Abstract
Objective
We and numerous others have long used sheep as a predictive model system in which to explore stem cell transplantation. Unfortunately, while numerous markers are available to identify and isolate human HSC, no reagents exist that allow HSC/progenitors from sheep to be identified or purified, greatly impeding the application of this well-established large animal model to the study of autologous or allogeneic HSC transplantation. The current studies were undertaken to create a monoclonal antibody (MoAb) to sheep CD34 that would enable isolation and study of sheep HSC/progenitors.
Methods
A partial cDNA to the extracellular domain of the sheep CD34 antigen was PCR cloned, characterized, and used to genetically immunize mice and create hybridomas.
Results
The resultant MoAb to sheep CD34 allows flow cytometric detection of sheep HSC/progenitors present within bone marrow, cord blood, and mobilized peripheral blood. Moreover, this antibody can be used to enrich for HSC/progenitors with enhanced in vitro colony-forming potential, and also identifies endothelial cells in situ within paraffin-embedded tissue sections, in similarity to antibodies to human CD34.
Conclusions
The availability of this monoclonal antibody recognizing the stem cell antigen CD34 in sheep will greatly facilitate the study of autologous and allogeneic HSC transplantation using this clinically relevant large animal model.
Keywords: CD34, hematopoietic stem cells, sheep model
Introduction
Sheep have long been used as a predictive model system in which to study development, disease, and physiology [1-10]. As a result of this physiologic similarity, since 1979, we and others have used the sheep model to explore stem cell transplantation [3, 10-28]. The large size and long life span of the sheep make it well-suited for the study of stem cell transplantation, since they allow evaluation of donor cell activity in the same animal for years after transplant and enable the investigator to obtain sufficient donor cells from the primary recipients to perform serial transplantation. Furthermore, by transplanting early in gestation, prior to immune maturation, it is possible to study enriched populations of putative human hematopoietic stem cells (HSC) in a healthy physiologically normal environment. Indeed, successful engraftment and multilineage differentiation of human HSC derived from fetal liver, fetal bone marrow, cord blood, adult bone marrow, and mobilized adult peripheral blood has now been observed in primary, secondary, and tertiary recipients using this model system [12, 15, 29-32].
However, while this model is ideal for studying the potential and behavior of human stem cells, as a xenogeneic model, events observed may not entirely reproduce what would be seen in a clinical setting. Unfortunately, while numerous markers are available to identify and isolate primitive human HSC, no reagents exist that identify or purify HSC/progenitors from sheep for transplantation studies, greatly impeding the application of this large animal model system to the study of autologous or allogeneic HSC transplantation.
Numerous markers are present on human HSC, but to date, CD34 has been the most widely used for HSC identification and isolation. CD34 is an integral membrane glycoprotein whose precise function is largely unknown [33, 34]. CD34 was first identified using the early human myeloblastic cell line KG-1a [35, 36], and CD34+ cells represent roughly 1-3% of bone marrow mononuclear cells (BMMNC) in a normal adult [33, 34]. Recent studies have now demonstrated that CD34 expression by HSC is a reversible process influenced by cell activation, and that some of the most primitive quiescent HSC may in fact be CD34- [37-41]. Nevertheless, the demonstration that autologous BM CD34+ were able to durably engraft baboons [42], led to the testing of human CD34+ cells for both autologous and allogeneic transplantations. This enriched cell population has produced durable hematopoietic reconstitution in both settings, providing evidence that CD34 is expressed on at least some of the most primitive long-term engrafting HSC, and establishing the rationale for widespread use of CD34+ cells for clinical transplantations.
Although we and others have used the fetal sheep model extensively to study the potential and behavior of human HSC, there are no antibodies which allow identification or purification of sheep HSC/progenitors, hindering the development of experimental HSC transplantation strategies in this model. Therefore, in the present studies, we developed monoclonal antibodies to ovine CD34. We PCR cloned and sequenced an 858bp cDNA corresponding to the extracellular domain of sheep CD34, genetically immunized mice, and created monoclonal antibodies. One antibody (8D11) was selected for all subsequent studies. Using flow cytometry, 8D11 identified a small, discrete population of CD45+ cells within sheep BM and cord blood (CB). This population comprised 1.1±0.4% of the total sheep BMMNC and 3.7±0.4% in CB, proportions in close accord with the incidence of CD34+ cells in human BM and CB. The ability of 8D11 to enrich for sheep hematopoietic progenitors was demonstrated by magnetically sorting 8D11+ cells and showing that these CD34+ cells were roughly 100-fold enriched for colony-forming potential (CFU) and 10-fold for CAFC as compared with BMMNC, whereas CD34-negative cells were devoid of progenitors with colony-forming potential. Further evidence of the utility of 8D11 as a marker of primitive hematopoietic cells in the sheep model came from studies in which gene-marked HSC/progenitors were identified in vivo with 8D11 2.5 years after in utero gene transfer, and studies which showed that G-CSF mobilization resulted in a 56-fold increase in the absolute levels of circulating CD34+ cells on day 2 of mobilization. In addition to its ability to identify sheep HSC/progenitors, 8D11 also robustly labeled the lining of blood vessels in sheep tissues, further extending the utility of this antibody.
In conclusion, this first successful generation of a monoclonal antibody to sheep CD34 will greatly facilitate using the sheep as a large animal model to study allogeneic HSC transplantation both in utero and in post-natal recipients using BM, CB, and mobilized PB as cell sources.
Materials and Methods
Cloning of extracellular domain of sheep CD34
To obtain a cDNA clone for the extracellular domain of the sheep CD34 molecule, RNA purified from freshly isolated sheep dermal fibroblasts was provided by Dr. Paul Simmons and used to synthesize cDNA using the SuperScript® II First-Strand Synthesis System (Invitrogen, Carlsbad, CA). These resultant cDNAs were employed as templates to amplify an 858bp fragment of the extracellular domain of sheep CD34 by PCR (30 cycles) using the following primers: sense primer: 5′-atgctgggccgcaggggcgcg-3′; antisense primer: 5′-ggtcttccgggaatagctctggtg -3′. Since the sheep CD34 gene had not been sequenced, these primers were designed based on the NCBI sequence for bovine CD34. The resultant PCR product was analyzed on a 0.8% agarose gel to confirm that the correct size product had been obtained. The 858bp band corresponding to the extracellular domain of the sheep CD34 gene was excised from the gel, purified using the Qiaex II kit (Qiagen, Inc., Valencia, CA) and cloned into the pGEM-T Easy vector (Promega, San Luis Obispo, CA), according to manufacturer's instructions. The recombinant plasmid was propagated in MAX Efficiency® DH5α™ Competent Cells (Invitrogen, Carslbad, CA) and purified using the QiaPrep Midiprep system (Qiagen). Sequencing was performed using SP6 and T7 sequencing primers at the Nevada Genomics Center. Once the DNA sequence had been analyzed and aligned with bovine and goat sequences for CD34 to confirm its identity, the recombinant pGEM-T Easy plasmid was provided to Genovac AG (Freiburg, Germany).
Genetic immunization and production of monoclonal antibody to sheep CD34
The recombinant sheep CD34 plasmid (pGEM-T Easy) was employed by Genovac AG (Freiburg, Germany) for the commercial production of custom monoclonal antibodies using a proprietary procedure. Briefly, the sheep CD34 cDNA was subcloned into a proprietary expression vector and used to genetically immunize mice by repeated intra-dermal injections, thus stimulating an immune response. Lymphocytes harvested from the mice were fused to mouse myeloma cells to establish CD34-specific hybridomas. The hybridomas were screened at Genovac using proprietary recombinant cells transfected in vitro with a sheep CD34-expression vector. We then tested positive hybridoma supernatants on primary sheep hematopoietic cells and tissue sections. All sheep were a Rambouillet/Merino cross. One clone (8D11) was chosen for subsequent subcloning, yielding a pure IgG1-producing hybridoma, the supernatant of which was used for all subsequent studies.
Collection and isolation of sheep BMMNC
BM was aspirated into heparinized syringes from the posterior iliac crest of healthy control adult sheep following the procedure detailed in an IACUC-approved protocol, and BMMNC isolated by Ficoll-Hypaque (Sigma Chemicals) density gradient centrifugation, washed twice and resuspended in IMDM, 10% FCS.
Collection and isolation of sheep CBMNC
Cord blood was aspirated into heparinized syringes from the umbilical vein of healthy control sheep during delivery following the procedure detailed in an IACUC-approved protocol. CBMNC were isolated as detailed above for BMMNC.
Methylcellulose colony assays
BMMNC were cultured at a concentration of 5×105/ml in methylcellulose (CFU-Mix, CFU-GM, BFU-E) using erythropoietin–containing MethoCult 4330 supplemented with recombinant ovine IL-3 (100U/ml), GM-CSF (100U/ml), SCF (1000U/mL) and sheep leukocyte-derived PHA-stimulated leukocyte conditioned medium (PHA-LCM) (5% vol/vol), as previously described [13, 43]. The plates were incubated at 37°C in a humidified atmosphere of 5% CO2 in air for 9-12 days. Hematopoietic colonies were then enumerated in situ [13, 43]. In the case of magnetically sorted CD34+ BM cells, cells obtained after MiniMacs sorting were cultured at 1×105cells/ml.
CAFC Assays
CAFC assays were performed essentially as previously described for mouse [44-46] with minor modifications. In short, a sheep stromal layer was grown in 96-well microtiter plates (Costar, Cambridge, MA) in IMDM supplemented with 10% fetal calf serum, 5% horse serum, 10-5M hydrocortisone, 3.3mM L-glutamine, 80U/ml penicillin, 80μg/ml streptomycin, and 10-4M β–mercaptoethanol. These layers were then grow, to confluence, treated with Mitomycin C, and seeded with either whole unfractionated sheep BM or 8D11-selected BM cells at 20000, 10000, 5000, 2500, 1250, 625, 312, 156, 78, and 39 cells per well. The cells were maintained at 37°C and 5% CO2. Half of the medium was replaced weekly, and wells were evaluated at days 28 and 35 for cobblestone areas growing underneath the stromal layer, since it is well established that day 28 and 35 CAFC represent the most primitive HSC with long-term repopulating ability.
Flow cytometric analysis of sheep peripheral blood and bone marrow
1×106 sheep PB, CB, or BM MNC were transferred to separate tubes, pelleted, and resuspended in PBS, 0.1% sodium azide. Cells were fixed in 2% paraformaldehyde, blocked for 15 minutes with 10% normal goat serum in TBS, and stained for 30 minutes at room temperature with a 1:10 dilution of supernatant containing monoclonal anti-sheep CD34. CD34 staining was then visualized with a PE-labeled anti-mouse IgG secondary antibody (Southern Biotech, Birmingham, AL) diluted 1:20, washed, and the cells stained with a 1:10 dilution of FITC-labeled antibody to sheep CD45 (AbD Serotec, Raleigh, NC). Following a final wash, cells were analyzed on a FACScan (Becton Dickinson Immuno-Systems, San Jose, CA). Negative controls consisted of PB, CB, and BM MNC stained with IgG1 control antibody and the identical PE-labeled secondary antibody used for CD34 visualization.
Mobilization of chimeric and control sheep with granulocyte–colony-stimulating factor
Human recombinant granulocyte–colony-stimulating factor (G-CSF) Neupogen (Amgen, Inc., Thousand Oaks, CA) was administered for 4 days once a day subcutaneously in the morning, weighing animals before injection, and using a dose of 4.8-5.7μg/kg. PB was drawn daily prior to injection and analyzed by flow cytometry to evaluate the total white blood cell (WBC) count and CD34+ progenitor cell content within the circulation.
Magnetic sorting of sheep CD34+ cells
BMMNC were washed twice with MiniMacs (Miltenyi Biotec, Inc., Auburn, CA) wash buffer, counted, and brought to a concentration of 108 cells per 300μl of buffer. 300μl of cell suspension was stained for 30min at 4°C with 50μl of monoclonal anti-ovine CD34. Samples were washed with MiniMacs buffer and the cell pellet resuspended in 300ul of MiniMacs buffer and incubated with 50μl of Goat anti-mouse IgG microbeads for 15min at room temperature. Samples were again washed and resuspended in 500μl of MiniMacs buffer. CD34+ cells were then separated on a MiniMacs magnetic column as per the manufacturer's instructions (Miltenyi Biotec, Inc.).
Immunohistochemical labeling of endothelial cells with anti-ovine CD34
Paraffin-embedded sections were de-waxed in xylene and rehydrated through a graded ethanol series to diH2O. Sections were blocked for 15-minutes with serum-free Protein Block (Dako, Carpentaria, CA) and incubated at room temperature for 1 hour with a 1:10 dilution of 8D11 supernatant. Residual unbound primary antibody was removed by three washes in TBS, containing 0.05% Tween 20. Sections were then incubated for 30 minutes at room temperature in the dark with PE-conjugated anti-mouse IgG secondary antibody (Southern Biotech) diluted 1:20. After three 1 minute washes, sections were counterstained with DAPI/Antifade solution (Vector Laboratories, Burlingame, CA) and coverslipped. Slides were viewed on an Olympus BX60 microscope. Sections processed in the absence of primary antibody served as controls.
Western Blotting
To characterize the hematopoietic protein recognized by clone 8D11, sheep BMMNC were magnetically sorted with clone 8D11. The resultant cell pellet was resuspended in 200μl of PBS and 200μl of 2X protein sample buffer (0.5M Tris-HCl pH 6.8, 5% glycerol, 2% SDS and 100 mM DTT). Samples were boiled for 5 minutes and centrifuged at 16,000g for 15 min to remove insoluble material. Samples were loaded onto a pre-cast 7.5% Ready gel with a 4.5% stacker (BioRad Laboratories, Hercules, CA) and run for 1 hour at 200 volts. After electrophoresis, the proteins were transferred to nitrocellulose membranes in a Bio-Rad Electroblotter Apparatus. Membranes were blocked in TBST (TBS, 0.05% Tween 20) containing 5% milk for 1 h at room temperature and then reacted overnight at 4°C with 8D11 diluted 1:50 in TBST-milk. After three washes in TBST, the membranes were treated with affinity-purified horse radish peroxidase (HRP)-conjugated goat anti-mouse IgG (Promega, Madison, WI) diluted 1:1,000. Membranes were washed three times in TBST, and then developed with a DAB-based HRP detection kit (Vector Laboratories, Burlingame, CA).
Digital Image Acquisition
All images were captured with an Olympus DP70 CCD camera attached to an Olympus BX60 microscope, using Olympus DP Controller Software version 2.1.1.183 (Olympus America, Melville, NY, USA). Images were then subjected to minimal global processing, such as brightness and contrast adjustment and color balance in Adobe Photoshop CS.
Results
PCR cloning and Sequence Analysis of the Extracellular Domain of Sheep CD34
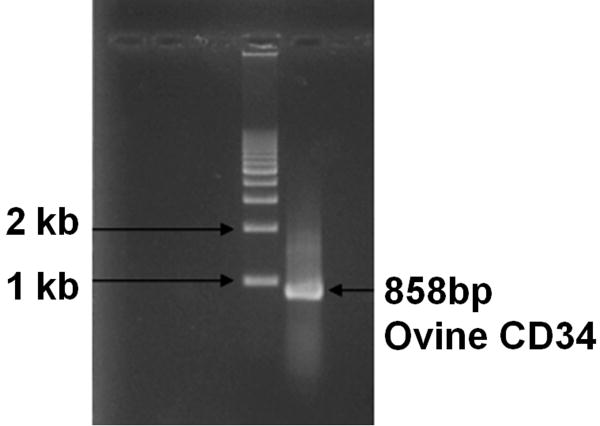
RNA isolated from sheep dermal fibroblasts was reverse transcribed to cDNA and subjected to PCR with primers designed to amplify the extracellular domain of the sheep CD34 molecule, as described in Materials and Methods. Since the sheep sequence was unknown at the time of initiating these studies, we designed the primers using the bovine CD34 sequence based on the rationale that, as a general rule, sheep and cow homology at the nucleotide level is greater than 90% within the coding regions. PCR amplification yielded an 858bp product (Figure 1). The full nucleotide sequence of this cDNA is shown in Figure 2A. This sequence was then subjected to BLAST sequence alignment search against the cow and human databases at NCBI, which revealed that the extracellular domain of the sheep CD34 molecule shares roughly 92% homology with the bovine counterpart. When aligned to the human CD34 sequence, sheep CD34 shared 90% homology for the first 84 nucleotides and roughly 78% homology from nucleotides 453-858, while exhibiting minimal homology between nucleotides 85 and 452. We next analyzed the amplified sequence using NCBI's ORF Finder and confirmed that we had obtained a single open reading frame, the first three nucleotides of which were the ATG start codon. The resultant amino acid sequence for the extracellular domain of sheep CD34 appears in Figure 2B. When aligned to the cow and human protein sequences using NCBI's Protein Blast alignment tool, we found that the extracellular portion of sheep CD34 was roughly 85% homologous to bovine CD34 and 53% homologous to human CD34 at the amino acid level.
Figure 1. PCR Cloning of the Extracellular Domain of Ovine CD34.
PCR with primers based on the known bovine CD34 sequence were used to amplify the previously uncharacterized 858bp extracellular domain of the ovine CD34 mRNA using cDNA from freshly isolated sheep dermal fibroblasts as a template, as described in Materials and Methods.
Figure 2.
A) Nucleotide Sequence of the Extracellular Domain of Ovine CD34. The 858bp PCR product (see Fig. 1) was excised from the gel, purified, and cloned into the pGEM–T Easy vector (Promega), as described in Materials and Methods. Sequencing was performed using T7 and SP6 primers at the Nevada Genomics Center. The sequences of the primers used to amplify the ovine CD34 extracellular domain are indicated with underlined text, and the transcription start codon is indicated in red. B) Amino Acid Sequence of the Extracellular Domain of Ovine CD34. Ovine CD34 nucleotide sequence was analyzed using NCBI's ORF Finder. This analysis confirmed the presence of a single open reading frame, the first three nucleotides of which were the ATG start codon. The resultant amino acid sequence for the extracellular domain of sheep CD34 is shown here.
Successful Generation of Monoclonal Antibody to the Extracellular Domain of Ovine CD34
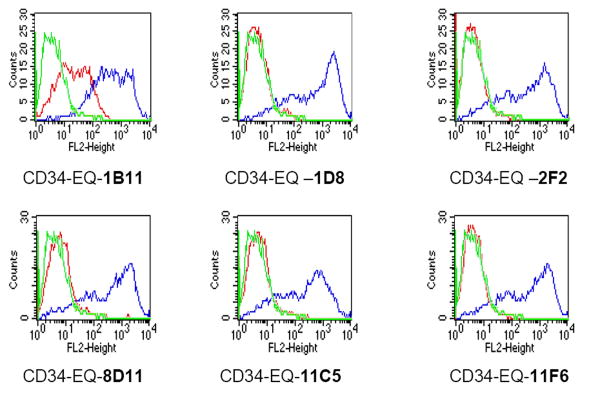
The recombinant pGEM-T Easy plasmid clone harboring the sheep CD34 extracellular domain was employed for the commercial production of custom monoclonal antibodies at Genovac AG (please see Materials and Methods for details). The production of CD34-specific hybridomas is described in detail in the Materials and Methods. The hybridoma supernatants were initially screened at Genovac by flow cytometry using cells transfected in vitro either with the CD34-expression vector, which was used to immunize the mice or with the identical “empty” vector containing no CD34 insert. Using this procedure, 6 potential clones, producing ovine CD34-specific monoclonal antibodies, were identified. As can be seen in Figure 3A, which is a representative FACS plot of these six most promising clones, this procedure resulted in the generation of hybridomas that yielded good specificity for ovine CD34. Subsequently, the hybridoma supernatants from these positive clones were further tested on primary sheep hematopoietic cells and sheep tissue sections at the University of Nevada, Reno, as detailed below.
Figure 3. Specificity of anti-ovine CD34 Monoclonal Antibodies.
The supernatant of the hybridomas, created as described in Materials and Methods, were employed to stain cells that were transiently transfected with either the empty expression vector (red) or with the expression vector containing the cDNA to the extracellular domain of ovine CD34 (blue). Cells stained with an irrelevant antibody are shown in green.
Ability of anti-ovine CD34 monoclonal antibodies to identify sheep hematopoietic stem/progenitor cells in cord blood and bone marrow and allow their purification
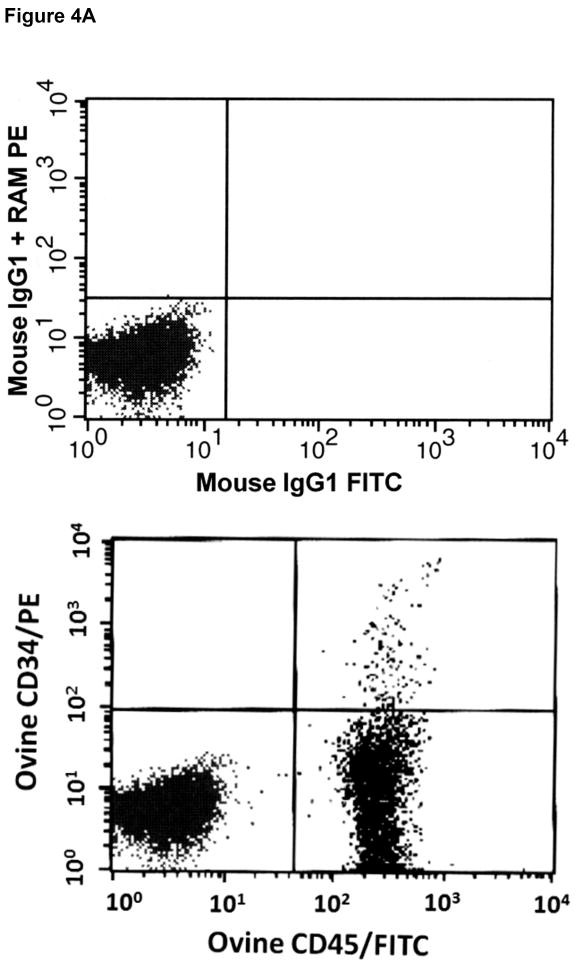
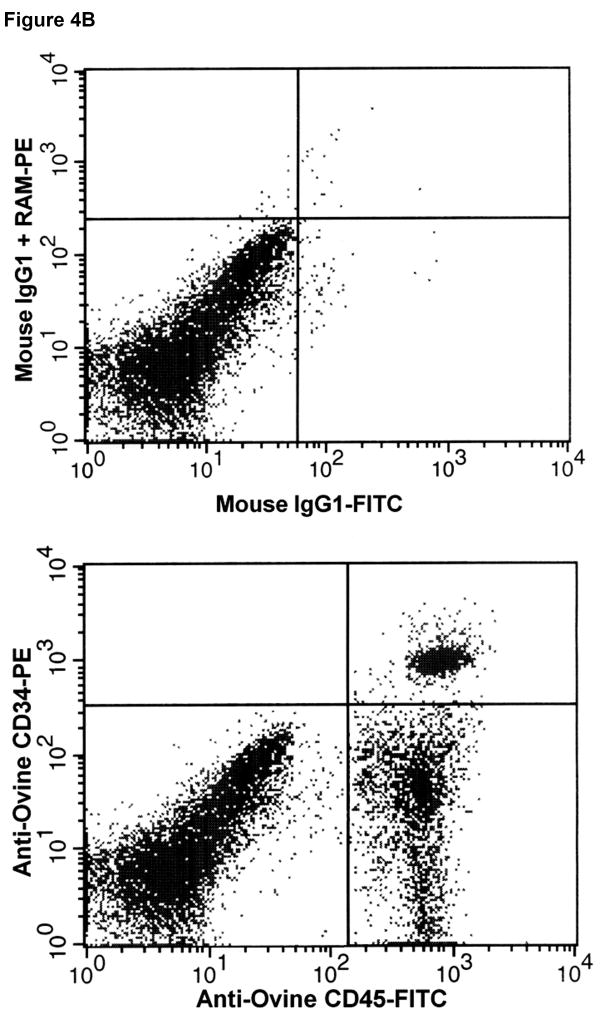
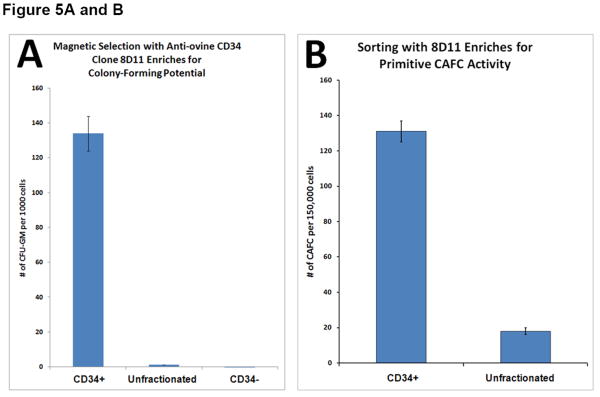
Having established that the 6 hybridoma clones potentially recognized the extracellular domain of ovine CD34, based on the screening employing transiently transfected cells (see above), we next attempted to determine whether these antibodies were capable of identifying and ultimately enriching for hematopoietic stem/progenitor cells. To accomplish this, we collected cord blood from a panel of normal healthy sheep at birth (n=12) and bone marrow aspirates from a panel of normal healthy adult sheep (n=13), and mononuclear cells were obtained by Ficoll-Hypaque density gradient centrifugation. These cells were then stained with anti-ovine CD45-FITC and each of the six candidate anti-ovine CD34 monoclonal antibodies. The anti-ovine CD34 antibodies were then detected with anti-mouse IgG-PE and the cells analyzed by flow cytometry. Each of these clones was able to detect a small, discrete population of CD45+CD34+ cells within the cord blood and bone marrow of each of the sheep. However, clone 8D11 consistently gave the best results with all 10 sheep in the panel, for this reason, clone 8D11 was used for the remaining studies in this manuscript. Figure 4A shows a representative FACS plot obtained when BMMNC from one of the 13 control sheep were stained with clone 8D11, and Figure 4B shows a representative FACS plot obtained when CBMNC from one of the 12 control sheep were stained with clone 8D11. The CD45+CD34+ population identified with this clone comprised 1.1±0.4% of the total marrow nucleated cells (n=13) and 3.7±0.4% of the total cord blood nucleated cells (n=12). These percentages are in agreement with the levels of CD34 cells found in human cord blood and adult human bone marrow, suggesting that the antibodies were identifying the appropriate cell population. To confirm this experimentally, we used clone 8D11 to perform magnetic cell sorting (MiniMacs) of either fresh (n=4) or frozen (n=3) adult sheep bone marrow mononuclear cells and assessed the methylcellulose colony-forming potential of the cells prior to and following magnetic sorting to assess the ability of this antibody to enrich for primitive sheep stem/progenitor cells. As can be seen in Figure 5A, which displays the results obtained with fresh sheep bone marrow, magnetic selection with clone 8D11 resulted in a roughly 100-fold enrichment for cells with colony-forming potential (p<0.001, 2-tailed t-test), since the CD34+ fraction generated 134±10 colonies per 103 cells plated, while the unfractionated BM cells generated only 1.36±0.06 colonies per 103 cells plated. Moreover, when the CD34- flow through fraction from the MiniMacs column was plated in methylcellulose, this population only generated 0.1±0.08 colonies/103 plated cells, strongly supporting the conclusion that the primitive stem/progenitor cells with colony-forming potential were found almost exclusively within the CD34+ fraction identified with clone 8D11. Three additional experiments were then performed with frozen sheep bone marrow, again assessing the ability of the 8D11 clone to identify and enrich for cells with primitive colony-forming potential. As we saw with the fresh bone marrow, essentially all cells with colony-forming potential were present within the CD34+ cell fraction producing an average of 177±138, 98±17, and 223±39 colonies per 103 cells plated in each experiment, respectively. In contrast, the CD34- fraction did not produce any colonies in any of the three experiments conducted with frozen sheep BM, even if plated at a 50-fold higher cell number (5×104 cells).
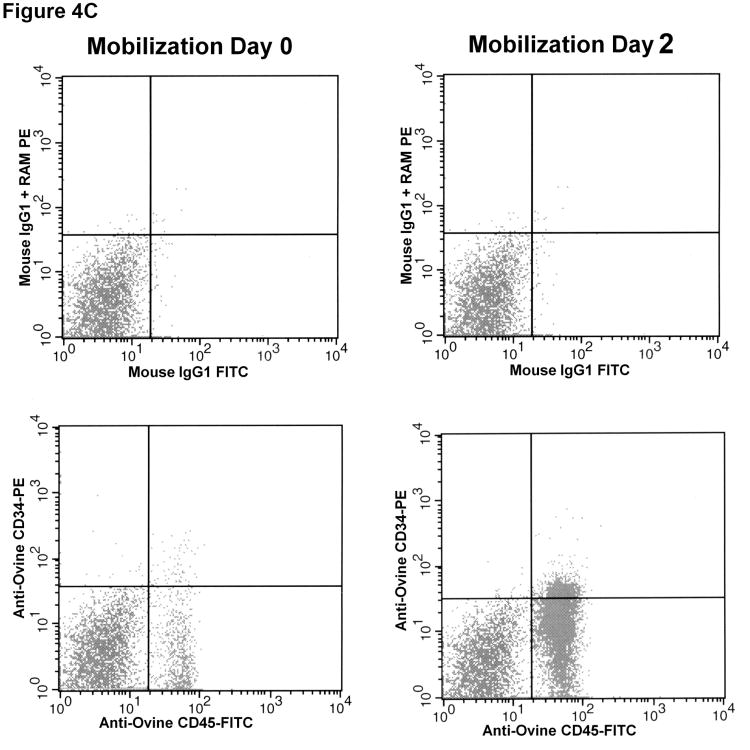
Figure 4. Identification of sheep hematopoietic progenitor cells in sheep bone marrow (A), cord blood (B), and mobilized peripheral blood (C) using the anti-ovine CD34 antibody.
Mononuclear cells obtained from the bone marrow (A), cord blood (B), and G-CSF mobilized peripheral blood (C) of normal healthy sheep were stained with anti-ovine CD45-FITC and each of the panel of six anti-ovine CD34 antibodies, which were then detected with anti-mouse IgG-PE and analyzed by flow cytometry. Shown are representative results obtained with one of the monoclonal antibodies (clone 8D11), with the upper panel of each figure showing the background staining obtained with an isotype-matched (IgG1) control primary antibody and the identical anti-mouse IgG-PE secondary used to detect anti-ovine CD34, and the lower panel showing the staining obtained with anti-sheep CD34 clone 8D11.
Figure 5.
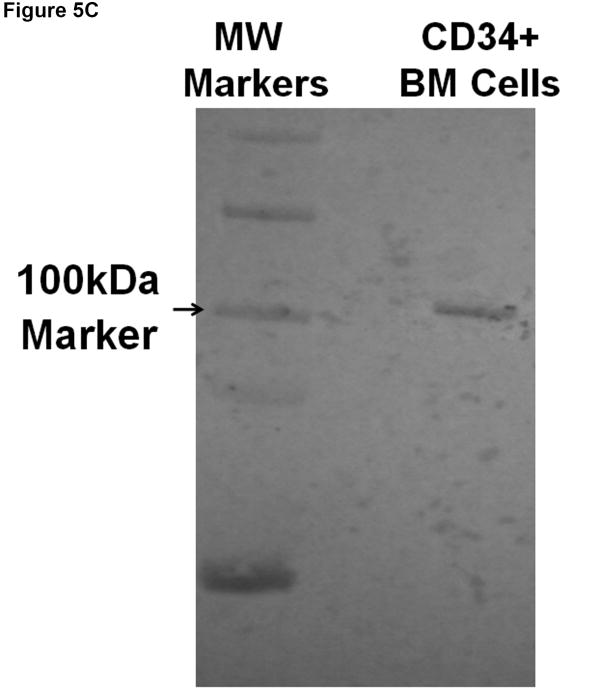
A) Anti-ovine CD34 antibody enables enrichment of hematopoietic cells with colony-forming potential. Bone marrow mononuclear cells were subjected to magnetic cell sorting with one of the anti-ovine CD34 antibodies (clone 8D11) and tested for their in vitro colony forming potential in methylcellulose assays. B) Anti-ovine CD34 antibody enables enrichment of primitive hematopoietic cobblestone area forming cells (CAFC). Bone marrow mononuclear cells were subjected to magnetic cell sorting with one of the anti-ovine CD34 antibodies (clone 8D11) and tested for the frequency of CAFC. C) Anti-Ovine CD34 antibody recognizes a single protein with predicted size. Bone marrow mononuclear cells were subjected to magnetic cell sorting with one of the anti-ovine CD34 antibodies (clone 8D11) and subsequently analyzed by Western Blotting to assess the specificity of the anti-CD34 antibody.
Further evidence for the ability of clone 8D11 to identify and allow isolation of sheep hematopoietic cells with primitive colony-forming potential came from studies in which we compared the frequency of cobblestone are-forming cells (CAFC) in unfractionated sheep bone marrow to that in the cells isolated based on 8D11 positivity. The results of these analyses (n=3) can be seen in Figure 5B. As shown in this figure, which is data from experiments in which 10,000 cells were plated per well, magnetic sorting with the 8D11 clone resulted in a roughly 10-fold enrichment for cells with CAFC potential (p<0.001, 2-tailed t-test). As we progressed down the limiting dilution curve, CAFC continued to be observed in the majority of wells with 8D11-selected cells down to our lowest dilution of only 39 cells per well. In contrast, when looking at the unfractionated marrow, CAFC were only observed in a fraction of the wells that had 312 cells, and were not observed in any of the wells in which fewer than 312 cells were plated. These data thus demonstrate that 8D11 labels hematopoietic cells with very primitive in vitro colony-forming potential, strongly suggesting it identifies HSC/progenitors present within sheep bone marrow.
To confirm the specificity of clone 8D11 for the CD34 antigen expressed on ovine HSC/progenitors, bone marrow mononuclear cells from a control sheep were subjected to magnetic cell sorting with clone 8D11. Protein extracts from the resultant CD34+ cells were then analyzed by Western blotting as detailed in the materials and methods, using 8D11 as the primary antibody. As can be seen in Figure 5C, this antibody recognizes a single protein near the anticipated molecular weight of roughly 100-110kDa[47], thus confirming its specificity for the ovine CD34 antigen.
8D11 Identifies Primitive Long-Lived HSC/progenitors In Vivo
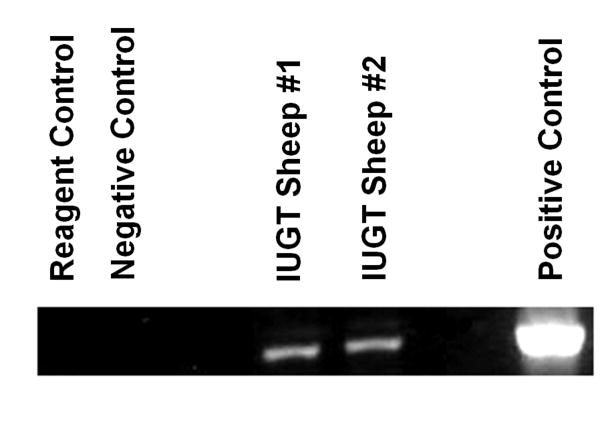
To further test the ability of 8D11 to identify primitive HSC and establish whether the identified cells possessed in vivo functionality, we used this antibody in other ongoing in utero gene transfer (IUGT) studies in our laboratory in which retroviral vectors were injected into the peritoneal cavity of early gestational sheep with the goal of achieving transduction of long-lived HSC in vivo [48-50]. These animals have exhibited long-term transduction within their hematopoietic system, some for over 5 years after in utero treatment, and we have previously reported that transplantation of unfractionated marrow from these sheep into secondary fetal sheep recipients results in long-term transgene presence/expression within the hematopoietic system of these secondary recipients [48-50]. In the present study, we used 8D11 to sort CD34+ cells from the bone marrow of several of the sheep from these studies to assess the presence of vector DNA within the HSC/progenitor compartment. As shown in Figure 6, which is a representative PCR from 2 of these animals (amplified as we have previously described [48-50]), CD34+ cells sorted from the BM of these animals at over 2.5 years after IUGT were still positive for the vector-encoded NeoR gene, confirming that the long-term persistence of transduced cells within the hematopoietic system of these sheep was indeed due to transduction of primitive CD34+ HSC/progenitors, and providing compelling, albeit somewhat indirect, evidence that the 8D11 antibody allows identification of cells with long-term repopulating ability in vivo.
Figure 6. Anti-ovine CD34 antibody detects HSC/progenitors with long-term repopulating ability in vivo.
Bone marrow cells from sheep that had received in utero gene transfer (IUGT) were stained with one of the anti-ovine CD34 antibodies (clone 8D11) at 2.5 years after IUGT and isolated by magnetic sorting. The resultant CD34+ cells were then subjected to PCR with primers specific to the vector-encoded NeoR gene to assess whether the long-term presence of transduced hematopoietic cells was the result of transduction of HSC/progenitors and to establish whether this antibody could identify long-term repopulating HSC/progenitors in vivo.
Mobilization of chimeric and control sheep with granulocyte–colony-stimulating factor
We have previously demonstrated that administration of human G-CSF (Neupogen) to sheep results in effective mobilization, as evidenced by a rapid rise on white blood cell count [12]. In the present studies, we wished to assess whether administration of G-CSF would mobilize HSC/progenitors identifiable with 8D11. As in our prior studies, administration of G-CSF resulted in a rapid rise in total white blood cell count from 1.4×106 cells/ml at day 0 to 12.8×106 cells/ml by day 2. While low levels of CD45+CD34+ cells (as identified by 8D11) were observed in the peripheral blood at day 0 (2.24%), by day 2 of mobilization, the levels of CD45+CD34+ cells had risen 6.2-fold to 13.83%. Even more striking was the rise in absolute numbers of CD45+CD34+ cells in the circulation as a result of G-CSF mobilization. After taking into account the rise in total white blood cell count, the levels of CD45+CD34+ cells in circulation increased from 31×104 cells/ml at day 0 to 1.77×106 cells/ml by day 2 of mobilization, representing a 56-fold increase in the absolute numbers of CD45+CD34+ cells in circulation. By day 4, levels had decreased back to 4.79% which, due to the increased levels of total white blood cells, still represented roughly 27-fold higher absolute numbers of CD34+ cells/ml of PB than at day 0 prior to mobilization. In similarity to what is seen in clinical patients undergoing mobilization with G-CSF, we also observed a modest 1.7-fold increase in the percentage of CD34+ cells within the bone marrow of sheep receiving G-CSF, with this rise occurring at day 4. These studies thus demonstrate that G-CSF efficiently mobilizes CD34+ HSC/progenitors in sheep, and provides further confirmation that clone 8D11 allows identification of primitive HSC/progenitors in vivo. Figure 4C shows a representative dot plot obtained after staining normal sheep peripheral blood with clone 8D11 prior to and at two days following mobilization with G-CSF.
Ability of anti-ovine CD34 monoclonal antibodies to identify endothelial cells within sheep tissue sections
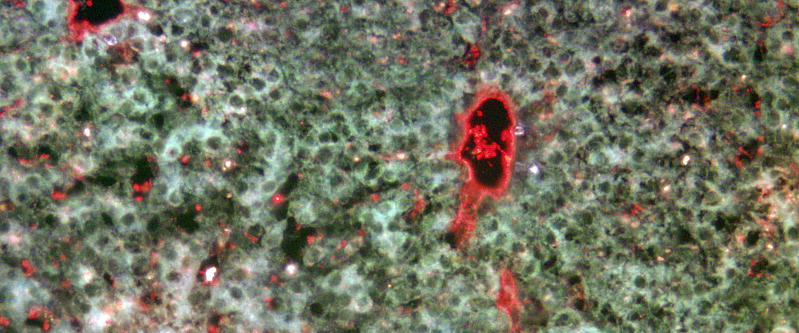
In addition to hematopoietic stem/progenitor cells, microvascular endothelial cells are one of the main cell types within the body that express significant levels of the CD34 antigen[51]. Having demonstrated that the anti-ovine CD34 monoclonal antibodies were able to identify and allow selection of hematopoietic stem/progenitor cells, we next investigated whether these antibodies could also identify microvascular endothelial cells within formalin-fixed, paraffin-embedded sheep tissue blocks. To accomplish this objective, tissue sections were prepared from the liver of normal healthy adult sheep and stained with one of the anti-ovine CD34 antibodies (clone 8D11). The signal was then detected by incubating the sections with anti-mouse IgG-PE to assess whether this antibody could also be used to identify endothelial cells within this context. As can be seen in Figure 7, this antibody robustly labeled the lining of the hepatic vessels, and some hematopoietic cells within the vessels as well, providing compelling evidence that the epitope recognized by this clone is preserved upon formalin fixation and paraffin embedding, and further extending the utility of this antibody to include analysis of paraffin-embedded tissue sections.
Figure 7. Anti-ovine CD34 antibody detects endothelial cells in formalin-fixed paraffin-embedded tissue sections.
In order to assess whether anti-ovine CD34 antibodies could also be used to identify endothelial cells, tissue sections prepared from the liver of normal healthy adult sheep were stained with the anti-ovine CD34 antibody, clone 8D11, and the signal was detected with anti-mouse IgG-PE. Low level autofluorescence in the FITC channel was used to view tissue morphology/architecture. Image captured with 60X objective.
Discussion
Sheep possess several characteristics that make them well-suited as a model system in which to explore stem cell transplantation including their large size, their relatively long life span, the fact that they are outbred, allowing far more genetic variation, much like what is seen within human populations, and the fact that they share many important physiological and developmental characteristics with humans [1-10], [12, 15, 29, 30, 32, 39]. Unfortunately, while numerous markers are available to identify and isolate primitive human HSC, no reagents exist that allow hematopoietic stem and progenitor cells from sheep to be identified or purified for transplantation studies. This dearth of HSC/progenitor reagents has greatly impeded the application of this valuable large animal model system to the study of autologous or allogeneic HSC transplantation. In the present studies, we used PCR to clone a portion of the extracellular domain of the ovine CD34 molecule, using primers based on the publicly available bovine sequence, given the lack of sequence data on the sheep homolog. Sequence analysis at the nucleotide and protein level revealed a fair degree of homology to the human CD34 molecule at both levels, but, not surprisingly, a much greater level of homology to the bovine molecule.
Having established the degree of homology at the sequence level, we next determined whether the ovine CD34 molecule possessed homology to its human counterpart at the functional level. Our in vitro colony-forming assays revealed that sorting for the CD34+ fraction with the 8D11 clone resulted in a substantial enrichment for primitive hematopoietic cells with colony-forming potential at both the CFU-GM level and at the level of much more primitive CAFC. Furthermore, these analyses demonstrated that essentially all of the cells with colony-forming potential were contained within the fraction of cells labeling positively with 8D11, since the negative flow-through fraction obtained from either fresh or frozen sheep BM was incapable of forming colonies in vitro.
Further confirmation of the ability of this clone to identify functional HSC/progenitors came from our studies in which this antibody was used to sort CD34+ cells from the bone marrow of animals that had received in utero gene transfer by direct intraperitoneal vector injection prior to commencing the present studies [48-50]. These animals had consistently exhibited transgene positivity within the bone marrow and peripheral blood throughout the course of these other ongoing studies. In the present study, we used 8D11 to sort CD34+ cells from these sheep and analyze this population of cells for the presence of the vector-encoded NeoR gene by PCR. These studies demonstrated that the two animals analyzed both contained NeoR+ cells within the CD34+ population recognized by 8D11, thus providing evidence that this clone does recognize HSC/progenitors with long-term in vivo engraftment capability. This was further confirmed by studies in which we showed that G-CSF mobilization of sheep resulted in a 56-fold increase in the absolute levels of circulating CD34+ cells by day 2 of mobilization. It is important to note that the true gold standard for proving that our CD34 antibody recognizes true long-term engrafting HSC in sheep would be to perform experiments in which CD34+ cells are transplanted into sheep recipients and assessed for their ability to provide durable hematopoietic repopulation. Unfortunately, while experiments of this nature are simple and can be performed quickly in small animal models such as mice, in the sheep model, in similarity to other large animals or humans, long-term repopulation experiments would likely require an additional 8-12 months. Thus, for this initial report on the creation and characterization of the antibody to sheep CD34, we used the retroviral marking study as an indirect means of assessing the ability of this antibody to identify HSC, or at least very long-lived progenitors in vivo.
Antibodies to the human CD34 antigen have been classified into class I, II and III antibodies based on CD34 glycosylation. Unfortunately, we do not yet know whether this same class system will apply to the sheep CD34 molecule and, if so, into which class our antibody will fall, although we do know that our antibody does not recognize the human CD34 antigen (data not shown). This lack of species cross-reactivity is perhaps not surprising, given that commercially available antibodies to human CD34 do not recognize the ovine counterpart. With respect to classification of the anti-ovine CD34 antibody, we presume that since the cDNA was injected into mice, it is most likely that our antibody would fall into class III, since glycosylation in mouse is likely different from sheep. However, to answer this question definitively, future studies will be required in which the ovine CD34 antigen is deglycosylated with various enzymes and the mAb tested for retention or loss of reactivity.
In conclusion, this first successful generation of a monoclonal antibody to the sheep CD34 antigen and proof that this antibody labels primitive HSC/progenitors in vivo engrafting potential will greatly facilitate the use of the well established sheep model to study allogeneic HSC transplantation both in utero and in post-natal recipients using bone marrow, mobilized peripheral blood, and cord blood as cell sources.
Acknowledgments
This work was supported by Grant Nos. HD043038, HL52955 and HL49042 from the National Institutes of Health.
Footnotes
Author Contributions:
CDP: Conception and design, financial support, collection, analysis, and interpretation of data, and writing of manuscript
DDH-F and PJS: Provision of study materials and collection/interpretation of data
CS and VV: Collection and assembly of data
DT: Other: Provided veterinary care for sheep
GA-P: Conception and design, financial support, collection, analysis, and interpretation of data
EDZ: Financial support, administrative support, final approval of manuscript
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barry JS, Anthony RV. The pregnant sheep as a model for human pregnancy. Theriogenology. 2008;69:55–67. doi: 10.1016/j.theriogenology.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter AM. Animal models of human placentation--a review. Placenta. 2007;28 Suppl A:S41–47. doi: 10.1016/j.placenta.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Donahue RE, Kuramoto K, Dunbar CE. Large animal models for stem and progenitor cell analysis. In: Coligan John E, et al., editors. Current protocols in immunology. Chapter 22. 2005. p. Unit 22A 21. [DOI] [PubMed] [Google Scholar]
- 4.Jenkin G, Young IR. Mechanisms responsible for parturition; the use of experimental models. Animal reproduction science. 2004:82–83. 567–581. doi: 10.1016/j.anireprosci.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Jenne CN, Kennedy LJ, Reynolds JD. Antibody repertoire development in the sheep. Developmental and comparative immunology. 2006;30:165–174. doi: 10.1016/j.dci.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Pearce AI, Richards RG, Milz S, Schneider E, Pearce SG. Animal models for implant biomaterial research in bone: a review. European cells & materials. 2007;13:1–10. doi: 10.22203/ecm.v013a01. [DOI] [PubMed] [Google Scholar]
- 7.Rabbani S, Ahmadi H, Fayazzadeh E, et al. Development of an ovine model of myocardial infarction. ANZ journal of surgery. 2008;78:78–81. doi: 10.1111/j.1445-2197.2007.04359.x. [DOI] [PubMed] [Google Scholar]
- 8.Scheerlinck JP, Snibson KJ, Bowles VM, Sutton P. Biomedical applications of sheep models: from asthma to vaccines. Trends in biotechnology. 2008;26:259–266. doi: 10.1016/j.tibtech.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Wallace JM, Regnault TR, Limesand SW, Hay WW, Jr, Anthony RV. Investigating the causes of low birth weight in contrasting ovine paradigms. The Journal of physiology. 2005;565:19–26. doi: 10.1113/jphysiol.2004.082032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noia G, Romano D, Terzano GM, et al. Ovine fetal growth curves in twin pregnancy: ultrasonographic assessment. Clinical and experimental obstetrics & gynecology. 2002;29:251–256. [PubMed] [Google Scholar]
- 11.Almeida-Porada G, Flake AW, Glimp HA, Zanjani ED. Cotransplantation of stroma results in enhancement of engraftment and early expression of donor hematopoietic stem cells in utero. Experimental hematology. 1999;27:1569–1575. doi: 10.1016/s0301-472x(99)00090-9. [DOI] [PubMed] [Google Scholar]
- 12.Almeida-Porada G, Porada C, Gupta N, Torabi A, Thain D, Zanjani ED. The human-sheep chimeras as a model for human stem cell mobilization and evaluation of hematopoietic grafts' potential. Experimental hematology. 2007;35:1594–1600. doi: 10.1016/j.exphem.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roodman GD, Zanjani ED. Endogenous erythroid colony-forming cells in fetal and newborn sheep. The Journal of laboratory and clinical medicine. 1979;94:699–707. [PubMed] [Google Scholar]
- 14.Troeger C, Surbek D, Schoberlein A, et al. In utero haematopoietic stem cell transplantation. Experiences in mice, sheep and humans. Swiss Med Wkly. 2006;136:498–503. doi: 10.4414/smw.2006.11380. [DOI] [PubMed] [Google Scholar]
- 15.Zanjani ED, Almeida-Porada G, Flake AW. The human/sheep xenograft model: a large animal model of human hematopoiesis. International journal of hematology. 1996;63:179–192. doi: 10.1016/0925-5710(96)00445-8. [DOI] [PubMed] [Google Scholar]
- 16.Jones DR, Anderson EM, Liu DT, Walker RM. Tolerance induction following in utero stem cell transplantation. Ernst Schering Research Foundation workshop. 2001:187–196. doi: 10.1007/978-3-662-04469-8_12. [DOI] [PubMed] [Google Scholar]
- 17.Liechty KW, MacKenzie TC, Shaaban AF, et al. Human mesenchymal stem cells engraft and demonstrate site-specific differentiation after in utero transplantation in sheep. Nature medicine. 2000;6:1282–1286. doi: 10.1038/81395. [DOI] [PubMed] [Google Scholar]
- 18.Mackenzie TC, Flake AW. Human mesenchymal stem cells persist, demonstrate site-specific multipotential differentiation, and are present in sites of wound healing and tissue regeneration after transplantation into fetal sheep. Blood cells, molecules & diseases. 2001;27:601–604. doi: 10.1006/bcmd.2001.0424. [DOI] [PubMed] [Google Scholar]
- 19.Mackenzie TC, Flake AW. Multilineage differentiation of human MSC after in utero transplantation. Cytotherapy. 2001;3:403–405. doi: 10.1080/146532401753277571. [DOI] [PubMed] [Google Scholar]
- 20.Michelini M, Papini S, Rosellini A, et al. Prolonged human/sheep cellular chimerism following transplantation of human hemopoietic stem cells into the ewe celomic cavity. The International journal of developmental biology. 2008;52:365–370. doi: 10.1387/ijdb.072362mm. [DOI] [PubMed] [Google Scholar]
- 21.Noia G, Ligato MS, Cesari E, et al. Source of cell injected is a critical factors for short and long engraftment in xeno-transplantation. Cell proliferation. 2008;41 Suppl 1:41–50. doi: 10.1111/j.1365-2184.2008.00481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noia G, Pierelli L, Bonanno G, et al. A novel route of transplantation of human cord blood stem cells in preimmune fetal sheep: the intracelomic cavity. Stem cells (Dayton, Ohio) 2003;21:638–646. doi: 10.1634/stemcells.21-6-638. [DOI] [PubMed] [Google Scholar]
- 23.Noia G, Pierelli L, Bonanno G, et al. The intracoelomic route: a new approach for in utero human cord blood stem cell transplantation. Fetal diagnosis and therapy. 2004;19:13–22. doi: 10.1159/000074254. [DOI] [PubMed] [Google Scholar]
- 24.Schoeberlein A, Holzgreve W, Dudler L, Hahn S, Surbek DV. In utero transplantation of autologous and allogeneic fetal liver stem cells in ovine fetuses. American journal of obstetrics and gynecology. 2004;191:1030–1036. doi: 10.1016/j.ajog.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 25.Schoeberlein A, Holzgreve W, Dudler L, Hahn S, Surbek DV. Tissue-specific engraftment after in utero transplantation of allogeneic mesenchymal stem cells into sheep fetuses. American journal of obstetrics and gynecology. 2005;192:1044–1052. doi: 10.1016/j.ajog.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 26.Surbek DV, Holzgreve W. Prenatal transplantation of hematopoietic stem cells: overview. Ernst Schering Research Foundation workshop. 2001:115–122. doi: 10.1007/978-3-662-04469-8_8. [DOI] [PubMed] [Google Scholar]
- 27.Surbek DV, Holzgreve W, Nicolaides KH. Haematopoietic stem cell transplantation and gene therapy in the fetus: ready for clinical use? Human reproduction update. 2001;7:85–91. doi: 10.1093/humupd/7.1.085. [DOI] [PubMed] [Google Scholar]
- 28.Young AJ, Holzgreve W, Dudler L, Schoeberlein A, Surbek DV. Engraftment of human cord blood-derived stem cells in preimmune ovine fetuses after ultrasound-guided in utero transplantation. American journal of obstetrics and gynecology. 2003;189:698–701. doi: 10.1067/s0002-9378(03)00716-6. [DOI] [PubMed] [Google Scholar]
- 29.Civin CI, Almeida-Porada G, Lee MJ, Olweus J, Terstappen LW, Zanjani ED. Sustained, retransplantable, multilineage engraftment of highly purified adult human bone marrow stem cells in vivo. Blood. 1996;88:4102–4109. [PubMed] [Google Scholar]
- 30.Shimizu Y, Ogawa M, Kobayashi M, Almeida-Porada G, Zanjani ED. Engraftment of cultured human hematopoietic cells in sheep. Blood. 1998;91:3688–3692. [PubMed] [Google Scholar]
- 31.Giesert C, Almeida-Porada G, Scheffold A, Kanz L, Zanjani ED, Buhring HJ. The monoclonal antibody W7C5 defines a novel surface antigen on hematopoietic stem cells. Annals of the New York Academy of Sciences. 2001;938:175–183. doi: 10.1111/j.1749-6632.2001.tb03587.x. [DOI] [PubMed] [Google Scholar]
- 32.Lewis ID, Almeida-Porada G, Du J, et al. Umbilical cord blood cells capable of engrafting in primary, secondary, and tertiary xenogeneic hosts are preserved after ex vivo culture in a noncontact system. Blood. 2001;97:3441–3449. doi: 10.1182/blood.v97.11.3441. [DOI] [PubMed] [Google Scholar]
- 33.Krause DS, Fackler MJ, Civin CI, May WS. CD34: structure, biology, and clinical utility. Blood. 1996;87:1–13. [PubMed] [Google Scholar]
- 34.Stella CC, Cazzola M, De Fabritiis P, et al. CD34-positive cells: biology and clinical relevance. Haematologica. 1995;80:367–387. [PubMed] [Google Scholar]
- 35.Civin CI, Strauss LC, Brovall C, Fackler MJ, Schwartz JF, Shaper JH. Antigenic analysis of hematopoiesis. III. A hematopoietic progenitor cell surface antigen defined by a monoclonal antibody raised against KG-1a cells. J Immunol. 1984;133:157–165. [PubMed] [Google Scholar]
- 36.Koeffler HP, Billing R, Lusis AJ, Sparkes R, Golde DW. An undifferentiated variant derived from the human acute myelogenous leukemia cell line (KG-1) Blood. 1980;56:265–273. [PubMed] [Google Scholar]
- 37.Matsuoka S, Ebihara Y, Xu M, et al. CD34 expression on long-term repopulating hematopoietic stem cells changes during developmental stages. Blood. 2001;97:419–425. doi: 10.1182/blood.v97.2.419. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa M, Tajima F, Ito T, Sato T, Laver JH, Deguchi T. CD34 expression by murine hematopoietic stem cells. Developmental changes and kinetic alterations. Annals of the New York Academy of Sciences. 2001;938:139–145. doi: 10.1111/j.1749-6632.2001.tb03583.x. [DOI] [PubMed] [Google Scholar]
- 39.Verfaillie CM, Almeida-Porada G, Wissink S, Zanjani ED. Kinetics of engraftment of CD34(-) and CD34(+) cells from mobilized blood differs from that of CD34(-) and CD34(+) cells from bone marrow. Experimental hematology. 2000;28:1071–1079. doi: 10.1016/s0301-472x(00)00506-3. [DOI] [PubMed] [Google Scholar]
- 40.Zanjani ED, Almeida-Porada G, Livingston AG, Flake AW, Ogawa M. Human bone marrow CD34- cells engraft in vivo and undergo multilineage expression that includes giving rise to CD34+ cells. Experimental hematology. 1998;26:353–360. [PubMed] [Google Scholar]
- 41.Zanjani ED, Almeida-Porada G, Livingston AG, Zeng H, Ogawa M. Reversible expression of CD34 by adult human bone marrow long-term engrafting hematopoietic stem cells. Experimental hematology. 2003;31:406–412. doi: 10.1016/s0301-472x(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 42.Berenson RJ, Andrews RG, Bensinger WI, et al. Antigen CD34+ marrow cells engraft lethally irradiated baboons. The Journal of clinical investigation. 1988;81:951–955. doi: 10.1172/JCI113409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kantoff PW, Flake AW, Eglitis MA, et al. In utero gene transfer and expression: a sheep transplantation model. Blood. 1989;73:1066–1073. [PubMed] [Google Scholar]
- 44.Ploemacher RE, van der Sluijs JP, van Beurden CA, Baert MR, Chan PL. Use of limiting-dilution type long-term marrow cultures in frequency analysis of marrow-repopulating and spleen colony-forming hematopoietic stem cells in the mouse. Blood. 1991;78:2527–2533. [PubMed] [Google Scholar]
- 45.Ploemacher RE, van der Sluijs JP, Voerman JS, Brons NH. An in vitro limiting-dilution assay of long-term repopulating hematopoietic stem cells in the mouse. Blood. 1989;74:2755–2763. [PubMed] [Google Scholar]
- 46.van der Sluijs JP, de Jong JP, Brons NH, Ploemacher RE. Marrow repopulating cells, but not CFU-S, establish long-term in vitro hemopoiesis on a marrow-derived stromal layer. Experimental hematology. 1990;18:893–896. [PubMed] [Google Scholar]
- 47.Fina L, Molgaard HV, Robertson D, et al. Expression of the CD34 gene in vascular endothelial cells. Blood. 1990;75:2417–2426. [PubMed] [Google Scholar]
- 48.Porada CD, Park P, Almeida-Porada G, Zanjani ED. The sheep model of in utero gene therapy. Fetal diagnosis and therapy. 2004;19:23–30. doi: 10.1159/000074255. [DOI] [PubMed] [Google Scholar]
- 49.Porada CD, Tran N, Eglitis M, et al. In utero gene therapy: transfer and long-term expression of the bacterial neo(r) gene in sheep after direct injection of retroviral vectors into preimmune fetuses. Human gene therapy. 1998;9:1571–1585. doi: 10.1089/hum.1998.9.11-1571. [DOI] [PubMed] [Google Scholar]
- 50.Tran ND, Porada CD, Zhao Y, Almeida-Porada G, Anderson WF, Zanjani ED. In utero transfer and expression of exogenous genes in sheep. Experimental hematology. 2000;28:17–30. doi: 10.1016/s0301-472x(99)00133-2. [DOI] [PubMed] [Google Scholar]
- 51.Pusztaszeri MP, Seelentag W, Bosman FT. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J Histochem Cytochem. 2006;54:385–395. doi: 10.1369/jhc.4A6514.2005. [DOI] [PubMed] [Google Scholar]