Abstract
Recent work has demonstrated considerable benefits of intracolonial genetic diversity for the productivity of honeybee colonies: single-patriline colonies have depressed foraging rates, smaller food stores and slower weight gain relative to multiple-patriline colonies. We explored whether differences in the use of foraging-related communication behaviour (waggle dances and shaking signals) underlie differences in foraging effort of genetically diverse and genetically uniform colonies. We created three pairs of colonies; each pair had one colony headed by a multiply mated queen (inseminated by 15 drones) and one colony headed by a singly mated queen. For each pair, we monitored the production of foraging-related signals over the course of 3 days. Foragers in genetically diverse colonies had substantially more information available to them about food resources than foragers in uniform colonies. On average, in genetically diverse colonies compared with genetically uniform colonies, 36% more waggle dances were identified daily, dancers performed 62% more waggle runs per dance, foragers reported food discoveries that were farther from the nest and 91% more shaking signals were exchanged among workers each morning prior to foraging. Extreme polyandry by honeybee queens enhances the production of worker–worker communication signals that facilitate the swift discovery and exploitation of food resources.
Keywords: dance communication, extreme polyandry, genetic diversity, shaking signal, recruitment, waggle dance
1. Introduction
The evolution of polyandry in phylogenetically isolated taxa of social insects is an intriguing phenomenon owing to high costs of this behaviour for the fitness of queens. When a queen mates with multiple males, she risks predation and exposure to sexually transmitted diseases, incurs energetic costs and—with presumable consequences for selective forces that maintain sociality—she decreases the average relatedness among her offspring, the cooperating members of her colony. For these reasons, the occurrence of polyandry is expected to be infrequent among social bees, wasps and ants (order: Hymenoptera). Surveys of the level of polyandry across taxa largely support this prediction, but nevertheless, polyandry, although rare, is found consistently across a wide range of biologically diverse social insects (reviewed by Boomsma & Ratnieks 1996; Strassmann 2001). Honeybees (genus Apis) stand out among polyandrous groups because, without exception, queens of all species mate with a strikingly high number of males (mean 7–41 effective mating frequency per queen across all species; reviewed by Tarpy & Nielsen (2002)).
Such a strong bias towards extreme polyandry in honeybees suggests that, despite potential costs to fitness, multiple mating by queens generates a substantial selective advantage for colonies. Many hypotheses have been proposed to explain the benefits of polyandry for social groups, and available evidence indicates that several of them are germane to honeybees (reviewed by Boomsma & Ratnieks 1996; Crozier & Fjerdingstad 2001). Multiple mating by honeybee queens can reduce the fitness load of producing diploid (sterile) drones, which result when fertilized eggs that are meant to yield workers are homozygous at the sex-determination locus (Tarpy & Page 2002). Multiple-patriline worker populations also have greater resistance to disease (honeybees: Tarpy & Seeley 2006, Seeley & Tarpy 2007; bumble-bees: Baer & Schmid-Hempel 1999, 2003; leaf-cutting ants: Hughes & Boomsma 2004) and are better at stabilizing within-nest conditions as the environment fluctuates (Jones et al. 2004) than worker populations comprising a single patriline. Often heralded as a strong explanation for these observations and supported by the demonstration that mating frequency is linked to colony growth (harvester ants: Cole & Wiernasz 1999), intracolonial genetic diversity is hypothesized to improve colony efficiency by introducing into colonies genetically based variability in the response thresholds for tasks that workers perform (Robinson & Page 1989). The link between worker genotype and the probability of task performance has been demonstrated repeatedly in honeybees (reviewed by Crozier & Fjerdingstad 2001), but short-term studies that compared the development of genetically uniform colonies against colonies with unnaturally low numbers of patrilines have not provided persuasive evidence that polyandry increases the collective productivity of honeybee workforces (Oldroyd et al. 1992; Fuchs & Schade 1994; Page et al. 1995).
Recently, Mattila & Seeley (2007) conducted a year-long study that revealed unambiguous differences in the long-term productivity and fitness of genetically diverse (15 patrilines) and genetically uniform (one patriline) colonies when new nests are founded after a swarming event. During the first months of colony founding, polyandrous colonies built comb faster, stockpiled more food and reared greater numbers of workers and drones than monandrous colonies (worker populations were similar in size at the start of the study). Inequalities in productivity accumulated over time and, with the onset of cold weather, all of the genetically uniform colonies exhausted their food reserve and subsequently froze to death by mid-winter, whereas 25% of the genetically diverse colonies survived to the next spring. The impressive fitness gains bestowed on colonies by multiple paternity were attributable in part to large differences in foraging effort between colony types; on the majority of days, given worker populations of equal size and with identical foraging opportunities, genetically diverse colonies had foraging rates that were 27–78% higher than those of genetically uniform colonies (Mattila & Seeley 2007).
We speculated that differences in the foraging effort of colonies would result if robust advertisement of food resources by waggle-dancing foragers was hampered by a loss of genetic diversity in single-patriline colonies. Workers of different patrilines vary in the extent to which they express key behaviours that contribute to the rapid discovery and exploitation of food resources, such as their likelihood of waggle dancing (Arnold et al. 2002) or scouting (Dreller 1998) or the distance at which they forage (Oldroyd et al. 1993). Accordingly, the presence of multiple patrilines in a colony may broaden a workforce's foraging effort in a variety of complex ways that would be reflected in the waggle dances of foragers as they report food discoveries to their nestmates (e.g. more foragers, more dances, a greater range of food resources). Moreover, workers in genetically diverse colonies may perform more shaking signals each morning to activate the workforce. The shaking signal, when a worker grips another worker with her forelegs and vigorously shakes both herself and her nestmate, is a modulating signal that conveys the broad meaning ‘prepare for greater activity’ (Seeley et al. 1998) or ‘reallocate labour to different activities’ (Nieh 1998). Shaking signals can have a variety of effects on a recipient, one of which is to increase her activity level and movement towards the dance floor, the area near the nest's entrance where foragers dance and unload nectar (reviewed by Schneider & Lewis 2004). Shaking signals are performed primarily by experienced foragers (Biesmeijer 2003) and are observed most frequently prior to each morning's flight if preceded by a day of good forage (Schneider et al. 1986; Seeley et al. 1998). Peaks in shaking signals each morning are positively correlated with dance activity later in the day (Schneider et al. 1986).
To determine whether polyandrous colonies benefit from genetic diversity by an increased amount of information available to foragers about food resources, we compared the production by workers of foraging-related communication signals in three pairs of genetically diverse and genetically uniform colonies. We observed each pair of colonies in the same setting and over the course of 3 days, and by deciphering workers' waggle dances throughout the day and recording the frequency of shaking signals each morning, we compared between colony types (i) the number of dances performed throughout the day, (ii) the number of waggling signals each dancer produced to report her find, (iii) the distance from the hive of advertised food sites and (iv) the production of shaking signals prior to flight each day.
2. Material and methods
(a) Controlling queen mating
Supersister queens, daughters of a single Carniolan queen mated with one drone (r=0.75), were instrumentally inseminated to create genetically diverse and genetically uniform colonies for our study. Each genetically diverse colony had a ‘polyandrous’ queen that was inseminated with a mix of semen obtained from 15 different drones. For each queen, a unique set of drones was selected randomly from a pool of over 1000 drones (11 different drone source colonies, approx. 100 drones colony−1); their semen was collected, pooled, stirred gently and 0.8 μl of the mixed ejaculate was used for insemination. Worker populations derived from queens that have been instrumentally inseminated in this manner contain workers representing offspring of each drone father (Haberl & Moritz 1994). Each genetically uniform colony had a ‘monandrous’ queen that had been inseminated with 0.8 μl of semen from a single drone (a unique drone for each queen). Each drone was selected from the same pool that was used for multiple-drone inseminations. Further details regarding the insemination process and queen rearing procedures are described by Mattila & Seeley (2007, electronic supplementary material); a separate group of queens was used for this study, although drones were drawn from the same pool.
Queens were reared and inseminated by a queen breeder (Glenn Apiaries, Fallbrook, California) during March 2006. Inseminated queens were labelled by the breeder as belonging to group ‘A’ or ‘B’ (five colonies group−1); none of the authors knew the insemination status of queens in either group, and this information was not revealed until all behavioural data had been collected.
(b) Establishing genetically diverse and uniform colonies
Inseminated queens were shipped to Ithaca, New York, where each one was installed in a queenless colony in a five-frame hive on 20 April 2006. Data collection did not begin until 28 June 2006, which allowed ample time for worker populations to be replaced by the offspring of the introduced queens. After queen introduction, colonies were examined weekly to remove supersedure cells and to ensure that queens continued to lay eggs consistently; colonies that had queens with poor egg-laying patterns were removed from the study. Furthermore, because genetically uniform colonies are more susceptible to disease than genetically diverse colonies (Seeley & Tarpy 2007), all colonies were medicated to minimize the potential for an effect of treatable diseases on the activity of workers. Treatments included those for American and European foul brood (oxytetracycline, Terramycin; Pfizer, Inc., Exton, Pennsylvania) and varroa mites (fluvalinate, Apistan; Wellmark International, Schaumburg, Illinois). Colonies that showed signs of untreatable diseases (e.g. chalkbrood) were removed from the study. After excluding three unsuitable colonies from the study, three pairs of colonies (one genetically diverse colony and one genetically uniform colony per pair) derived from the 10 queens shipped by the breeder were used for the study.
(c) Documenting activity on the dance floor
Throughout the summer, we videotaped waggle dances performed by foragers in our paired study colonies. Each pair of colonies was observed over the course of 3 days and in the same setting: the first pair was observed during 28–30 June 2006 (inclusive), the second during 13–15 July 2006 and the third during 9–11 August 2006.
To document activity on the dance floor over these periods, each colony had its queen, an empty frame and one frame covered with bees (approx. 2000 workers), brood and food transferred to a two-frame observation hive. For each pair of colonies, the contents of the brood/food frame and the number of workers were carefully matched by measuring the area covered by each with a 2.5×2.5 cm grid; thus each colony had similar amounts of bees, resources and stages of brood to tend. The entrance of each observation hive was designed to force returning foragers to enter on only one side of the comb; thus all dances were performed on a single dance floor, which was recorded by a digital video camera (Sony, DCR-HC90) positioned on one side of the hive (Seeley 1995). Hives in each pair were installed side by side at the Liddell Field Station (Cornell University campus) at least 2 days before observations began, so that workers had time to adjust to their new hive, entrance and surroundings. Once videotaping began, the side of the comb on which workers entered the hive was recorded continually from 07.00 to 19.00 to ensure that all activity on the dance floor was captured on tape.
(d) Analysing waggle dances, shaking signals and relative foraging activity
We counted and translated the waggle dances performed by foragers over the course of each day to compare between genetically diverse and genetically uniform colonies: the number of dances performed throughout the day, the number of waggle signals workers produced when they returned to colonies, the distance to food sites advertised by dances and the start of dancing each day. Thus, to characterize the foraging effort of each colony on each day through waggle dances, we watched the first minute of every 5-min interval of videotape, we identified any dancer that performed at least four waggle runs, and then for each dancer, we measured the angle of four runs relative to vertical (direction to food relative to the Sun's azimuth) and run duration (distance to food). These values were averaged for each dancer, and the mean values were used to determine the site advertised by the dance relative to the position of the colony. Videotapes were played back on a digital video editor (Sony, DSR-30); dance angles were estimated using a rotating compass that was affixed to the video monitor and the duration of each waggle run was measured to 1/30 of a second. The specific locations of food sites in space, as gleaned from dances, were estimated according to von Frisch's (1967, p. 100) standard curve of distance versus waggle-run duration for a Carniolan colony and by adjusting compass direction to account for the movement of the Sun throughout the day (USA Naval Observatory, http://aa.usno.navy.mil/data/docs/altaz.php). We felt that assessing the dance activity of foragers during one in every 5 min of videotape provided a reasonable comparison of waggle-dance activity between genetically diverse and genetically uniform colonies, although it undoubtedly underestimated the total number of times returning foragers danced in each colony throughout the day.
We also observed individual foragers on the dance floor to determine whether there was a difference between genetically diverse and genetically uniform colonies in the quantity of information advertised by a single forager. On each day that colonies were videotaped, we selected 20 waggle-dancing foragers from each colony and quantified their dance behaviour after they returned to the hive and before they left on another foraging trip. Each dancer was selected by identifying the first worker to waggle at the start of a 1-min interval of tape; each 1-min interval was separated by nine intervening minutes of tape and the same time period (12.00 to 15.20) was analysed each day. Once a waggle dancer was identified, we rewound the tape to the point where she entered the hive and traced her movements until she left the dance floor (either above to empty comb or below to leave the hive). While the dancer was in sight, we recorded (i) the number of waggle runs she performed and (ii) the number of ‘dances’ she completed (at least eight consecutive waggle runs in the same location, which is the mean number of runs followed by dance followers that successfully find an advertised food site; Judd 1995).
We wondered how the activation of the workforce each morning with shaking signals differed between genetically diverse and genetically uniform colonies. Thus, each morning that colonies were videotaped, we documented the number of shaking signals recorded within the field of view of the camera, as well as the number of workers that produced the signals. For each colony, shaking signals and shaking bees were counted during the first minute of every 5-min interval of tape during the first hour of the day (07.00 to 08.00).
Finally, we calculated relative foraging rates for colonies in each pair to determine whether the level of communication among workers (waggle dances and shaking signals) reflected foraging activity. On the days that colonies were observed (except 29 June), we counted the number of workers that left each hive per minute between 12.00 and 12.30, from which we calculated the mean number of bees exiting per minute.
(e) Data analyses
Data are reported as mean±s.e. (with one standard deviation (s.d.) provided). The level of significance for all statistical tests was set at α=0.05. We used one-tailed tests to improve the power of our analyses because we could predict in advance that genetically diverse colonies would outperform genetically uniform colonies in foraging-related tasks (Mattila & Seeley 2007). When measures were made on multiple workers from the same colony (i.e. distance to food advertised by a waggle dance, number of waggles per dancing worker), each measure was treated as a subsample of that colony in a one-way ANOVA, with data pooled across 3 days of observation (Proc Mixed; SAS Institute 2004). For data that included one estimate made on a colony per day (i.e. number of dances performed daily, number of shaking signals performed during the first hour of foraging, relative foraging activity), we calculated means for each colony across the 3-day observation period and then used paired t-tests to compare the mean performance of genetically diverse and genetically uniform colonies (Proc Univariate; SAS Institute 2004). Although our data probably did not meet the assumption of normality for parametric tests, the use of paired t-tests permitted us to keep α=0.05 as our level of significance, whereas the equivalent non-parametric Wilcoxon signed-rank test could not resolve significant differences below α=0.125 for n=3 pairs. We provided raw data for all colonies whenever paired t-tests were employed (table 1).
Table 1.
Comparison of signal production and foraging activity in three pairs of genetically diverse and genetically uniform colonies, over 3 days of observation per pair. (Total number of signals produced per day were determined by counting signals observed on the dance floor of hives during the first minute of every 5-min interval of observation (waggle dances were counted from 07.00 to 19.00; shaking signals were counted from 07.00 to 08.00). Relative foraging rates were determined from 12.00 to 12.30. Means, s.e. and s.d. were calculated using 3-day colony means; percentage of increase=((genetically diverse−genetically uniform)/genetically uniform)×100%.)
| pair | day | total no. of waggle dances | total no. of shaking signals | relative foraging rates (mean no. workers exiting min−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| genetically uniform | genetically diverse | increase (%) | genetically uniform | genetically diverse | increase (%) | genetically uniform | genetically diverse | increase (%) | ||
| 1 | 28 Jun | 395 | 329 | −17 | 322 | 781 | 143 | 10.0 | 10.6 | 6 |
| 29 Jun | 267 | 296 | 11 | 205 | 889 | 337 | — | — | — | |
| 30 Jun | 153 | 206 | 35 | 220 | 559 | 154 | 9.8 | 31.5 | 221 | |
| 2 | 13 Jul | 90 | 196 | 118 | 208 | 400 | 92 | 8.5 | 13.9 | 64 |
| 14 Jul | 172 | 334 | 94 | 284 | 284 | 0 | 7.3 | 15.4 | 111 | |
| 15 Jul | 155 | 262 | 69 | 111 | 171 | 54 | 6.5 | 13.7 | 111 | |
| 3 | 9 Aug | 113 | 227 | 101 | 143 | 205 | 43 | 3.9 | 9.1 | 133 |
| 10 Aug | 182 | 270 | 48 | 351 | 283 | −19 | 7.7 | 11.5 | 49 | |
| 11 Aug | 107 | 108 | 1 | 241 | 403 | 67 | 4.7 | 6.7 | 43 | |
| mean±s.e. (s.d.) | 181.6±45.1 (78.1) | 247.6±23.2 (40.3) | 231.7±15.4 (26.6) | 441.7±150.7 (261.0) | 7.9±1.3 (2.2) | 14.8±3.5 (5.9) | ||||
3. Results
(a) Waggle-dance activity in colonies
One way that we compared dance communication in genetically diverse and genetically uniform colonies was to count the number of waggle dances that were performed in colonies during the first minute of each 5-min interval of videotape that we recorded as colonies foraged. This sampling regimen yielded a total of 3862 dances from which we decoded direction and distance information. The difference in waggle-dance activity between the two colony types varied within pairs and over 9 days of observation, but we found that, on average, the number of waggle dances performed over a day was 36% greater in genetically diverse colonies compared with genetically uniform colonies (table 1). On 7 of 9 days of observation, workers in genetically diverse colonies performed appreciably more waggle dances than workers in genetically uniform colonies (10–118% increase in dancing), and on 3 days, the difference in the daily total of dances approached or was greater than two times in favour of genetically diverse colonies (table 1). Although there were more dances per day on average in genetically diverse versus genetically uniform colonies, we could detect only a highly suggestive trend towards increased dancing with greater diversity owing to our small sample size and variability in the degree to which genetically diverse colonies outdanced uniform colonies, i.e. greater difference in relative performance between colonies in pair 2 compared with pairs 1 and 3, which inflates standard error of the mean and drives down the size of the t-statistic (table 1; paired t-test of mean daily dance total per colony: t2=1.9, p=0.09).
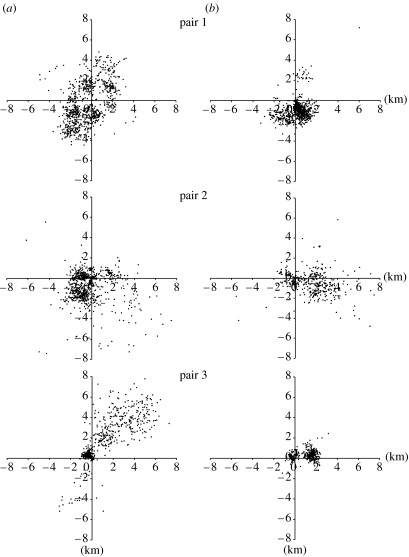
Dancing foragers in genetically diverse colonies advertised food sites that were a mean of 800 m farther away from the colony than sites reported by foragers in genetically uniform colonies (mean 2.4±0.2 km (s.d. 2.0) versus 1.6±0.3 km (s.d. 1.2); ANOVA with subsampling—effect of genetic diversity: F1,4=8.5, p=0.02, subsampling effect: F4,3715=26.9, p<0.0001). Plots of the location of food discoveries surrounding each pair of colonies, as indicated by dances over 3 days, confirm that foragers in genetically diverse colonies reported food finds over greater distances than foragers in genetically uniform colonies (figure 1).
Figure 1.
Foraging maps for three pairs of (a) genetically diverse and (b) genetically uniform colonies as both colonies in each pair foraged in the same environment over the course of 3 days. Maps show estimated locations of food advertised by the dances of workers throughout the 3 days that each pair of colonies was observed. Estimations of locations are based on von Frisch's (1967) standard curves of waggle duration versus distance from the hive for Carniolan workers. The intersection of the axes marks the location of the pair of hives.
There was no difference between pairs of genetically diverse and genetically uniform colonies in the time each morning at which foragers started to dance (paired t-test of difference in start time: t2=1.3, p=0.16). On five of nine mornings, the first dance was observed in genetically diverse colonies (10–60 min sooner than in genetically uniform colonies), and on three of nine mornings, genetically uniform colonies had the first dance (5–20 min before genetically uniform colonies). One morning, foragers began to dance in both colonies almost simultaneously.
(b) Behaviour of individual waggle dancers
We wondered whether the amount of information available to foragers about food resources was increased by the behaviour of an individual waggle dancer as she danced. By observing a subset of waggle dancers in each colony, we found that dancers in genetically diverse colonies performed 62% more waggle runs than dancers in genetically uniform colonies (mean 39.8±2.9 (s.d. 5.3) runs worker−1 versus 24.6±2.9 (s.d. 6.7) runs worker−1; ANOVA with subsampling—effect of genetic diversity: F1,4=14.1, p=0.01, subsampling effect: F4,354=2.1, p=0.04). Furthermore, dancers in genetically uniform colonies completed a mean of only 1.3±0.2 (s.d. 0.4) dances (at least eight consecutive waggle runs in the same location on the comb), fewer than the mean of 2.2±0.2 (s.d. 0.3) dances that were performed by dancers in genetically diverse colonies (ANOVA with subsampling—effect of genetic diversity: F1,4=17.1, p=0.005, subsampling effect: F4,354=1.9, p=0.06).
(c) Activation of workforce each morning
We counted the number of shaking signals observed during one in every 5 min of videotape throughout the first hour that colonies were monitored each morning. Within the field of view of the camera, an average of 91% more shaking signals were counted in genetically diverse colonies compared with genetically uniform colonies between 07.00 and 08.00. The occurrence of shaking signals was 43–337% greater in genetically diverse colonies relative to genetically uniform colonies on 7 of 9 days; signals were equally frequent on 1 day and 19% more numerous in uniform colonies on the remaining day (table 1). Moreover, during each 1-min interval, 50% more individuals were observed giving shaking signals to other workers in genetically diverse colonies (mean 10.2±1.6 (s.d. 4.9) workers) than in genetically uniform colonies (mean 6.8±1.0 (s.d. 2.9) workers). Strong trends towards an increase in the number of shaking signals and the number of shaking bees were observed across all pairs of colonies, although t-tests were not significant owing to a small sample size and a large difference in the production of shaking signals between colonies in pair 1 contrasted with pairs 2 and 3 (paired t-tests; mean total number of shakes per colony: t2=1.5, p=0.13; mean number of shaking workers per colony: t2=1.7, p=0.11).
(d) Relative foraging activity
Between 12.00 and 12.30 each day, more workers exited the hives of genetically diverse colonies, presumably to forage, compared with genetically uniform colonies on every day of observation; foraging rates were 87% greater on average when colony workforces were genetically diverse (table 1; paired t-test of mean foraging rate per colony: t2=3.3, p=0.04).
4. Discussion
We found persuasive evidence that the production of foraging-related communication signals (waggle dances and shaking signals) is enhanced by intracolonial genetic diversity—a consequence of polyandry by queens—and that an increase in the availability of information about foraging conditions is associated with greater foraging rates in genetically diverse colonies relative to genetically uniform colonies. Our study required us to make detailed measurements within each colony, which limited our ability to examine a large number of colonies; despite these constraints, we found strong and consistent trends towards increased expression of signalling behaviour with greater intracolonial genetic diversity. On average, the presence of multiple patrilines in a honeybee colony reliably increased the breadth of information shared among foragers about the location of food resources in several conspicuous ways and, in almost every measure, differences between genetically diverse and genetically uniform colonies were considerable. First, over the course of the day, foragers were observed waggle dancing 36% more often in genetically diverse colonies compared with genetically uniform colonies. Second, waggle-dancing foragers in genetically diverse colonies performed a mean of 62% more waggles after a foraging trip than dancers in genetically uniform colonies. Finally, forager workforces that comprised multiple patrilines reported food discoveries that were almost 1 km farther from the nest than single-patriline workforces, which implies that foragers exploited food sources over a larger area. Evidence suggests that workers in different patrilines vary in the distance at which they prefer to forage (Oldroyd et al. 1993); here, we see this preference manifested in a more expansive foraging effort by polyandrous colonies, which could allow for a more robust response to sudden changes in food availability. Thus, given similarly sized populations and foraging opportunities, workers in genetically diverse colonies had access to better information about potential food resources than workers in genetically uniform colonies because there were greater numbers of dances, each dancing worker conveyed more information as she danced and dancers advertised finds farther afield from their nest. Furthermore, the frequency of shaking signals was substantially (nearly two times) higher each morning in genetically diverse versus genetically uniform colonies, which has been correlated previously with higher numbers of dances later in the day (Schneider et al. 1986). Indeed, we found that increased utilization of waggle dances and shaking signals mirrored increased foraging rates of genetically diverse colonies relative to genetically uniform colonies.
This study is the first real step towards documenting the reasons why a lack of genetic diversity affects the organization of a colony's forager workforce, specifically, how it is registered in the expression of foraging-related communication among workers. Moreover, we have demonstrated that a reduction in communication among workers, as a consequence of a monandry, is linked with low foraging activity at the colony level. The results of this study reveal, at least in part, the behavioural differences of workers that can account for depressed levels of foraging in colonies headed by monandrous queens relative to those led by polyandrous queens (reported by Mattila & Seeley 2007). And importantly, a reduction in foraging effort contributes to severe fitness consequences for genetically uniform colonies that experience reduced productivity of the forager workforce (exacerbated by slow colony development) to the point where founding colonies cannot store sufficient food reserves to survive their first winter (Mattila & Seeley 2007).
Presently, it is not clear why the use of dance communication is stifled by a lack of diversity in colonies, but it is probably related to genetic differences among patrilines in thresholds for expressing foraging- and recruitment-related signalling behaviour. For example, we know that waggle dancing and food scouting are not performed evenly among patrilines in colonies (Dreller 1998; Arnold et al. 2002). It is probable that the presence of patrilines with low thresholds for these behaviours boosted the number of dances that were observed in our genetically diverse colonies. Furthermore, we know that foragers from the same colony differ in the rate at which they increase their dance response (number of waggle runs) in reaction to increases in resource profitability (Seeley 1994). If the nature of this stimulus–response function is linked to genotype, then we would expect some foragers to respond to a resource with more waggle runs than other workers and, in general, to see more ‘vigorous’ responders as patriline numbers rise, as we saw in this study. A reduction in the amount of information exchanged among workers in single-patriline colonies may stem from or be further exacerbated by workers that have a genetic tendency for low lifetime participation in foraging (Guzman-Novoa & Gary 1993) or a low probability of adopting different forager roles (e.g. inspector, scout, recruit, reactivated forager, dancer). In other words, we do not know the degree to which signalling is impaired by a poor foraging effort or vice versa. Truly, our results hint that intracolonial genetic diversity could have far-reaching consequences for improving the productivity, flexibility and efficiency of the forager workforce. Yet, it is difficult to establish concrete ideas about how multiple paternity divides labour within colonies because almost nothing is understood about response thresholds of workers and the strength of worker response once thresholds are exceeded, or the relationship of either to patriline membership. Careful experiments in this vein are required to unravel these mysteries.
A comparison of the production of waggle-dance signals in the second pair of colonies was of particular interest to us. This pair showed the largest and most consistent difference in the daily frequency of waggle dances over the 3 days of observation (table 1) and it was the only one that was studied during the summer's single intense honey flow (see fig. 3 in Mattila & Seeley (2007) for seasonal weight gain of colonies in the same location). Despite an abundance of natural forage—other colonies at Liddell were foraging busily and filling their hives with nectar for the first time that summer, as was the genetically diverse colony in the pair—the genetically uniform colony had a relatively low number of dances and a shocking lack of activity on the dance floor throughout each day of observation. We wonder whether this indicates that single-patriline colonies are particularly ineffectual at capitalizing on food resources that become plentiful suddenly, thus underutilizing opportunities to make net gains in weight. With rather limited opportunities to gain weight annually (only 6–8 weeks of the year in our area; Seeley & Visscher 1985, Mattila & Seeley 2007), an inability to activate the forager workforce when foraging conditions become excellent would be selected against strongly. Conversely, traits that improved the response of colonies to rapidly shifting conditions, such as polyandry, would be expected to increase in frequency accordingly. The results from this pair of colonies are suggestive and would explain large differences in the rate of weight gain between genetically diverse and genetically uniform colonies studied over the same period and in the same location (Mattila & Seeley 2007), although further experimentation is required to explore the relationship between genetic diversity and colony response to the sudden availability of food resources.
Hence, when the weight of evidence is considered, polyandry by queens most probably evolved in and is maintained in honeybees because it has plural selective advantages for colonies, advantages that outweigh its costs and result in greater fitness gains than would be realized in its absence. Intracolonial genetic diversity improves a colony's resistance to disease (Tarpy & Seeley 2006; Seeley & Tarpy 2007), decreases the probability of a high percentage of inviable worker brood (Tarpy & Page 2002) and enhances the productivity of a colony's workforce (Mattila & Seeley 2007), shown here by—one of potentially numerous mechanisms—increasing the amount of information shared among foragers about lucrative food resources. The amplifying effect that multiple paternity has on the use of dance communication among workers may play a very significant role in the pervasive and extreme nature of polyandry across Apis species, all of which share a complex communication system that facilitates the exploitation of patchy and ephemeral resources for the survival of energy-demanding colonies. Here, an increase from 1 to 15 patrilines in a colony caused a considerable rise in the frequency of the sophisticated communication signals that are produced to inform nestmates about foraging conditions. In turn, the accompanying increase in foraging rate undoubtedly enhances the long-term survival and fitness of colonies (Mattila & Seeley 2007). Synergism between dance communication and multiple mating in honeybees may be one of the main selective forces propelling the evolution of the extreme polyandry that characterizes this monophyletic group. Where extreme polyandry is found in other highly derived eusocial insect taxa, such as harvester and army ants (Denny et al. 2004; Kronauer et al. 2004; Rheindt et al. 2004; Wiernasz et al. 2004), intracolonial genetic diversity may play a similar role in their complex and well-organized foraging efforts.
Acknowledgments
Funding was provided by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada to H.R.M. and by a grant from the National Research Initiative of the US Department of Agriculture Cooperative State Research, Education and Extension Service to T.D.S. (no. 2003-35302-13387). We thank two anonymous reviewers for careful comments that sharpened our interpretation of the study.
References
- Arnold G, Quenet B, Papin C, Masson C, Kirchner W.H. Intra-colonial variability in the dance communication in honeybees (Apis mellifera) Ethology. 2002;108:751–761. doi:10.1046/j.1439-0310.2002.00809.x [Google Scholar]
- Baer B, Schmid-Hempel P. Experimental variation in polyandry affects parasite loads and fitness in a bumble-bee. Nature. 1999;397:151–154. doi:10.1038/16451 [Google Scholar]
- Baer B, Schmid-Hempel P. Bumblebee workers from different sire groups vary in susceptibility to parasite infection. Ecol. Lett. 2003;6:106–110. doi:10.1046/j.1461-0248.2003.00411.x [Google Scholar]
- Biesmeijer J.C. The occurrence and context of the shaking signal in honey bees (Apis mellifera) exploiting natural food sources. Ethology. 2003;109:1009–1020. doi:10.1046/j.0179-1613.2003.00939.x [Google Scholar]
- Boomsma J.J, Ratnieks F.L.W. Paternity in eusocial Hymenoptera. Phil. Trans. R. Soc. B. 1996;351:947–975. doi:10.1098/rstb.1996.0087 [Google Scholar]
- Cole B.J, Wiernasz D.C. The selective advantage of low relatedness. Science. 1999;285:891–893. doi: 10.1126/science.285.5429.891. doi:10.1126/science.285.5429.891 [DOI] [PubMed] [Google Scholar]
- Crozier R.H, Fjerdingstad E.J. Polyandry in social Hymenoptera—disunity in diversity? Ann. Zool. Fenn. 2001;38:267–285. [Google Scholar]
- Denny A.J, Franks N.R, Powell S, Edwards K.J. Exceptionally high levels of multiple mating in an army ant. Naturwissenschaften. 2004;91:396–399. doi: 10.1007/s00114-004-0546-4. doi:10.1007/s00114-004-0546-4 [DOI] [PubMed] [Google Scholar]
- Dreller C. Division of labor between scouts and recruits: genetic influence and mechanisms. Behav. Ecol. Sociobiol. 1998;43:191–196. doi:10.1007/s002650050480 [Google Scholar]
- Fuchs S, Schade V. Lower performance in honeybee colonies of uniform paternity. Apidologie. 1994;25:155–168. doi:10.1051/apido:19940204 [Google Scholar]
- Guzman-Novoa E, Gary N.E. Genotypic variability of components of foraging behavior in honey bees (Hymenoptera: Apidae) J. Econ. Entomol. 1993;86:715–721. [Google Scholar]
- Haberl M, Moritz R.F.A. Estimation of intracolonial worker relationship in a honey bee colony (Apis mellifera L.) using DNA fingerprinting. Insect. Soc. 1994;41:263–272. doi:10.1007/BF01242297 [Google Scholar]
- Hughes W.O.H, Boomsma J.J. Genetic diversity and disease resistance in leaf-cutting ant societies. Evolution. 2004;58:1251–1260. doi: 10.1554/03-546. doi:10.1554/03-546 [DOI] [PubMed] [Google Scholar]
- Jones J.C, Myerscough M.R, Graham S, Oldroyd B.P. Honey bee nest thermoregulation: diversity promotes stability. Science. 2004;305:402–404. doi: 10.1126/science.1096340. doi:10.1126/science.1096340 [DOI] [PubMed] [Google Scholar]
- Judd T.M. The waggle dance of the honey bee: which bees following a dancer successfully acquire the information? J. Insect. Behav. 1995;8:343–354. doi:10.1007/BF01989363 [Google Scholar]
- Kronauer D.J.C, Schöning C, Pedersen J.S, Boomsma J.J, Gadau J. Extreme queen-mating frequency and colony fission in African army ants. Mol. Ecol. 2004;13:2381–2388. doi: 10.1111/j.1365-294X.2004.02262.x. doi:10.1111/j.1365-294X.2004.02262.x [DOI] [PubMed] [Google Scholar]
- Mattila H.R, Seeley T.D. Genetic diversity in honey bee colonies enhances productivity and fitness. Science. 2007;317:362–364. doi: 10.1126/science.1143046. doi:10.1126/science.1143046 [DOI] [PubMed] [Google Scholar]
- Nieh J.C. The honey bee shaking signal: function and design of a modulatory communication signal. Behav. Ecol. Sociobiol. 1998;42:23–36. doi:10.1007/s002650050408 [Google Scholar]
- Oldroyd B.P, Rinderer T.E, Harbo J.R, Buco S.M. Effects of intracolonial genetic diversity on honey bee (Hymenoptera: Apidae) colony performance. Ann. Entomol. Soc. Am. 1992;85:335–343. [Google Scholar]
- Oldroyd B.P, Rinderer T.E, Buco S.M, Beaman L.D. Genetic variance in honey bees for preferred foraging distance. Anim. Behav. 1993;45:323–332. doi:10.1006/anbe.1993.1037 [Google Scholar]
- Page R.E, Jr, Robinson G.E, Fondrk M.K, Nasr M.E. Effects of worker genotypic diversity on honey bee colony development and behavior (Apis mellifera L.) Behav. Ecol. Sociobiol. 1995;36:387–396. doi:10.1007/s002650050161 [Google Scholar]
- Rheindt F.E, Gadau J, Strehl C.-P, Hölldobler B. Extremely high mating frequency in the Florida harvester ant (Pogonomyrmex badius) Behav. Ecol. Sociobiol. 2004;56:472–481. doi:10.1007/s00265-004-0808-3 [Google Scholar]
- Robinson G.E, Page R.E., Jr . Genetic basis for division of labor in an insect society. In: Breed M.D, Page R.E Jr, editors. The genetics of social evolution. Westview Press; Boulder, CO: 1989. pp. 61–80. [Google Scholar]
- SAS Institute. SAS/STAT 9.1 user's guide. SAS Institute, Inc; Cary, NC: 2004. [Google Scholar]
- Schneider S.S, Lewis L.A. The vibration signal, modulatory communication and the organization of labor in honey bees, Apis mellifera. Apidologie. 2004;35:117–131. doi:10.1051/apido:2004006 [Google Scholar]
- Schneider S.S, Stamps J.A, Gary N.E. The vibration dance of the honey bee. I. Communication regulating foraging on two time scales. Anim. Behav. 1986;34:377–385. doi:10.1016/S0003-3472(86)80105-1 [Google Scholar]
- Seeley T.D. Honey bee foragers as sensory units of their colonies. Behav. Ecol. Sociobiol. 1994;34:51–62. doi:10.1007/s002650050018 [Google Scholar]
- Seeley T.D. Harvard University Press; Cambridge, MA: 1995. The wisdom of the hive. [Google Scholar]
- Seeley T.D, Tarpy D.R. Queen promiscuity lowers disease within honeybee colonies. Proc. R. Soc. B. 2007;274:67–72. doi: 10.1098/rspb.2006.3702. doi:10.1098/rspb.2006.3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley T.D, Visscher P.K. Survival of honeybees in cold climates: the critical timing of colony growth and reproduction. Ecol. Entomol. 1985;10:81–88. [Google Scholar]
- Seeley T.D, Weidenmüller A, Kühnholz S. The shaking signal of the honey bee informs workers to prepare for greater activity. Ethology. 1998;104:10–26. [Google Scholar]
- Strassmann J. The rarity of multiple mating by females in the social Hymenoptera. Insect. Soc. 2001;48:1–13. doi:10.1007/PL00001737 [Google Scholar]
- Tarpy D.R, Nielsen D.I. Sampling error, effective paternity, and estimating the genetic structure of honey bee colonies (Hymenoptera: Apidae) Ann. Entomol. Soc. Am. 2002;95:513–528. doi:10.1603/0013-8746(2002)095[0513:SEEPAE]2.0CO;2 [Google Scholar]
- Tarpy D.R, Page R.E., Jr Sex determination and the evolution of polyandry in honey bees (Apis mellifera) Behav. Ecol. Sociobiol. 2002;52:143–150. doi:10.1007/s00265-002-0498-7 [Google Scholar]
- Tarpy D.R, Seeley T.D. Lower disease infections in honeybee (Apis mellifera) colonies headed by polyandrous vs monandrous queens. Naturwissenschaften. 2006;93:195–199. doi: 10.1007/s00114-006-0091-4. doi:10.1007/s00114-006-0091-4 [DOI] [PubMed] [Google Scholar]
- von Frisch K. Harvard University Press; Cambridge, MA: 1967. The dance language and orientation of bees. [Google Scholar]
- Wiernasz D.C, Perroni C.L, Cole B.J. Polyandry and fitness in the western harvester ant, Pogonomyrmex occidentalis. Mol. Ecol. 2004;13:1601–1606. doi: 10.1111/j.1365-294X.2004.02153.x. doi:10.1111/j.1365-294X.2004.02153.x [DOI] [PubMed] [Google Scholar]