Summary
Biomembranes are complex, heterogeneous, dynamic systems playing essential roles in numerous processes such as cell signaling and membrane trafficking. Model membranes provide simpler platforms for studying biomembrane dynamics under well-controlled environments. Here we present a modified polymer lift-off approach to introduce chemical complexity into biomimetic membranes by constructing domains of one lipid composition (here, didodecylphosphatidylcholine) that are surrounded by a different lipid composition (e.g., dipentadecylphosphatidylcholine), which we refer to as patterned backfilled samples. Fluorescence microscopy and correlation spectroscopy were used to characterize this patterning approach. We observe two types of domain populations: one with diffuse boundaries and a minor fraction with sharp edges. Lipids within the diffuse domains in patterned backfilled samples undergo anomalous diffusion, which results from nonideally mixed clusters of gel phase lipid within the fluid domains. No lateral diffusion was observed within the minor population of domains with well-defined borders. These results suggest that, while membrane patterning by a variety of approaches is useful for biophysical and biosensor applications, a thorough and systematic characterization of the resulting biomimetic membrane, and its unpatterned counterpart, is essential.
Keywords: patterning, lipid bilayers, fluorescence correlation spectroscopy, lateral diffusion
Introduction
Microdomains (such as “lipid rafts”[1]) in biological membranes have been proposed to participate in numerous functions, such as signal transduction[2, 3] and membrane trafficking[4]. Given the inherent complexity of cell membranes, model systems (i.e., giant unilamellar vesicles[5], supported planar lipid bilayers and monolayers[6–8] and black lipid membranes[9]) have been developed to provide a simpler platform for step-by-step studies in controlled environments. Domains of different lipid phases can form spontaneously when two or more lipids are mixed together[8, 10–12], where segregation of lipids into different phases occurs mainly due to the inherent properties of the participating lipid molecules. Due to this spontaneous segregation, these model systems do not allow us to probe the dynamics of microdomains in a spatially and temporally well-controlled manner. As a result, newly developed micropatterning approaches such as microcontact printing[13–15], agarose stamping[16], UV photolysis[17–20] and a polymer-based dry lift-off approach[21, 22] may allow formation of spatially organized microdomains of one lipid composition within the context of another lipid composition, thereby allowing us to extend beyond the phase diagram. Such spatially organized model systems could then act as building blocks for integrated biosensors to facilitate studies for a variety of cellular processes[22].
Planar supported membranes consist of a continuous bilayer of lipids that have been deposited onto a solid or a polymer-based support[23] as a result of vesicle fusion[12] or Langmuir-Blodgett/Langmuir-Schaefer deposition[6]. Due to the presence of thin film of water between the bilayer and substrate, supported bilayers retain many of the physical characteristics of naturally occurring cell membranes and are amenable to a variety of measurement and surface modification techniques, such as micropatterning, due to their planar geometry. To date, arrays of single composition lipid domains on solid supports has been accomplished using microcontact printing[13], UV photolysis[17, 19, 20], agarose stamping[16] and a polymer-based dry lift-off approach[21, 22, 24]. We focused in this work on the latter patterning method, in which a parylene film is patterned via microlithography, the biomolecule (here, lipids) deposited, and the parylene film removed to reveal arrays of biomolecule on the substrate. To date, parylene lift-off has been used to deposit domains of haptenated bilayers for antibody recognition[24] and for cell stimulation[22, 25]; however, introducing further chemical complexity within the bilayer itself by surrounding the domains with another lipid composition had not yet been done.
In this work, we describe a combination of polymer lift-off and backfilling to construct chemically more complex supported bilayers. For our initial studies, we used lipids that are immiscible: didodecylphosphatidylcholine (di12:0 PC) which is fluid at room temperature and dipentadecylphosphatidylcholine (di15:0 PC) which is in the gel phase. We find that the backfilled interdomain regions of gel phase spatially restrict the fluid domains, and two types of domains exist on patterned backfilled samples, which we characterize with fluorescence microscopy and fluorescence correlation spectroscopy (FCS). We find that the two types of domains vary in properties (e.g., melting behavior, diffusion), which we pin down to their respective sources. We determined that the backfilling step introduces gel phase lipids inside fluid patterns that cluster and act as obstacles leading to anomalous diffusion of fluid lipids. Further, these results suggest that if backfilling is used to introduce complexity into patterned biomimetic membranes, it will be critical to optimize lipid concentrations on unpatterned bilayers first to determine concentrations that lead to minimal intercalation of the backfilling lipid. These results lead us to suggest that a thorough, quantitiative characterization of patterned bilayers, regardless of the patterning method used, is essential for any study in which the membrane properties must be known and well-defined.
Materials and methods
Materials
1,2-didodecyl-sn-glycerol-3-phosphocholine (di12:0 PC) and 1,2-dipenta-decyl-sn-glycerol-3-phosphocholine (di15:0 PC) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). The fluorescent lipid analogs, 1,1′-dihexadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine (diI-C16) and 2-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)-1-hexadecanoyl-sn-glycero-3-phosphocholine (bodipy PC), were purchased from Invitrogen (Carlsbad, CA, USA). Lipids and fluorescent analogs were used without additional purification.
Preparation of supported planar bilayers
The day prior to an experiment, small unilamellar vesicles (SUVs) were formed as described previously[26, 27]. Lipids at a desired composition, e.g., di12:0 PC ± 0.5 mol% bodipy PC or di15:0 PC ± 0.5 mol% diI-C16, were added to a test tube, previously cleaned in ethanolic potassium hydroxide, and dried under nitrogen. Dried lipids were then resuspended at a 2 mM concentration in 50 mM Tris, pH 7.4, 100 mM NaCl (TBS), probe sonicated until clarified and subjected to ultracentrifugation (Airfuge, 30 psi, 1 h). The top quarter of the supernatant was collected and stored overnight at room temperature and used within 24 hours. On the day of an experiment, the SUV solution (75 μL) was applied to a sandwich made of a detergent-cleaned 3″ × 1″ glass slide and a 22 mm × 22 mm glass coverslip that had been cleaned in oxygen plasma immediately prior to application of the SUV suspension. SUVs spontaneously fuse to form uniform lipid bilayers, and after a 30 min incubation in a humidified chamber, samples were rinsed with TBS to remove unfused vesicles. Bilayer samples were sealed with VALAP (Vaseline:lanolin:paraffin [2:1:1, wt/wt]) and measurements carried out immediately. Samples formed in this manner will be referred to as unpatterned bilayers. For some control experiments, unpatterned di12:0 PC ± 0.5 mol% bodipy PC bilayers were incubated with 75 μL of either di15:0 PC ± 0.5 mol% diI-C16 SUVs or TBS for 30 min. These bilayers were then extensively rinsed with TBS, sealed with VALAP and used for imaging and FCS measurements.
Preparation of patterned surfaces via polymer lift-off
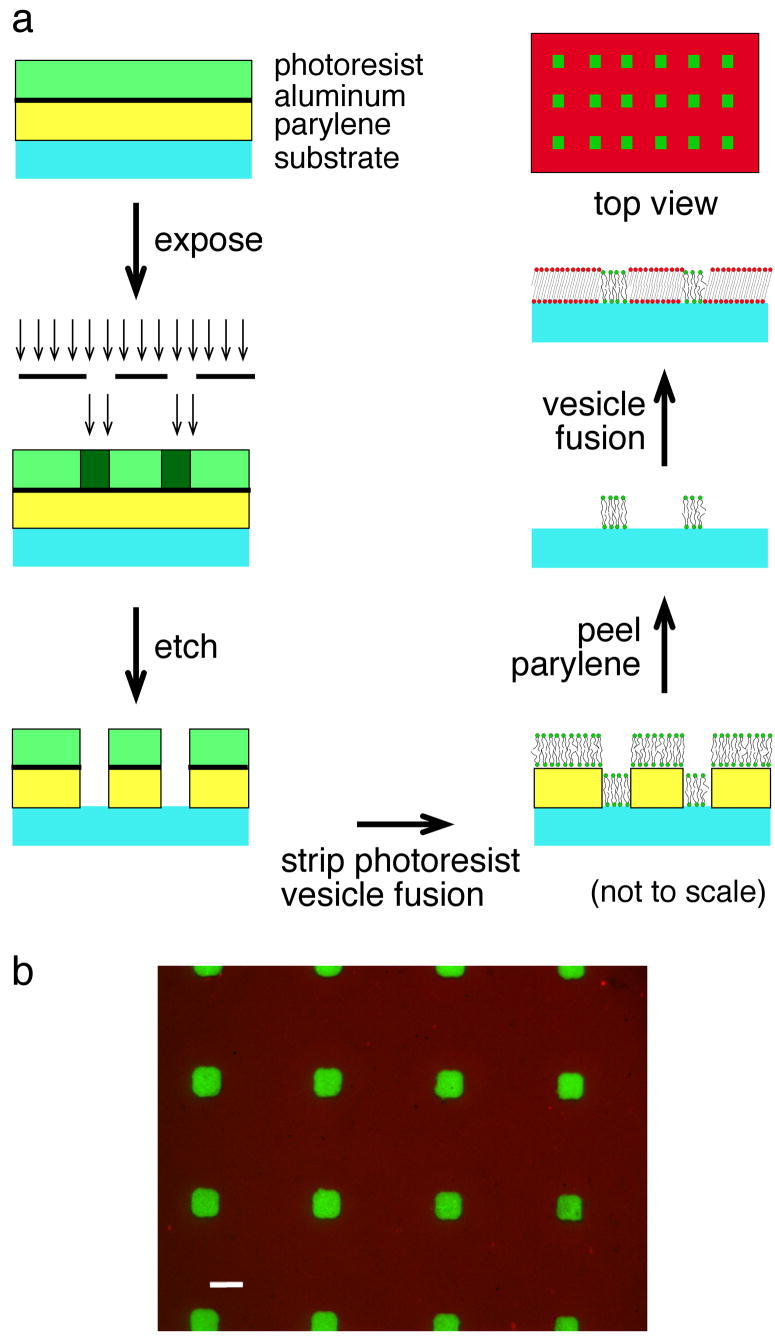
Parylene C (Parylene Deposition System 2010, Specialty Coating Systems; Indianapolis, IN, USA) was vapor deposited onto a detergent cleaned glass substrate (22 mm × 22 mm). After parylene deposition (Fig. 1), a 100 nm film of aluminum metal (Thermionics VE-90 thermal evaporator; Port Townsend, WA, USA) was evaporated onto the parylene coated surface, followed by application of a 1.2 μm-thick film of Shipley 1827 positive photoresist. The substrate was baked for 90 s at 105°C. The pattern from a quartz photomask was transferred to the photoresist via UV exposure in a contact aligner. The photoresist and the underlying aluminum were developed in CD26 base. The pattern was then dry-etched in a reactive ion etcher (Plasma Therm 720 RIE Reactive Ion Etch System; St. Petersburg, FL, USA) under the following conditions: 5 sccm argon, 90 sccm oxygen and 10 sccm carbon tetrafluoride at 400 W and 50 mTorr. As a result of dry-etching, exposed parylene was etched to the bare glass surface (Fig. 1), as assessed by X-ray photoelectron spectroscopy (not shown). Samples were rinsed with acetone and isopropyl alcohol, the aluminum was dissolved in CD26, and finally the sample was rinsed with deionized water and dried under nitrogen.
Figure 1.
Schematic illustration of lipid micropatterning via a modified polymer lift-off approach and a representative image. (a) Parylene, aluminum and photoresist were deposited on a glass substrate. Following exposure to ultraviolet light through a photomask, the photoresist and aluminum layers were developed and removed, and parylene was dry-etched in an oxygen-rich plasma. Substrates were cleaned and incubated with SUVs (e.g., bodipy PC-labeled di12:0 PC) to form domain arrays after removal of parylene. The exposed substrate was then coated with a bilayer of another lipid composition (e.g., di15:0 PC labeled with diI-C16) that had formed via vesicle fusion. (b) A representative image of patterned di12:0 PC containing 0.5 mol% bodipy PC (green) and backfilled di15:0 PC with 0.5 mol% diI-C16 (red). Images were captured sequentially with appropriate emission filters to minimize bleedthrough between the two fluorescence signals. Bar, 10 μm.
Patterned bilayer formation
The patterned substrate was first cleaned in an oxygen plasma. SUVs (75 μL) of fluid lipids (e.g., di12:0 PC labeled with bodipy PC) were fused onto this freshly cleaned surface in a humidified chamber for 30 min, resulting in bilayer assembly on the exposed glass surface. After 30 min, the surface was extensively rinsed with TBS, and the parylene layer was removed from the substrate, leaving behind patterned arrays of bilayers. After forming a sandwich with a detergent cleaned 3″ × 1″ glass slide, SUVs (75 μL) of gel phase lipid (e.g., di15:0 PC labeled with diI-C16) were injected, incubated for 30 min, washed with TBS and sealed. These samples will be referred to as patterned backfilled samples henceforth. Positive and negative controls consisted of unpatterned bilayers of either di12:0 PC or di15:0 PC with and without fluorescent dye, respectively. An additional negative control for background fluorescence was a TBS only sample.
Fluorescence imaging
All samples were imaged with a Photometrics CoolSnap HQ CCD detector (Tucson, AZ, USA) on a Nikon TE2000U inverted microscope with a 60× 1.2 NA objective (Nikon PlanApo, Melville, NY, USA) at room temperature (~25°C), unless otherwise noted. For bodipy PC detection, a 485/15 excitation filter, 520/20 emission filter and 505 DRLP dichroic were used, and for diI-C16 detection, a 555/10 excitation filter, 650/20 emission filter and 560 DRLP dichroic were used; these filter combinations eliminated bleedthrough. Excitation and emission filter wheels, x-y translation stage, (Ludl Electronic Products, Hawthorne, NY, USA), and image acquisition were driven by ISee Imaging software (Raleigh, NC, USA) on a Pentium class PC running Linux[27]. Samples were illuminated with mercury lamp excitation and exposure times were kept constant for a given day of experiments. All images were background and flatfield corrected. An objective heater (Bioptechs, Butler, PA, USA) was used for the bilayer melting experiments.
FCS characterization of lateral diffusion on patterned bilayers
Confocal FCS was used to characterize the lateral mobility of bodipy PC in di12:0 PC bilayers. FCS experiments were carried out on a Nikon TE2000U inverted microscope as described by Kyoung et al.[27]. A laser beam (either the 488 nm line from a Coherent Innova 90C-6 argon ion laser [Santa Clara, CA, USA] or 543 nm HeNe laser from Meredith Instruments [Glendale, AZ, USA]) was focused through the epi-port of the microscope and projected onto the sample by overfilling the back aperture of a Nikon PlanApo 60× 1.2 NA objective. Typical excitation powers ranged from 4.4–30 μW at the sample plane with negligible photobleaching. A pinhole (50 μm diameter) was placed immediately in front of a GaAsP photomultiplier tube (Hamamatsu H7421-40; Bridgewater, NJ, USA) in an image plane conjugate to the sample. Correlation curves were acquired with a Hamamatsu M9003 correlation card in a Pentium class PC running Windows XP.
For a given day of FCS experiments, the structure parameter, ωo, was determined using 0.1–1 nM rhodamine green following Kyoung et al.[27], and lateral diffusion measurements were carried out on bilayer samples. Normalized data were fit to one and two component diffusion in two dimensions with Igor Pro (WaveMetrics; Lake Oswego, OR, USA) according to[28, 29]
| (1) |
where G(τm) is the normalized autocorrelation function for two-dimensional diffusion, τ is the time interval, and τDi is the characteristic diffusion times for each fraction ( ). For fluorescence fluctuations due to restricted non-Brownian diffusion in two dimensions, the normalized correlation function, G(τ) can be modified such that diffusion time of a molecule, τD, can be calculated following[30]
| (2) |
where α is the anomalous diffusion exponent that is less than unity[30, 31]. For all cases, χ2 was calculated to determine which model best described the data. Diffusion coefficients (D) reported in Tables 1 and 2 were calculated as .
Table 1.
Lateral diffusion of bodipy PC in patterned or unpatterned di12:0 PC supported membranes.
| Sample | D ± SD (× 10−9 cm2 s−1) | αa |
|---|---|---|
| unpatterned di12:0 PC + 0.5 mol% bodipy PC (n = 83) | 6.7 ± 2.1b,d | — |
| patterned di12:0 PC + 0.5 mol% bodipy PC (n = 42) | 1.9 ± 0.5c | 0.74 ± 0.13 |
| patterned di12:0 PC + 0.5 mol% bodipy PC at 37°C (n = 20) | 7.4 ± 2.5b,d | — |
| unpatterned di12:0 PC + 1 × 10−6 mol% bodipy PC (n = 54) | 7.7 ± 3.4b,d | — |
| unpatterned di12:0 PC + 1 × 10−5 mol% bodipy PC (n = 53) | 8.1 ± 2.3b,d | — |
Table 2.
Lateral diffusion of bodipy PC in unpatterned di12:0 PC supported membranes incubated with SUVs composed of di15:0 PC for 30 min followed by extensive washing with buffer.
| Unpatterned supported bilayer composition | SUV composition | Temperature (°C) | D ± SD (× 10−9 cm2 s−1) | αa |
|---|---|---|---|---|
| di12:0 PC + 0.5 mol% bodipy PC | di15:0 PC + 0.5 mol% diI-C16 | 25 (n = 20) | 4.6 ± 0.9b | 0.79 ± 0.06 |
| 37 (n = 18) | 7.9 ± 2.9c,d | — | ||
| 25 (n = 18) | 4.5 ± 1.8b | 0.78 ± 0.09 | ||
|
| ||||
| di12:0 PC + 0.5 mol% bodipy PC | di15:0 PC − 0.5 mol% diI-C16 | 25 (n = 22) | 5.8 ± 1.6b | 0.81 ± 0.08 |
| 37 (n = 20) | 6.8 ± 2.8c,d | — | ||
| 25 (n = 17) | 6.1 ± 1.7b | 0.79 ± 0.08 | ||
|
| ||||
| di12:0 PC + 0.5 mol% bodipy PC | —e | 25 (n = 22) | 7.1 ± 2.2c | — |
| 37 (n = 20) | 11.1 ± 1.9c,d | — | ||
| 25 (n = 17) | 7.9 ± 2.1c | — | ||
|
| ||||
| di12:0 PC + 0.5 mol% bodipy PC | —f | 25 (n = 83) | 6.7 ± 2.1c | — |
| 37 (n = 17) | 8.9 ± 1.7c,d | — | ||
| 25 (n = 20) | 6.7 ± 2.3c | — | ||
α is the anomalous diffusion exponent.
Fit to Eq. 2.
Fit to Eq. 1, where m = 1.
F > Fcrit, as determined by single factor ANOVA, indicating a statistically significant difference from rest of the cases at 95% confidence limit.
30 min incubation with TBS buffer only, in place of SUV suspension, followed by extensive washing.
Planar bilayers processed normally with no 30 min incubation.
Bilayer melting
Samples patterned with di12:0 PC (melting temperature, Tm = −1°C) and backfilled with di15:0 PC (Tm = 33°C) were heated beyond the phase transition temperature of di15:0 PC to 37°C for 30 min to confirm bilayer continuity across the entire patterned sample. FCS was performed on unpatterned di15:0 PC planar supported lipid bilayers at 37°C to confirm their fluidity.
Atomic force microscopy
Glass substrates that were uniformly coated with parylene were etched using reactive ion etching (RIE) as described above. These substrates were imaged in air using tapping-mode AFM with a MultiMode Scanning Probe Microscope and Nanoscope IIIa controller (Digital Instruments; Edina, MN, USA) using silicon doped tips (Veeco Nanoprobe; Camarillo, CA, USA) having a spring constant of 0.5 N m−1. For comparison as a control, detergent cleaned glass substrates, otherwise used for making supported lipid bilayers, were also imaged under similar conditions.
Results and discussion
In addition to microcontact printing[13, 14, 32–34], agarose stamping[16] and UV photolysis[17–20], a polymer lift-off approach has been developed to form arrays of lipid microdomains on a variety of surfaces[22, 24, 25]. Fig. 1a describes the modified polymer lift-off approach in detail. We initially followed the patterning approach described by Craighead et al.[22, 24, 35]; that is, simply using photoresist and parylene to transfer the desired patterns of 5–10 μm features to the underlying glass substrate, following dry-etching. However, when we inspected the patterned substrate after lipid vesicle fusion, we observed that lipids were deposited in a nonspecific manner, in addition to the desired pattern, indicating the presence of defects in the substrate prior to bilayer preparation. These defects likely result from the photoresist and parylene polymers having similar etch selectivity and rates by the oxygen plasma. Also because photoresists are known to have defects in them[36], oxygen plasma is able to attack the parylene under the resist. To prevent the formation of these pinhole defects, we modified our process to introduce a hard mask of aluminum[37], which would then be chemically wet-etched in CD26 developer (tetramethylammonium hydroxide in water) during processing, thereby allowing us to develop simultaneously the photoresist and aluminum. SUVs of defined lipid composition were then introduced to form self-assembled bilayers on the exposed glass substrate and parylene film (Fig. 1). Arrays of lipid domains formed after the parylene film was removed. To backfill the resulting interdomain regions, we introduced SUVs of another chemically well-defined composition, and we then characterized these samples using quantitative fluorescence microscopy and correlation spectroscopy.
Although the 100 nm aluminum film is completely removed from the patterned substrate prior to lipid bilayer deposition, we carried out control studies to ensure that the addition of aluminum does not interfere with lipid bilayer organization and diffusion on the substrate. We compared the lateral diffusion of fluid bilayers that result when substrates are prepared with parylene and photoresist in the presence and absence of the intermediate aluminum film. For these experiments, we used a clear photomask (i.e., one with no pattern) to completely expose the photoresist, effectively preparing unpatterned bilayers of fluorescently labeled di12:0 PC. In both cases, the diffusion of di12:0 PC was one-component simple Brownian diffusion of similar magnitude to that of conventionally prepared unpatterned di12:0 PC bilayers (~10−9 cm2 s−1; data not shown and Table 1).
In our initial characterization studies on patterned backfilled samples, we chose to use a combination of lipid compositions that are immiscible at room temperature: di12:0 PC (Tm = −1°C), which is fluid at room temperature, ± bodipy PC (as a fluorescent label); and di15:0 PC (Tm = 33°C), which is in the gel phase at room temperature, ± diI-C16 (as a fluorescent label). To facilitate quantitative imaging, we used 5–10 μm features, which are easily resolvable optically. Typically, we patterned arrays of a particular lipid bilayer (most commonly, the fluid di12:0 PC) and then introduced another degree of complexity by subsequent incubation with a second lipid component (e.g., di15:0 PC). Using epifluorescence microscopy (Fig. 1b), we found that the complex bilayers retain their patterns when di12:0 PC was patterned and di15:0 PC was used for backfilling, and the feature size matched those on the photomask used in lithography. These results indicate that on these patterned substrates, the outside gel phase lipid bilayer can spatially confine fluid phase lipid domains on a planar support. Similar results have been reported earlier where PDMS stamping[15] and UV photolysis[18, 20] was used to kinetically trap components of a binary mixture to form stable lipid patterns.
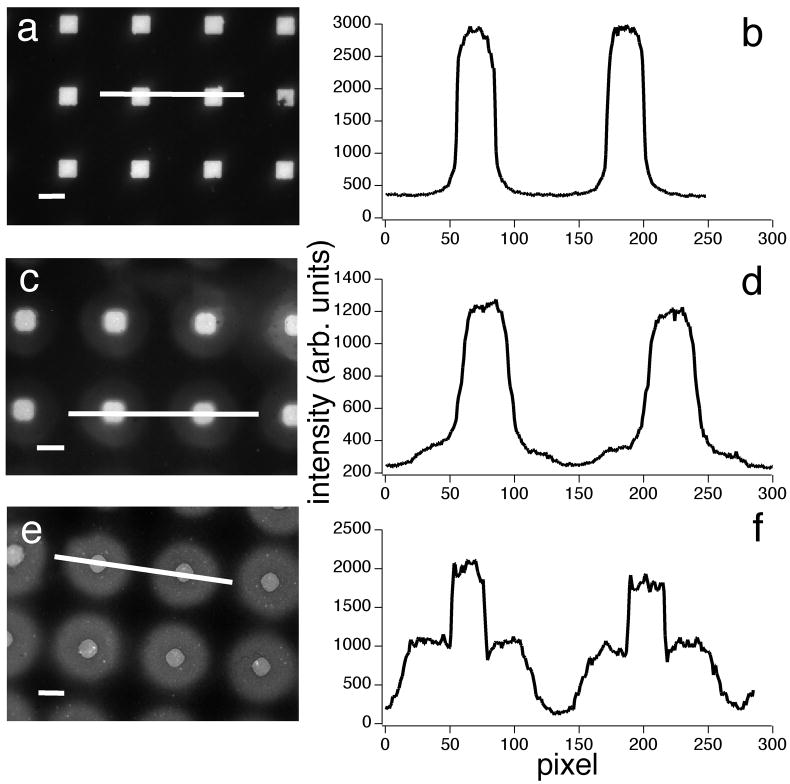
Upon inspection of the patterned substrates, we observed two types of arrays on samples formed from the deposition of fluorescently labeled di12:0 PC into the patterned features, followed by backfilling with di15:0 PC (Fig. 2). Nearly (19 ± 3)% (n = 10, where n is the number of patterned substrates) of the total number of patterns had sharply defined edges (Fig. 2a), which remained stable for at least 6 h. A line scan of the fluorescence intensity shows that the fluid lipid is restricted to the patterned features by the surrounding gel phase (Fig. 2b). The majority of the patterned arrays, however, have more diffuse boundaries (Fig. 2c and e), in which the fluid lipid diffuses into the gel phase. Interestingly, these two types of domains were randomly distributed throughout the surface of a given substrate, suggesting that these differences likely do not result from the location of the substrate within the RIE chamber where we would expect a systematic distribution.
Figure 2.
Three examples of patterned domains present on a single substrate. Bodipy PC-labeled domains of di12:0 PC were surrounded by di15:0 PC. Two populations of domains are observed: one with sharply defined boundaries (a), and the other with diffuse borders (c, e). Panels (b, d and f) show the corresponding intensity line profiles of panels (a, c and e). Bar, 10 μm.
We then hypothesized that surface differences lead to the two domain types. To test this possibility, we examined patterned substrates with fluorescence microscopy and tapping-mode AFM. In one experiment, we incubated substrates with SUVs of bodipy PC-labeled di12:0 PC to form planar bilayers after the patterned parylene was removed; this sample preparation contrasts with our usual procedure in which vesicle fusion is carried out prior to parylene removal. If no surface modification occurred, we would expect to see uniform bilayers; however, we observed faint patterns even though patterned parylene was completely removed from these substrates prior to lipid vesicle fusion (not shown), leading us to conclude that the substrates may be modified during the fabrication process, possibly during reactive ion etching. Such plasma-induced surface roughness of glass has been previously reported, particularly when oxygen plasma is used at high power (i.e., >100 W) and/or long exposure times[38]. To test this idea further, we imaged plasma-etched substrates using tapping-mode AFM. As shown in Fig. 3, some areas of surfaces were found to be non-uniform with roughness. The observed roughness was not due to residual parylene, which we confirmed using X-ray photoelectron spectroscopy. Polymerized parylene C is a poly-paraxylylene with a chlorine substitution of one of the aromatic hydrogens. The absence of a chlorine signal confirms that parylene is completely removed under our etching conditions. In contrast, controls of detergent-cleaned substrates that had been plasma cleaned had negligible surface roughness (not shown). These results suggest that vesicle fusion on etched, pitted surfaces might lead to nonuniform lipid bilayers that may exhibit hindered or nonexistent lateral diffusion.
Figure 3.
AFM image of a plasma-etched glass surface (a) and the corresponding line scan (b). Unpatterned parylene deposited onto glass was etched completely prior to imaging in air to assess surface roughness resulting from fabrication. Bar, 2.5 μm.
To measure the lateral diffusion of fluid domains on backfilled samples, we used confocal FCS, which not only yields the diffusion coefficient of the species in a system[30, 39–43] but also can be used to identify whether anomalous diffusion occurs; that is, when the diffusing species encounters obstacles or is hindered in some way[31, 44]. Patterned, fluorescently labeled fluid phase arrays surrounded by di15:0 PC exhibit anomalous diffusion (1.9 ± 0.5) × 10−9 cm2 s−1 (n = 42, where n is the number of measurements), with an anomalous exponent α = 0.72 ± 0.13 (Table 1; Fig. 4, open circles). Interestingly, lateral diffusion measurements (Fig. 4) could only be performed on diffuse patterns (e.g., similar to those shown in Fig. 2c and e). Domains with sharp borders (Fig. 2a) did not undergo lateral diffusion on our experimental time scale (not shown). Unpatterned supported bilayers yielded one-component Brownian motion, where D = (6.7 ± 2.1) × 10−9 cm2 s−1 (n = 83) (Table 1; Fig. 4, closed circles), as previously observed for other fluid supported bilayers[40, 45].
Figure 4.
Representative lateral diffusion measurements of bodipy PC in di12:0 PC bilayers. Open circles represent diffusion within patterned di12:0 PC domains, which is best described as anomalous diffusion (Eq. 2). Diffusion in an unpatterned di12:0 PC bilayer (closed circles) exhibits one-component Brownian motion (Eq. 1, m = 1). Results are shown in Table 1.
We hypothesized that the anomalous diffusion observed for the diffuse domains of fluid lipids may be due to surface roughness (Fig. 3), a possible experimental artifact, or gel phase lipid clusters within the domains. We tested the first possibility in the following experiment. As described above for the AFM study, we completely etched unpatterned parylene that was deposited on glass substrates. Bilayers of di12:0 PC labeled with bodipy PC were formed, and FCS was used to measure the lateral diffusion. If surface roughness was the primary cause, we would expect anomalous diffusion on these unpatterned, but etched, substrates. However, we observe that the lipids exhibit one-component Brownian motion, thereby ruling out this possible explanation.
FCS is inherently a single molecule technique and requires low concentrations (~nM) of fluorophores to obtain sufficiently large fluctuations that allow correlation[46–48]. However, these low fluorophore concentrations preclude imaging to identify a region of interest for an FCS experiment, due to the very low signal-to-noise. As noted previously, we used 0.5 mol% fluorescent lipid, which allowed us to strategically position the laser within a domain as well as characterize the domain type (Fig. 2). For FCS experiments, we reduced the concentration via photobleaching, with the mercury arc lamp, to obtain an intensity equivalent to 1 × 10−6–1 × 10−5 mol%. To test whether photobleaching in this manner may affect the validity of our FCS measurements, we prepared unpatterned di12:0 PC bilayers that contain either 1 × 10−6 or 1 × 10−5 mol% bodipy PC and compared these results to unpatterned bilayers containing 0.5 mol% bodipy PC that had been photobleached. As shown in Table 1, all three samples yielded one-component Brownian motion, and we observed no statistically significant difference in the lateral diffusion coefficient, as assessed by single-factor ANOVA. These results suggest that anomalous diffusion likely does not result from our experimental preparation for reducing the fluorophore intensity.
We next turned our attention to whether anomalous diffusion could be attributed to partial mixing of the backfilled gel phase lipids in the fluid domains or whether they are completely excluded from the domains. To test this idea, we prepared samples that were fluorescently labeled in both the fluid domains (with bodipy PC) and in the backfilled interdomain region (with diI-C16) and imaged sequentially, with appropriate filters to minimize bleedthrough (Fig. 5a and c). From intensity profiles (Fig. 5 b and d), we observe that the fluorescent intensity due to diI-C16 in the backfilled gel phase is low but not completely excluded from the domains, indicating that some mixing occurs during the backfilling step. Despite this mixing of the two phases, good contrast is observed. This result agrees with another reported work on patterned backfilled samples in which two chemically different thiol monolayers in the patterned and backfilled areas were found to mix at room temperature[49]. Given our results, we hypothesized that the anomalous behavior of lipid is due to the presence of gel phase clusters inside the patterned fluid phase regions. To test this hypothesis further, we carried out FCS diffusion measurements on patterned, fluorescently labeled fluid lipid arrays that were surrounded by di15:0 PC. When these membranes are heated beyond the phase transition temperature of di15:0 PC and maintained at 37°C for 30 min, we found that the anomalous component disappears and the lateral diffusion becomes Brownian with D = (7.4 ± 2.5) × 10−9 cm2 s−1 (n = 20) (Table 1). Because the gel and fluid phase lipids used in this report mix nonideally and are immiscible at room temperature, these results suggest that di15:0 PC lipids likely form clusters inside fluid domains, which act as diffusion obstacles.
Figure 5.
Representative paired epifluorescence images of domain arrays. Domains of fluid di12:0 PC containing bodipy PC (a) is surrounded by di15:0 PC labeled with diI-C16 (c) that had been backfilled. Panels (b and d) are the corresponding fluorescence intensity profiles for panels (a and c), respectively. Images were captured sequentially with appropriate filters to minimize bleedthrough. Bar, 10 μm.
Although we are using saturating concentrations of lipids during model membrane formation based on the work of Thompson [e.g., 26, and NL Thompson and ED Sheets, unpublished results, 50, 51], we wanted to test further whether di15:0 PC could intercalate into pre-formed di12:0 PC membranes. We prepared unpatterned supported bilayers composed of di12:0 PC ± bodipy PC; however, prior to sealing with VALAP, we incubated these bilayers with an additional 75 μL of SUVs composed of di15:0 PC ± diI-C16, in the same manner that we prepared patterned backfilled bilayers. We then measured the lateral diffusion of bodipy PC in the fluid bilayers to determine whether gel phase lipids intercalate and lead to anomalous diffusion and compared these results to di12:0 PC bilayers that had not been incubated with di15:0 PC. Upon “backfilling” (i.e., incubation with the di15:0 PC SUVs), these samples experience anomalous diffusion at room temperature ([4.6 ± 0.9] × 10−9 cm2 s−1, α = 0.79 ± 0.06, n = 20; Table 2; Fig. 6, open circles). Upon heating to 37°C (above the Tm of di15:0 PC), anomalous diffusion disappears and the data fit to one-component Brownian diffusion where D = (7.9 ± 2.9) × 10−9 cm2 s−1 (n = 18; Table 2; Fig. 6, closed circles). When these samples cool to room temperature, bodipy PC once again exhibits anomalous diffusion (Table 2). The appearance of temperature-dependent anomalous diffusion in these samples, where no surface roughness is present due to the absence of etching, suggests that intercalation of gel phase lipids is likely the main source of obstacles that lead to anomalous diffusion. Thus, we suggest that it will be essential to initially optimize lipid concentrations on unpatterned bilayers to determine concentrations that lead to minimal intercalation of the backfilling lipid. Further, these results also suggest caution in interpreting the biophysical behavior of backfilled biomimetic membranes, and we propose a thorough quantitative characterization of such membranes.
Figure 6.
Representative lateral diffusion measurements of bodipy PC in di12:0 PC bilayers incubated with SUVs composed of di15:0 PC for 30 min followed by extensive washing with buffer. Open circles represent diffusion in unpatterned di12:0 PC supported membranes at room temperature, which is best described as anomalous diffusion (Eq. 2). Diffusion in unpat-terned di12:0 PC supported membranes at 37°C (closed circles) exhibits one-component Brownian motion (Eq. 1, m = 1). Results are shown in Table 2.
Because our long-term goal is to investigate molecular dynamics occurring in complex patterned bilayers, we also wanted to determine whether backfilled samples formed continuous bilayers. We therefore carried out melting experiments in which di15:0 PC underwent a phase transition at 33°C. As shown in Fig. 7, fluid domains were labeled with bodipy PC and the interdomain gel phase was labeled with diI-C16. In these experiments, the temperature was increased from room temperature (Fig. 7a) to 37°C and maintained for 30 min to ensure mixing (Fig. 7b). The lipids on the patterned backfilled samples remain well mixed after cooling to room temperature (Fig. 7c), indicating that domain pinning that has been observed for some other supported bilayers does not occur[6, 7]. However, when we examine the melting behavior of arrays with sharply defined features (e.g., Fig. 2a), we observe that the patterned and backfilled lipids do not mix (Fig. 7d–f), supporting the FCS measurements in which fluid lipids were incapable of lateral diffusion which possibly results from a physical barrier (i.e., due to surface roughness; Fig. 3). The reason why surface roughness affects only a minor subset of domains is not clear from the present data, and further study is needed to clarify this issue.
Figure 7.
Melting behavior of the two types of patterned domains. Fluid domains of bodipy PC-labeled di12:0 PC (green) with diffuse edges (a–c) or with sharp boundaries (d–f) are surrounded by diI-C16-labeled di15:0 PC (red). The temperature was increased from room temperature (a, d) through the Tm of di15:0 PC (33°C) to 37°C for 30 min (b, e) then decreased to room temperature (c, f). Images were captured sequentially with appropriate filters to minimize bleedthrough. Bar, 10 μm.
Conclusions
Here we report the systematic characterization of spatially arranged, two-phase supported lipid bilayers obtained via a modified polymer lift-off approach. We patterned arrays of fluid domains that were surrounded by a backfilled gel phase. These domains can generally be described as a population with diffuse boundaries, in which the fluid lipids undergo anomalous diffusion and form continuous bilayers with the gel phase, and as a minor population with sharp borders, in which the fluid lipids do not diffuse laterally or form continuous bilayers. Anomalous diffusion likely results from small, nonideally mixed clusters of gel phase lipids within the domains, which act as obstacles to diffusion. Given these results on a relatively simple complex bilayer (that is, immiscible fluid and gel lipids), it is essential to fully and quantitatively characterize biomimetic membrane systems as additional complexity (e.g., miscible lipids, cholesterol) is introduced or when choosing among various patterning approaches. Further, these results suggest that if backfilling is used to introduce complexity into patterned biomimetic membranes, it will be critical to optimize lipid concentrations on unpatterned bilayers first to determine concentrations that lead to minimal intercalation of the backfilling lipid. We propose that the work reported here represents an important cautionary tale for the growing field of patterned biomimetic membranes, regardless of the particular patterning approach. Keeping these caveats in mind, however, patterned, complex bilayers with spatially addressable domains will likely provide useful building blocks for the development of integrated biosensors and critical infrastructure for cell signaling studies in vitro.
Acknowledgments
We thank Prof. Ahmed A. Heikal (Penn State Bioengineering) and Angel M. Davey (Penn State Chemistry) for helpful comments on this manuscript. This work was supported, in part, by The Pennsylvania State University, the Penn State Materials Research Institute, the Penn State MRSEC under NSF grant DMR 0213623, and the Center for Optical Technologies, which is supported by the Commonwealth of Pennsylvania. This publication was also supported by the Pennsylvania State University Materials Research Institute Nano Fabrication Network and the National Science Foundation Cooperative Agreement No. 0335765, National Nanotechnology Infrastructure Network, with Cornell University. Additional acknowledgment is made to the Donors of the American Chemical Society Petroleum Research Fund and Avanti Polar Lipids, Inc. for partial support of this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edidin M. The state of lipid rafts: from model membranes to cells. Annu Rev Biophys Biomol Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 2.Sheets ED, Holowka D, Baird B. Membrane organization in immunoglobulin E receptor signaling. Curr Opin Chem Biol. 1999;3:95–99. doi: 10.1016/s1367-5931(99)80017-9. [DOI] [PubMed] [Google Scholar]
- 3.Janes PW, Ley SC, Magee AI, Kabouridis PS. The role of lipid rafts in T cell antigen receptor (TCR) signalling. Semin Immunol. 2000;12:23–34. doi: 10.1006/smim.2000.0204. [DOI] [PubMed] [Google Scholar]
- 4.Alonso MA, Millan J. The role of lipid rafts in signalling and membrane trafficking in T lymphocytes. J Cell Sci. 2001;114:3957–3965. doi: 10.1242/jcs.114.22.3957. [DOI] [PubMed] [Google Scholar]
- 5.Feigenson GW, Buboltz JT. Ternary phase diagram of dipalmitoyl-PC/dilauroyl-PC/cholesterol: nanoscopic domain formation driven by cholesterol. Biophys J. 2001;80:2775–2788. doi: 10.1016/S0006-3495(01)76245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stottrup BL, Veatch SL, Keller SL. Nonequilibrium behavior in supported lipid membranes containing cholesterol. Biophys J. 2004;86:2942–2950. doi: 10.1016/S0006-3495(04)74345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietrich C, Bagatolli LA, Volovyk ZN, Thompson NL, Levi M, Jacobson K, Gratton E. Lipid rafts reconstituted in model membranes. Biophys J. 2001;80:1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietrich C, Volovyk ZN, Levi M, Thompson NL, Jacobson K. Partitioning of Thy-1, GM1, and cross-linked phospholipid analogs into lipid rafts reconstituted in supported model membrane monolayers. Proc Natl Acad Sci USA. 2001;98:10642–10647. doi: 10.1073/pnas.191168698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samsonov AV, Mihalyov I, Cohen FS. Characterization of cholesterol-sphingomyelin domains and their dynamics in bilayer membranes. Biophys J. 2001;81:1486–1500. doi: 10.1016/S0006-3495(01)75803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw JE, Slade A, Yip CM. Simultaneous in situ total internal reflection fluorescence/Atomic force microscopy studies of DPPC/d POPC microdomains in supported planar bilayers. J Am Chem Soc. 2003;125:11838–11839. doi: 10.1021/ja0370894. [DOI] [PubMed] [Google Scholar]
- 11.Burns AR. Domain structure in model membrane bilayers investigated by simultaneous atomic force microscopy and fluorescence imaging. Langmuir. 2003;19:8358–8363. [Google Scholar]
- 12.Kalb E, Frey S, Tamm LK. Formation of supported planar bilayers by fusion of vesicles to supported phospholipid monolayers. Biochim Biophys Acta. 1992;1103:307–316. doi: 10.1016/0005-2736(92)90101-q. [DOI] [PubMed] [Google Scholar]
- 13.Hovis JS, Boxer SG. Patterning and composition arrays of supported lipid bilayers by microcontact printing. Langmuir. 2001;17:3400–3405. [Google Scholar]
- 14.Sapuri-Butti AR, Li QL, Groves JT, Parikh AN. Nonequilibrium patterns of cholesterol-rich chemical heterogenieties within single fluid supported phospholipid bilayer. Langmuir. 2006;22:5374–5384. doi: 10.1021/la052248d. [DOI] [PubMed] [Google Scholar]
- 15.Jung SY, Holden MA, Cremer PS, Collier CP. Two-component membrane lithography via backfilling. ChemPhysChem. 2005;6:423–426. doi: 10.1002/cphc.200400540. [DOI] [PubMed] [Google Scholar]
- 16.Majd S, Mayer M. Hydrogel stamping of arrays of supported lipid bilayers with various lipid compositions for the screening of drug-membrane and protein-membrane interactions. Angew Chem Int Ed. 2005;44:6697–6700. doi: 10.1002/anie.200502189. [DOI] [PubMed] [Google Scholar]
- 17.Howland MC, Supri-Butti AR, Dixit SS, Dattelbaum AM, Shreve AP, Parikh AN. Phospholipid morphologies on photochemically patterned silane monolayers. J Am Chem Soc. 2005;127:6752–6765. doi: 10.1021/ja043439q. [DOI] [PubMed] [Google Scholar]
- 18.Yee CK, Amweg ML, Parikh AN. Direct photochemical patterning and refunctionalization of supported phospholipid bilayers. J Am Chem Soc. 2004;126:13962–13972. doi: 10.1021/ja047714k. [DOI] [PubMed] [Google Scholar]
- 19.Yu C, Parikh AN, Groves JT. Direct patterning of membrane derivatized colloids using in-situ UV-ozone photolithography. Adv Mater. 2005;17:1477–1480. [Google Scholar]
- 20.Yee CK, Amweg ML, Parikh AN. Membrane photolithography: direct micropatterning and manipulation of fluid phospholipid membranes in the aqueous phase using deep-UV light. Adv Mater. 2004;16:1184–1189. [Google Scholar]
- 21.Ilic B, Craighead HG. Topographical patterning of chemically sensitive biological materials using a polymer-based dry lift off. Biomed Microdev. 2000;2:317–322. [Google Scholar]
- 22.Orth RN, Wu M, Holowka DA, Craighead HG, Baird B. Mast cell activation on patterned lipid bilayers of subcellular dimensions. Langmuir. 2003;19:1599–1605. [Google Scholar]
- 23.Naumann CA, Prucker T, Lehmann T, Ruhe J, Knoll W, Bank CW. The polymer-supported phospholipid bilayer: tethering a new approach to substrate membrane stabilization. Biomacromol. 2002;3:27–35. doi: 10.1021/bm0100211. [DOI] [PubMed] [Google Scholar]
- 24.Orth RN, Kameoka J, Zipfel WR, Ilic B, Webb WW, Clark TG, Craighead HG. Creating biological membranes on the micron scale: forming patterned lipid bilayers using a polymer lift-off technique. Biophys J. 2003;85:3066–3073. doi: 10.1016/S0006-3495(03)74725-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu M, Holowka D, Craighead HG, Baird B. Visualization of plasma membrane compartmentalization with patterned lipid bilayers. Proc Natl Acad Sci USA. 2004;101:13798–13803. doi: 10.1073/pnas.0403835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearce KH, Hiskey RG, Thompson NL. Surface binding kinetics of prothrombin fragment 1 on planar membranes measured by total internal reflection fluorescence microscopy. Biochem. 1992;31:5983–5995. doi: 10.1021/bi00141a005. [DOI] [PubMed] [Google Scholar]
- 27.Kyoung M, Karunwi K, Sheets ED. A versatile multi-mode microscope to probe and manipulate nanoparticles and biomolecules. J Microsc. 2007;225:136–146. doi: 10.1111/j.1365-2818.2007.01725.x. [DOI] [PubMed] [Google Scholar]
- 28.Hac AE, Seeger HM, Fidora M, Heimburg T. Diffusion in two-component lipid membranes: a fluorescence correlation spectroscopy and Monte Carlo simulation. Biophys J. 2005;88:317–333. doi: 10.1529/biophysj.104.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bacia K, Scherfeld D, Kahya N, Schwille P. Fluorescence correlation spectroscopy relates rafts in model and native membranes. Biophys J. 2004;87:1034–1043. doi: 10.1529/biophysj.104.040519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wawrezinieck L, Rigneault H, Marguet D, Lenne P. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys J. 2005;89:4029–4042. doi: 10.1529/biophysj.105.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheets ED, Lee GM, Simson R, Jacobson K. Transient confinement of a glycosylphosphatidylinositol-anchored protein in plasma membrane. Biochem. 1997;36:12449–12458. doi: 10.1021/bi9710939. [DOI] [PubMed] [Google Scholar]
- 32.Kung LA, Kam L, Hovis JS, Boxer SG. Patterning hybrid surfaces of proteins and supported lipid bilayers. Langmuir. 2000;16:6773–6776. [Google Scholar]
- 33.Hovis JS, Boxer SG. Patterning barriers to lateral diffusion in supported lipid bilayer membranes by blotting and stamping. Langmuir. 2000;16:894–897. [Google Scholar]
- 34.Groves JT, Ulman N, Boxer SG. Micropatterning fluid lipid bilayers on solid supports. Science. 1997;275:651–653. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 35.Moran-Mirabal JM, Tan CP, Orth RN, Williams EO, Craighead HG, Lin DM. Controlling microarray spot morphology with polymer liftoff arrays. Anal Chem. 2007;79:1109–1114. doi: 10.1021/ac061898z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maclver BA, Puzio LC. A method for observing micrometer-size defects in resists. J Electrochem Soc. 1982;129:2384–2385. [Google Scholar]
- 37.Atsuta K, Suzuki H, Takeuchi S. A parylene lift-off process with microfluidic channels for selective protein patterning. J Micromech Microeng. 2007;17:496–500. [Google Scholar]
- 38.Choi SW, Choi WB, Lee YH, Ju BK, Sung MY, Kim BH. The analysis of oxygen plasma pretreatment for improving anodic bonding. J Electrochem Soc. 2002;149:G8–G11. [Google Scholar]
- 39.Burns AR, Frankel DJ, Buranda T. Local mobility in lipid domains of supported bilayer characterized by atomic force microscopy and fluorescence correlation spectroscopy. Biophys J. 2005;89:1081–1093. doi: 10.1529/biophysj.105.060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Garnick S. Lipid diffusion compared in outer and inner leaflets of planar supported lipid bilayer. J Chem Phys. 2005;123:211104–211114. doi: 10.1063/1.2138699. [DOI] [PubMed] [Google Scholar]
- 41.Scherfeld D, Kahya N, Schwille P. Lipid dynamics and domain formation in model membranes composed to ternary mixtures of unsaturated and saturated phosphatidylcholines and cholesterol. Biophys J. 2003;85:3758–3768. doi: 10.1016/S0006-3495(03)74791-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwille P, Korlach J, Webb WW. Fluorescence correlation spectroscopy with single molecule sensitivity on cell and model membranes. Cytometry. 1999;36:176–182. doi: 10.1002/(sici)1097-0320(19990701)36:3<176::aid-cyto5>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 43.Benda A, Benes M, Marecek V, Lhotsky A, Hermens WT, Hof M. How to determine diffusion coefficients in planar phospholipid systems by confocal fluorescence correlation spectroscopy. Langmuir. 2003;19:4120–4126. [Google Scholar]
- 44.Saxton MJ. Anomalous diffusion due to obstacles: A Monte Carlo study. Biophys J. 1994;66:394–401. doi: 10.1016/s0006-3495(94)80789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saxton MJ, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 46.Kahya N, Scherfeld D, Bacia K, Schwille P. Lipid domain formation and dynamics in giant unilamellar vesicles explored by fluorescence correlation spectroscopy. J Struc Bio. 2004;147:77–89. doi: 10.1016/j.jsb.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 47.Kahya N, Schwille P. Fluorescence correlation studies of lipid domains in model membranes. Mol Mem Biol. 2006;23:29–39. doi: 10.1080/09687860500489099. [DOI] [PubMed] [Google Scholar]
- 48.Hess ST, Huang S, Heikal AA, Webb WW. Biological and chemical applications of fluorescence correlation spectroscopy: a review. Biochem. 2002;41:697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- 49.Cimatu K, Baldelli S. Sum frequency generation microscopy of microcontact-printed mixed self-assembled monolayers. J Phys Chem B. 2006;110:1807–1813. doi: 10.1021/jp0562779. [DOI] [PubMed] [Google Scholar]
- 50.Huang Z, Pearce K, Thompson N. Translational diffusion of bovine prothrombin fragment 1 weakly bound to supported planar membranes: measurement by total internal reflection with fluorescence pattern photobleaching recovery. Biophys J. 1994;67:1754–1766. doi: 10.1016/S0006-3495(94)80650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan T, Yang B, Thompson NL, Maekawa S, Epand R, Jacobson K. Binding of NAP-22, a calmodulin binding neuronal protein, to raft like domains in model membranes. Biochemistry. 2003;42:4780–4786. doi: 10.1021/bi0265877. [DOI] [PubMed] [Google Scholar]