous monitoring of a population over a long time scale, are required. Greenwood et al. (1978) explored the relationship between inbreeding depression and natal dispersal using 11 years of data from the long-term study of great tits in Wytham Woods. However, their study did not formally test relationships between inbreeding and dispersal, and only a limited number of inbreeding pairs were identified for which natal dispersal distances could be investigated. The aim of the present study was to use a much more extensive dataset for the same population, involving over 5000 dispersal events recorded over 44 years, to test the relationship between dispersal and inbreeding at different levels. In a previous study, we showed that close inbreeding in this population reduces fitness by 55%, calculated in terms of the number of fledged grand-offspring relative to an outbred pair (Szulkin et al. 2007). Here, we demonstrate a strong effect of limited natal dispersal on the likelihood of inbreeding, and suggest that dispersal should be considered a fundamental mechanism of inbreeding avoidance in many vertebrate species.
2. Material and methods
(a) Study population
The great tit Parus major is a small hole nesting passerine bird that has been studied at Wytham Woods (Oxfordshire, UK, 1°20′ W 51°46′ N) since 1947 (Perrins 1979). The population breeds almost exclusively in over 1000 artificial nest-boxes, scattered at variable densities across ca 380 ha of semi-natural deciduous woodland. The number and location of nest-boxes throughout the site, as well as the breeding protocol, have remained fairly constant since 1964; the exact coordinates of all nest-boxes were digitally mapped in 2005 (Wilkin et al. 2006). Great tit breeding events are identified by performing regular checks of all nest-boxes in the study area throughout the breeding season. Nestlings are ringed 15 days after hatching, and parents caught and identified while feeding young at the nest. Immigration rates into the population are relatively high, as on average 40% of males and 47% of females breeding in any year within Wytham are born outside the forest (McCleery et al. 2004); immigrants are assumed to be unrelated to each other for the purposes of this study. Establishing parental identity, combined with offspring ringing at the nest allowed us to build a pedigree of all breeding birds in Wytham, as well as to determine natal dispersal distances of birds born and breeding in the site. The pedigree relies on the assumption that social fathers are also genetic fathers; extra-pair fertilization, however, can occur, and is estimated to be of the order of 14–19% (Blakey 1994; S. C. Patrick 2006, unpublished work) in this population. But while EPP can change the genetic relationship of an offspring to its social father, with the exception of daughters mating with their social father, it can at most only halve the relatedness of pairs of inbreeding birds, and it does not interfere in our estimates of natal dispersal distance. More detailed information on the collection of life-history data can be found in McCleery et al. (2004), and discussion on pedigree construction in Szulkin et al. (2007).
(b) Natal dispersal distances
We defined natal dispersal distance as the Euclidean distance between the natal nest-box (or box of rearing in cases where cross-fostering occurred) and the nest-box of an individual breeding for the first time. While some individuals bred more than once in their lifetime (75.4% birds bred only with one partner, 18.2% bred with two, 5% bred with three, 1.1% with four and 0.1% with five different partners), only their natal dispersal distance was taken into account for these analyses. Because dispersal between breeding events is very limited in this population (Harvey et al. 1979), the natal dispersal distance of a bird effectively reflects its lifetime dispersal, and analyses conducted on all dispersal events yield identical trends as those presented in the study.
(c) Defining inbreeding
Identities of parents and their offspring allowed us to build a pedigree covering the period from 1958 to 2007; the dispersal data are only estimated for the years 1964–2007 as all boxes were not in position until 1964. In rare cases where experiments such as cross-fostering were carried out on a nest and it was impossible to determine the biological identity of nestlings, such individuals were classified as having unknown parents. Similarly, in breeding events where one or both parents were unknown, the offspring were assigned a unique virtual sire and dam identity, which allowed siblings to be identified despite unknown parental identity.
Inbreeding occurs when relatives mate, and offspring born from such matings are more likely to carry genes that are identical by descent (Lynch & Walsh 1998); the proportion of genes identical by descent will increase the greater relatedness between its parents. The pedigree of all parents and offspring allows coefficients of inbreeding f (Wright 1922; Keller & Waller 2002) to be estimated for each nestling or breeding event. In this study, we define ‘inbreeding individuals’ as those mating with a relative, and siring inbred offspring. The highest inbreeding that can occur in birds and mammals within one generation is often described as ‘close’ inbreeding, and yields offspring with an inbreeding coefficient of f=0.25. Such inbreeding arises, assuming previously outbred individuals, in the case of brother–sister or parent–offspring mating. We used the software Pedigree Viewer (available at http://www.personal.une.edu.au/∼bkinghor/pedigree.htm) to estimate inbreeding coefficients of all nestlings ringed during the breeding season between 1958 and 2007. In order to facilitate comparison, all breeding events were assigned a particular inbreeding class: if an individual bred only with unrelated partners in its lifetime, its status was defined as ‘outbred’ (f=0.0), and the partners were thus ‘outbreeding’. Owing to the substantial pedigree depth in some cases (see Szulkin et al. (2007) for more details), we could detect some low values of inbreeding, which were likely to be the result of high pedigree resolution. We therefore assigned all inbreeding events with non-null inbreeding and f lower than 0.03125, to an inbreeding class defined as f=0.0002, as the median value of inbreeding for this class was 0.0002. Four classes with successively increasing inbreeding followed: f=0.03125; f=0.06125 (first-cousin mating); f=0.125 (grandparent–grand-offspring/double first-cousin mating); and finally the highest inbreeding class, f=0.25. Thus, even if an individual once mated with an unrelated mate but then mated with its sibling in the following breeding event, the related partners were qualified as inbreeding (f=0.25) in their lifetime.
(d) Statistical analysis
In general, because natal dispersal distances tend to be right skewed, we used square-root transformed data for all analyses where natal dispersal was used as a response variable, but we report untransformed values together with the median and the interquartile range (IQR) in figures and tables as measures of central tendency and variability.
(i) Natal dispersal distance in different inbreeding classes
To infer differences in natal dispersal distances between inbreeding classes, we used linear mixed models to control for the potential non-independence of breeding siblings by fitting the breeding event from which the bird originated as a random effect in the models. For the close inbreeding category (f=0.25), we excluded values of natal dispersal distances of parents involved in parent–offspring matings, because parental natal dispersal occurred before their offspring were born. In the case of brother–sister matings, we also included only one sibling at random of a pair to avoid pseudoreplication. We fitted the effect of inbreeding on dispersal using a continuous variable with several levels of inbreeding (f=0.0, 0.002, 0.03125, 0.0625, 0.125 and 0.25). We additionally fitted a sex×inbreeding interaction, the birth year of the breeding individual (categorical variable), its standardized egg-laying date (calculated as ‘egg-laying date’−‘average egg-laying date for a given year’) averaged over an individual's lifetime, and the distance from the bird's first breeding nest-box to the forest edge as additional explanatory variables influencing natal dispersal. Variables where p>0.1 were dropped from the model. The amount of variance explained by our model was estimated as a proportion of the difference between residual variance when only random effects were included, and the residual variance of the model where both random and all fixed effects taken into account. Natal dispersal distances of different types of close inbreeding (parent–offspring, brother–sister matings) and of outbreeding male and female individuals were further explored using matched pair tests.
(ii) Matched pair tests
A comparison of all birds mating with related and unrelated kin is potentially vulnerable to environmental and demographic effects on inbreeding, dispersal or both. For example, both inbreeding and dispersal might be greater when population density is high or in poorer habitats. To take into account such environmental heterogeneity, we matched focal nests where related individuals bred with the closest nest-box where unrelated individuals bred in the same year. Because, in some cases, the closest breeding event yielded information on natal dispersal distance of only one of the two parents (when one individual was born outside Wytham), the closest matched pair nest-box could differ for males and females at a focal nest.
Nest-box coordinates and information on nest-box occupancy for each year were used to generate a matrix of nest-box distances for each year. We used Matlab v. 7.1 to generate sequences of active nest-boxes (i.e. where a breeding event occurred) that matched focal nest-boxes where f≥0.03125 according to the following criteria: (i) male and/or female natal dispersal distances were known, (ii) pair members of matched nest-boxes were unrelated to each other (thus siring offspring with f=0.0), and (iii) the nest-box was the closest to the focal nest-box satisfying criteria (i) and (ii).
We compared natal dispersal distances of inbreeding and outbreeding individuals from matched breeding events using matched pair t-tests of square-root transformed natal dispersal distances. Because natal dispersal distances of brother–sister pairs are potentially pseudoreplicated (all sibling pairs but one were reared together), we also compared differences in natal dispersal distances between inbreeding siblings and outbreeding individuals using a randomly chosen member of a pair of siblings, maintaining equal proportions of males and females (27 cases of brother–sister matings; 14 males and 13 females included).
(iii) Likelihood of inbreeding
The fact that inbreeding individuals may differ in their natal dispersal distances relative to outbreeding individuals does not necessary tell us how the likelihood of inbreeding changes with dispersal distance. We therefore analysed differences in the likelihood of inbreeding with respect to dispersal distance using generalized linear models (GLMs) where inbreeding (f≥0.03125) was fitted as a binomial variable (inbred or not). Additional explanatory variables fitted in the model were sex, the interaction between sex and dispersal distance, an individual's birth year, distance of the first breeding nest-box to the forest edge and standardized egg-laying date. Variables where p>0.1 were dropped from the model. In the case of parent–offspring matings, parental natal dispersal distance occurred before its offspring was born; we therefore excluded parental dispersal distances from the dataset when parents mated with their offspring. When males and females were analysed together, only one sibling of a brother–sister mating pair (chosen at random) was included in the analysis. All statistical analyses were carried out using Genstat v. 8.1 (VSN International Ltd).
3. Results
We analysed 44 years of great tit (P. major) breeding data from Wytham Woods, UK, which yielded 5289 male and female records of natal dispersal events for which we could estimate relatedness. Natal dispersal distances, as measured between the natal site and the first breeding event within a pair of individuals, differed between the sexes, as expected, with females dispersing 49% further than males on average (median and IQR: males 528 m (298–931, n=2772); females 788 m (456–1338, n=2517)). We identified 78 cases of natal dispersal distances where individuals were inbreeding at f=0.25 in their lifetime (out of which 54, 8 and 16 individuals were involved in brother–sister, father–daughter and mother–son matings, respectively), and 25, 45, 29 and 554 cases where individuals were inbreeding at f=0.125, 0.0625, 0.03125 and 0.0002, respectively.
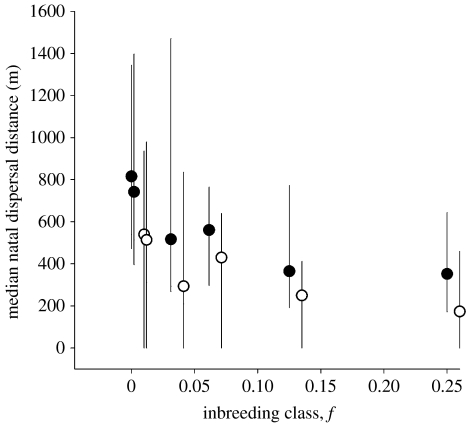
Individuals that mated with kin and sired inbred offspring dispersed shorter distances than individuals who bred with unrelated mates (figure 1; table 1). Although the trend for a relationship between inbreeding and short dispersal distances appears to be stronger for females, the interaction between sex and inbreeding was not significant and was thus removed from the final model (table 1).
Figure 1.
Natal dispersal distances (median and IQR) for male and female great tits (open and filled circles, respectively) with respect to six different inbreeding classes. Parent–offspring matings are excluded from this analysis. The values for males have been offset on the abscissa to facilitate presentation.
Table 1.
Individuals mating with related kin disperse short distances. Mixed model with normal errors and birth origin (unique birthplace) fitted as random effects; natal dispersal distances of parents involved in parent–offspring matings are excluded, and in the case of brother–sister matings, only one sibling of a pair is included. Square-rooted values of natal dispersal distance were used as response variable in order to achieve normality. Variance explained by the model, 14%. Residual d.f., 5091.
| d.f. | wald | p | regression coefficient | s.e. | |
|---|---|---|---|---|---|
| sex | 1 | 236.38 | <0.001 | ♂: −4.34 | 0.28 |
| inbreeding | 1 | 72.04 | <0.001 | −47.91 | 5.64 |
| birth year | 47 | 86.23 | <0.001 | ||
| egg-laying date | 1 | 3.03 | 0.082 | 0.044 | 0.025 |
| distance to forest edge | 1 | 3.81 | 0.051 | −0.0025 | 0.0013 |
| rejected term | |||||
| sex×inbreeding | 1 | 0.95 | 0.329 | ||
(a) Matching inbreeding and outbreeding pairs living at close proximity
Because recorded dispersal distances can be influenced by small- and large-scale environmental heterogeneity, we used a matched pairs approach to compare each inbreeding pair with the nearest outbreeding pair in the same year. The median distance between matched nest-boxes was of 96 m (68–140 m IQR) for males and 103 m (70–157 m IQR) for females (see table 2 for distances between matched nest-boxes in different inbreeding classes). For each inbreeding class but the lowest one (f=0.0002), we found a significant effect of mate relatedness on individual natal dispersal distance (table 2); the mean natal dispersal distance was significantly shorter for both males and females that mated with kin compared with outbreeding birds.
Table 2.
Natal dispersal distances (m) of individuals mating with related and unrelated partners (median+IQR). (The matched pair t-test uses square-rooted values of natal dispersal distances.)
| inbreeding class | d.f. | t | p | mean difference (95% CI) | dispersal distance of inbreeding individuals | matched dispersal distance of outbreeding individuals | n | distance between matched nest-boxes |
|---|---|---|---|---|---|---|---|---|
| f=0.25 | ||||||||
| type of close inbreeding | ||||||||
| only one of the two siblings (randomly selected) | 26 | 2.82 | 0.009 | 7.20 (1.94–12.45) | ♂: 377 (160–542) | ♂: 681 (381–1069) | 27 | ♂: 115 (44–181) |
| ♀: 349 (231–664) | ♀: 702 (379–1199) | 27 | ♀: 146 (71–193) | |||||
| parents | 7 | 1.65 | 0.144 | 9.29 (−4.06 to 22.64) | ♂: 553 (313–1812) | ♂: 449 (300–634) | 4 | ♂: 113 (91–129) |
| ♀: 671 (618–883) | ♀: 276 (150–470) | 4 | ♀: 224 (179–241) | |||||
| offspring | 15 | 6.14 | <0.001 | 22.63 (14.77–30.48) | ♂: 66 (9–107) | ♂: 834 (405–1696) | 12 | ♂: 102 (64–139) |
| ♀: 161 (0–338) | ♀: 682 (307–1685) | 4 | ♀: 91 (71–113) | |||||
| brothers | 26 | 1.83 | 0.079 | 4.60 (0.56–9.76) | 352 (172–570) | 608 (301–1049) | 27 | 121 (54–178) |
| sisters | 26 | 3.59 | 0.001 | 11.42 (4.88–17.96) | 377 (188–644) | 1002 (557–1712) | 27 | 129 (71–223) |
| f=0.125 | 24 | 5.37 | <0.001 | 14.65 (9.02–20.27) | ♂: 250 (142–413) | ♂: 562 (421–1980) | 12 | ♂: 109 (73–137) |
| ♀: 365 (192–773) | ♀: 1466 (627–1912) | 13 | ♀: 130 (75–151) | |||||
| f=0.0625 | 44 | 2.33 | 0.024 | 4.56 (0.63–8.57) | ♂: 430 (303–641) | ♂: 701 (266–956) | 22 | ♂: 72 (53–117) |
| ♀: 561 (297–765) | ♀: 776 (382–1345) | 23 | ♀: 109 (67–130) | |||||
| f=0.03125 | 28 | 2.31 | 0.029 | 5.35 (0.60–10.11) | ♂: 245 (207–745) | ♂: 509 (353–785) | 14 | ♂: 83 (63–150) |
| ♀: 517 (268–1471) | ♀: 1183 (918–1590) | 15 | ♀: 95 (79–149) | |||||
| f=0.0002 | 553 | 0.08 | 0.937 | 0.05 (−1.24 to 1.14) | ♂: 514 (312–980) | ♂: 540 (308–940) | 293 | ♂: 97 (69–139) |
| ♀: 742 (396–1398) | ♀: 829 (412–1378) | 271 | ♀: 99 (68–156) | |||||
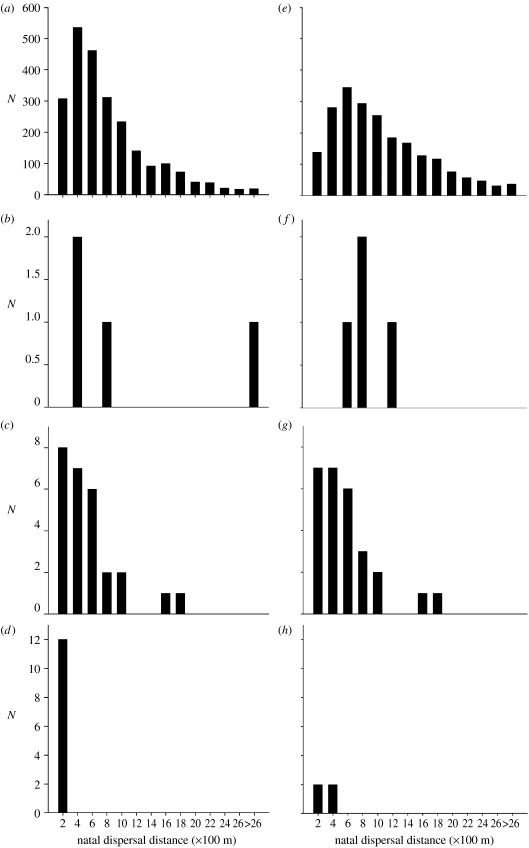
Dispersal distances of individuals involved in different types of close inbreeding, defined here as siring offspring with f=0.25, differed considerably from each other. Parent–offspring matings typically resulted from offspring hardly dispersing at all from their natal site, while parental natal dispersal distances did not differ from dispersal distances of outbreeding individuals (table 2; figure 2). Siblings that mated with each other, in contrast, dispersed shorter distances than would be expected for either males or females (table 2; figure 2). For individuals that mated with both related and unrelated kin in their lifetimes, there was no clear evidence that inbreeding occurred closer to their natal site (paired t-test =1.82, d.f.=59, p=0.074), as the median dispersal distance while inbreeding was of 308 m (136–430, n=32) for males and 410 m (195–692, n=28) for females; median dispersal distance while outbreeding: 363 m (193–508, n=32) for males and 383 m (223–692, n=28) for females.
Figure 2.
Frequency distributions of natal dispersal distances for (a–d) males and (e–h) females that either (a,e) outbred or were involved in close inbreeding of the following types: (b) fathers from father–daughter matings, (c) brothers from brother–sister matings, (d) sons from mother–son matings, (f) mothers from mother–son matings, (g) sisters from brother–sister matings and (h) daughters from father–daughter matings.
(b) Likelihood of inbreeding when breeding close to home
Individuals that mated with a related partner dispersed shorter distances relative to birds mating with unrelated partners; this, in turn, led to a significantly increased likelihood of inbreeding for those individuals that dispersed short distances (table 3; figure 3).
Table 3.
Natal dispersal influences the likelihood of inbreeding (coded as ‘0’/‘1’, and equivalent to ‘1’ whenever f≥0.03125) in males and females. (GLM with binomial error distribution and logit link. Parents from parent–offspring matings are excluded from the analysis, and only one randomly chosen sibling from brother–sister pairs is included when both sexes are analysed together. Variance explained by the model: 10.8%.)
| dataset | d.f. | deviance | deviance ratio | p | regression coefficient | s.e. |
|---|---|---|---|---|---|---|
| dispersal distance | 1 | 60.4 | 90.4 | <0.001 | −0.0015 | 0.0002 |
| sex | 1 | 3.2 | 3.16 | 0.075 | −0.307 | 0.172 |
| parental birth year | 47 | 61.2 | 1.30 | 0.08 | ||
| residual | 5204 | 1025.5 | ||||
| rejected terms | ||||||
| sex×dispersal distance | 1 | 0.2 | 0.2 | 0.655 | ||
| distance to forest edge | 1 | 0.5 | 0.51 | 0.474 | ||
| egg-laying date | 1 | 0.0008 | 0.0006 | 0.978 | ||
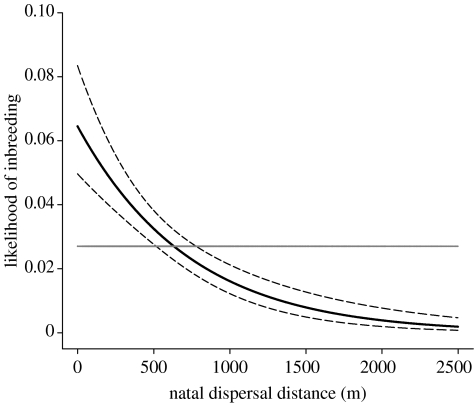
Figure 3.
The likelihood of mating with kin decreases with respect to natal dispersal distance (m). The bold line shows the fitted values from a GLM with binomial errors where natal dispersal distance was fitted as a predictor to inbreeding (f≥0.03125); dashed lines show the 95% CI for the fit. The horizontal line represents the overall population average likelihood of inbreeding.
The likelihood of inbreeding at f≥0.03125 exceeded the population average when birds dispersed less than 632 m (515–778 m, 95% CI), while the likelihood of close inbreeding (f=0.25) increased 3.4-fold (2.3–5, 95% CI) relative to the population average when birds dispersed less than 200 m. The importance of natal dispersal distance on the likelihood of inbreeding was equally large when males and females were analysed separately (table 3).
4. Discussion
Using a large and long-term dataset that allowed the estimation of many dispersal events for wild great tits, we found that birds mating with kin disperse shorter distances than those who outbreed. Moreover, males and females that do not disperse far are substantially more likely to inbreed than individuals dispersing longer distances. Hence, we show that there is a direct relationship between natal dispersal distances and the likelihood of inbreeding, for both males and females. Below, we discuss the implications of these findings for understanding the operation and evolution of inbreeding avoidance mechanisms and dispersal in general.
Hansson et al. (2007) suggested that, in most situations, dispersal should be a sufficient inbreeding avoidance mechanism; however, direct evidence for a link between dispersal and inbreeding is scarce. Greenwood et al. (1978) showed a trend for reduced natal dispersal distances of birds mating with kin using a dataset of Wytham great tit breeding data spanning from 1964 to 1975, yet the results were not always clear-cut, nor were they formally tested. Using a long-term dataset spanning from 1964 to 2007 of the same great tit population, we could test the relationship between dispersal and inbreeding with greater power and resolution. We are aware of only one other study that has directly tested the relationship between dispersal and inbreeding. Schiegg et al. (2006) showed that in the cooperatively breeding red-cockaded woodpecker Picoides borealis, the offspring of females that dispersed short distances from their natal site were more likely to be inbred. There was no evidence that this was the case for dispersal of their fathers and, interestingly, remaining on the natal territory was not associated with an elevated risk of inbreeding at all.
While dispersal distances vary depending on the inbreeding group considered in our study, it is noteworthy that birds involved in inbreeding with other than first-order relatives (e.g. f=0.03125) were also found to disperse shorter distances than outbreeding individuals. These classes of inbreeding differ from those involving first-order relatives because the inbreeding individuals need not necessarily have encountered each other previously, and the dispersal events can be considered to be independent; in the case of first-order relatives breeding, limited dispersal might result from non-independent dispersal. Sex differences in dispersal tended to be more marked with lower inbreeding coefficients, and there are probably two reasons for this: first, short dispersal distances are bounded by zero, while long dispersal distances are limited only by the size of the study site; second, the dispersal distance must converge when the majority of close inbreeding events are for siblings.
Individuals involved in close inbreeding (f=0.25) dispersed shorter distances than outbreeding individuals, yet this category of inbreeding was not homogenous in terms of patterns of natal dispersal: in parent–offspring matings, parents had an average dispersal distance, whereas offspring dispersal was very limited. Previous work on this population has shown that dispersal by adults between breeding attempts is very limited (Harvey et al. 1979); hence these parent–offspring matings seem to result from limited dispersal of only the offspring. Siblings that mated with each other dispersed further than offspring in parent–offspring matings, yet the distances were much shorter than those of outbreeding individuals, and overall contributed to the greater likelihood of inbreeding in short dispersal classes. However, it is notable that some cases of close inbreeding resulted even though a pair of siblings had dispersed further than the median for outbreeding birds (figure 2c,g: two cases of sibling pairs inbreeding having dispersed further than the median). While it has been found that sibling tits often disperse in similar directions, but not necessarily the same distances (Matthysen et al. 2005), more research is needed into the patterns of pair formation, dispersal and the break-up of family groups after fledging. We do not yet know whether brother–sister mating, the most frequent form of close inbreeding in our study population, is mainly the result of random mating between individuals that dispersed short distances or whether unusually strong pair bonds were formed while in the original family group, which may lead to restricted dispersal. The effect of natal dispersal distances in sib matings seems to vary between species, as a long-term study of blue tits Parus caeruleus in The Netherlands does not show reduced dispersal in cases of brother–sister mating (A. J. van Noordwijk 2007, personal communication).
Natal dispersal is notorious for its vulnerability to bias: when populations breeding within a limited area are studied, particularly when breeding sites are provided as here, the recorded ‘dispersal’ trait is limited by a finite number of possible observations, dependent on the position of the natal nest-box in the study plot, the size and shape of the study plot and the distance moved by a bird between its natal and breeding site (van Noordwijk 1984). While a matched pairs approach has been used in other studies to infer fitness costs resulting from different patterns of dispersal in siblings (as in Pärt 1991), to test for random mating (Keller & Arcese 1998) or to compare survival relative to sibling heterozygosity (Hansson et al. 2001), we suggest that it is also an effective way to incorporate environmental heterogeneity, including factors that are unmeasured. While methods aimed at establishing the true nature of the natal dispersal distribution are currently being developed (van Noordwijk 2006; unpublished manuscript), the use of matched pairs of observations (this study) provides a simple yet powerful alternative to more complex models aiming to control for environmental heterogeneity.
Depending on initial assumptions, conclusions of theoretical models investigating factors shaping the evolution of dispersal vary, and give varying weight to the importance of inbreeding to dispersal. For example, Guillaume & Perrin (2006) suggested that inbreeding should have a limited impact on the evolution of dispersal; by contrast, Roze & Rousset (2005) find that inbreeding may substantially influence the evolution of dispersal, while Perrin & Mazalov (1999, 2000) suggest that inbreeding avoidance cannot be the only selective pressure shaping dispersal, and define its influence on dispersal as ‘complex’. A clearly lacking element is empirical testing of the relationship between inbreeding and dispersal. While our study is of the current association between inbreeding and dispersal, it suggests that selection may still act to favour increased dispersal due to the risk of inbreeding, as the likelihood of inbreeding increased particularly steeply for very short distances. For example, individuals that dispersed less than 200 m have a likelihood of 2.8% (1.9–4.1%, 95% CI) of close (f=0.25) inbreeding. Given a 55% reduction in terms of fledged grand-offspring caused by this level of inbreeding (Szulkin et al. 2007) in this population, the expected reduction in fitness due to inbreeding, for individuals dispersing less than 200 m, is of the order of 1.5% (1.0–2.3 95% CI). Although selection differentials against limited dispersal due to inbreeding alone are currently relatively weak, it must be emphasized that our calculations are based on the current situation where dispersal is already operating as inbreeding avoidance behaviour. As stated by Pärt (1996), current frequencies of inbreeding inherently include dispersal as an inbreeding avoidance mechanism, and it is therefore noteworthy that inbreeding apparently still causes selection against short distance dispersal even at its current state. Of course, a full assessment of selection on dispersal behaviour is very challenging, and would require experimental manipulation of dispersal over a large scale, with the concomitant problem that fitness estimates for different classes of dispersers may be biased.
While we have shown evidence for a link between inbreeding and dispersal, we are not in favour of ‘unifactorial’ hypotheses (Lambin et al. 2001) such as those that argue that dispersal functions primarily to avoid close inbreeding. Several other factors are very likely to influence the evolution of dispersal, such as competition for mates and resources (Hamilton & May 1977; Greenwood 1980) or the temporal variability of the environment (Olivieri et al. 1995; Gandon & Michalakis 1999). Our findings, however, simply strongly support the fundamental importance of dispersal as a mean of inbreeding avoidance, and suggest that inbreeding avoidance is an important element in the shaping of the evolution of dispersal.
An increasing number of studies show no evidence for active inbreeding avoidance through kin discrimination, as shown by tests of random mating (van Noordwijk et al. 1985; van Tienderen & van Noordwijk 1988; Keller & Arcese 1998; Hansson et al. 2007) or of increased rates of EPP in inbred broods (Kempenaers et al. 1996; Foerster et al. 2003; Foerster et al. 2006), and there are only a few studies suggesting the reverse, which have generally used marker-based estimates of inbreeding (Blomqvist et al. 2002; Eimes et al. 2005; see Griffith & Montgomerie 2003). Our results are consistent with what would be expected if mating occurred at random, as the likelihood of inbreeding is a function of distance of an individual's breeding site relative to its natal nest. While tests for active inbreeding avoidance through increased rates of EPP, or via direct assessment of kin recognition, have not been performed for this population, their presence, or absence, should not decrease the importance that natal dispersal has on inbreeding. In agreement with the suggestions of Hansson et al. (2007), this study shows that in this great tit population, natal dispersal distance is a key factor facilitating inbreeding avoidance, which coupled with marked inbreeding depression, suggests that inbreeding avoidance should have played a major role in the shaping of the evolution of dispersal.
Acknowledgments
The work was funded by the Christopher Welch Trust and The Queen's College. Work carried out in this study complies with current UK regulations. We thank the many generations of fieldworkers who collected data for this study. We are very grateful to Arie van Noordwijk and Robin McCleery for their discussions on dispersal, and in particular for sharing their ideas and suggestions for this manuscript. We thank Przemysław Żelazowski for help in spatial analyses and Teddy Wilkin for providing nest-box coordinates from Wytham Woods.
Footnotes
One contribution of 18 to a Special Issue ‘Evolutionary dynamics of wild populations’.
References
- Bengtsson B.O. Avoiding inbreeding—at what cost? J. Theor. Biol. 1978;73:439–444. doi: 10.1016/0022-5193(78)90151-0. doi:10.1016/0022-5193(78)90151-0 [DOI] [PubMed] [Google Scholar]
- Blakey J.K. Genetic evidence for extra-pair fertilizations in a monogamous passerine, the great tit Parus major. Ibis. 1994;136:457–462. [Google Scholar]
- Blomqvist D, et al. Genetic similarity between mates and extra-pair parentage in three species of shorebirds. Nature. 2002;419:613–615. doi: 10.1038/nature01104. doi:10.1038/nature01104 [DOI] [PubMed] [Google Scholar]
- Bulmer M.G. Inbreeding in the great tit. Heredity. 1973;30:313–925. doi: 10.1038/hdy.1973.41. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The genetic basis of inbreeding depression. Genet. Res. 1999;74:329–340. doi: 10.1017/s0016672399004152. doi:10.1017/S0016672399004152 [DOI] [PubMed] [Google Scholar]
- Clobert J, Danchin E, Dhondt A.A, Nichols J.D. Oxford University Press; Oxford, UK: 2001. Dispersal. [Google Scholar]
- Eimes J.A, Parker P.G, Brown J.L, Brown E.R. Extrapair fertilization and genetic similarity of social mates in the Mexican jay. Behav. Ecol. 2005;16:456–460. doi:10.1093/beheco/ari010 [Google Scholar]
- Foerster K, Delhey K, Johnsen A, Lifjeld J.T, Kempenaers B. Females increase offspring heterozygosity and fitness through extra-pair matings. Nature. 2003;425:714–717. doi: 10.1038/nature01969. doi:10.1038/nature01969 [DOI] [PubMed] [Google Scholar]
- Foerster K, Valcu M, Johnsen A, Kempenaers B. A spatial genetic structure and effects of relatedness on mate choice in a wild bird population. Mol. Ecol. 2006;15:4555–4567. doi: 10.1111/j.1365-294X.2006.03091.x. doi:10.1111/j.1365-294X.2006.03091.x [DOI] [PubMed] [Google Scholar]
- Gandon S. Kin competition, the cost of inbreeding and the evolution of dispersal. J. Theor. Biol. 1999;200:345–364. doi: 10.1006/jtbi.1999.0994. doi:10.1006/jtbi.1999.0994 [DOI] [PubMed] [Google Scholar]
- Gandon S, Michalakis Y. Evolutionarily stable dispersal rate in a metapopulation with extinctions and kin competition. J. Theor. Biol. 1999;199:275–290. doi: 10.1006/jtbi.1999.0960. doi:10.1006/jtbi.1999.0960 [DOI] [PubMed] [Google Scholar]
- Gandon S, Michalakis Y. Multiple causes of the evolution of dispersal. In: Clobert J, Danchin E, Dhondt A.A, Nichols J.D, editors. Dispersal. Oxford University Press; Oxford, UK: 2001. pp. 155–188. [Google Scholar]
- Greenwood P.J. Mating systems, philopatry and dispersal in birds and mammals. Anim. Behav. 1980;28:1140–1162. doi:10.1016/S0003-3472(80)80103-5 [Google Scholar]
- Greenwood P.J, Harvey P.H, Perrins C.M. Inbreeding and dispersal in the great tit. Nature. 1978;271:52–54. doi:10.1038/271052a0 [Google Scholar]
- Griffith S.C, Montgomerie R. Why do birds engage in extra-pair copulation? Nature. 2003;422:833. doi: 10.1038/422833a. doi:10.1038/422833a [DOI] [PubMed] [Google Scholar]
- Guillaume F, Perrin N. Joint evolution of dispersal and inbreeding load. Genetics. 2006;173:497–509. doi: 10.1534/genetics.105.046847. doi:10.1534/genetics.105.046847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W.D, May R.M. Dispersal in stable habitats. Nature. 1977;269:578–581. doi:10.1038/269578a0 [Google Scholar]
- Hansson B, Bensch S, Hasselquist D, Åkesson M. Microsatellite diversity predicts recruitment of sibling great reed warblers. Proc. R. Soc. B. 2001;268:1287–1291. doi: 10.1098/rspb.2001.1640. doi:10.1098/rspb.2001.1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson B, Jack L, Christians J.K, Pemberton J.M, Akesson M, Westerdahl H, Bensch S, Hasselquist D. No evidence for inbreeding avoidance in a great reed warbler population. Behav. Ecol. 2007;18:157–164. doi:10.1093/beheco/arl062 [Google Scholar]
- Harvey P.H, Greenwood P.J, Perrins C.M. Breeding area fidelity of great tits (Parus major) J. Anim. Ecol. 1979;48:305–313. doi:10.2307/4115 [Google Scholar]
- Howard W.E. Innate and environmental dispersal of individual vertebrates. Am. Midl. Nat. 1960;63:152–161. doi:10.2307/2422936 [Google Scholar]
- Johnson M.L, Gaines M.S. Evolution of dispersal—theoretical models and empirical tests using birds and mammals. Annu. Rev. Ecol. Syst. 1990;21:449–480. doi:10.1146/annurev.es.21.110190.002313 [Google Scholar]
- Keller L.F, Arcese P. No evidence for inbreeding avoidance in a natural population of song sparrows (Melospiza melodia) Am. Nat. 1998;152:380–392. doi: 10.1086/286176. doi:10.1086/286176 [DOI] [PubMed] [Google Scholar]
- Keller L.F, Waller D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002;17:230–241. doi:10.1016/S0169-5347(02)02489-8 [Google Scholar]
- Kempenaers B, Adriaensen F, Van Noordwijk A.J, Dhondt A.A. Genetic similarity, inbreeding and hatching failure in blue tits: are unhatched eggs infertile? Proc. R. Soc. B. 1996;263:179–185. doi:10.1098/rspb.1996.0029 [Google Scholar]
- Lambin X, Aars J, Poertney S.B. Dispersal, intraspecific competition and kin facilitation: a review of the empirical evidence. In: Clobert J, Danchin E, Dhondt A.A, Nichols J.D, editors. Dispersal. Oxford University Press; Oxford, UK: 2001. pp. 110–122. [Google Scholar]
- Lynch M, Walsh B. Sinauer Associates, Inc.; Sunderland, MA: 1998. Genetics and analysis of quantitative traits. [Google Scholar]
- Matthysen E, Van de Casteele T, Adriaensen F. Do sibling tits (Parus major, P. caeruleus) disperse over similar distances and in similar directions? Oecologia. 2005;143:301–307. doi: 10.1007/s00442-004-1760-7. doi:10.1007/s00442-004-1760-7 [DOI] [PubMed] [Google Scholar]
- McCleery R.H, Pettifor R.A, Armbruster P, Meyer K, Sheldon B.C, Perrins C.M. Components of variance underlying fitness in a natural population of the great tit Parus major. Am. Nat. 2004;164:E62–E72. doi: 10.1086/422660. doi:10.1086/422660 [DOI] [PubMed] [Google Scholar]
- Motro U. Avoiding inbreeding and sibling competition—the evolution of sexual dimorphism for dispersal. Am. Nat. 1991;137:108–115. doi:10.1086/285148 [Google Scholar]
- Olivieri I, Michalakis Y, Gouyon P.H. Metapopulation genetics and the evolution of dispersal. Am. Nat. 1995;146:202–228. doi:10.1086/285795 [Google Scholar]
- Pärt T. Philopatry pays: a comparison between collared flycatcher sisters. Am. Nat. 1991;138:790–796. doi:10.1086/285252 [Google Scholar]
- Pärt T. Problems with testing inbreeding avoidance: the case of the collared flycatcher. Evolution. 1996;50:1625–1630. doi: 10.1111/j.1558-5646.1996.tb03934.x. doi:10.2307/2410898 [DOI] [PubMed] [Google Scholar]
- Perrin N, Goudet J. Inbreeding, kinship, and the evolution of natal dispersal. In: Clobert J, Danchin E, Dhondt A.A, Nichols J.D, editors. Dispersal. Oxford University Press; Oxford, UK: 2001. pp. 123–154. [Google Scholar]
- Perrin N, Mazalov V. Dispersal and inbreeding avoidance. Am. Nat. 1999;154:282–292. doi: 10.1086/303236. doi:10.1086/303236 [DOI] [PubMed] [Google Scholar]
- Perrin N, Mazalov V. Local competition, inbreeding, and the evolution of sex-biased dispersal. Am. Nat. 2000;155:116–127. doi: 10.1086/303296. doi:10.1086/303296 [DOI] [PubMed] [Google Scholar]
- Perrins C.M. Collins; London, UK: 1979. British tits. [Google Scholar]
- Roze D, Rousset F. Inbreeding depression and the evolution of dispersal rates: a multilocus model. Am. Nat. 2005;166:708–721. doi: 10.1086/497543. doi:10.1086/497543 [DOI] [PubMed] [Google Scholar]
- Schiegg K, Daniels S.J, Walters J.R, Priddy J.A, Pasinelli G. Inbreeding in red-cockaded woodpeckers: effects of natal dispersal distance and territory location. Biol. Conserv. 2006;131:544–552. doi:10.1016/j.biocon.2006.03.001 [Google Scholar]
- Szulkin M, Garant D, McCleery R.H, Sheldon B.C. Inbreeding depression along a life-history continuum in the great tit. J. Evol. Biol. 2007;20:1531–1543. doi: 10.1111/j.1420-9101.2007.01325.x. doi:10.1111/j.1420-9101.2007.01325.x [DOI] [PubMed] [Google Scholar]
- van Noordwijk A.J. Problems in the analysis of dispersal and a critique on its heritability in the great tit. J. Anim. Ecol. 1984;53:533–544. doi:10.2307/4532 [Google Scholar]
- van Noordwijk A.J. Measuring natal dispersal as distance-related recruitment rates. J. Ornithol. 2006;147:45. doi:10.1007/s10336-006-0093-1 [Google Scholar]
- van Noordwijk A.J, Van Tienderen P.H, de Jong G. Genealogical evidence for random mating in a natural population of the great tit (Parus major L.) Naturwissenschaften. 1985;72:104–106. doi:10.1007/BF00508149 [Google Scholar]
- van Tienderen P.H, van Noordwijk A.J. Dispersal, kinship and inbreeding in an island population of the great tit. J. Evol. Biol. 1988;1:117–137. doi:10.1046/j.1420-9101.1988.1020117.x [Google Scholar]
- Waser P.M, Austad S.N, Keane B. When should animals tolerate inbreeding. Am. Nat. 1986;128:529–537. doi:10.1086/284585 [Google Scholar]
- Wilkin T.A, Garant D, Gosler A.G, Sheldon B.C. Density effects on life-history traits in a wild population of the great tit Parus major: analyses of long-term data with GIS techniques. J. Anim. Ecol. 2006;75:604–615. doi: 10.1111/j.1365-2656.2006.01078.x. doi:10.1111/j.1365-2656.2006.01078.x [DOI] [PubMed] [Google Scholar]
- Wright S. Coefficients of inbreeding and relationship. Am. Nat. 1922;56:330–338. doi:10.1086/279872 [Google Scholar]