Abstract
Metabolic fuels act on hypothalamic neurons to regulate feeding behavior and energy homeostasis, but the signaling mechanisms mediating these effects are not fully clear. Rats placed on a low-protein diet (10% of calories) exhibited increased food intake (P < 0.05) and hypothalamic Agouti-related protein (Agrp) gene expression (P = 0.002). Direct intracerebroventricular injection of either an amino acid mixture (RPMI 1640) or leucine alone (1 μg) suppressed 24-h food intake (P < 0.05), indicating that increasing amino acid concentrations within the brain is sufficient to suppress food intake. To define a cellular mechanism for these direct effects, GT1–7 hypothalamic cells were exposed to low amino acids for 16 h. Decreasing amino acid availability increased Agrp mRNA levels in GT1–7 cells (P < 0.01), and this effect was attenuated by replacement of the amino acid leucine (P < 0.05). Acute exposure to elevated amino acid concentrations increased ribosomal protein S6 kinase phosphorylation via a rapamycin-sensitive mechanism, suggesting that amino acids directly stimulated mammalian target of rapamycin (mTOR) signaling. To test whether mTOR signaling contributes to amino acid inhibition of Agrp gene expression, GT1–7 cells cultured in either low or high amino acids for 16 h and were also treated with rapamcyin (50 nM). Rapamycin treatment increased Agrp mRNA levels in cells exposed to high amino acids (P = 0.01). Taken together, these observations indicate that amino acids can act within the brain to inhibit food intake and that a direct, mTOR-dependent inhibition of Agrp gene expression may contribute to this effect.
Keywords: Hypothalamus, food intake, neuropeptide, mammalian target of rapamycin
Hypothalamic neurons regulate energy homeostasis by responding to changes in a variety of circulating factors and in turn projecting to downstream sites regulating feeding behavior and autonomic outflow (8, 10, 24). Although much work has focused on the regulation of these neuronal populations by hormones such as leptin, several lines of evidence indicate that nutrient fuels also directly influence the function of specific populations of hypothalamic neurons (20, 32).
Included within this nutrient-sensing system is the ability to detect changes in protein or amino acid availability. A large body of literature indicates that changes in dietary protein influence feeding behavior (1, 3, 17, 25, 28, 33–35). In addition, variations in individual amino acids are also detected by a central nutrient-sensing system, as illustrated by the rapid aversion that develops to diets that are severely imbalanced for an essential amino acid (12, 16, 29, 31) and by the fact that rats will preferentially self-select between two imbalanced diets to meet amino acid requirements (11). Taken together, the above observations support the existence of neural mechanisms that sense and respond to changes in protein or amino acid availability.
Recent work by Cota et al. (5) suggests that hypothalamic neurons participate in this protein-sensing system and that the serine/threonine kinase mammalian target of rapamycin (mTOR) is a cellular mediator of this effect. mTOR is an intracellular signaling molecule sensitive to both amino acid and growth factor signaling and is classically described as a “metabolic sensor” (7, 9). mTOR is expressed within hypothalamic Agrp and Pomc-expressing neurons and is necessary for acute suppression of food intake following central leucine administration (5). These observations are therefore consistent with a model in which hypothalamic mTOR contributes to the regulation of food intake by amino acids.
The current work focuses on the ability of amino acids to directly influence hypothalamic cells. This work demonstrates that amino acids inhibit Agrp gene expression via an mTOR-dependent mechanism, and suggests that this direct effect may contribute to alterations in food intake that occur in response to variations in amino acid availability.
MATERIALS AND METHODS
Animals
All procedures were performed in accordance with the National Institutes of Health guidelines for the care and use of animals and were approved by the Animal Care and Use Committee of Pennington Biomedical Research Center. Male Sprague Dawley rats (Harlan Laboratories, Indianapolis, IN; Charles River Laboratories, Wilmington, MA) were housed singly and maintained on a 12:12-h light-dark cycle with ad libitum access to standard rat chow and water unless otherwise noted.
Effects of a low-protein diet on food intake and hypothalamic Agrp gene expression
Male rats weighing 262 ± 1.52 g were randomly assigned to semipurified, isocaloric diets containing protein at either 20% of total energy (20% protein; control) or 10% of total energy (7–9 rats/group). Diet composition is provided in Table 1. Powdered diets were placed within food cups that were weighed daily, and rats were maintained on wire-bottomed cages for accurate measures of spillage. Daily food intake and body weight was monitored for 7 days, and, on day 7, rats were killed and brains collected and stored. Mediobasal hypothalamic punches were subsequently collected, and mRNA was isolated using Tri-Reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer’s instructions. Expression of Agrp was determined using real-time PCR as described below.
Table 1.
Diet composition
| Protein, %
|
||
|---|---|---|
| 10 | 20 | |
| Ingredients, % | ||
| Casein | 10 | 20 |
| Carbohydrate | 55 | 55 |
| Corn oil | 14.4 | 10 |
| Fiber | 15.6 | 10 |
| Minerals | 3.7 | 3.7 |
| Vitamins | 1.0 | 1.0 |
| Methionine | 0.3 | 0.3 |
| Total calories, % | ||
| Protein | 10.2 | 20.5 |
| Carbohydrate | 56.4 | 56.4 |
| Fat | 33.3 | 23.1 |
Effects of acute, central administration of amino acids on food intake
Male Sprague Dawley rats (300–350 g) were surgically implanted with indwelling third-ventricular cannulas (23). Using aseptic techniques, the skull was exposed, head screws were inserted, and a 22-gauge stainless steel cannula was implanted at coordinates 2.2 posterior from bregma and 7.5 ventral from dura. After anchoring the cannula and sealing all skull openings with dental acrylic, the incision was sutured, and a 28-gauge obdurator was placed in the cannula. After surgery, animals were treated with analgesics and allowed to recover at least 1 wk before further study. Following surgical recovery, rats were fasted for 24 h. Before food replacement (24 h), rats were treated intracerebroventricular with either vehicle (PBS; 2 μl), RPMI 1640 Amino Acids Solution (Sigma-Aldrich, St. Louis, MO), or 1 μg of leucine (7 rats/group). RPMI 1640 contains a mixture of amino acids appropriate for cell culture and comes as a 50× stock. This stock was diluted to 10× in PBS, and a subsequent injection of 2 μl of this 10× solution delivers 1 μg of leucine plus a balance of 18 additional amino acids. After intracerebroventricular injection (2 h), food was replaced, and 4- and 24-h food intake was recorded. To more closely examine the effects of leucine on food intake, rats bearing intracerebroventricular cannulas were fasted for 24 h as above. Before food replacement (2 h), rats were treated intracerebroventricularly with either vehicle (PBS; 2 μl) or three doses of leucine (1, 3, or 10 μg; 7–9 rats/group). Food was replaced 2 h later, and 4- and 24-h food intake was recorded.
In vitro culture of GT1–7 hypothalamic cell line in varying levels of amino acids
All in vitro experiments used the GT1–7 hypothalamic cell line, which was a generous gift from Dr. Pamela Mellon (University of California, San Diego, CA). Before study, cells were cultured in DMEM (with amino acids) containing 25 mM glucose, 10% FBS, 50 units of penicillin, and 50 μg/ml streptomycin at 37°C in 5% CO2 until becoming confluent. For subsequent studies in which amino acid concentrations were varied, DMEM with typical amino acids (100%) and DMEM without amino acids (0%) were mixed to create appropriate ratios for culture. All media contained 10 mM glucose. Chronic (16 h) experiments to assess changes in Agrp gene expression also included 1% FBS, whereas serum was absent in more acute studies of mTOR signaling.
Effects of reduce amino acid availability on Agrp gene expression in vitro
To test the effects of reduced amino acid availability on Agrp gene expression, GT1–7 cells were cultured for 16 h in media containing five levels of amino acids (5, 10, 25, 40, and 100%), with 100% being standard DMEM. After 16 h of culture, cells were collected, RNA was extracted, and levels of Agrp mRNA were determined using real-time PCR.
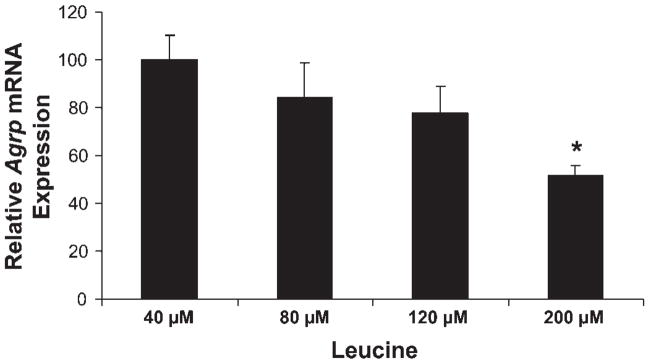
To determine whether changes in leucine concentration influence Agrp gene expression, cells were cultured for 16 h in DMEM containing no amino acids but supplemented with 40, 80, 120, and 200 μM leucine. Leucine at 40 μM represents the concentration in 5% amino acids. After 16 h of culture, cells were harvested, and Agrp mRNA levels were determined.
Effects of amino acids on mTOR signaling in hypothalamic cells
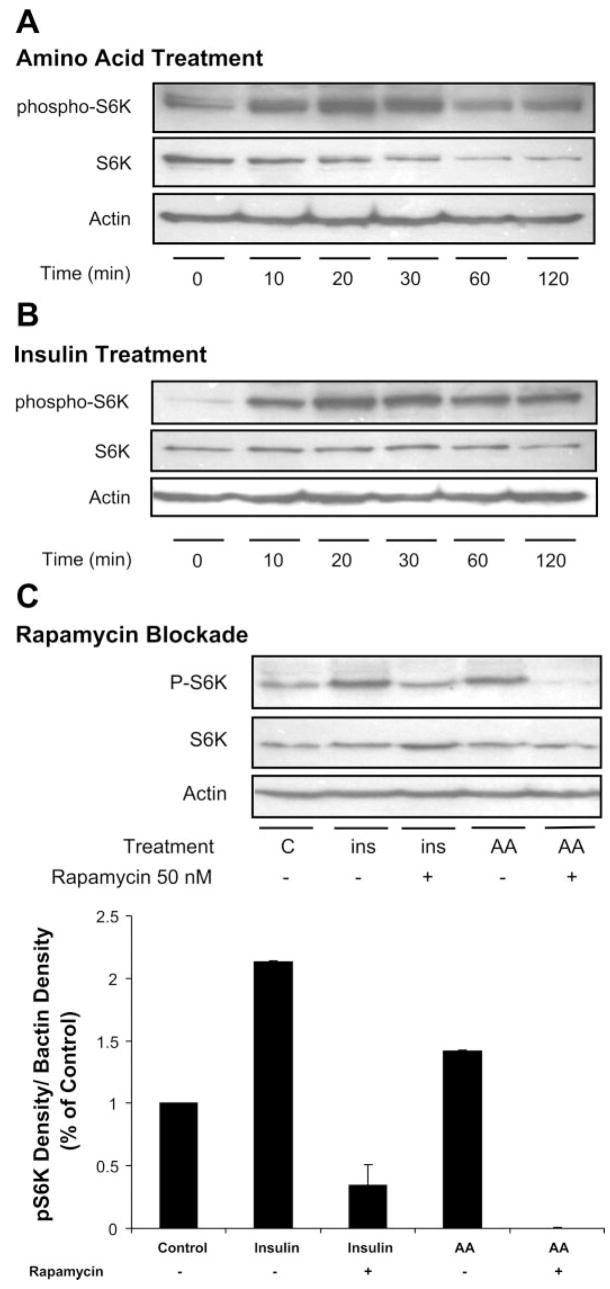
To determine whether amino acids directly impact mTOR signaling within hypothalamic cells, GT1–7 cells were preincubated in 5% amino acids for 7 h. Media were then removed and replaced with media containing 100% amino acids, and cells were collected 0, 10, 20, 30, 60, and 120 min following amino acid stimulation for protein extraction and Western Blot analysis of phosphoribosomal protein S6 kinase (S6K1), as described below. To serve as a positive control, a separate experiment was conducted as above, except that cells were treated with insulin (10 mM) and then collected at the above time points for protein extraction and Western Blot analysis of phospho-S6K1.
To determine whether inhibition of mTOR signaling would block the effects of amino acids or insulin on phosphorylation of S6K1, cells were preincubated in low (5%) amino acids for 7 h as above. During the last half-hour of incubation, some groups were pretreated with rapamycin (50 nM), a pharmacological inhibitor of mTOR. Cells were then transferred to the following four conditions: 100% amino acids, insulin (10 mM), 100% amino acids plus rapamycin, or insulin plus rapamycin. After exposure to high amino acids or insulin (20 min), cells were harvested for protein extraction and Western Blot analysis of phospho-S6K1.
Effect of pharmacological inhibition of mTOR on amino acid-dependent inhibition of Agrp gene expression
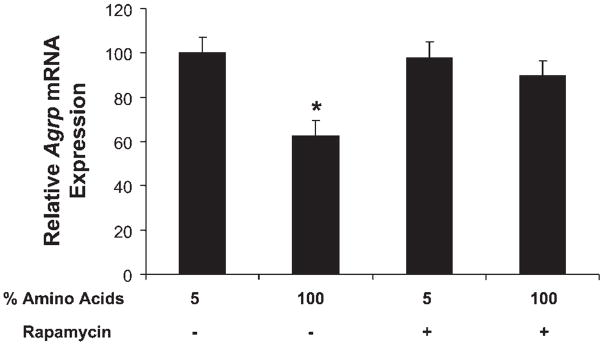
GT1–7 cells were cultured under four different conditions consisting of basal conditions (DMEM, 1% FBS, and 10 mM glucose) with 5% amino acids or 100% amino acids. Within each group, cells were treated with either vehicle or rapamycin (50 nM) for 16 h. Subsequently, cells were harvested for RNA extraction and assessment of Agrp gene expression via real-time PCR.
Real-time PCR analysis
Total RNA was isolated with Tri-Reagent (Molecular Research Center) and quantified spectrophotometrically. Confirmation of intact 18S and 28S RNA bands was achieved by ethidium bromide staining after electrophoresis of samples in 1% agarose/formaldehyde gels. Taqman probes and primers for genes of interest were designed using PrimerExpress Software (Applied Biosystems, Branchburg, NJ) and were synthesized by Biosearch Technologies (Novato, CA). Real-time RT-PCR reactions were prepared with a Taqman 100Rxn Core Reagent Kit (Applied Biosystems). PCR amplification was carried out in optical 96-well plates in an ABI PRISM 7700 sequence detector (Applied Biosystems) under the following conditions: 48°C for 30 min, then 95°C for 10 min for one cycle, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. All expression data were normalized to 18S rRNA expression levels. Primers and probes are as follows: mouse Agrp: forward GTACCGCCACGAACCTCTGT, reverse TCCCCTGCCTTTCCCAAA, probe TCGCACCTAGCCAATGGATGTT; rat Agrp: forward TTGGCAGAGGTGCTAGATCCA, reverse AGGACTCGTGCAGCCTTACAC, probe CGAGTCTCGTTCTCCGCGTCGC; 18S rRNA: forward ACGGCCGGTACAGTGAAACT, reverse GAGCGAGCGACCAAAGGA, probe CCATAACTGATTTAATGAGCCATTCG.
Protein extraction and Western blot
GT1–7 cells were harvested and placed in ice-cold lysis buffer containing 62.5 mM Tris · HCl pH 6.8, 1% SDS, and 1% protease inhibitor, and cells were homogenized via sonification. Cell lysate was then centrifuged at 10,000 g for 10 min, and supernate was collected as total protein lysates. Total protein (40 μg) was separated via SDS-PAGE and transferred to polyvinyl-idene difluoride membranes. Membranes were washed in TBS plus 0.1% Tween 20 (TBS-T) and blocked in TBS-T plus 5% BSA/nonfat dry milk for 1 h before incubation with primary antibodies (Cell Signaling Technology, Beverly, MA) to either phospho-S6K1 (1: 2,000 dilution), total S6K1 (1:2,000 dilution), or β-actin (1:5,000 dilution) Membranes were then washed and incubated with corresponding secondary antibodies. Positive signal was imaged using a chemiluminescent substrate (ECL Reagent; Amersham Biosciences, Piscataway, NJ) and exposure to autoradiograph film. After being probed with the phospho-specific antibody, the membrane was stripped using 0.5 N NaOH for 20 min and reprobed for total antibody followed by β-actin antibody.
Statistical analysis
Data were analyzed using the SAS software package (SAS V8; SAS Institute, Cary NC) using a two-tailed t-test or ANOVA using the general linear model (GLM) procedure. Repeated-measures analysis of changes in food intake and body weight over time was conducted using the GLM procedure of SAS. When experiment-wide tests were significant, post hoc comparisons were made using the LSMEANS statement with the PDIFF option and thus represent least-significant difference tests for preplanned comparisons. Gene expression was normalized to 18S rRNA expression levels before analysis. Densitometric analysis of the Western blot results was conducted on scanned images using the QuantityOne 1-D Analysis software (Bio-Rad). Pixel density for phosphorylated S6K was determined and normalized to levels of β-actin. Data are presented as means ± SE.
RESULTS
Low-protein diets increase food intake and hypothalamic Agrp gene expression
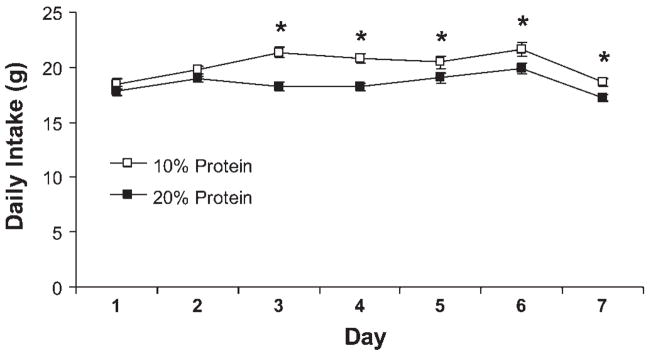
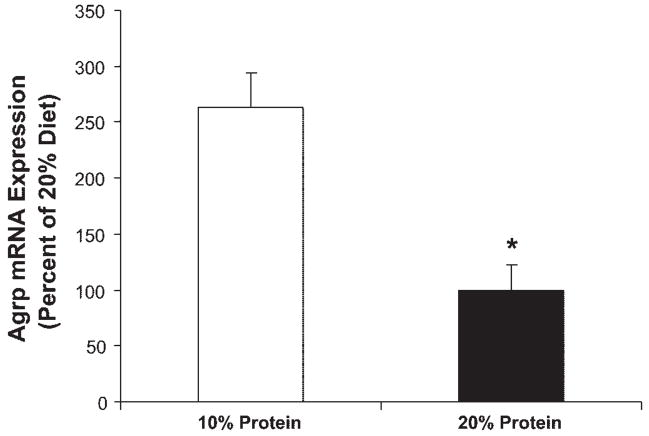
Male Sprague Dawley rats were placed on a purified, isocaloric diet providing protein at either 20% of total energy (20% protein; control) or 10% of total energy (Table 1), and changes in food intake and body weight were assessed over 7 days. Rats consuming the low-protein diet exhibited a significant increase in daily and cumulative food intake within 3 days, and this difference persisted for the remainder of the study (P < 0.05; Fig. 1). In conjunction with increased food intake, hypothalamic Agrp mRNA levels were increased by day 7 in low-protein rats compared with controls (P = 0.002; Fig. 2). These observations are consistent with previous evidence that alterations in dietary protein levels influence food intake (3, 17, 34, 35) and suggest that increases in Agrp may contribute to this hyperphagia.
Fig. 1.
Effects of a low-protein diet on food intake. Male Sprague Dawley rats were placed on a purified, isocaloric diet providing protein at either 20% of total energy (20% protein; control) or 10% of total energy (7–9 rats/group), and changes in food intake and body weight were assessed over 7 days. Exposure to the low-protein diet resulted in a significant increase in daily food intake beginning on day 3, and this increase persisted for the remainder of the study (*P < 0.05).
Fig. 2.
Effects of a low-protein diet on hypothalamic Agrp gene expression. After 7 days on the low-protein diet, brains were collected, small punches of mediobasal hypothalamus were collected, and total RNA was extracted. Levels of Agrp mRNA were assessed via real-time PCR. Hypothalamic Agrp mRNA expression was significantly increased in rats exposed to 7 days of a low-protein diet compared with controls (*P = 0.002).
Acute, central administration of amino acids reduces food intake
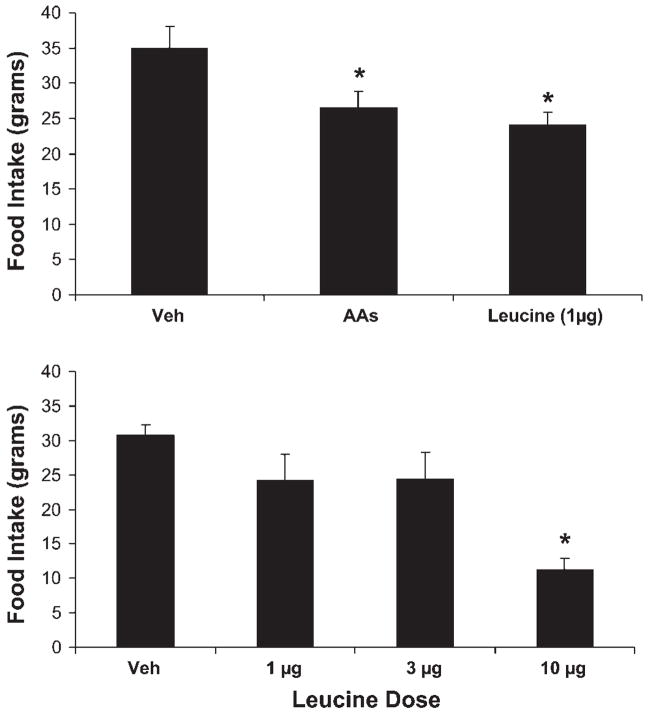
To test whether amino acids are capable of directly acting within the brain to inhibit feeding behavior, 24-h-fasted rats were injected with an amino acid mixture (10× RPMI 1640; 2 μl) or the amino acid leucine. The amount of leucine injected (1 μg) was chosen to match that contained within the amino acid mixture, although the amino acid mixture also delivered other essential amino acids. Injection of both the amino acid mixture and leucine (1 μg) significantly suppressed 24-h food intake (P < 0.05; Fig. 3A). To more closely assess the effects of leucine on feeding behavior, a separate group of cannulated rats was treated with increasing doses of leucine (1, 3, and 10 μg), and food intake during the subsequent 24 h was determined. Intracerebroventricular injection of leucine dose dependently suppressed 24-h food intake, with 10 μg leucine inducing a particularly robust decrease in food intake (P < 0.001; Fig. 3B).
Fig. 3.
Effect of acute, central administration of amino acids on food intake. A: rats bearing 3rd-ventricular cannulas were fasted for 24 h and subsequently injected (icv; 7 rats/group) with vehicle, 1 μg of leucine, or a cell culture-based amino acid mixture (10× RPMI; 2 μl) that contained 1 μg of leucine in addition to other essential amino acids. Injection of both the amino acid mixture and leucine significantly suppressed 4- and 24-h food intake (*P < 0.05). B: a separate group of icv-cannulated rats was fasted as above and injected icv with increasing doses of leucine (1, 3, and 10 μg; 7–9 rats/group), and food intake during the subsequent 24 h was determined. Icv injection of leucine dose-dependently suppressed 4 and 24-h food intake, with 1 μg leucine exerting a modest anorectic effect at 24 h (P = 0.06) and 10 μg leucine robustly suppressing intake (*P < 0.01).
Reduced amino acid availability increases Agrp gene expression in vitro
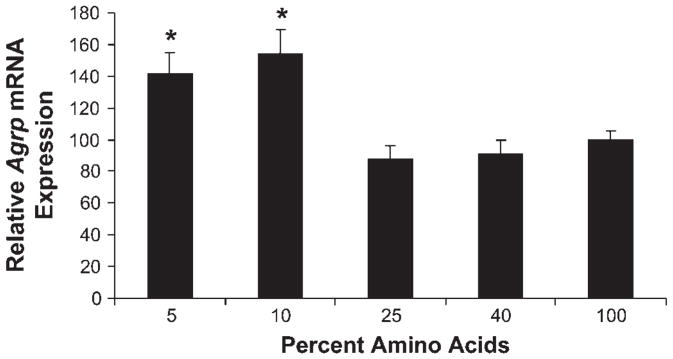
To determine whether changes in amino acid availability could exert direct effects on Agrp gene expression, the GT1–7 hypothalamic cell line was used. Cells were cultured in varying levels of amino acids for 16 h and then harvested to assess gene expression via real-time PCR. Compared with typical levels of amino acids used in cell culture (standard DMEM = 100%), decreasing amino acid levels to 10% or 5% of basal resulted in significant increases in Agrp mRNA levels (P < 0.001; Fig. 4). To test whether leucine alone could directly regulate Agrp gene expression, cells cultured in 5% amino acids for 16 h were supplemented with increasing concentrations of leucine. Leucine replacement resulted in a dose-dependent, linear decrease in Agrp mRNA levels in cells cultured in low amino acid conditions (P < 0.01; Fig. 5).
Fig. 4.
Effects of reduced amino acid availability on Agrp gene expression in vitro. The GT1–7 hypothalamic cell line was used to assess the effects of reduced amino acids on Agrp gene expression in vitro. Cells were cultured in 5, 10, 25, 40, and 100% amino acids for 16 h, with 100% representing standard DMEM. Following exposure to low amino acids, cells were harvested to assess Agrp gene expression via real-time PCR. Decreasing amino acid levels to 10 or 5% of basal resulted in significant increases in Agrp mRNA levels (*P < 0.001).
Fig. 5.
Effect of leucine on Agrp gene expression in vitro. GT1–7 cells were cultured in 5% amino acids for 16 h and supplemented with 40, 80, 120, or 200 μM leucine. After 16 h, cells were harvested, total RNA was extracted, and Agrp mRNA levels were determined via real-time PCR. Leucine supplementation to cells cultured in low amino acids resulted in a dose-dependent, linear decrease in Agrp mRNA levels in cells cultured in low amino acid conditions (P < 0.01).
Amino acids activate mTOR signaling in GT1–7 cells
To define a potential cellular mechanism for amino acid regulation of food intake and Agrp gene expression, phosphorylation of S6K1 was assessed, since S6K1 is a downstream substrate for mTOR and thus a marker of mTOR signaling. GT1–7 cells were cultured at 5% of basal amino acid concentrations for 7 h to increase basal Agrp expression. Cells were then placed in media containing 100% amino acids, and, at defined time points, cells were collected for Western blot analysis of S6K1 phosphorylation. In this paradigm, amino acid stimulation increased S6K1 phosphorylation in a time-dependent manner, peaking at 20–30 min after initial exposure (Fig. 6A). As expected, insulin treatment also rapidly induced S6K1 phosphorylation within GT1–7 cells (Fig. 6B).
Fig. 6.
Effect of amino acids and insulin on mammalian target of rapamycin (mTOR) signaling in hypothalamic cells. A: GT1–7 cells were cultured at 5% of basal amino acid concentrations for 7 h and then placed in media containing 100% amino acids. At defined time points following exposure to high amino acids, cells were collected for Western blot analysis of ribosomal protein S6 kinase (S6K1) phosphorylation. B: in similar fashion, GT1–7 cells cultured in normal levels of amino acids were treated with insulin (10 mM), cells were collected at similar time points, and total protein was used to assess phosphorylation of S6K1. C: to test whether amino acid-dependent activation of S6K1 requires mTOR signaling, GT1–7 cells incubated in low amino acid concentrations were treated for 20 min with either 100% amino acids or insulin (10 mM) in the presence or absence of the mTOR inhibitor rapamycin. Rapamycin treatment (50 nM) attenuated both amino acid- and insulin-induced increases in S6K1 phosphorylation (*P < 0.05).
To test whether amino acid-dependent activation of S6K1 requires mTOR signaling, GT1–7 cells were preincubated in low amino acid concentrations and then treated for 20 min with either 100% amino acids or insulin (10 mM) in the presence or absence of the mTOR inhibitor rapamycin. As expected, both insulin and amino acids increased phosphorylation of S6K1 (Fig. 6C; P < 0.05). Treatment with rapamycin (50 nM) prevented both the amino acid- and insulin-induced increases in S6K1 phosphorylation, consistent with a model in which mTOR acts a sensor of amino acids within hypothalamic cells.
Activation of mTOR is necessary for amino acid-dependent inhibition of Agrp gene expression
To test whether mTOR signaling contributes to amino acid-dependent inhibition of Agrp gene expression, GT1–7 cells were cultured in either 5 or 100% amino acids in the presence or absence of rapamycin (50 nm). As before, Agrp mRNA levels were significantly higher in cells cultured in 5% amino acids verses those cultured in 100% (P = 0.002; Fig. 7). Treatment with rapamycin had no effect on the already elevated Agrp gene expression in cells cultured in 5% amino acids. However, rapamycin pretreatment increased Agrp mRNA levels in cells cultured in 100% amino acids such that Agrp levels were significantly higher than those of cells cultured in 100% amino acids without rapamycin (P = 0.01) and similar to those cells cultured in 5% amino acids. Taken together, these observations suggest that mTOR signaling is necessary for the inhibition of Agrp gene expression by amino acids.
Fig. 7.
Activation of mTOR is necessary for amino acid-dependent inhibition of Agrp gene expression. GT1–7 cells were cultured in either 5 or 100% amino acids in the presence or absence of rapamycin (50 nm), generating four treatment groups. After 16 h of culture, cells were harvested, and RNA was extracted to assess Agrp gene expression via real-time PCR. Treatment with rapamycin had no effect on the already elevated Agrp mRNA levels in cells cultured in 5% amino acids but increased Agrp mRNA levels in cells cultured in 100% amino acids such that levels were significantly higher than those of cells cultured in 100% amino acids without rapamycin (*P = 0.01).
DISCUSSION
Nutrient fuels such as glucose and fatty acids act locally within the brain to regulate feeding behavior and energy homeostasis (19, 22, 27, 30). Work presented herein provides support for a model in which amino acids also act within the brain to suppress food intake and Agrp gene expression. Work by Cota et al. (5) indicates that mTOR signaling components are expressed within hypothalamic AgRP neurons and that mTOR signaling is necessary for leucine-dependent inhibition of feeding (5). The current data replicate the suppression of food intake in response to intracerebroventricular administration of amino acids and, in addition, demonstrate that amino acids directly inhibit Agrp expression within a hypothalamically derived cell line and that this inhibition requires the activation of mTOR signaling. Considering that changes in dietary amino acid availability also altered hypothalamic Agrp gene expression, these observations support a model in which hypothalamic AgRP neurons function as nutrient sensors, linking changes in hormonal and nutrient (amino acid) availability to changes in food intake and body weight.
It is well-accepted that the manipulation of dietary protein can exert significant effects on feeding behavior (1, 25, 28, 33). Low-protein diets increase food intake (34, 35), whereas high-protein diets tend to suppress food intake (3, 17). If simultaneously offered a choice of a low- and high-protein diet, animals tend to selectively balance the consumption of each diet such that adequate protein is consumed (14). These observations highlight the significant effects that protein can exert on feeding behavior and suggest that physiological systems exist that monitor and regulate protein consumption. Although it is possible that protein status is sensed generally, several lines of observation suggest that mechanisms exist for the sensing of individual amino acids. For instance, it is well-established that diets that are severely amino acid imbalanced induce aversive responses, leading to decreased food intake (16, 26). However, if offered a choice between two diets that are imbalanced for different amino acids, rats will effectively self-select between these two diets to meet their amino acid requirements, even though each diet would be aversive if offered alone (11). Clearly these observations indicate that animals can sense changes in dietary protein, as well as variations in individual amino acids. A large body of work implicates the anterior piriform cortex in the aversive response to amino acid imbalances, and infusion of the limiting amino acid directly into the piriform cortex is sufficient to block the aversive response to amino acid imbalance (12). However, although it is clear that the anterior piriform cortex plays a critical role in the aversive response to amino acid imbalance, whether this brain area is also involved in the homeostatic regulation of feeding behavior is less clear.
One brain area that is widely accepted to play a key role in the homeostatic regulation of food intake is the hypothalamus (8). Hypothalamic neurons are sensitive to a wide range of hormones and nutrients, and thus a logical hypothesis is that these neurons might also play a role in protein sensing (20, 32). In particular, hypothalamic neurons containing the orexigenic neuropeptides AgRP and neuropeptide Y (NPY) respond to a variety of hormones and nutrients. Feeding a low-protein diet resulted in an increase in food intake, consistent with previous work (34, 35). This increased intake was accompanied by a significant increase in hypothalamic Agrp gene expression. It is unlikely that the increase in Agrp expression is secondary to the increased food intake, since increases in food intake and body weight would be predicted to suppress Agrp mRNA levels. Considering that Npy gene expression is also increased by low-protein diets (34), these data are most consistent with a model in which decreases in dietary protein activate NPY/AgRP neurons and lead to increases in food intake.
Whereas the above observations suggest that dietary amino acids influence central mechanisms regulating energy balance, these experiments do not delineate whether the detection mechanism exists within the brain or instead within peripheral tissues such as the liver or gastrointestinal tract. Brain amino acid concentrations are sensitive to changes in dietary amino acid availability as well as the concentrations of amino acids within the circulation, which supports the possibility that the brain could be directly sensitive to changes in amino acid availability (4, 6, 13). To discriminate between a direct effect of amino acids on the brain vs. an indirect effect within the periphery, third-ventricular cannulas were used to directly deliver amino acids in the brain, thus bypassing any peripheral sensing mechanism. In this model, central delivery of amino acids significantly decreased food intake (5). This observation does not rule out contributions from peripheral mechanisms, such as recent evidence that gut-derived PYY contributes to the reduction in food intake following high-protein diets (2). However, these observations do indicate that central sensing mechanisms are sufficient to mediate the effects of amino acids on feeding behavior.
A second question regarding the mechanism of central protein sensing is the nature of the signal itself. One possibility is that the brain tracks the availability of many amino acids simultaneously. However, an alternate possibility is that specific amino acids serve as markers for total protein availability. In support of this latter alternative, multiple observations suggest that the branched-chain amino acids, and particularly leucine, are well-suited to serve as nutrient signals (5, 15, 21). To test whether leucine alone was sufficient to alter feeding behavior, the effects of direct brain delivery of leucine were determined and compared with the amino acid mixture. In this paradigm, leucine alone decreased food intake similarly to the amino acid mixture. This comparison is significant considering that the amount of leucine delivered (1 μg) was the same in both treatments, but the amino acid mixture contained a large variety of additional amino acids. The efficacy of an amino acid mixture to suppress food intake was therefore similar to that of the same dose of leucine alone. In addition, subsequent experiments using higher leucine doses produced more robust decreases in food intake. Work recently published by Cota et al. (5) supports a key role of leucine, with intracerebroventricular administration of leucine significantly suppressing food intake but a similar dose of the branched-chain amino acid valine having no effect. Taken together with the current results, these observations indicate that amino acids can act locally within the brain to suppress food intake and that leucine in particular may function as a key amino acid signal within the brain, much as it does within peripheral tissues (18).
The above observations indicate that dietary amino acids suppress food intake and Agrp gene expression and that a direct action of specific amino acids on the brain may contribute to this effect. To more closely examine the cellular mechanisms mediating amino acid regulation of Agrp gene expression, we focused on the GT1–7 hypothalamic cell line. GT1–7 cells express Agrp and respond to hormonal and nutrient cues in a manner similar to hypothalamic neurons in vivo (19). Consistent with the in vivo effects of a low-protein diet, reducing amino acids levels to 10 or 5% of baseline (basal DMEM is considered 100% amino acids) resulted in a significant increase in Agrp mRNA levels in GT1–7 cells. This observation not only complements the in vivo observations reported here, they also indicate that changes in amino acid availability can have direct effects on Agrp gene expression.
Our previous in vivo observations indicate that leucine may act as key signal of amino acid availability. To test this hypothesis in vitro, cells cultured in low amino acid conditions were supplemented with increasing levels of leucine. Leucine replacement dose-dependently decreased Agrp gene expression in a setting of reduced amino acids, suggesting that leucine alone is sufficient to inhibit Agrp gene expression in vitro, much as it regulated feeding behavior in vivo.
If amino acids act directly to inhibit food intake and Agrp gene expression, what is the cellular mechanism mediating these effects? A large body of work suggests that amino acids, and particularly leucine, act intracellularly on the serine-threonine kinase mTOR (15, 21). mTOR is classically associated with cellular nutrient sensing and is proposed to link nutrient and growth factor signaling to cellular protein metabolism, particularly protein translation (7, 9). mTOR phosphorylates a variety of downstream signaling molecules, but S6K1 and eukaryotic initiation factor 4E-binding protein 1 are established mediators of its cellular effects on protein metabolism. Recent work by Cota et al. (5) implicates mTOR in leucine-dependent inhibition of feeding and demonstrates that mTOR signaling is active within AgRP neurons (5). Available evidence therefore suggests that mTOR is a cellular mediator of amino acid action.
To test whether amino acids directly activate mTOR signaling within a hypothalamic cell line, GT1–7 cells were exposed to an acute change from low to high amino acids. In this paradigm, exposure to increased amino acids increased S6K1 phosphorylation. In addition, this induction was blocked by pretreatment with rapamycin, indicating that the effects were dependent on mTOR signaling. Insulin is also a well-known activator of mTOR signaling. As with amino acids, insulin treatment also rapidly induced phosphorylation of S6K, and this effect was also sensitive to rapamycin pretreatment.
The above observations indicate that amino acids activate mTOR signaling in GT1–7 cells, thus raising the possibility that mTOR may mediate the effects of amino acids on Agrp gene expression. To test this hypothesis, GT1–7 cells were exposed to either 5 or 100% amino acids as before but were also treated with the mTOR inhibitor rapamycin. As expected, cells exposed to 5% amino acids had significantly higher levels of Agrp mRNA compared with those cultured in 100% amino acids. Rapamycin treatment had no effect in cells cultured in low amino acids, which was expected considering that mTOR signaling is reduced in situations of low amino acids. Contrastingly, rapamycin treatment significantly increased Agrp mRNA levels in cells cultured in high amino acids, effectively blocking the amino acid-dependent inhibition of Agrp expression. This observation suggests that the activation of mTOR signaling is a necessary component for amino acid inhibition of Agrp gene expression in a hypothalamic cell line.
The work presented herein represents a series of experiments supporting a direct effect of amino acids on Agrp gene expression. Changes in dietary protein exert significant effects on feeding behavior, and, based on the current work, it appears likely that at least some of these effects are mediated by a direct effect of amino acids on hypothalamic AgRP neurons. In addition, these observations also specifically implicate leucine as an important amino acid signal, since many of the effects of amino acids were replicated by leucine alone. Finally, these observations support recent work implicating mTOR as an intracellular mediator of amino acid action within the hypothalamus and indicate that amino acids can exert direct effects on Agrp gene expression via a mechanism that requires intact mTOR signaling.
Acknowledgments
GRANTS
This work was supported by the Pennington Medical Foundation (C. D. Morrison and R. J. Martin) and National Institutes of Health Grants NS-051570 (C. D. Morrison) and RR-021945 (C. D. Morrison).
References
- 1.Anderson GH, Moore SE. Dietary proteins in the regulation of food intake and body weight in humans. J Nutr. 2004;134:974S–979S. doi: 10.1093/jn/134.4.974S. [DOI] [PubMed] [Google Scholar]
- 2.Batterham RL, Heffron H, Kapoor S, Chivers JE, Chandarana K, Herzog H, Le Roux CW, Thomas EL, Bell JD, Withers DJ. Critical role for peptide YY in protein-mediated satiation and body-weight regulation. Cell Metab. 2006;4:223–233. doi: 10.1016/j.cmet.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Bensaid A, Tome D, L’Heureux-Bourdon D, Even P, Gietzen D, Morens C, Gaudichon C, Larue-Achagiotis C, Fromentin G. A high-protein diet enhances satiety without conditioned taste aversion in the rat. Physiol Behav. 2003;78:311–320. doi: 10.1016/s0031-9384(02)00977-0. [DOI] [PubMed] [Google Scholar]
- 4.Choi YH, Fletcher PJ, Anderson GH. Extracellular amino acid profiles in the paraventricular nucleus of the rat hypothalamus are influenced by diet composition. Brain Res. 2001;892:320–328. doi: 10.1016/s0006-8993(00)03267-4. [DOI] [PubMed] [Google Scholar]
- 5.Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, Seeley RJ. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- 6.Currie PJ, Chang N, Luo S, Anderson GH. Microdialysis as a tool to measure dietary and regional effects on the complete profile of extracellular amino acids in the hypothalamus of rats. Life Sci. 1995;57:1911–1923. doi: 10.1016/0024-3205(95)02178-l. [DOI] [PubMed] [Google Scholar]
- 7.Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 8.Elmquist JK, Coppari R, Balthasar N, Ichinose M, Lowell BB. Identifying hypothalamic pathways controlling food intake, body weight, and glucose homeostasis. J Comp Neurol. 2005;493:63–71. doi: 10.1002/cne.20786. [DOI] [PubMed] [Google Scholar]
- 9.Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 10.Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004;116:337–350. doi: 10.1016/s0092-8674(03)01081-x. [DOI] [PubMed] [Google Scholar]
- 11.Fromentin G, Nicolaidis S. Rebalancing essential amino acids intake by self-selection in the rat. Br J Nutr. 1996;75:669–682. doi: 10.1079/bjn19960172. [DOI] [PubMed] [Google Scholar]
- 12.Gietzen DW, Rogers QR. Nutritional homeostasis and indispensable amino acid sensing: a new solution to an old puzzle. Trends Neurosci. 2006;29:91–99. doi: 10.1016/j.tins.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Gustafson JM, Dodds SJ, Burgus RC, Mercer LP. Prediction of brain and serum free amino acid profiles in rats fed graded levels of protein. J Nutr. 1986;116:1667–1681. doi: 10.1093/jn/116.9.1667. [DOI] [PubMed] [Google Scholar]
- 14.Harper AE, Peters JC. Protein intake, brain amino acid and serotonin concentrations and protein self-selection. J Nutr. 1989;119:677–689. doi: 10.1093/jn/119.5.677. [DOI] [PubMed] [Google Scholar]
- 15.Kimball SR, Jefferson LS. Signaling pathways and molecular mechanisms through which branched-chain amino acids mediate translational control of protein synthesis. J Nutr. 2006;136:227S–231S. doi: 10.1093/jn/136.1.227S. [DOI] [PubMed] [Google Scholar]
- 16.Koehnle TJ, Stephens AL, Gietzen DW. Threonine-imbalanced diet alters first-meal microstructure in rats. Physiol Behav. 2004;81:15–21. doi: 10.1016/j.physbeh.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 17.L’Heureux-Bouron D, Tome D, Rampin O, Even PC, Larue-Achagiotis C, Fromentin G. Total subdiaphragmatic vagotomy does not suppress high protein diet-induced food intake depression in rats. J Nutr. 2003;133:2639–2642. doi: 10.1093/jn/133.8.2639. [DOI] [PubMed] [Google Scholar]
- 18.Layman DK, Walker DA. Potential importance of leucine in treatment of obesity and the metabolic syndrome. J Nutr. 2006;136:319S–323S. doi: 10.1093/jn/136.1.319S. [DOI] [PubMed] [Google Scholar]
- 19.Lee K, Li B, Xi X, Suh Y, Martin RJ. Role of neuronal energy status in the regulation of adenosine 5′-monophosphate-activated protein kinase, orexigenic neuropeptides expression, and feeding behavior. Endocrinology. 2005;146:3–10. doi: 10.1210/en.2004-0968. [DOI] [PubMed] [Google Scholar]
- 20.Levin BE, Routh VH, Kang L, Sanders NM, Dunn-Meynell AA. Neuronal glucosensing: what do we know after 50 years? Diabetes. 2004;53:2521–2528. doi: 10.2337/diabetes.53.10.2521. [DOI] [PubMed] [Google Scholar]
- 21.Meijer AJ. Amino acids as regulators and components of nonproteinogenic pathways. J Nutr. 2003;133:2057S–2062S. doi: 10.1093/jn/133.6.2057S. [DOI] [PubMed] [Google Scholar]
- 22.Minokoshi Y, Alquier T, Furukawa N, Kim YB, Lee A, Xue B, Mu J, Foufelle F, Ferre P, Birnbaum MJ, Stuck BJ, Kahn BB. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature. 2004;428:569–574. doi: 10.1038/nature02440. [DOI] [PubMed] [Google Scholar]
- 23.Morrison CD, White CL, Wang Z, Lee SY, Lawrence DS, Cefalu WT, Zhang ZY, Gettys TW. Increased hypothalamic protein tyrosine phosphatase 1B contributes to leptin resistance with age. Endocrinology. 2007;148:433–440. doi: 10.1210/en.2006-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 25.Musten B, Peace D, Anderson GH. Food intake regulation in the weanling rat: self-selection of protein and energy. J Nutr. 1974;104:563–572. doi: 10.1093/jn/104.5.563. [DOI] [PubMed] [Google Scholar]
- 26.Naito-Hoopes M, McArthur LH, Gietzen DW, Rogers QR. Learned preference and aversion for complete and isoleucine-devoid diets in rats. Physiol Behav. 1993;53:485–494. doi: 10.1016/0031-9384(93)90142-3. [DOI] [PubMed] [Google Scholar]
- 27.Obici S, Feng Z, Morgan K, Stein D, Karkanias G, Rossetti L. Central administration of oleic acid inhibits glucose production and food intake. Diabetes. 2002;51:271–275. doi: 10.2337/diabetes.51.2.271. [DOI] [PubMed] [Google Scholar]
- 28.Peters JC, Harper AE. Influence of dietary protein level on protein self-selection and plasma and brain amino acid concentrations. Physiol Behav. 1984;33:783–790. doi: 10.1016/0031-9384(84)90048-9. [DOI] [PubMed] [Google Scholar]
- 29.Picard ML, Uzu G, Dunnington EA, Siegel PB. Food intake adjustments of chicks: short term reactions to deficiencies in lysine, methionine and tryptophan. Br Poult Sci. 1993;34:737–746. doi: 10.1080/00071669308417632. [DOI] [PubMed] [Google Scholar]
- 30.Ronnett GV, Kim EK, Landree LE, Tu Y. Fatty acid metabolism as a target for obesity treatment. Physiol Behav. 2005;85:25–35. doi: 10.1016/j.physbeh.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Russell MC, Koehnle TJ, Barrett JA, Blevins JE, Gietzen DW. The rapid anorectic response to a threonine imbalanced diet is decreased by injection of threonine into the anterior piriform cortex of rats. Nutr Neurosci. 2003;6:247–251. doi: 10.1080/1028415031000151567. [DOI] [PubMed] [Google Scholar]
- 32.Seeley RJ, York DA. Fuel sensing and the central nervous system (CNS): implications for the regulation of energy balance and the treatment for obesity. Obes Rev. 2005;6:259–265. doi: 10.1111/j.1467-789X.2005.00193.x. [DOI] [PubMed] [Google Scholar]
- 33.Westerterp-Plantenga MS. The significance of protein in food intake and body weight regulation. Curr Opin Clin Nutr Metab Care. 2003;6:635–638. doi: 10.1097/00075197-200311000-00005. [DOI] [PubMed] [Google Scholar]
- 34.White BD, He B, Dean RG, Martin RJ. Low protein diets increase neuropeptide Y gene expression in the basomedial hypothalamus of rats. J Nutr. 1994;124:1152–1160. doi: 10.1093/jn/124.8.1152. [DOI] [PubMed] [Google Scholar]
- 35.White BD, Porter MH, Martin RJ. Effects of age on the feeding response to moderately low dietary protein in rats. Physiol Behav. 2000;68:673–681. doi: 10.1016/s0031-9384(99)00229-2. [DOI] [PubMed] [Google Scholar]