Abstract
Epidemiological models generally assume that the number of susceptible individuals that become infected within a unit of time depends on the density of the hosts and the concentration of parasites (i.e. mass-action principle). However, empirical studies have found significant deviations from this assumption due to biotic and abiotic factors, such as seasonality, the spatial structure of the host population and host heterogeneity with respect to immunity and susceptibility. In this paper, we examine the effect of the dose level of the bacterial endoparasite Pasteuria ramosa on the infection rate of its host, the water flea Daphnia magna. Using seven host clones and two parasite isolates, we measure the fraction of infected hosts after exposure to eight different parasite doses to determine whether there is variation in the infection process across different host clone–parasite isolate combinations. In five combinations, a pronounced dose-dependent infection pattern was found. Using a likelihood approach, we compare the infection data of these five combinations to the fit of three mathematical models: a mass-action model, a parasite antagonism model (i.e. an increase in the parasite dose leads to an under-proportionate increase in the infection rate per host) and a heterogeneous host model. We found that the host heterogeneity model, in which we assumed the existence of non-inherited phenotypic differences in host susceptibilities to the parasite, provides the best fit. Our analysis suggests that among 5 out of the 14 host clone–parasite isolate combinations that resulted in appreciable infections, non-genetic host heterogeneity plays an important role.
Keywords: Daphnia magna, dose–response curve, frailty model, mass-action infection, Pasteuria ramosa
1. Introduction
Parasites and pathogens play a vital role in the regulation (Anderson & May 1979) and coevolution (Jaenike 1978; Hamilton 1980) of their host populations. Understanding and modelling the infection dynamics of host–parasite systems will consequently be of utter importance, if we are to make predictions about the spread of infectious diseases. Although the transmission term (i.e. the number of susceptible individuals that become infected per time unit) is often assumed to depend on the density of the hosts and the concentration of the parasites (mass-action principle of chemical kinetics, Hamer 1906), many studies have found significant deviations from the mass-action assumption due to, for example, differential susceptibility to the pathogen (Knell et al. 1998) and behavioural and physiological changes at high host density (D'amico et al. 1996).
In an attempt to reconcile modelled transmission term data and real data, several non-exclusive explanations have been suggested (reviewed in Deredec & Courchamp 2003). For instance, in the case of vector-borne and sexually transmitted diseases, the number of conspecifics encountered by a contagious individual may depend on the proportion of infected hosts in the population, yielding a frequency-dependent transmission term (also called proportionate mixing, e.g. Dietz & Schenzle 1985). Even if the transmission of a given parasite is consistent with the mass-action principle, certain biotic (e.g. genetic, physiological) and abiotic (e.g. seasonality, Altizer et al. 2006) aspects of the host may influence the interaction. For example, the host population dynamics and spatial structure (e.g. habitat heterogeneity, metapopulation) substantially affect parasite transmission, thereby violating the assumption of a homogeneously (i.e. randomly) mixed system (Grosholz 1993; Burdon et al. 1995; Fromont et al. 1998; Lopez et al. 2005). Additionally, the host's immune system may buffer against the initial increase in parasite abundance up to a certain point, depending on the (genetically based) immunological heterogeneity of the host and the initial parasite inoculum (Antia et al. 1994; Antia & Lipsitch 1997; Leite et al. 2000).
In host–parasite systems with free parasite stages, one can test the dependence of the infection rate on the density of the free parasite stages by exposing hosts to different doses of the parasite. In most models of these systems, parasite dose density is a key variable, as evident from infection experiments involving both macroparasites (Ashworth et al. 1996; Osnas & Lively 2004) and microparasites (Ebert et al. 2000b; Little & Ebert 2000; Regoes et al. 2003; Hughes et al. 2004; Brunner et al. 2005). In a previous study, we investigated how the infection rate of Daphnia magna depends on the dose of its parasite Pasteuria ramosa (Regoes et al. 2003). We found that, although the infection process largely conformed to the mass-action principle, there was a statistically significant deviation from a mass-action infection term that might be explained by inter-individual, non-genetic variation in the hosts' susceptibility. In that first study, however, we focused on one clone of D. magna and one isolate of P. ramosa, thereby controlling for genetic variation in host susceptibility.
In the present study, we extend our previous analysis to the combinations of seven D. magna clones and two P. ramosa isolates to determine whether there is a significant difference in the infection process across different host clone–parasite isolate combinations. We hypothesize that genetic variations among hosts and parasites may result in further significant deviations from a mass-action infection term.
2. Material and methods
(a) Host–parasite system
The cladoceran D. magna Straus is a cyclical parthenogenetic zooplankter that inhabits eutrophic shallow ponds in Eurasia. Sexual reproduction is triggered environmentally (Kleiven et al. 1992), but maternal and environmental effects can be controlled in the laboratory. Daphnia magna is naturally infected by a wide variety of bacterial, microsporidial and fungal parasites (Green 1974), with detrimental impact on host fitness (Stirnadel & Ebert 1997; Ebert et al. 2000b; Ebert 2005). Age at first reproduction is approximately 10 days, and the lifespan of uninfected Daphnia is up to 100 days, whereas parasitized individuals have considerably shorter lifespans (Ebert et al. 2000a).
Pasteuria ramosa Metchnikoff 1888 is a bacterial obligate endoparasite of Daphnia (Ebert et al. 1996; Ebert 2005). Transmission occurs through contact between the host and the water-borne spores, and probably requires the filter-feeding host to ingest the endospores. Following infection, the bacterium grows in the body cavity of its host, causing sterilization; ultimately, millions of endospores are released from the decaying cadaver of an infected host (Ebert et al. 1996). Castration can occur before the host produces any clutches, and mature spores are found in the host approximately 20 days following infection.
(b) Experimental setup
The Daphnia clones used in the experiment were isolated from several ponds in Europe: Belgium (B2 and M10), Gaarzerfeld in North Germany (DG), Hungary (HO1 and HO2) and Munich, Germany (Mu10 and Mu11). Each clone was generated by isolating parthenogenetic eggs from the brood chamber of an adult female and raising the clonal offspring in isolation. Following isolation and prior to the experiment, all clones were kept in the laboratory under standardized conditions. In preparation for the experiment, seven Daphnia clones were stock cultured in 400 ml glass beakers (12 beakers per clone each containing eight individuals) with artificial medium (Klüttgen et al. 1994; modified as per Ebert et al. 1998), where they were fed daily with approximately 1.5×105 cells ml−1 medium of the chemostat-cultured unicellular algae Scenedesmus gracilis. We used offspring from the third generation of these isofemale lines.
We placed 4-day-old juveniles singly in 100 ml jars with 20 ml of artificial medium in combination with seven Daphnia clones×two Pasteuria isolates×eight dose levels as well as a control treatment without parasites for each Daphnia clone. We used 14 replicates per treatment, making a total of 1666 individuals for the experiment. The temperature was 20±0.5°C and the individuals were kept with a cycle of 16 hours L : 8 hours D. All the treatments were evenly distributed across two incubators and randomly shuffled within the drawers of each incubator.
The Pasteuria isolates used to infect the Daphnia were originally obtained from infected hosts from Gaarzerfeld, Germany (P1) and Belgium (P4). In the case of P1, we started with one single infected female and in the case of P4 with 10 infected females. These hosts were well fed until they died, upon which their parasite spores were used to propagate infection via the generally susceptible host clone HO2. This process was repeated three times until there were enough spore-carrying cadavers for infection. We prepared spore solutions by grinding approximately 180 and 200 HO2 cadavers infected with P1 and P4, respectively. All cadavers were carefully homogenized, and the density of the initial spore concentrations was determined using a haemocytometer (Thoma ruling). The highest spore concentration for a dose treatment was 6 250 000 spores in 20 ml. This dose was subsequently diluted seven times by a factor of five to produce each of the next lower concentrations, resulting in eight dose levels from 80 to 6 250 000 spores (4–312 500 spores ml−1).
All animals except controls were exposed to their respective Pasteuria dose on day 5 and were initially fed approximately 1×106 algae cells per Daphnia per day. A week after infection we added 80 ml of fresh medium to each jar; thereafter, medium was replaced on a weekly basis. To accommodate the animal's growing demand for food, we increased the daily food level on days 10, 14, 17, 22, 27 and 32 to 2×106, 3×106, 5×106, 6×106, 7.5×106 and 10×106 algae cells per individual per day, respectively. Dead animals were recorded daily, but only animals that died after day 14 were dissected and checked for disease. Animals that died earlier could not be reliably checked for infection and were thus excluded from the analysis. On day 44, all remaining animals were scored for infection based on colour (infected animals lose their typical transparency and are brownish-reddish) and lack of eggs. When in doubt, we dissected the animal and checked for infection under a phase contrast microscope, but we found no discrepancies with our initial diagnosis.
(c) Mathematical models
We fit three mathematical models to the infection data of our experiment using a likelihood approach. We transformed previous mathematical models (Regoes et al. 2003) into standard survival problems, where ‘survival’ means to remain uninfected. We derived expressions for the expected relationship between the fraction of uninfected hosts and the parasite dose to which the hosts were exposed. This approach is similar to the one adopted by mathematical epidemiologists who investigate vaccine efficacies (e.g. Halloran et al. 1996; Longini & Halloran 1996).
In the mass-action infection model, we assume that the hazard λ of ‘dying’, or becoming infected, is proportional at any moment to the infection rate b and the parasite concentration P at that time,
| (2.1) |
The cumulative hazard Λ to become infected during the exposure time texp is
| (2.2) |
The integral simplifies because we assume that the parasite concentration is constant during the time of exposure texp. We ignore the death rate of the parasite during exposure, since P. ramosa spores can survive several decades (Decaestecker et al. 2004).
The population survival function is given by S=e−Λ, which, for the dose level j, expands to
| (2.3) |
A similar derivation shows the population survival function for the parasite antagonism model
| (2.4) |
An antagonism effect of the parasite spores implies that k<1, that is, an increase in the parasite inoculum will lead to an under-proportionate increase in the infection rate per host. Similarly, k>1 would describe a synergistic relationship among the parasite spores, whereby an increase in the parasite dose causes an over-proportionate increase in the infection rate per host; for k=1, equation (2.4) is equivalent to the survival function of the mass-action model in equation (2.3).
Although individuals from each D. magna clone are genetically identical, they may differ phenotypically in their susceptibility to infection. As a result, highly susceptible hosts will be infected first, that is, at low parasite doses, leading to disproportionately more infections at low doses. On the other hand, as the parasite dose increases, the pool of susceptible hosts is exhausted, and it becomes more difficult to infect the remaining hosts. These dynamics are described by the following population survival function:
| (2.5) |
This survival function for the heterogeneous host model assumes that the susceptibilities of the hosts are Γ-distributed with a mean and a variance . This is a standard extension of simple survival models referred to as frailty models (Hougaard 1986; Aalen 1988).
The likelihood function for all three models can be defined as,
| (2.6) |
Here, Sj denotes the survival function given by equations (2.3), (2.4) or (2.5). The binomial coefficients are omitted because they only scale the likelihood in a manner independent of the parameters to be estimated. The optimization of the likelihood is technically easier done on its logarithm, the log likelihood,
| (2.7) |
The likelihood functions defined above were programmed in the R language (R Development Core Team 2005), and maximum-likelihood estimators of the model parameters were obtained using the functions optimize (for the mass-action infection model) and optim with the default method ‘Nelder–Mead’ (for the parasite antagonism and heterogeneous host models). Standard errors of the maximum-likelihood estimators were obtained using the Fisher information
| (2.8) |
Hereby, p denotes any model parameter. The derivatives of the log likelihood, l, were calculated using the computer algebra system Maxima (de Souza et al. 2004).
3. Results
(a) Infection experiment
We exposed each of seven D. magna clones to two P. ramosa isolates in eight different dose levels and recorded infection prevalence. During the first two weeks, we observed approximately 10% unexplained host mortality across all treatments including controls. This mortality, however, did not differ between treatments and controls. In particular, there was no increase in the mortality rate with the parasite dose.
The outcome of the infection experiment with the Daphnia–Pasteuria combinations can be grouped into three categories (table 1). First, some Daphnia clones were found to be uninfected, even when exposed to the highest parasite spore concentration. For instance, B2, HO1, Mu10 and Mu11 never became infected by P1, and DG and Mu11 never became infected by P4. Second, other Daphnia clones were found to be relatively resistant to all but the two or three highest doses, e.g. B2, HO1 and Mu10 were largely resistant to P4. However, even at these high doses, the percentages of infection were mostly low. Therefore, these specific Daphnia–Pasteuria combinations could not be further analysed due to insufficient data points. Third, the remaining five combinations resulted in appreciable infections in all but the lowest dose level. These five combinations were used to fit the mass-action infection model, the parasite antagonism model and the heterogeneous host model.
Table 1.
Percentage of infected individuals at each dose level. (All control treatments remained uninfected for all host clone–parasite isolate combinations. Cells marked in italics represent entries for those combinations which were included into the mathematical analysis.)
| Daphnia clone | |||||||
|---|---|---|---|---|---|---|---|
| dose | HO2 | M10 | DG | Mu10 | B2 | HO1 | Mu11 |
| P1 | |||||||
| 80 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 400 | 29 | 0 | 22 | 0 | 0 | 0 | 0 |
| 2000 | 27 | 23 | 33 | 0 | 0 | 0 | 0 |
| 10 000 | 11 | 46 | 90 | 0 | 0 | 0 | 0 |
| 50 000 | 75 | 92 | 83 | 0 | 0 | 0 | 0 |
| 250 000 | 83 | 93 | 83 | 0 | 0 | 0 | 0 |
| 1 250 000 | 100 | 92 | 91 | 0 | 0 | 0 | 0 |
| 6 250 000 | 100 | 100 | 92 | 0 | 0 | 0 | 0 |
| P4 | |||||||
| 80 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 400 | 9 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2000 | 43 | 36 | 0 | 0 | 0 | 0 | 0 |
| 10 000 | 64 | 54 | 0 | 0 | 0 | 0 | 0 |
| 50 000 | 85 | 86 | 0 | 0 | 0 | 0 | 0 |
| 250 000 | 100 | 79 | 0 | 9 | 0 | 0 | 0 |
| 1 250 000 | 93 | 100 | 0 | 38 | 15 | 0 | 0 |
| 6 250 000 | 100 | 100 | 0 | 85 | 14 | 10 | 0 |
(b) Fitting of infection models
First, we fit our three models to the pooled data of the five successful combinations, disregarding variation across different host–parasite combinations. By fitting our models to the pooled data, we do not presuppose that the specific host–parasite combinations are irrelevant. On the contrary, fitting the data pooled across host–parasite combinations is a necessary null model that allows us to determine whether there are differences between combinations in the infection parameters (see below). We found that the log likelihood of the mass-action infection model is −374.5, whereas this value is much higher for the parasite antagonism and host heterogeneity models (table 2). We compared the fits statistically with a chi-squared test. In this test, one calculates the statistic −2(l(Model 1)−l(Model 2)) and tests if this statistic conforms with a chi-squared distribution, whose degrees of freedom are equal to the difference in the number of parameters of the two models being compared (d.f.=1 in our case). We found that both the parasite antagonism model (likelihood=−185.1) and the host heterogeneity model (likelihood=−173.0) fit significantly better than the mass-action infection model (Pr<10−16 in both cases). The likelihood of the host heterogeneity model is the highest, but we cannot compare it with the parasite antagonism model using a chi-squared test because neither model is an extension of the other (because the log likelihood of the host heterogeneity model is higher by 12 than that of the parasite antagonism model, and both the models have the same number of parameters, alternative methods of comparison, such as the Akaike Information Criterion, would favour the host heterogeneity model).
Table 2.
Model fits.
| infection model | parameter estimatesa | log likelihood, l |
|---|---|---|
| mass-action | b=(2.12±0.17)×10−5 ml d−1 | −374.5 |
| parasite antagonism | −185.1 | |
| heterogeneous host | −173.0 |
Maximum-likelihood estimates±s.e.
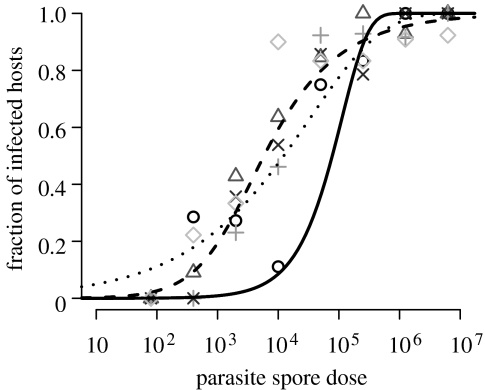
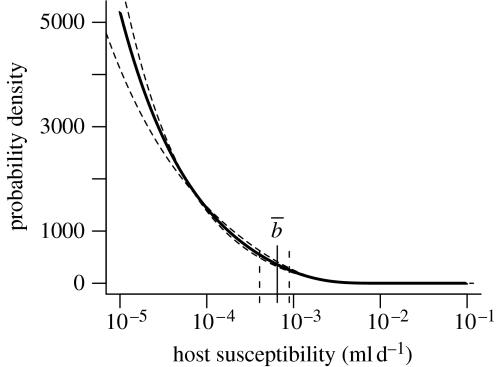
The maximum-likelihood estimates of the parameters and their standard errors are listed in table 2. The estimated infection probability is lower than the one we found previously (Regoes et al. 2003), a difference we elaborate on below. We can calculate the ID50 (i.e. the infectious dose at which 50% of the animals become infected) from the estimates of the heterogeneous host model, . In figure 1, we show the fit of the host heterogeneity model to the data. In figure 2, we plot the distribution of host susceptibilities which is most consistent with our data. The mean susceptibility of the host population is the probability that a given D. magna clone becomes infected by either Pasteuria isolates (P1 or P4), as long as this Daphnia clone is susceptible. Figure 2 also provides a 95% CI for the parameters and v.
Figure 1.
Plot of the proportion of infected hosts in relation to the parasite spore dose used. Each symbol represents a host–parasite combination (circles, HO2+P1; triangles, HO2+P4; pluses, M10+P1; crosses, M10+P4 and diamonds, DG+P1). The solid, dotted and dashed lines represent the fits of the mass-action infection, parasite antagonism and heterogeneous host models, respectively.
Figure 2.
Estimated susceptibility distribution of the population solid line. marks the mean susceptibility of the host population. The dashed lines show the distribution for 95% confidence limits of the parameters and v.
To investigate whether the infection dynamics differ among various host–parasite combinations, we fit our three models to each combination separately, thus allowing the parameter estimates to differ between combinations. We then compared the sum of the log likelihoods of the resulting model fits to the fits of the pooled dataset with a chi-squared test.
Whereas the likelihood of the mass-action infection model increased significantly from −374.5 to −340.7 (Pr=7×10−14), if we allowed the infection probability to differ between host–parasite combinations, the fit of the parasite antagonism and the host heterogeneity models showed no significant improvement (table 3). However, the fit of the mass-action infection model, which allows for differences in the infection probability, is still much worse than the parasite antagonism or the host heterogeneity models.
Table 3.
Comparisons of models allowing for differences between host–parasite combinations to basal models with a chi-squared test.
| Model 1: Model 2 | −2(l(Model 1)−l(Model 2)) | d.f. | Pr-value |
|---|---|---|---|
| mass-action: mass-action+combinations | 67.8 | 4 | 7×10−14 |
| parasite antagonism: parasite antagonism+combinations | 11.2 | 8 | 0.19 |
| heterogeneous host: heterogeneous host+combinations | 7.8 | 8 | 0.45 |
In summary, we conclude that our data strongly deviates from a simple mass-action infection model either in the form of host heterogeneity or parasite antagonism, but we find no evidence for variation of the infection dynamics across combinations of susceptible hosts and parasites.
4. Discussion
In the present study, we examined what effect the dose level of the bacterial endoparasite P. ramosa had on the infection rate of the water flea D. magna. We developed three mathematical models and fit them to infection data from five different host–parasite combinations using a log-likelihood approach. First, we analysed the infection dynamics neglecting the existence of different combinations of host clones and parasite isolates in our dataset. Our analysis shows significant deviation from a mass-action assumption model. Although the goodness-of-fit of our two alternative models, parasite antagonism and host heterogeneity, is significantly better, the heterogeneous host model with its particular Γ distribution of host susceptibilities gives the best fit.
This result differs from our previous study (Regoes et al. 2003), in which the goodness-of-fit of the mass-action infection model was not so strikingly worse than that of the other two models (parasite antagonism and host heterogeneity). This difference stems mainly from the difference in our mathematical methods: in the previous study we used a least-squares approach, whereas in the present study we used a likelihood approach, chosen because it provides a more appropriate and exact statistical description by which to estimate infection probabilities, and because it is also most consistent with mathematical studies in epidemiology (Halloran et al. 1996; Longini & Halloran 1996). Since the likelihood approach is more exact, it is also more sensitive to statistical signals in the data that indicate deviation from the model. In the case of the mass-action infection model, the ‘signal’ indicating a deviation is the fact that even at the highest doses, not every host becomes infected. Given this increased sensitivity, the mass-action infection model fit is worse than the fits of the two other models (figure 1, solid line), and the estimate of the infection probability is lower than in our previous study.
The main goal of the present study was to investigate potential differences in the infection process across host clone and parasite isolate combinations. We anticipated that host genetics would strongly influence infectivity, given the high variation for resistance of D. magna to its endoparasite P. ramosa (Ebert et al. 1998; Little & Ebert 2000). Our results show that there are very strong differences among host–parasite combinations, ranging from combinations without any infections to highly compatible combinations (table 1). For further analysis, we focused on those combinations where enough infections occurred to make the analysis possible. Interestingly, the relationship between parasite dose and infection probability did not differ significantly across the five compatible host–parasite combinations in our mathematical analysis. This result is contrary to our expectation that the infection probability would vary across host–parasite combinations.
A picture—albeit rather speculative—that emerges from our analysis is that infection probability is a ‘binary’ rather than a continuous parameter, meaning that any isolate of P. ramosa will either be unable to infect a given clone of D. magna or infect it with a probability of approximately 6×10−4 ml−1. Such a binary infectivity will serve as a barrier against the adaptation of the parasite to new host clones. This ‘binary infection hypothesis’ ignores the fact that some of the resistant combinations contained a few hosts that became infected at very high doses from one of the two parasite lines. It is possible that the P4 P. ramosa used here contained a mixture of different parasite strains. In this case, the low infection probability of P4 in Mu10, B2 and HO1 (table 1) may be the result of a parasite population that contains a high proportion of non-infectious strains and a low proportion of an infectious strain—in accordance with a binary infection. A follow-up experiment (data not shown) confirmed that these infections were indeed caused by a third parasite line being present in very low abundance in the P4 isolate suspensions (F. Ben-Ami & D. Ebert 2006, unpublished data). Although the purity of the parasite isolates is an important issue for a binary interpretation of the infection process, we would like to emphasize that it does not play a role in deviance from the mass-action model that we observed here. In a previous study, we showed that the observed deviations from the mass-action infection model cannot be explained by parasite heterogeneity or stochastic effects (Regoes et al. 2003).
Out of 14, 6 host clone–parasite isolate combinations did not result in infection even at the highest dose. On the other hand, P1 was highly infective in the clone DG from where it was isolated, which is consistent with patterns of local adaptation. The remaining four combinations with appreciable infection were allopatric. Although infectivity is often contingent on pre-adaptation of specific combinations, the lack of variation in the infection probability across the five combinations with appreciable infection in our experiment suggests that local adaptation does not influence the dynamics of the actual infection process. This is consistent with an earlier study that tested local adaptation in infectivity among populations of D. magna and P. ramosa (Ebert et al. 1998).
Serial passage of D. magna clones by P. ramosa lines may increase infectivity in the host of passage during five passages (Little et al. 2006). If our binary infection hypothesis holds, this finding may be interpreted as selection for infective and against non-infective parasite genotypes within a mixed-isolate infection. The question arises whether such selection, which may have happened during culturing of our P. ramosa isolates before the actual experiment, caused the here reported findings. We believe the answer is no. Our main finding is that among the compatible combinations non-genetic effects (within-host clones) cause a substantial deviation from a mass-action model. Genetic effects of the parasite, which would be expected as a result of selection, are less likely to explain our finding. Furthermore, if selection of the parasite had taken place, it would have increased genetic differences among combinations, rather than making them small and non-significant. We cannot rule out, however, that the low infectivity observed in certain combinations was caused by selection against variants, which had been previously (i.e. before selection) able to infect in these apparently incompatible combinations.
The best model in the present study is the one in which we assume that host individuals of the same genotype have different susceptibilities to the parasite. What could the nature of this variation be? Studies of gypsy moth larvae (Dwyer et al. 1997), the Indian meal moth (Knell et al. 1998) and their viruses suggested earlier that host heterogeneity among individuals leads to deviation from the usual mass-action infection model. However, the hosts in these studies reproduce sexually, making it hard to disentangle the nature of the variation in susceptibility. In our study, variation is most probably caused by non-inherited, phenotypic differences among individual hosts of the same clone. Such non-genetic effects on resistance (e.g. physiological and environmental influences) have been observed before in this system (Duncan et al. 2006) and were attributed to the host's immune response. In summary, these findings suggest that although infectivity per se depends on the genetic specificity of a host clone–parasite isolate combination, it is further dependent on individual differences among hosts, which may be influenced by external factors (e.g. micro-environmental variation among our experimental jars) or by internal factors, such as molecular mechanisms of the immune system (e.g. alternative splicing, Watson et al. 2005) or within-clone variation in life-history traits (e.g. differences in size at birth). This forms a testable hypothesis, which can be disentangled experimentally.
Acknowledgments
We thank three anonymous referees for valuable comments, M. Ackermann and A. Yates for fruitful discussions on statistical issues, J. Hottinger for laboratory assistance, and N. Boileau, D. Brites, I. Colson and L. Zimmermann for helping with the experimental set-up. This study was supported by an EU Marie Curie Intra-European Fellowship (contract no. MEIF-CT-2005-010690) to F.B.A.
References
- Aalen O.O. Heterogeneity in survival analysis. Stat. Med. 1988;7:1121–1137. doi: 10.1002/sim.4780071105. doi:10.1002/sim.4780071105 [DOI] [PubMed] [Google Scholar]
- Altizer S, Dobson A, Hosseini P, Hudson P, Pascual M, Rohani P. Seasonality and the dynamics of infectious diseases. Ecol. Lett. 2006;9:467–484. doi: 10.1111/j.1461-0248.2005.00879.x. doi:10.1111/j.1461-0248.2005.00879.x [DOI] [PubMed] [Google Scholar]
- Anderson R.M, May R.M. Population biology of infectious diseases: part I. Nature. 1979;280:361–366. doi: 10.1038/280361a0. doi:10.1038/280361a0 [DOI] [PubMed] [Google Scholar]
- Antia R, Lipsitch M. Mathematical models of parasite responses to host immune defenses. Parasitology. 1997;115:S155–S167. doi: 10.1017/s003118209700200x. doi:10.1017/S003118209700200X [DOI] [PubMed] [Google Scholar]
- Antia R, Levin B.R, May R.M. Within-host population-dynamics and the evolution and maintenance of microparasite virulence. Am. Nat. 1994;144:457–472. doi:10.1086/285686 [Google Scholar]
- Ashworth S.T, Kennedy C.R, Blanc G. Density-dependent effects of Anguillicola crassus (Nematoda) within and on its copepod intermediate hosts. Parasitology. 1996;113:303–309. doi: 10.1017/s003118200008207x. [DOI] [PubMed] [Google Scholar]
- Brunner J.L, Richards K, Collins J.P. Dose and host characteristics influence virulence of ranavirus infections. Oecologia. 2005;144:399–406. doi: 10.1007/s00442-005-0093-5. doi:10.1007/s00442-005-0093-5 [DOI] [PubMed] [Google Scholar]
- Burdon J.J, Ericson L, Müller W.J. Temporal and spatial changes in a metapopulation of the rust pathogen Triphragmium ulmariae and its host, Filipendula ulmaria. J. Ecol. 1995;83:979–989. doi:10.2307/2261179 [Google Scholar]
- D'amico V, Elkinton J.S, Dwyer G, Burand J.P, Buonaccorsi J.P. Virus transmission in gypsy moths is not a simple mass action process. Ecology. 1996;77:201–206. doi:10.2307/2265669 [Google Scholar]
- Decaestecker E, Lefever C, De Meester L, Ebert D. Haunted by the past: evidence for resting stage banks of microparasites and epibionts of Daphnia. Limnol. Oceanogr. 2004;49:1355–1364. [Google Scholar]
- Deredec A, Courchamp F. Extinction thresholds in host–parasite dynamics. Ann. Zool. Fenn. 2003;40:115–130. [Google Scholar]
- de Souza, P. N., Fateman, R. J., Moses, J. & Yapp, C. 2004 The maxima book. See http://maxima.sourceforge.net/docs/maximabook/maximabook-19-Sept-2004.pdf
- Dietz K, Schenzle D. Proportionate mixing models for age-dependent infection transmission. J. Math. Biol. 1985;22:117–120. doi: 10.1007/BF00276550. doi:10.1007/BF00276550 [DOI] [PubMed] [Google Scholar]
- Duncan A, Mitchell S.E, Little T.J. Parasite-mediated selection in Daphnia: the role of sex and diapause. J. Evol. Biol. 2006;19:1183–1189. doi: 10.1111/j.1420-9101.2006.01085.x. doi:10.1111/j.1420-9101.2006.01085.x [DOI] [PubMed] [Google Scholar]
- Dwyer G, Elkinton J.S, Buonaccorsi J.P. Host heterogeneity in susceptibility and disease dynamics: tests of a mathematical model. Am. Nat. 1997;150:685–707. doi: 10.1086/286089. doi:10.1086/286089 [DOI] [PubMed] [Google Scholar]
- Ebert, D. 2005 Ecology, epidemiology, and evolution of parasitism in Daphnia [Internet]. National Library of Medicine (US), National Center for Biotechnology Information: Bethesda, MD. See http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books
- Ebert D, Rainey P, Embley T.M, Scholz D. Development, life cycle, ultrastructure and phylogenetic position of Pasteuria ramosa Metchnikoff 1888: rediscovery of an obligate endoparasite of Daphnia magna Straus. Phil. Trans. R. Soc. B. 1996;351:1689–1701. doi:10.1098/rstb.1996.0151 [Google Scholar]
- Ebert D, Zschokke-Rohringer C.D, Carius H.J. Within- and between-population variation for resistance of Daphnia magna to the bacterial endoparasite Pasteuria ramosa. Proc. R. Soc. B. 1998;265:2127–2134. doi:10.1098/rspb.1998.0549 [Google Scholar]
- Ebert D, Lipsitch M, Mangin K.L. The effect of parasites on host population density and extinction: experimental epidemiology with Daphnia and six microparasites. Am. Nat. 2000a;156:459–477. doi: 10.1086/303404. doi:10.1086/303404 [DOI] [PubMed] [Google Scholar]
- Ebert D, Zschokke-Rohringer C.D, Carius H.J. Dose effects and density-dependent regulation of two microparasites of Daphnia magna. Oecologia. 2000b;122:200–209. doi: 10.1007/PL00008847. doi:10.1007/PL00008847 [DOI] [PubMed] [Google Scholar]
- Fromont E, Pontier D, Langlais M. Dynamics of a feline retrovirus (FeLV) in host populations with variable spatial structure. Proc. R. Soc. B. 1998;265:1097–1104. doi: 10.1098/rspb.1998.0404. doi:10.1098/rspb.1998.0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. Parasites and epibionts of Cladocera. Trans. Zool. Soc. Lond. 1974;32:417–515. [Google Scholar]
- Grosholz E.D. The influence of habitat heterogeneity on host–pathogen population dynamics. Oecologia. 1993;96:347–353. doi: 10.1007/BF00317504. doi:10.1007/BF00317504 [DOI] [PubMed] [Google Scholar]
- Halloran M.E, Longini I.M, Struchiner C.J. Estimability and interpretation of vaccine efficacy using frailty mixing models. Am. J. Epidemiol. 1996;144:83–97. doi: 10.1093/oxfordjournals.aje.a008858. [DOI] [PubMed] [Google Scholar]
- Hamer W.H. Epidemic disease in England. Lancet. 1906;1:733–739. [Google Scholar]
- Hamilton W.D. Sex versus non-sex versus parasite. Oikos. 1980;35:282–290. doi:10.2307/3544435 [Google Scholar]
- Hougaard P. A class of multivariate failure time distributions. Biometrika. 1986;73:671–678. [Google Scholar]
- Hughes W.O.H, Petersen K.S, Ugelvig L.V, Pedersen D, Thomsen L, Poulsen M, Boomsma J.J. Density-dependence and within-host competition in a semelparous parasite of leaf-cutting ants. BMC Evol. Biol. 2004;4:45. doi: 10.1186/1471-2148-4-45. doi:10.1186/1471-2148-4-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J. A hypothesis to account for the maintenance of sex within populations. Evol. Theory. 1978;3:191–194. [Google Scholar]
- Kleiven O.T, Larsson P, Hobaek A. Sexual reproduction in Daphnia magna requires three stimuli. Oikos. 1992;65:197–206. doi:10.2307/3545010 [Google Scholar]
- Klüttgen B, Dümler U, Engels M, Ratte H.T. ADaM, an artificial freshwater for the culture of zooplankton. Water Res. 1994;28:743–746. doi:10.1016/0043-1354(94)90157-0 [Google Scholar]
- Knell R.J, Begon M, Thompson D.J. Transmission of Plodia interpunctella granulosis virus does not conform to the mass action model. J. Anim. Ecol. 1998;67:592–599. doi:10.1046/j.1365-2656.1998.00219.x [Google Scholar]
- Leite M.B.F, Bassanezi R.C, Yang H.M. The basic reproduction ratio for a model of directly transmitted infections considering the virus charge and the immunological response. IMA J. Math. Appl. Med. Biol. 2000;17:15–31. doi:10.1093/imammb/17.1.15 [PubMed] [Google Scholar]
- Little T.J, Ebert D. The cause of parasitic infection in natural populations of Daphnia (Crustacea: Cladocera): the role of host genetics. Proc. R. Soc. B. 2000;267:2037–2042. doi: 10.1098/rspb.2000.1246. doi:10.1098/rspb.2000.1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little T.J, Watt K, Ebert D. Parasite–host specificity: experimental studies on the basis of parasite adaptation. Evolution. 2006;60:31–38. [PubMed] [Google Scholar]
- Longini I.M, Halloran M.E. A frailty mixture model for estimating vaccine efficacy. Appl. Stat. 1996;45:165–173. doi:10.2307/2986152 [Google Scholar]
- Lopez J.E, Gallinot L.P, Wade M.J. Spread of parasites in metapopulations: an experimental study of the effects of host migration rate and local host population size. Parasitology. 2005;130:323–332. doi: 10.1017/s0031182004006602. doi:10.1017/S0031182004006602 [DOI] [PubMed] [Google Scholar]
- Osnas E.E, Lively C.M. Parasite dose, prevalence of infection and local adaptation in a host–parasite system. Parasitology. 2004;128:223–228. doi: 10.1017/s0031182003004360. doi:10.1017/S0031182003004360 [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing; Vienna, Austria: 2005. R: a language and environment for statistical computing. [Google Scholar]
- Regoes R.R, Hottinger J.W, Sygnarski L, Ebert D. The infection rate of Daphnia magna by Pasteuria ramosa conforms with the mass-action principle. Epidemiol. Infect. 2003;131:957–966. doi: 10.1017/s0950268803008793. doi:10.1017/S0950268803008793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnadel H.A, Ebert D. Prevalence, host specificity and impact on host fecundity of microparasites and epibionts in three sympatric Daphnia species. J. Anim. Ecol. 1997;66:212–222. doi:10.2307/6023 [Google Scholar]
- Watson F.L, Püttmann-Holgado R, Thomas F, Lamar D.L, Hughes M, Kondo M, Rebel V.I, Schmucker D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. doi:10.1126/science.1116887 [DOI] [PubMed] [Google Scholar]