Abstract
Copper is essential for proper functioning of cytochrome c oxidases, and therefore for cellular respiration in eukaryotes and many bacteria. Here we show that a new periplasmic protein (PCuAC) selectively inserts Cu(I) ions into subunit II of Thermus thermophilus ba3 oxidase to generate a native CuA site. The purported metallochaperone Sco1 is unable to deliver copper ions; instead, it works as a thiol-disulfide reductase to maintain the correct oxidation state of the CuA cysteine ligands.
CuA is a dinuclear copper site within the soluble domain of subunit II (Cox2) of bacterial and eukaryotic cytochrome c oxidases (CcOs), whose function is to convey electrons from a soluble cytochrome c to the catalytic heme a3-CuB center of CcO (refs. 1,2). The proper assembly of the CuA site is essential for the catalytic machinery of a functional oxidase. Several proteins have been identified as key players in the delivery of metal ions to the CuA site3, but the detailed molecular mechanisms and the specific roles of each protein are poorly understood4. In prokaryotes two protein families have been proposed to be involved in CuA site formation. The first includes proteins that are able to bind Cu(I) through methionine and histidine residues arranged in a highly conserved H(M)X10MX21HXM motif 5 (referred to as periplasmic CuA chaperone (PCuAC) hereafter). The second consists of the Sco proteins, whose mechanism of action in CuA assembly as thioredoxins or metallochaperones is still debated6. These proteins (PCuAC and Sco) are often found in the same bacterial operon, and most of the identified operons that encode Sco also contain a gene for Cox2 (ref. 7). PCuAC and Sco proteins occur together in the Vibrio cholerae bacterium (http://string.embl.de).
The CuA-containing subunit II from Thermus thermophilus ba3 oxidase (Tt CuA hereafter), T. thermophilus PCuAC (Tt PCuAC) and T. thermophilus Sco1 (Tt Sco1), which are all located in the bacterial periplasm, were expressed as truncated soluble versions in which the transmembrane helices (and the signal peptide region in Tt PCuAC) were not included, thus resulting in constructs of 136, 172 and 121 amino acids, respectively (Supplementary Methods and Supplementary Fig. 1 online).
Tt PCuAC binds one equivalent of Cu(I), as also observed for the homologous protein DR1885 (ref. 5). The Cu(I) binding affinity, measured by competition experiments with DTT followed by 1H-15N HSQC, is (2.2 ± 0.1) × 10−13 M (Supplementary Fig. 2, Supplementary Data and Supplementary Methods online). Upon addition of Cu(I) up to a 1:1 metal/protein ratio, the most affected region includes residues His46, Met61, Met83 and His85 and the neighboring residues (Supplementary Fig. 2), thus defining the copper binding ligands (similar to the Met3His ligand set found for DR1885 in homologous positions).
Sco proteins, in contrast to the Tt PCuAC protein family, are able to bind both Cu(I) and Cu(II) ions8–10. Cu(I) binding followed by NMR confirmed the involvement of metal ligands conserved in all Sco1 proteins (Cys47, Cys51 and His137) (Supplementary Fig. 2 and Supplementary Data). Apo-Tt Sco1 showed weaker Cu(I) binding capabilities compared with its eukaryotic homologs (a lower limit of ~10−10 M was estimated for the Kd of Cu(I)–Tt Sco1). Cu(II)–Tt Sco1 presents spectroscopic features resembling those of the human and Bacillus subtilis homologs8,10,11 (Supplementary Data and Supplementary Fig. 3 online).
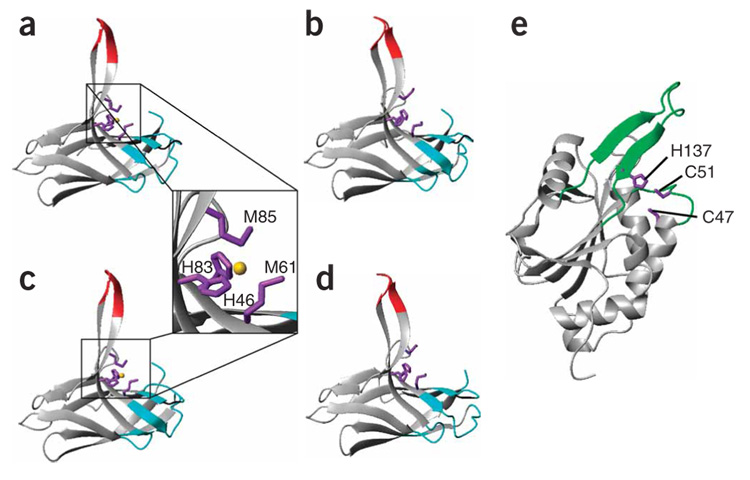
The solution structures of apo-Tt PCuAC, Cu(I)–Tt PCuAC and apo-Tt Sco1 were determined using NMR-derived restraints (Supplementary Methods). The NMR spectra of apo- and Cu(I)–Tt PCuAC variants show two sets of signals in a 65:35 ratio (Supplementary Fig. 3) for 23 and 24 residues, respectively, which originate from a cis-trans isomerization of the Gly13-Pro14 peptide bond. The two structures resulting from this isomerism in both apo- and Cu(I)–Tt PCuAC were obtained (Fig. 1a–d and Supplementary Tables 1 and 2 online). The structural differences induced by this isomerization are localized in loops 1 and 8 and do not affect Cu(I) binding (Fig. 1a–d). Tt PCuAC is arranged in a cupredoxin-like fold (Fig. 1a–d), except that strands µ4 and µ5 form an extended, flexible solvent-exposed µ-hairpin that is longer than the one reported for the homologous DR1885 protein5. The Cu(I) ion is coordinated in a tetrahedral arrangement to the sulfur atoms of Met61 and Met85, the Nδ 1 atom of His46 and the Nε2 atom of His83 (Fig. 1a,c). The structure of apo-Tt Sco1 in the reduced form adopts the thioredoxin-like fold already observed for all Sco1 homologs12–16 (Fig. 1e and Supplementary Table 3 online). Loop 8 (which includes the metal ligand His137) adopts an extended conformation in Tt Sco1 (Fig. 1e) resembling that observed for the human and yeast proteins, in contrast with the shorter (and less extended) loop 8 present in Bs Sco1. The cysteine metal ligands are solvent exposed (as in other Sco1 proteins), which is in agreement with the observation of rapid air oxidation.
Figure 1. Solution structures of apo-Tt PCuAC, Cu(I)–Tt PCuAC and apo-Tt Sco1.
(a–d) The trans (a,b) and cis (c,d) conformations resulting from Pro14 isomerization in Tt PCuAC are shown for each protein. The methionine and histidine residues that bind the metal are shown as purple sticks in Tt PCuAC. The Cu(I) ion is rendered as a golden sphere. The inset highlights the metal binding site of Cu(I)–Tt PCuAC. Residues experiencing double conformations in Tt PCuAC as a consequence of cis-trans proline isomerization are shown in cyan, and those displaying fast backbone motions at 298 K are shown in red. (e) The metal ligands Cys47, Cys51 and His137 of apo-Tt Sco1 are shown as purple sticks, and loops 3 and 8 are shown in green.
The availability of the NMR resonance assignments of the apo and the metallated forms of the two possible copper donors, as well as that of the CuA-containing soluble fragment of the T. thermophilus ba3 oxidase, allowed us to investigate, through NMR, copper uptake by the CuA fragment. We simultaneously monitored the occurrence of copper transfer and the formation of the correct metallated form, and also identified the copper donor protein, by detecting the resulting apo state of the protein that had transferred the copper ions. This strategy also allows the identification of possible transient intermediates.
Addition of Cu(I) to reduced 15N apo-Tt CuA under anaerobic conditions gives rise to the fully metallated protein in the reduced state (Supplementary Data and Supplementary Fig. 4 online). Exposure of this species to oxygen resulted in the formation of a purple species with the characteristic electronic spectrum and 1H NMR signals of the oxidized, mixed-valence CuA center (Supplementary Fig. 4)17. This indicates that the CuA center can be formed in vitro without the assistance of any protein when the cysteine residues of the CuA center are reduced. The affinity of both copper ions in Tt CuA is in the femtomolar range based on competition studies with DTT (Supplementary Methods) and is thus higher than that of Tt PCuAC and Tt Sco1. However, because Cu(I) is not freely available in the periplasmic space, a Cu(I) chaperone is needed to deliver two Cu(I) ions to the apo-CuA protein.
We initially explored the possible role of Sco1 as a Cu(I) or Cu(II) donor to apo-CuA. No evidence of copper uptake by the CuA protein or of metal depletion of Tt Sco1 was observed, which suggests that Tt Sco1 is not responsible for the direct delivery of Cu(I) or Cu(II) ions into apo-Tt CuA.
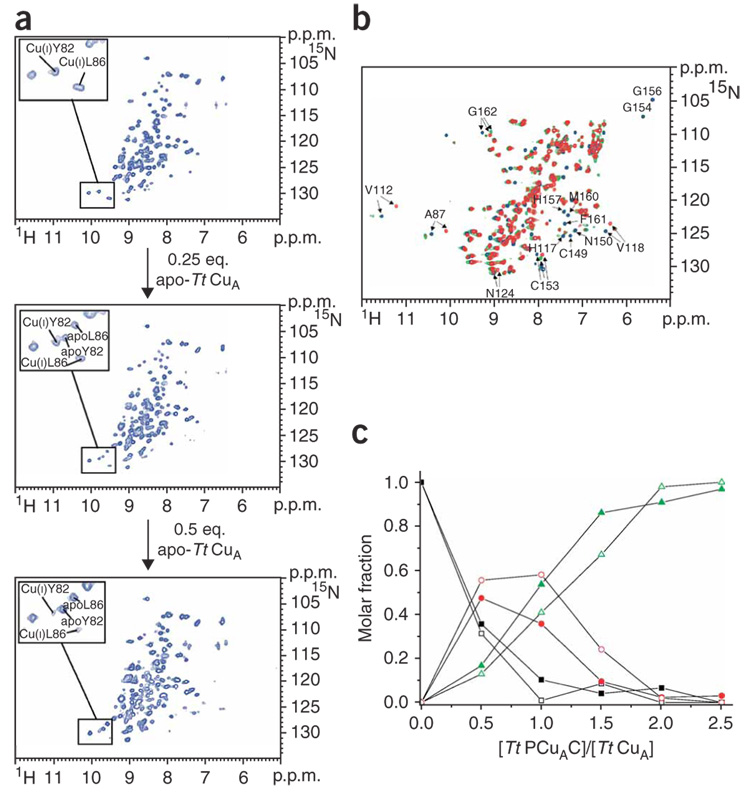
Genomic studies have suggested that Tt PCuAC homologs could be involved in copper transport in the periplasmic space of bacteria5,18, and we decided to test this previously unexplored hypothesis. When apo-Tt CuA was added to a solution of 15N Cu(I)–Tt PCuAC, a new set of resonances corresponding to apo-Tt PCuAC was observed (Fig. 2a). A step-wise titration led to the progressive disappearance of the signals corresponding to Cu(I)–Tt PCuAC, with the concomitant increase of signals from the apo form (Fig. 2a). This process was complete at a Tt CuA/Tt PCuAC ratio of 1:2 (Fig. 2a). The complementary experiment to characterize the metallated species was performed by adding two equivalents of unlabeled Cu(I)–Tt PCuAC to 15N apo-Tt CuA, which led to the typical 1H-15N HSQC pattern of native Cu(I)2–Tt CuA (Fig. 2b). A step-wise titration disclosed the formation of an intermediate species when less than one equivalent of Cu(I)–Tt PCuAC was added to the CuA domain (Fig. 2b,c). This species then converted into the dimetallated Cu(I)2–Tt CuA upon further addition of the donor protein (Fig. 2c). This intermediate species displayed resonances with chemical shifts differing from those of both the apo- and the fully metallated species (Fig. 2b), and it did not show any line broadening with respect to the other forms of the CuA domain. This behavior, together with the observation that no intermediate was identified by monitoring the copper release from Cu(I)–Tt PCuAC to apo-Tt CuA, allowed us to rule out the detection of a possible complex between the two proteins, and suggested that this intermediate corresponds to a singly metallated CuA species. We conclude that Tt PCuAC is capable of transferring two Cu(I) ions to the reduced apo-Tt CuA site sequentially, thereby eliciting the formation of the binuclear CuA center in the reduced state (Fig. 2c). Aerobic oxidation of this mixture leads to the mixed-valence, oxidized CuA center, as revealed by its characteristic electronic spectrum (Supplementary Fig. 4).
Figure 2. Cu(I) transfer reaction between Cu(I)–Tt PCuAC and apo-Tt CuA.
(a) Titration of 15N-labeled Cu(I)–Tt PCuAC with unlabeled apo-Tt CuA followed by 1H-15N HSQC experiments. In the insets, residues followed to map the metal exchange reaction are shown. (b) Overlay of the 1H-15N HSQC spectra of 15N apo-Tt CuA (red), and after addition of one (green) and two (blue) equivalents of unlabeled Cu(I)–Tt PCuAC. Some residues indicating the formation of Cu(I)2–Tt CuA and of a partially metallated CuA species are labeled. (c) Plot showing the relative concentrations of Cu(I)2–Tt CuA (green), the partially metallated CuA protein (red) and apo-Tt CuA (black) as a function of the Tt PCuAC/Tt CuA molar ratio. The signals of residues Cys153 (solid symbols) and Gly162 (open symbols) (whose 1H-15N chemical shifts substantially change depending on the metallation state of Tt CuA) have been selected to evaluate the molar fractions of Tt CuA forms.
Sco1 is able to bind Cu(II) ions. The possible direct transfer of Cu(II) ions was explored by following the titration of a sample of 15N-labeled Cu(II)–Tt Sco1 with unlabeled apo-Tt CuA. There was no evidence of the formation of apo-Tt Sco1 in the 1H-15N HSQC spectra, thus revealing the inability of this protein to transfer Cu(II) ions to the CuA domain. When 2 equivalents of unlabeled Cu(I)–Tt PCuAC were added to the mixture of 15N-labeled Cu(II)–Tt Sco1 and 15N-labeled apo-Tt CuA, the reduced Cu(I)2–Tt CuA center was formed (Supplementary Fig. 5 online). This result indicates that (i) the presence of Cu(II)–Tt Sco1 does not prevent Cu(I)–Tt PCuAC from transferring Cu(I), and (ii) as the final product is the reduced Cu(I)2–Tt CuA center, the mechanism cannot involve the concerted action of both Cu(II)–Tt Sco1 and Cu(I)–Tt PCuAC, which would give rise to a mixed-valence, oxidized CuA site.
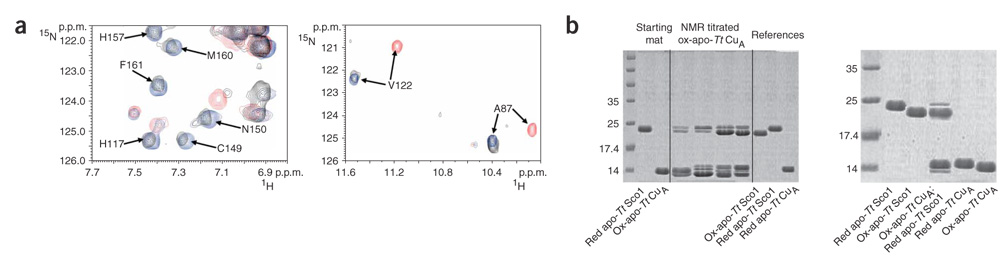
Sco1, which has already been shown to be essential for copper upload into the CuA site in B. subtilis19, is characterized by a thioredoxin-like fold. We therefore investigated the possible thiol-disulfide oxidoreductase activity of Tt Sco1 during copper uptake. Oxidized apo-Tt CuA, in which the two cysteine ligands had been aerobically oxidized to form a disulphide bond (ox-apo-Tt CuA hereafter), was not able to bind copper ions when Cu(I)–Tt PCuAC was added. However, when 15N-labeled ox-apo-Tt CuA was titrated with a 2:1 mixture of Cu(I)– Tt PCuAC and reduced apo-Tt Sco1, the backbone resonances of the native Cu(I)2–Tt CuA appeared (Fig. 3a). Exposure of the reaction sample to atmospheric oxygen allowed the development of the typical UV-vis spectrum of the oxidized, mixed-valence CuA center, which confirmed that the native site had formed. A 4-acetamide-4′- amleimidylstilbene-2,2′-disulfonic acid, disodium salt (AMS)-reacted SDS gel of the reaction mixture (run under nonreducing conditions, Supplementary Methods) indicated that, at a Tt CuA/Tt Sco1/Tt PCuAC ratio of 1:1:2, Tt Sco1 is mostly in the oxidized state (Fig. 3b). Similarly, in a 1:1 Tt CuA/Tt Sco1 mixture, Tt Sco1 is essentially in an oxidized state, whereas Tt CuA is in a reduced state (Fig. 3b). These experiments show that Tt Sco1 is able to reduce the disulfide bond of ox-apo-Tt CuA protein, which demonstrates that this protein behaves as a thiol-disulfide reductase in in vitro CuA assembly.
Figure 3. Cu(I) transfer from Cu(I)–Tt PCuAC to oxidized apo-Tt CuA in the presence of reduced apo-Tt Sco1.
(a) 1H-15N HSQC overlay of oxidized apo-Tt CuA (red), Cu(I)2–Tt CuA (blue) and the final mixture of the titration between 15N-labeled oxidized apo-Tt CuA and unlabeled, reduced apo-Tt Sco1 and Cu(I)–Tt PCuAC in a 1:1:2 ratio, in the absence of DTT (black). (b) Left panel: AMS-reacted, nonreducing SDS gel of different aliquots from the NMR titration between 15N-labeled oxidized apo-Tt CuA and a 1:2 mixture of unlabeled, reduced apo-Tt Sco1 and Cu(I)–Tt PCuAC (shown in a). Four steps of the NMR titration are reported. The band of Tt CuA protein at 14.8 kDa is very close to that of Tt PCuAC, which indeed has a very similar molecular weight (13.2 kDa). Starting materials and the Tt PCuAC and Tt CuA proteins in their different redox states are also reported as a reference. Right panel: AMS-reacted, nonreducing SDS gel of a protein mixture containing oxidized apo-Tt CuA and reduced apo-Tt Sco1 in a 1:1 ratio. The proteins in their different redox states are also reported as a reference. Molecular weight markers are shown in first lanes.
Although it is possible that the in vivo pathway is more complex, the structural characterization of Tt PCuAC and of its copper binding properties and its ability to selectively and sequentially deliver two Cu(I) ions to apo-Tt CuA, thus giving rise to the native Cu(I)2–Tt CuA site in vitro, strongly supports the annotation of this protein as a periplasmic Cu(I) chaperone. Our data also indicate that Tt Sco1 is able to reduce the disulfide bond of the CuA center, thus allowing the CuA site to accept Cu(I) ions from Tt PCuAC. The mechanism of bacterial CuA assembly therefore consists of a sequential insertion of two Cu(I) ions donated by a metallochaperone once the disulfide bond of the CuA center is reduced by a thioredoxin. In eukaryotes the assembly of the CuA site is different, as Scos have a larger affinity for Cu(I) and may act as both chaperones and thioredoxins12,20–22.
Supplementary Material
Note: Supplementary information is available on the Nature Chemical Biology website.
ACKNOWLEDGMENTS
This work was supported by the European Commission (European Network of Research Infrastructures for Providing Access and Technological Advancements in Bio-NMR contract n° 026145, SPINE2-Complexes contract n° LSHG-CT-2006-031220 and Marie Curie host fellowships for early stage research training n° MEST-CT-2004-504391, NMR in Inorganic Structural Biology) and by a grant from Ente Cassa di Risparmio di Firenze. Work in Rosario (Argentina) was supported by the US National Institutes of Health (R01-GM068682), the Howard Hughes Medical Institute and Agencia Nacional de Promoción Científica y Tecnoló gica (PME2003-0026 and PICT2002-01-11625) grants to A.J.V. L.A.A. thanks Consejo Nacional de Investigaciones Científícas y Técnicas for a doctoral fellowship. We thank D. Winge (Departments of Medicine and Biochemistry, University of Utah Health Sciences Center, University of Utah) for kindly providing the expression plasmid for Tt Sco1.
Footnotes
Published online at http://www.nature.com/naturechemicalbiology/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Ostermeier C, Iwata S, Michel H. Curr. Opin. Struct Biol. 1996;6:460–466. doi: 10.1016/s0959-440x(96)80110-2. [DOI] [PubMed] [Google Scholar]
- 2.Maneg O, Malatesta F, Ludwig B, Drosou V. Biochim Biophys. Acta. 2004;1655:274–281. doi: 10.1016/j.bbabio.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Carr HS, Winge DR. Acc. Chem. Res. 2003;36:309–316. doi: 10.1021/ar0200807. [DOI] [PubMed] [Google Scholar]
- 4.Cobine PA, Pierrel F, Winge DR. Biochim. Biophys. Acta. 2006;1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Banci L, et al. Proc. Natl. Acad. Sci. USA. 2005;102:3994–3999. doi: 10.1073/pnas.0406150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khalimonchuk O, Winge DR. Biochim. Biophys. Acta. 2008;1783:618–628. doi: 10.1016/j.bbamcr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banci L, Bertini I, Cavallaro G, Rosato A. J. Proteome Res. 2007;6:1568–1579. doi: 10.1021/pr060538p. [DOI] [PubMed] [Google Scholar]
- 8.Horng YC, et al. J. Biol. Chem. 2005;280:34113–34122. doi: 10.1074/jbc.M506801200. [DOI] [PubMed] [Google Scholar]
- 9.McEwan AG, et al. FEBS Lett. 2002;518:10–16. doi: 10.1016/s0014-5793(02)02532-2. [DOI] [PubMed] [Google Scholar]
- 10.Andruzzi L, Nakano M, Nilges MJ, Blackburn NJ. J. Am. Chem. Soc. 2005;127:16548–16558. doi: 10.1021/ja0529539. [DOI] [PubMed] [Google Scholar]
- 11.Imriskova-Sosova I, et al. Biochemistry. 2005;44:16949–16956. doi: 10.1021/bi051343i. [DOI] [PubMed] [Google Scholar]
- 12.Banci L, et al. Proc. Natl. Acad. Sci. USA. 2006;103:8595–8600. doi: 10.1073/pnas.0601375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams JC, et al. J. Biol. Chem. 2005;280:15202–15211. doi: 10.1074/jbc.M410705200. [DOI] [PubMed] [Google Scholar]
- 14.Balatri E, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Structure. 2003;11:1431–1443. doi: 10.1016/j.str.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Ye Q, Imriskova-Sosova I, Hill BC, Jia Z. Biochemistry. 2005;44:2934–2942. doi: 10.1021/bi0480537. [DOI] [PubMed] [Google Scholar]
- 16.Abajian C, Rosenzweig AC. J. Biol. Inorg. Chem. 2006;11:459–466. doi: 10.1007/s00775-006-0096-7. [DOI] [PubMed] [Google Scholar]
- 17.Bertini I, et al. J. Am. Chem. Soc. 1996;118:11658–11659. [Google Scholar]
- 18.Arnesano F, Banci L, Bertini I, Martinelli M. J. Proteome Res. 2005;4:63–70. doi: 10.1021/pr049862f. [DOI] [PubMed] [Google Scholar]
- 19.Mattatall NR, Jazairi J, Hill BC. J. Biol. Chem. 2000;275:28802–28809. doi: 10.1074/jbc.M002741200. [DOI] [PubMed] [Google Scholar]
- 20.Banci L, et al. Proc. Natl. Acad. Sci. USA. 2007;104:15–20. [Google Scholar]
- 21.Banci L, et al. Structure. 2007;15:1132–1140. doi: 10.1016/j.str.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Banci L, et al. Proc. Natl. Acad. Sci. USA. 2008;105:6803–6808. doi: 10.1073/pnas.0800019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Chemical Biology website.