Abstract
This paper reports the design, synthesis, and characterization of a family of cyclic peptides that mimic protein quaternary structure through β-sheet interactions. These peptides are 54-membered-ring macrocycles comprising an extended heptapeptide β-strand, two Hao β-strand mimics [JACS 2000, 122, 7654] joined by one additional α-amino acid, and two δ-linked ornithine β-turn mimics [JACS 2003, 125, 876]. Peptide 3a, as the representative of these cyclic peptides, contains a heptapeptide sequence (TSFTYTS) adapted from the dimerization interface of protein NuG2 [PDB ID: 1mio]. 1H NMR studies of aqueous solutions of peptide 3a show a partially folded monomer in slow exchange with a strongly folded oligomer. NOE studies clearly show that the peptide self-associates through edge-to-edge β-sheet dimerization. Pulsed-field gradient (PFG) NMR diffusion coefficient measurements and analytical ultracentrifugation (AUC) studies establish that the oligomer is a tetramer. Collectively, these experiments suggest a model in which cyclic peptide 3a oligomerizes to form a dimer of β-sheet dimers. In this tetrameric β-sheet sandwich, the macrocyclic peptide 3a is folded to form a β-sheet, the β-sheet is dimerized through edge-to-edge interactions, and this dimer is further dimerized through hydrophobic face-to-face interactions involving the Phe and Tyr groups. Further studies of peptides 3b–3n, which are homologues of peptide 3a with 1–6 variations in the heptapeptide sequence, elucidate the importance of the heptapeptide sequence in the folding and oligomerization of this family of cyclic peptides. Studies of peptides 3b–3g show that aromatic residues across from Hao improve folding of the peptide, while studies of peptides 3h–3n indicate that hydrophobic residues at positions R3 and R5 of the heptapeptide sequence are important in oligomerization.
Introduction
β-Sheet interactions occur widely in protein folding and in protein-protein interactions: While intramolecular β-sheet interactions are important in protein secondary and tertiary structures, intermolecular β-sheet interactions are important in protein quaternary structure, protein molecular recognition, and protein aggregation.1 Intermolecular interactions are involved in HIV2 and cancer,3 and in neurodegenerative diseases such as Alzheimer’s, Huntington’s, and prion protein diseases.4 These interactions are also essential for the activity of many normal mammalian proteins, such as the Met repressor5 and defensin HNP-3,6 which function as β-sheet dimers.
Studies of β-sheet interactions using model systems have mostly focused on mimicking the secondary and tertiary structures of proteins with peptides that fold into β-sheet structures. Studies on β-hairpins, as the simplest β-sheet structures, have enhanced the general knowledge about the kinetics and thermodynamics of β-sheet folding in proteins.7 Large peptides that fold into larger β-sheet structures have been created either through de novo design or by employing the lessons learned from studying β-hairpins.7,8 Peptides that fold into β-sheet structures have also been created by combining amino acids and peptidomimetic templates that mimic β-turns and β-sheets.7,9
The mimicry of β-sheet quaternary interactions with simple model systems has received relatively little attention, despite the importance of these interactions in protein quaternary structure and molecular recognition. The design of simple model systems that participate in well-defined intermolecular β-sheet interactions is particularly challenging, because the β-strands that comprise β-sheets can uncontrollably assemble. Each β-strand presents two edges bearing “sticky” hydrogen-bonding groups, as well as two faces bearing side chains that can participate in additional interactions. For this reason, contemporary studies of β-sheet interactions have generally focused on creating large supramolecular assemblies or aggregates involving extended edge-to-edge and face-to-face interactions.10,11
A number of peptidic model systems that mimic β-sheet quaternary structure through the formation of dimers have been developed.12,13 Most of these systems dimerize in noncompetitive organic solvents and are designed to allow hydrogen bonding from only one edge of the β-sheet, with the other edge being blocked to avoid uncontrolled aggregation. The development of systems that form β-sheet dimers or other small well-defined oligomers in water has proven more challenging. In aqueous solution, hydrogen bonding between water molecules and the main-chain amide groups of small peptides strongly competes with the hydrogen bonding involved in intermolecular β-sheet interactions. In contrast to proteins, small peptides lack the large contact areas that can assist dimerization or oligomerization through numerous stabilizing side-chain interactions. Larger peptides can form larger contact areas but often form aggregates, rather than well-defined quaternary structures.
The de novo designed 33-mer βpep peptides, developed by Mayo and coworkers, arguably provide the most successful mimicry of protein β-sheet quaternary structure.8d,14 These 33-mer peptides exhibit a monomer-dimer-tetramer equilibrium, in which hydrophobic interactions stabilize a tetrameric β-sheet sandwich. This tetramer has a long dimerization sequence and forms as a heterodimer of two dimers that have identical sequence but different monomer-monomer interface alignments. A 21-mer peptide developed by Imperiali and coworkers also mimics some elements of protein β-sheet quaternary structure.15 This peptide folds into an α-helix joined to a β-hairpin and forms a homotetramer in which the four helices create a stabilizing hydrophobic core. To better understand and control the β-sheet quaternary structure and interactions of proteins, smaller and simpler water-soluble peptides that participate in well-defined β-sheet interactions are needed.
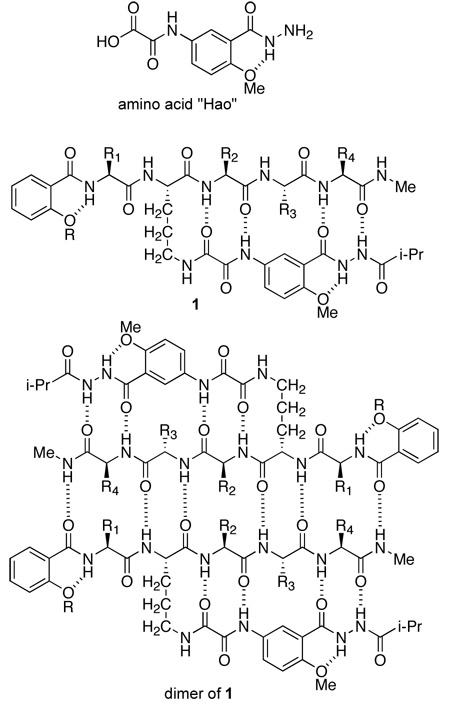
The Nowick research group’s efforts to mimic protein intermolecular β-sheet interactions have focused on developing β-sheet models that dimerize, because dimerization constitutes the simplest form of intermolecular β-sheet interaction and occurs widely among proteins.12,16 We have previously introduced the unnatural amino acid Hao, which behaves as a three-amino acid β-strand mimic and forms hydrogen bonds from only one edge.12b Peptides containing Hao, such as 1, fold into well-defined β-sheet structures and dimerize in weakly competitive organic solvents.12c We have used these peptides to probe sequence selectivity, enantioselectivity, and aromatic interactions in molecular recognition between β-sheets.12d–f
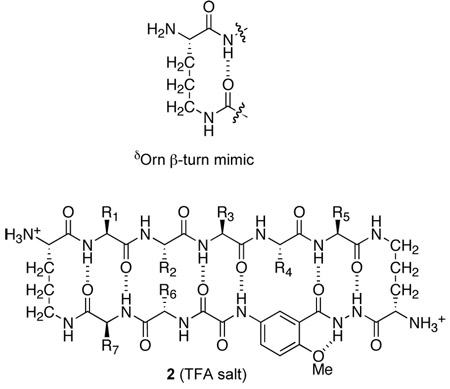
The Nowick research group has also introduced δ-linked ornithine (δOrn) as a new β-turn mimic.12c,17 We have recently used the Hao and δOrn turn units to create cyclic peptides 2 that form β-sheet structures in aqueous solution.18,19 These 42-membered-ring cyclic peptides consist of one Hao unit, two δOrn turn units, and a pentapeptide β-strand. These peptides do not form edge-to-edge β-sheet dimers in water, even though the related peptides 1, which also contain a pentapeptide β-strand, do dimerize in organic solvents. Cyclic peptides 2 appear to be too small to dimerize in water and may lack sufficient side-chain contacts and hydrogen-bonding groups to form well-defined dimers in aqueous solution.
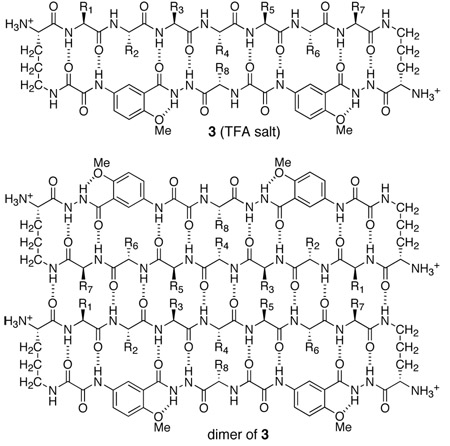
To achieve dimerization in water, we have now designed larger macrocyclic β-sheets 3 containing a heptapeptide β-strand, two Hao units joined by an additional α-amino acid, and two δOrn turn units. We hypothesized that cyclic peptides 3, which comprise a 54-membered ring, would dimerize through additional stabilizing interchain β-sheet interactions and hydrogen bonds.20 Here, we describe the design, synthesis, and evaluation of these β-sheets. Many of these cyclic peptides form tetramers composed of two edge-to-edge β-sheet dimers in water at millimolar concentrations. NOE studies show well-defined folding and dimer formation, while PFG NMR diffusion measurements and analytical ultracentrifugation studies establish the formation of tetramers, and in some cases higher oligomers.
Results and Discussion
This section describes the design, synthesis, and study of peptides 3. Peptide 3a constitutes the archetype of these 54-membered-ring cyclic β-sheets, while peptides 3b–3n constitute variants that permit the investigation of the effect of sequence on folding and oligomerization (Table 1). Subsection I describes studies of peptide 3a, while subsection II describes studies of peptides 3b–3n.
Table 1.
Amino acid sequence at positions R1–R8 for peptides 3a–3n.a
| Peptide | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 |
|---|---|---|---|---|---|---|---|---|
| 3a | Thr | Ser | Phe | Thr | Tyr | Thr | Ser | Lys |
| 3b | Thr | Ser | Phe | Glu | Tyr | Thr | Ser | Lys |
| 3c | Thr | Ser | Phe | Leu | Tyr | Thr | Ser | Lys |
| 3d | Thr | Ser | Phe | Tyr | Tyr | Thr | Ser | Lys |
| 3e | Thr | Leu | Phe | Thr | Tyr | Val | Ser | Lys |
| 3f | Thr | Tyr | Phe | Thr | Tyr | Phe | Ser | Lys |
| 3g | Thr | Tyr | Phe | Thr | Tyr | Tyr | Ser | Lys |
| 3h | Glu | Ser | Phe | Thr | Tyr | Thr | Lys | Lys |
| 3i | Thr | Ser | Leu | Thr | Val | Thr | Ser | Lys |
| 3j | Thr | Tyr | Ile | Thr | Val | Tyr | Ser | Lys |
| 3k | Thr | Tyr | Ile | Thr | Thr | Tyr | Ser | Lys |
| 3l | Thr | Tyr | Ser | Thr | Val | Tyr | Ser | Lys |
| 3m | Thr | Tyr | Ser | Thr | Thr | Tyr | Ser | Lys |
| 3n | Phe | Tyr | Ser | Thr | Thr | Phe | Tyr | Lys |
Peptides 3b–3n are homologues of peptide 3a with 1–6 variations in the amino acid sequence. The variations are shown in bold typeface.
I. Design, Synthesis, and Study of Macrocyclic Peptide 3a
Design of peptide 3a
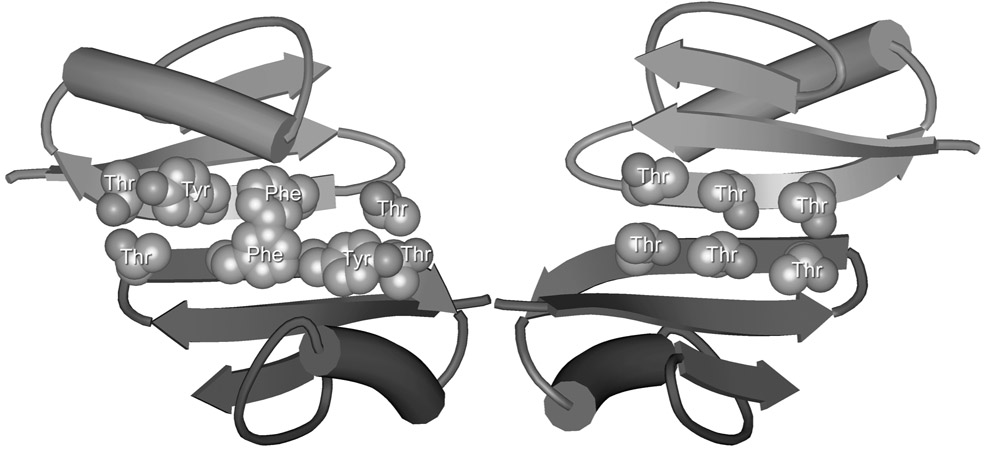
We designed macrocyclic peptide 3a with two Hao units connected by a lysine in the “lower” strand, an extended heptapeptide sequence in the “upper” strand, and two δOrn turn units connecting the two strands. We adapted the heptapeptide sequence (TSFTYTS) from the redesigned protein G variant NuG2 (TTFTYTT, Figure 1), which forms a dimer in the solid state.21 To simplify the NMR studies of the synthetic cyclic peptide, we replaced two of the Thr residues of NuG2 with Ser. We chose lysine as the amino acid connecting the two Hao units to enhance the solubility of the cyclic peptide in water.
Figure 1.
Views from the front and back sides of the crystallographic NuG2 dimer illustrating the heptapeptide β-strand dimerization interface TTFTYTT (PDB ID: 1mio).21
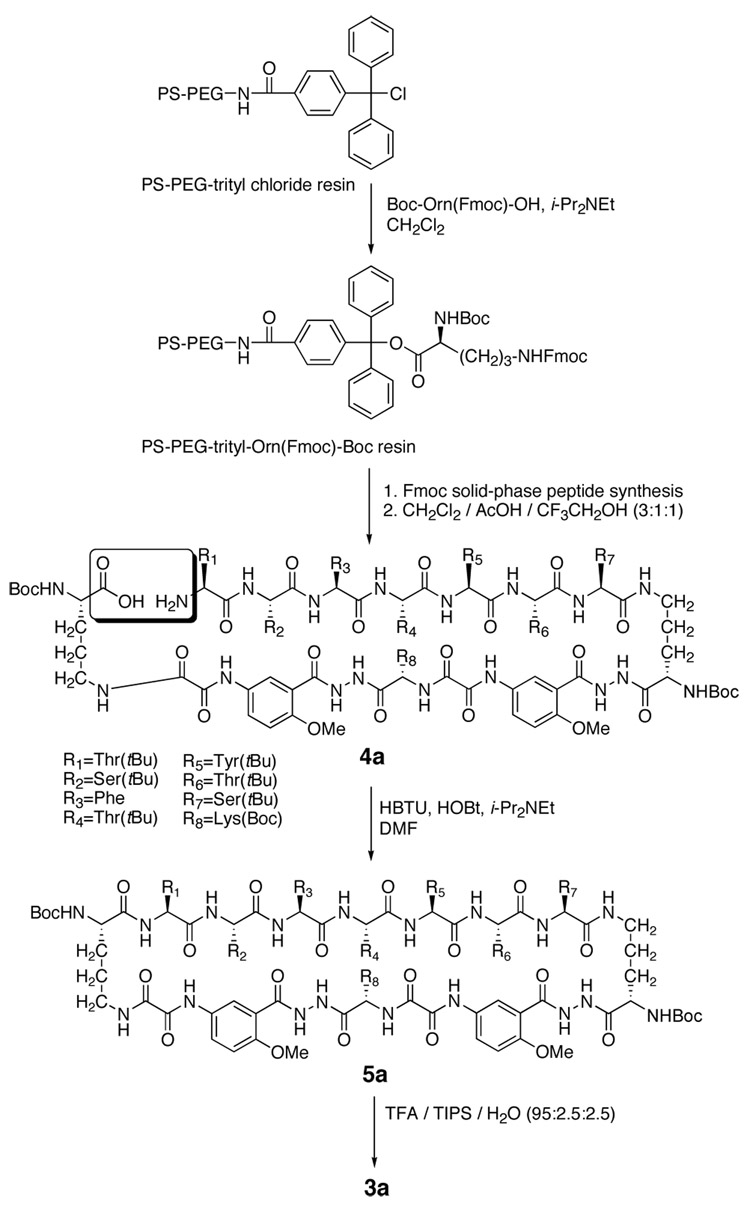
Synthesis of peptide 3a
Peptide 3a was easily prepared by synthesis of the corresponding linear peptide 4a on PS-PEG-trityl resin followed by solution-phase cyclization, side-chain deprotection, and purification. Scheme 1 summarizes the synthesis. The amino acid derivative Boc-Orn(Fmoc)-OH was first loaded on the PS-PEG-trityl chloride resin with i-Pr2NEt in CH2Cl2.22 Protected linear peptide 4a was then assembled on the resin by standard automated Fmoc solid-phase peptide synthesis and was cleaved from the resin under mildly acidic conditions. Peptide 4a was cyclized to protected cyclic peptide 5a by slow addition to HBTU and HOBt in dilute DMF solution. Deprotection with TFA followed by RP-HPLC purification gave peptide 3a as the trifluoroacetate salt in 15–30% overall yield, based on the loading of the resin with Boc-Orn(Fmoc)-OH. Typically, a 0.1-millimole-scale synthesis affords about 30–60 mg of pure cyclic peptide 3a. The peptide exhibits good solubility in water (> 10 mM), and solutions of the peptide are clear and do not form precipitate over time.
Scheme 1.
Synthesis of peptide 3a
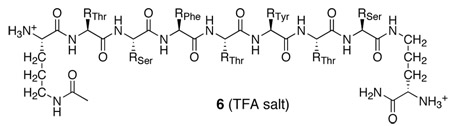
An acyclic control
To determine the effect of cyclicity and the Hao templates in 3a, we synthesized linear peptide 6 as a control. Peptide 6 may be thought of as a homologue of the “upper” peptide strand of 3a. It contains the heptapeptide sequence TSFTYTS and two δOrn units, but lacks the “lower” strand.
Folding and oligomerization of peptide 3a in water
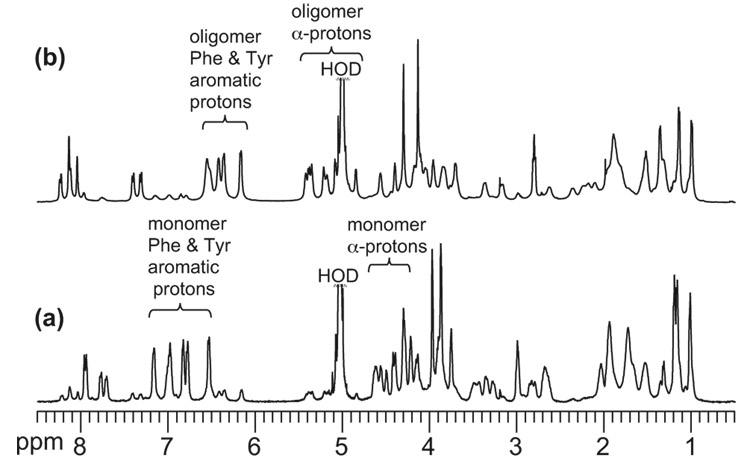
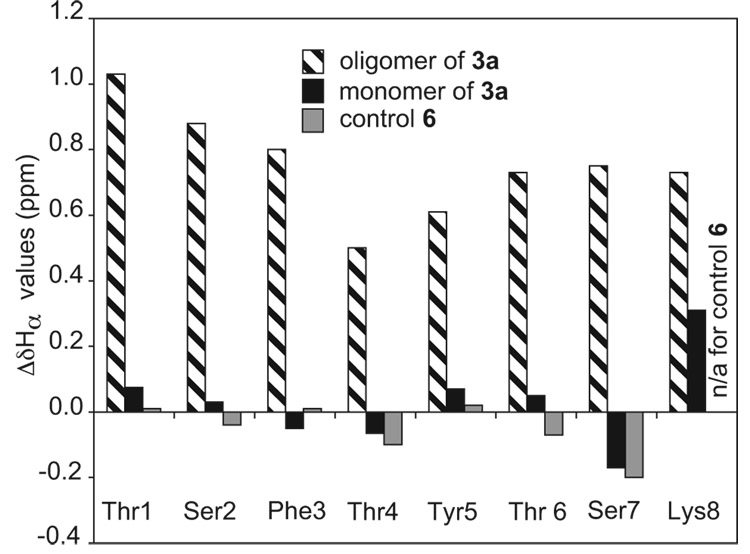
1H NMR spectra of aqueous solutions of peptide 3a show two sets of resonances in proportions that change with concentration (Figure 2). This result indicates the presence of at least two species that exchange slowly on the NMR time scale and suggests that the sets of resonances correspond to monomer and oligomer.23 The α-proton resonances of the oligomer exhibit substantial downfield shifting. While the chemical shifts of the α-proton resonances of the monomer and control peptide 6 are similar to those of amino acids in random coil conformations, those of the oligomer appear 0.5–1.0 ppm downfield. Figure 3 presents the deviation of the α-proton chemical shifts from published random coil values24 (ΔδHα = observed δHα - random coil δHα) for the oligomer, the monomer, and control peptide 6.25 The downfield shifting suggests that the oligomer adopts a β-sheet structure and that the monomer is largely unstructured. The aromatic resonances of the Phe and Tyr residues of the oligomer shift upfield relative to those of the monomer (Figure 2). The upfield shifting suggests aromatic interactions and possibly the formation of a hydrophobic core in the oligomer.
Figure 2.
1H NMR spectra of peptide 3a at concentrations of 1.0 mM (a) and 6.0 mM (b) in D2O at 500 MHz and 280 K.
Figure 3.
Deviation of the α-proton chemical shifts from published random coil values24 (ΔδHα = observed δHα - random coil δHα) for the oligomer, the monomer, and control peptide 6. 1H NMR studies were performed on 8.5 mM and 1.0 mM solutions of peptide 3a and on an 8.0 mM solution of control peptide 6 in D2O at 280 K.
The difference in the chemical shifts of the diastereotopic δ-protons of a δOrn turn unit (ΔδδOrn) reflects the degree of folding of the turn unit, and hence the peptide.17 We have previously observed that a ΔδδOrn value of about 0.6 ppm corresponds to complete folding in water for related peptides.17,18 Both δOrn turn units of peptide 3a exhibit ΔδδOrn values of 0.15 ppm for the monomer and 0.69 ppm for the oligomer. The small ΔδδOrn value for the monomer indicates only partial folding of the turn units, while the large value for the oligomer indicates predominant or complete folding. In contrast, control peptide 6 exhibits ΔδδOrn values of 0.00 and 0.05 ppm, indicating essentially no folding of these units.
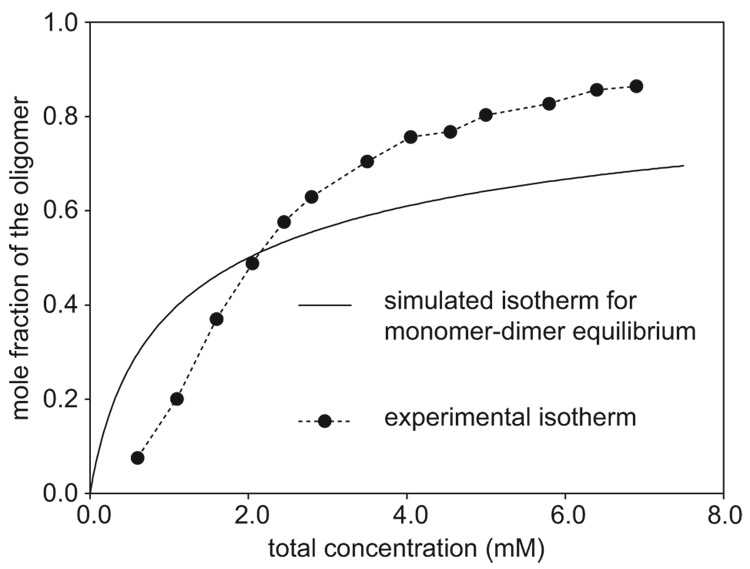
1H NMR concentration studies show a concentration dependence of oligomerization that is not consistent with a monomer-dimer system, but rather with a monomer and a higher oligomer. Figure 4 illustrates this concentration dependence. The relative concentrations of the peptide 3a monomer and oligomer were determined at 280 K by integrating the corresponding 1H NMR resonances. The mole fraction of the oligomer was then calculated and plotted against the total concentration of peptide 3a. The sigmoidal shape and inflection of the curve indicate that oligomerization is cooperative and that the oligomer must be at least a trimer. A simulated isotherm for a monomer-dimer equilibrium (also shown in Figure 4) lacks the sigmoidal shape and inflection and does not fit the data.
Figure 4.
Concentration dependence of oligomer formation of peptide 3a: mole fraction of oligomer vs. total peptide concentration. The relative concentrations of the monomer and oligomer were determined in D2O at 280 K by integrating the corresponding 1H NMR resonances. A simulated isotherm for a monomer-dimer equilibrium (K1,2 = 500 M−1), which is shown for comparison, does not fit the data.
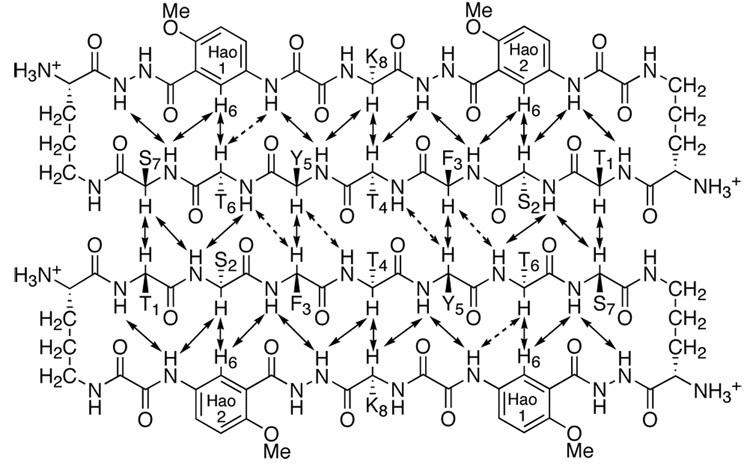
NOESY and ROESY studies of the oligomer in D2O and H2O–D2O (90:10) reveal an extensive network of NOEs associated with folding and edge-to-edge dimerization through antiparallel β-sheet formation. These experiments were carried out at 280 K and at high concentrations of peptide 3a (8.5 mM) to increase signal from the oligomer, reduce signal from the monomer, and minimize interference from overlap of these signals. Figure 5 summarizes the key NOEs between the protons that make up the backbone of the peptide chains (CHα, NH, and Hao-H6).
Figure 5.
Key NOEs involving main-chain inter-residue contacts in the β-sheet dimer of peptide 3a. Dashed double-headed arrows represent weak or ambiguous NOEs.
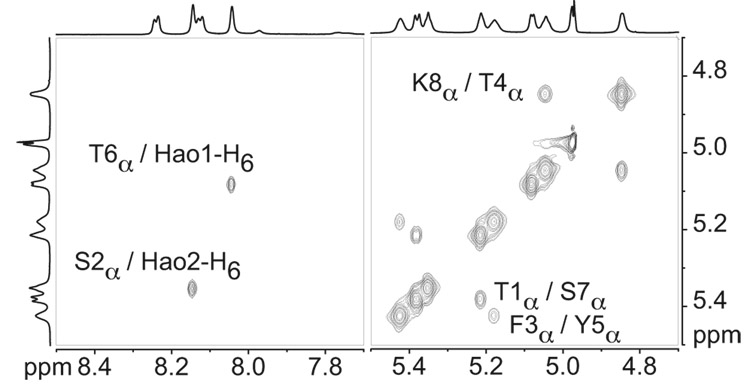
Figure 6 illustrates the five key NOE cross-peaks that are observed in D2O (those not involving NH groups). The intramolecular NOEs between the α-protons of Thr4 and Lys8 (T4α/K8α), between the α-proton of Ser2 and H6 of Hao2 (S2α/Hao2-H6), and between the α-proton of Thr6 and H6 of Hao1 (T6α/Hao1-H6) indicate folding. The intermolecular NOEs between the α-protons of Thr1 and Ser7 (T1α/S7α) and between the α-protons of Phe3 and Tyr5 (F3α/Y5α) indicate edge-to-edge dimerization. Additional NOEs involving NH groups, which further demonstrate folding and dimerization, are observed in H2O–D2O. (The Supporting Information provides additional details.) Collectively, these NOEs establish that macrocyclic peptide 3a folds to form β-sheets and that the β-sheets dimerize through edge-to-edge interactions.
Figure 6.
Selected expansions of the NOESY spectrum of the peptide 3a oligomer in D2O illustrating key NOEs resulting from folding and edge-to-edge dimerization in the oligomer. NOESY studies of an 8.5 mM solution of peptide 3a were performed at 800 MHz and 280 K with a 75-ms mixing time.
In contrast to the oligomer, the monomer of peptide 3a shows only weak long-range NOEs, indicating incomplete folding. 800 MHz NOESY studies of the monomer at submillimolar peptide concentrations show weak cross-peaks associated with several of the key intramolecular interresidue contacts in Figure 5 (e.g., S2α/Hao2-H6, T4α/K8α, and T6α/Hao1-H6). (The Supporting Information provides additional data.) A weak NOE between the α-proton of Tyr5 and H6 of Hao1 (Y5α/Hao1-H6) is also present, suggesting the presence of a minor alternate conformer. In conjunction with the small ΔδHα values and small ΔδδOrn values, the weak NOEs indicate partial β-sheet folding of the monomer.
The aromatic side chains of Phe3 and Tyr5 likely enhance folding of the monomer through interaction with the aromatic rings of the Hao units. Evidence for these aromatic interactions in the monomer of peptide 3a include upfield shifting of the aromatic protons of Phe3 and Tyr5 (0.25–0.35 ppm relative to control peptide 6) and interstrand NOEs between the aromatic protons (F3/Hao2 and Y5/Hao1).
Not surprisingly, control peptide 6 exhibits no long-range NOEs at concentrations up to 8 mM (Supporting Information). The absence of long-range NOEs establishes that control peptide 6 neither folds nor forms a distinct dimer.
Oligomerization state of peptide 3a
The NOE data clearly show that peptide 3a forms an oligomer composed of edge-to-edge dimers, but do not establish whether it is a simple dimer, a dimer of dimers (tetramer), a higher oligomer, or a mixture of oligomers.26 To elucidate the oligomeric state of peptide 3a and determine if the NMR resonances of the oligomer result from a single species or from multiple oligomeric species in rapid equilibrium on the NMR time scale, we performed pulsed-field gradient NMR (PFG NMR) diffusion measurements and analytical ultracentrifugation (AUC) experiments.
PFG NMR diffusion measurements.27,28
Diffusion coefficients (D) provide insight into the molecular weights and association states of molecules and oligomers in solution.28 PFG NMR diffusion studies are particularly well suited to measuring diffusion coefficients, because they permit the diffusion coefficients to be determined using the same NMR sample and conditions used for structural studies.28b In PFG NMR diffusion techniques, a magnetic field gradient is first encoded across the NMR tube, the molecules are then allowed to diffuse during a delay, and the magnetic field gradient is subsequently decoded. The diffusion coefficient is then determined by measuring the attenuation of the NMR signal, which results from diffusion.28c We measured the diffusion coefficients of the monomer and the oligomer of peptide 3a by using an sLED pulse sequence at 800 MHz and observing the attenuation of the signals from these species.28a (The Supporting Information provides experimental details.)
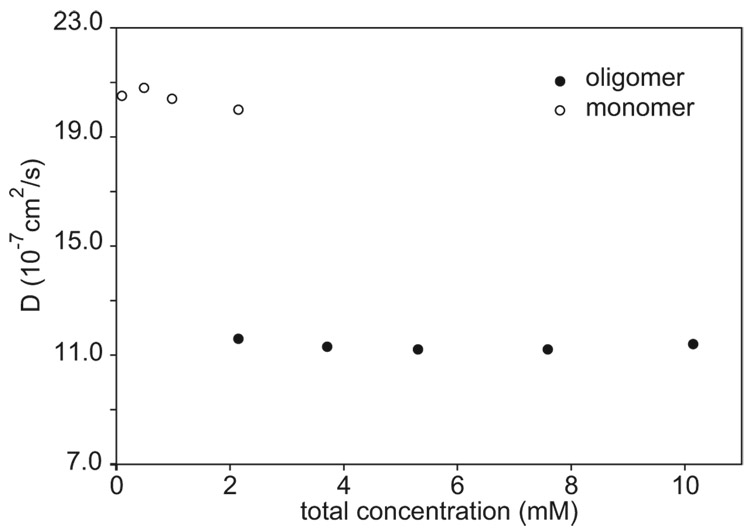
Measurement of the diffusion coefficients of peptide 3a as a function of concentration shows that the NMR resonances of the oligomer result from a single species and not from multiple oligomeric species in rapid chemical exchange. We carried out these measurements by varying the total concentration of peptide 3a from 0.1 to 10 mM in D2O solution at 298 K and were able to measure the diffusion coefficients of the monomer from 0.1–2 mM and the oligomer from 2–10 mM.29 The diffusion coefficients of the monomer and the oligomer do not vary significantly over the range of concentrations studied (Figure 7). If the oligomeric species consisted of smaller and larger oligomers in rapid exchange, increasing the concentration of peptide from 2 to 10 mM would shift the equilibrium toward the larger oligomers and would decrease the observed diffusion coefficient of the oligomeric species.
Figure 7.
The diffusion coefficients of the oligomer and the monomer of peptide 3a at various total concentrations of the peptide. The diffusion coefficients were measured in D2O at 298 K by using an sLED pulse sequence28a on an 800 MHz 1H NMR spectrometer.
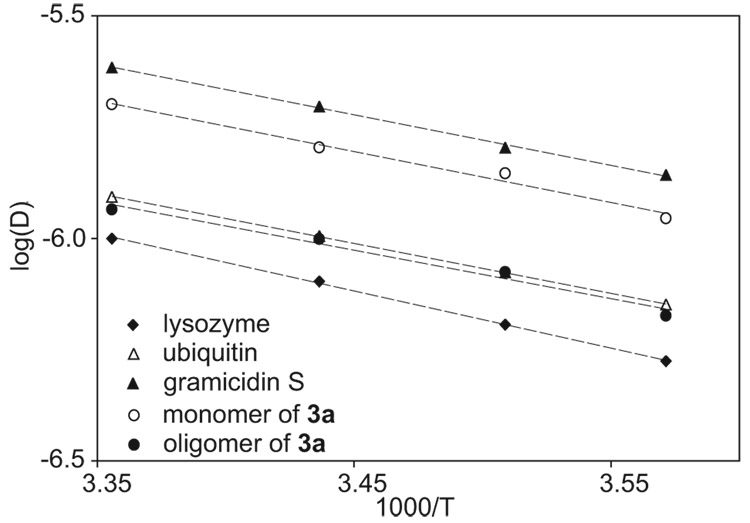
Measurement of the diffusion coefficients of peptide 3a as a function of temperature (T) reveals that the oligomerization states of the monomer and the oligomer do not vary over the temperature range of 280–298 K. In dilute solutions of proteins and peptides, in which the viscosity of the solution is almost identical to that of water, log(D) for a species with a single oligomerization state varies linearly with 1/T. The slope of the linear relationship is the same, regardless of the species, and reflects the activation energy of the self-diffusion of water.30,31 If the oligomerization state changes with temperature, the relationship between log(D) and 1/T deviates from linearity. The diffusion coefficients of the monomer and the oligomer of peptide 3a were respectively measured in 2 and 10 mM D2O solutions of the peptide from 280–298 K and compared to those of lysozyme, ubiquitin, and gramicidin S.30 Figure 8 illustrates the relationship between log(D) and 1/T for these species. The linear plots with similar slopes reflect that the oligomerization states of the monomer and the oligomer do not change significantly over the temperature range studied.30
Figure 8.
Comparison of the temperature dependence of the diffusion coefficients of the monomer and the oligomer of peptide 3a to those of lysozyme, ubiquitin, and gramicidin S in D2O. The diffusion coefficients were measured at 280, 285, 291, and 298 K by using an sLED pulse sequence28a on an 800 MHz 1H NMR spectrometer. The diffusion coefficients of these species were measured at the following concentrations: 3a monomer, 2 mM; 3a oligomer, 10 mM; lysozyme, 1.4 mM; ubiquitin, 1.0 mM; and gramicidin S, 1.1 mM.
The ratio of the diffusion coefficients of the oligomer and the monomer (Doligomer/Dmonomer) helps establish the oligomerization state.28a,32 If both the monomer, and the oligomer are envisioned as compact (spherical) structures, then Doligomer/Dmonomer should be 0.63 for a tetramer and 0.79 for a dimer.33 If, on the other hand, the tetramer is envisioned as either a tetrahedral or a square planar array of hard spheres, the ratio should be 0.62 or 0.58, respectively.32 With this hard-sphere model, the ratio should be 0.75 for a dimer. Over the temperature range of 280–298, the ratio of the diffusion coefficients for peptide 3a is 0.60 ± 0.03. This ratio of diffusion coefficients is consistent with a tetrameric oligomer.
Analytical ultracentrifugation studies34
To corroborate the tetramer formation of peptide 3a and to check for any aggregate invisible by NMR techniques (e.g., very large aggregates), we conducted analytical ultracentrifugation studies comprising both sedimentation velocity analysis and sedimentation equilibrium experiments. All studies were performed in 100 mM aqueous (H2O) NaCl solutions to avoid non-ideal behavior associated with repulsion between the charged peptides.35 The velocity experiments were performed at 60 000 rpm, while the equilibrium experiments were performed at 50 000, 55 000, and 60 000 rpm. Both sets of experiments were performed at 293 K. The partial specific volume of peptide 3a was estimated to be 0.706 cm3/g.36,37,38
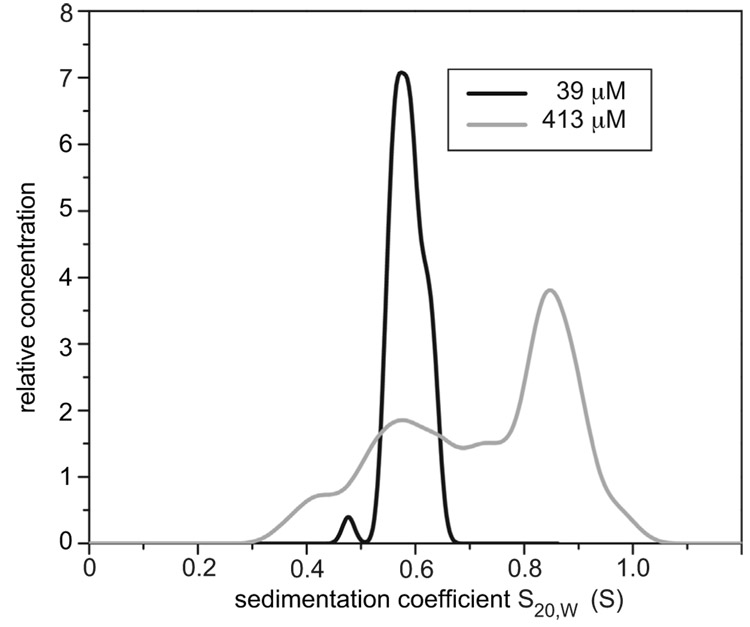
We conducted the sedimentation velocity experiments at 39 µM and 413 µM loading concentrations to observe both the monomer and the oligomer. The 39 µM experiment was performed using UV-absorbance at 280 nm; the 413 µM experiment was performed using Rayleigh interference. Enhanced van Holde-Weischet analysis39 of the data from the low-concentration experiment results in a single major peak (Figure 9).40 The high-concentration experiment gives two peaks with a diffuse reaction boundary: a smaller peak at the same position as the low-concentration experiment and a second larger peak from an oligomeric species (Figure 9). Collectively, these two experiments show a monomer and an oligomer in equilibrium and exclude the formation of high-molecular weight aggregates. Equilibration of the monomer and the oligomer on the time scale of the sedimentation velocity experiment may result in the poor resolution of the two species in the 413 µM experiment.
Figure 9.
Van Holde–Weischet distributions of sedimentation velocity experiments of peptide 3a measured with UV absorbance at 280 nm at 39 µM (black line) and with Rayleigh interference at 413 µM (grey line).
We performed the equilibrium analytical ultracentrifugation studies on 15 µM, 1.5 mM, and 9.0 mM solutions of peptide 3a to assure good signals from both the monomer and the oligomer. Data were collected by UV-absorbance scans at 15 µM and by Rayleigh interference measurements at 1.5 and 9.0 mM and were fit globally. Global fitting of data observed under multiple conditions, such as multiple rotor speeds and multiple loading concentrations enhances the confidence in each fitted parameter value.41 In such a fit, parameters such as the monomer molecular weight and the association constants are considered global parameters and are forced to be the same for all included data sets. Multiple models were attempted to fit the experimental data, including: (1) a single ideal species, (2) a two-component, non-interacting species model, (3) a fixed molecular weight distribution between 100–10,000 Dalton, (4) reversibly associating monomer-trimer, monomer-tetramer, and monomer-pentamer systems, and (5) a reversible monomer-dimer-tetramer model. Table 2 lists the results from these fits.
Table 2.
Global fitting results for various models to a combination of sedimentation equilibrium data from 15 µM, 1.5 mM, and 9.0 mM experiments.
| Model | Variance | MWmonomer | MWoligomer | K |
|---|---|---|---|---|
| single-component (15 µM data only) | 1.222 × 10−5 | 1.62 kD | n/a | n/a |
| single-component | 8.332 × 10−3 | 4.67 kD | n/a | n/a |
| two-component | 1.096 × 10−4 | 1.52 kD | 6.17 kD | n/a |
| fixed MW distribution | 1.022 × 10−4 | 1.67 kD a | 5.76 kDa | n/a |
| monomer-trimer | 4.021 × 10−4 | 2.37 kD | n/a | K1,3 = 1.04 × 106 M−2 |
| monomer-tetramer | 1.656 × 10−4 | 1.62 KD | n/a | K1,4 = 3.76 × 109 M−3 |
| monomer-pentamer | 2.586 × 10−4 | 1.25 kD | n/a | K1,5 = 9.05 × 1012 M−4 |
| monomer-dimer-tetramerb | 1.656 × 10−4 | 1.62 kD | n/a | K1,4 = 3.75 × 109 M−3 |
Weight-average measurements of each mode in a bimodal distribution. The Supporting Information provides further details.
Essentially no signal from a dimeric species is observed, and no monomer-dimer equilibrium constant (K1,2) can be determined.
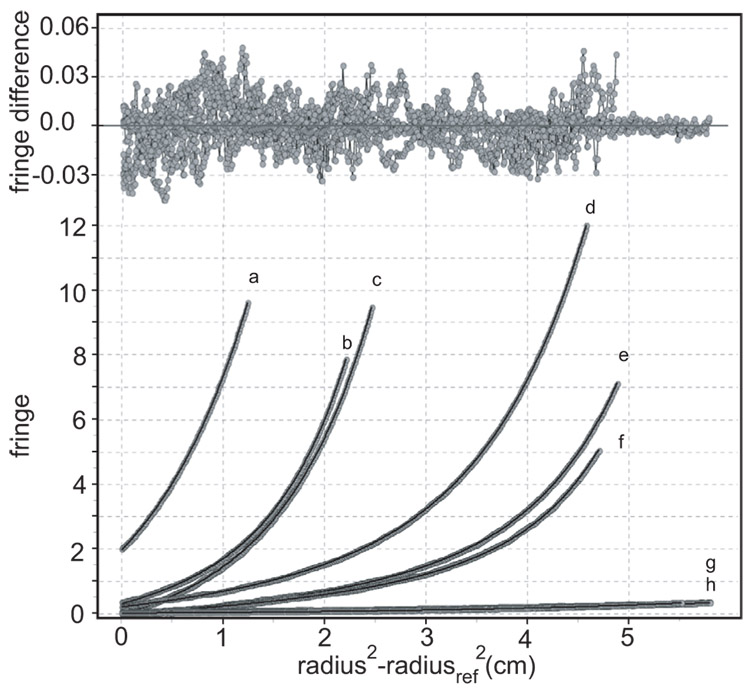
The data from the 15 µM experiment fit well to a single monomeric species, indicating that at low concentration peptide 3a is essentially all monomer. The data from the 1.5 and 9.0 mM experiments do not fit a single-component model, indicating the presence of a second component. Fitting with a two-component, ideal-species model suggests that the second component has the molecular weight of a tetramer. A fixed-molecular-weight-distribution model38a further confirms this analysis, giving a bimodal molecular weight distribution, with average molecular weight species consistent with a monomer-tetramer model (Supporting Information). This result is entirely consistent with the bimodal sedimentation distribution observed in the 413 µM velocity experiment shown in Figure 9. The model for the reversibly self-associating monomer-tetramer system produces a monomer molecular weight in excellent agreement with the actual monomer molecular weight (1.62 kD measured, 1.61 kD actual). Figure 10 shows a plot of the residuals and overlays for the monomer-tetramer fit.
Figure 10.
Sedimentation equilibrium data for peptide 3a fitted to a monomer-tetramer equilibrium model. Residuals of the fit are shown on the top, overlays on the bottom. Grey points represent experimental data at (a) 9 mM and 60 000 rpm, (b) 9 mM and 55 000 rpm, (c) 9 mM and 50 000 rpm, (d) 1.5 mM and 60 000 rpm, (e) 1.5 mM and 55 000 rpm, (f) 1.5 mM and 50 000 rpm, (g) 15 µM and 60 000 rpm, and (h) 15 µM and 55 000 rpm.42 Black curves represent the fitted model.
Other reversible self-association models that we tried also support the monomer-tetramer model by returning variances higher than the monomer-tetramer model and resulting in unreasonable monomer molecular weights. The monomer-trimer model results in a monomer molecular weight approximately 700 Dalton higher than the actual monomer molecular weight, while a monomer-pentamer model results in a monomer molecular weight about 400 Dalton lower than the actual monomer molecular weight. In both cases, the variance is 1.5–2.5 fold higher than the variance for the monomer-tetramer model. A reversible monomer-dimer-tetramer model produces identical results to the monomer-tetramer model, with essentially no signal from a monomer-dimer association, and a similar variance as the monomer-tetramer model.
Collectively, the AUC results corroborate that peptide 3a participates in a monomer-tetramer equilibrium. Monte Carlo analysis of the monomer-tetramer fit gives a molecular weight of 1.624 kD (95% confidence intervals: 1.619/1.629 kD) and a monomer-tetramer equilibrium constant (K1,4) of 3.76 × 109 M−3 (95% confidence intervals: 3.66 × 109 M−3/3.87 × 109 M−3). With this equilibrium constant, 50% of the peptide is tetramer at 0.8 mM and 293 K in 100 mM aqueous NaCl. The determined monomer molecular weight of 1.624 kD is in excellent agreement with the 1.614 kD actual molecular weight of peptide 3a.
Working model for the tetramer of peptide 3a
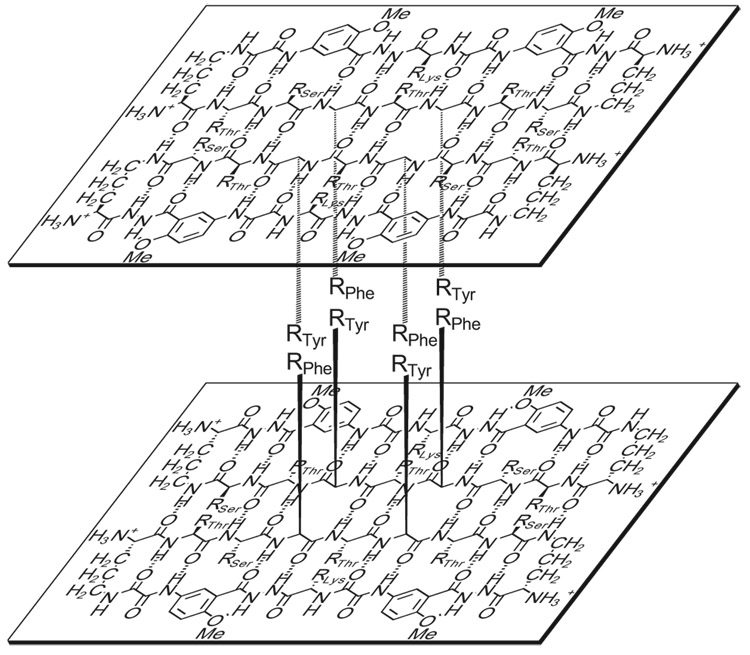
In conjunction with the NOE studies and the upfield shifting of the Phe and Tyr aromatic resonances, the PFG NMR experiments and analytical ultracentrifugation studies suggest a model in which the macrocyclic peptide 3a oligomerizes to form a dimer of β-sheet dimers (Figure 11). In this tetrameric β-sheet sandwich, the macrocyclic peptide 3a is folded to form a β-sheet, the β-sheet is dimerized through edge-to-edge interactions, and this dimer is further dimerized through hydrophobic face-to-face interactions involving the Phe and Tyr groups. (This description is just hierarchical and is not a chronological organization of events.) In this structure, the side chains of Thr1, Phe3, Tyr5, and Ser7 are on the interior, while those of Ser2, Thr4, Thr6, and Lys8 are on the exterior. The hydrophobic residues Phe3 and Tyr5 comprise a hydrophobic core.
Figure 11.
Illustration of the β-sheet sandwich tetramer of peptide 3a.
The self-assembly of peptide 3a into a tetrameric β-sheet sandwich significantly enhances the folding of the peptide. The ability of self-assembly to promote folding is common to a variety of systems. Examples include human transthyretin, which folds as a β-sheet tetramer but is less folded as the monomer,43 honeybee melittin, which folds as an α-helical tetramer but is unfolded as the monomer,44 and Mayo’s βpep peptides, which form well-structured β-sandwich tetramers, molten globule-like β-sandwich dimers, and unfolded monomers.14 The cooperativity between self-assembly and folding in these peptides and small proteins constitutes a fundamental natural principle for creating secondary and tertiary structures through quaternary structure and is akin to the highly cooperative folding of larger proteins.
II. Effect of the Heptapeptide Sequence on Folding and Oligomerization of the Monomer
The tetrameric β-sheet sandwich model shown in Figure 11 suggests that the nature and location of the side chains, and the face upon which they are displayed, should have profound effects on the folding and the oligomerization of the macrocyclic β-sheets. The side chains at positions R2, R4, and R6 of the heptapeptide sequence are on the exterior surface of the tetramer. The side chains at positions R2 and R6 are directly across from the aromatic rings of Hao units, while the side chain at position R4 is across from the side chain of Lys8. Interactions between these groups should affect the folding and oligomerization of the monomer. The side chains at positions R1, R3, R5, and R7 are on the interior of the tetramer, and those of R3 and R5 comprise a hydrophobic core. Interactions between the interior side chains should affect the hydrophobic core and thereby the stability of the tetramer.
We designed macrocyclic peptides 3b–3n to evaluate the roles of different interactions (e.g., polar, electrostatic, aromatic, and hydrophobic interactions) in folding and oligomerization. Peptides 3b–3n, which are homologues of peptide 3a with 1–6 variations in the heptapeptide sequence, were synthesized in an analogous fashion to peptide 3a. We studied the folding of the monomers of these peptides by 1H NMR spectroscopy in D2O solution at low total concentrations (0.4–1.5 mM) to minimize signal from the oligomers. In these studies of the monomers, we calculated the average of the ΔδHα values for the residues at positions R1–R7 and the average of the ΔδδOrn values for the two δOrn turn units.45 Figure 12 and Figure 13 present these average values; the individual α-proton chemical shifts and ΔδδOrn values are tabulated in the Supporting Information. We also studied the oligomerization properties of these peptides by 1H NMR spectroscopy in D2O solution at higher concentrations. Table 3 summarizes the folding of the monomers and the oligomerization properties of peptides 3a–3n.
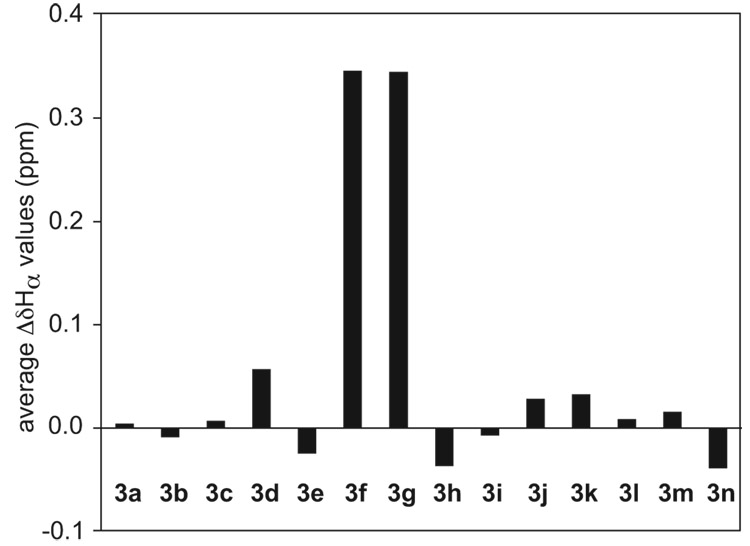
Figure 12.
Average ΔδHα values of the residues at positions R1–R7 for peptide 3a–3n monomers at 298 K in D2O.45
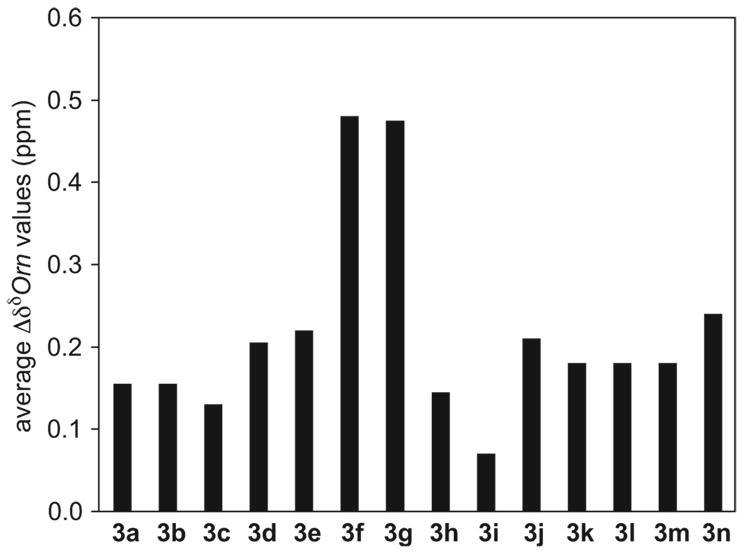
Figure 13.
Average ΔδδOrn values for peptide 3a–3n monomers at 298 K in D2O.45
Table 3.
Folding of the monomers and oligomerization properties of peptides 3a–3n.
| Peptide | Folding of the monomer | Oligomerization properties |
|---|---|---|
| 3a | partial | tetramera |
| 3b | partial | tetramerb |
| 3c | partial | tetramerb |
| 3d | partial | tetramerb |
| 3e | partial | tetramerb |
| 3f | good | tetramer (with a K1,4 higher than 3a)c |
| 3g | good | tetramer (with a K1,4 higher than 3a)d |
| 3h | partial | tetramer (with a K1,4 lower than 3a)e |
| 3i | partial | ____f |
| 3j | partial | hexamer or octamerg |
| 3k | partial | hexamer or octamerg |
| 3l | partial | ____f |
| 3m | partial | ____f |
| 3n | partial | ____f |
The oligomerization state was determined by PFG NMR diffusion measurements and AUC experiments.
The oligomerization state was inferred from similar oligomerization behavior as peptide 3a in concentration-dependent 1H NMR studies.
The oligomerization state was determined by PFG NMR diffusion measurements 46 and 1H NMR concentration studies.
The oligomerization state was inferred from similar oligomerization behavior as peptide 3f in concentration-dependent 1H NMR studies.
The oligomerization state was determined by PFG NMR diffusion measurements 47 and 1H NMR concentration studies.
Some self-association without distinct oligomer formation was observed by concentration-dependant 1H NMR studies.
The oligomerization state was determined by PFG NMR diffusion measurements.48
Variation in positions R2, R4, and R6
We synthesized and studied peptides 3b–3g to determine the effect of the side chains at positions R2, R4, and R6 of the heptapeptide sequence on the folding and oligomerization of the monomers. In peptides 3b, 3c, and 3d, we replaced the polar residue Thr4 of peptide 3a with the polar residue Glu, the hydrophobic residue Leu, and the aromatic residue Tyr. In peptide 3e, we replaced the polar residues Ser2 and Thr6 of peptide 3a with the hydrophobic residues Leu and Val. In peptide 3f, we replaced these residues with aromatic residues Tyr and Phe. In peptide 3g, we replaced these residues with Tyr.
The average ΔδHα values and the average ΔδδOrn values reveal substantially more folding in the monomers of peptides 3f and 3g than those of peptides 3a–3e (Figure 12 and Figure 13).46 The average ΔδHα values for the monomers of peptides 3f and 3g are large (0.35 ppm), while those for the monomers of peptides 3a–3e are essentially zero (−0.03–0.06 ppm).45 Consistent with this trend, the average ΔδδOrn values for the monomers of peptides 3f and 3g are large (0.48 ppm), while those for the monomers of peptides 3a–3e are small (0.13–0.22 ppm). The relatively large ΔδHα and ΔδHδOrn values for peptides 3f and 3g reflect the better folding of these peptides.
1H NMR studies indicate that the enhanced folding of the monomers of peptides 3f and 3g arises from additional aromatic interactions between the Hao units and the aromatic residues at positions R2 and R6.49 The aromatic resonances of the side chains at positions R2 and R6 of these peptides are shifted upfield by several tenths of a ppm relative to those in simple peptides.50 NOEs between these side chains and the Hao units provide additional evidence of interaction between these groups.
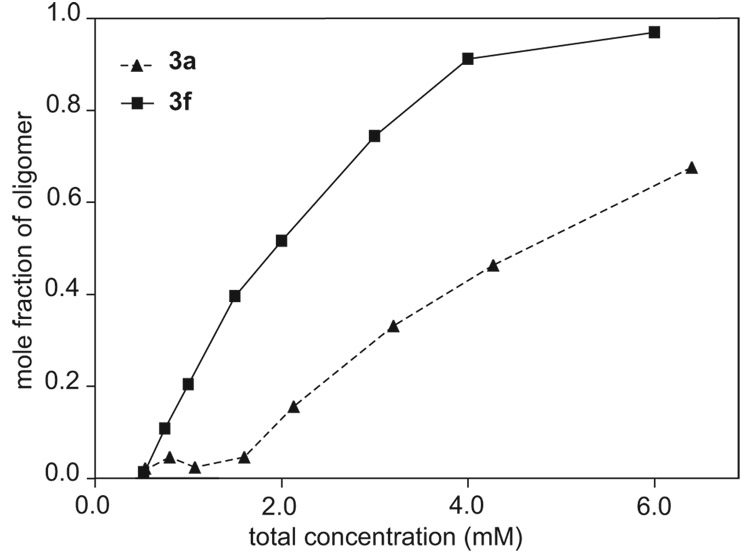
Peptides 3f and 3g form tetramers at substantially lower concentrations (higher K1,4) than peptides 3a–3e. Figure 14 compares the concentration-dependent oligomerization of peptides 3a and 3f at 298 K. At 2 mM total peptide concentration, less than 20% of 3a is tetrameric, while ca. 50% of 3f is tetrameric.46 Peptides 3b–3e show concentration-dependent oligomerization properties similar to peptide 3a, while peptide 3g shows concentration-dependent oligomerization properties similar to peptide 3f. The enhanced oligomerization of peptides 3f and 3g likely arise from their preorganization as well-folded monomers.
Figure 14.
Comparison of the concentration-dependent oligomerization of peptides 3a and 3f: mole fraction of oligomer vs. total peptide concentration. The relative concentrations of the monomer and oligomer were determined in D2O at 298 K by integrating the corresponding 1H NMR resonances.
Collectively, these studies of variation at positions R2, R4, and R6 suggest that aromatic residues at positions R2 and R6 improve folding of the monomer and that better-folded monomers oligomerize at lower concentrations (with higher association constants).
Variation in positions R1, R3, R5, and R7
We first synthesized and studied peptides 3h and 3i to determine the effect of the side chains at positions R1, R3, R5, and R7 of the heptapeptide sequence on oligomerization. In peptide 3h, we replaced the polar residues Thr1 and Ser7 of peptide 3a with the polar and charged residues Glu and Lys.45 In peptide 3i, we replaced the aromatic residues Phe3 and Tyr5 of peptide 3a with the hydrophobic residues Leu and Val. The results from these peptides subsequently prompted us to synthesize and study peptides 3j–3n.
Comparison of the ΔδHα and ΔδδOrn values of the monomers of peptides 3h and 3i to those of peptide 3a suggests that these variants have less β-strand character and less folded δOrn turns than peptide 3a (Figure 12 and Figure 13).45 At 298 K, the average ΔδHα of peptides 3h and 3i are −0.04 and −0.01 ppm, respectively, while that of peptide 3a is 0.00 ppm (Figure 12). The ΔδδOrn values for peptides 3h and 3i are 0.15 and 0.07 ppm, respectively, while that of peptide 3a is 0.16 ppm (Figure 13).
NMR studies of peptides 3h and 3i show little or no tetramer in water. Peptide 3h shows no tetramer at low concentrations and only traces of the tetramer at high concentration and low temperature (10 mM, 280 K).47 The poor folding of the monomer and the charged side chain of Lys7, which can destabilize the hydrophobic core of the tetramer, likely contribute to the decreased tetramer formation of this peptide. Peptide 3i does not show any oligomer at concentrations up to 8 mM and temperatures of 280–298 K. The weak folding and the absence of oligomerization in peptide 3i may result from the absence of aromatic residues at positions R3 and R5. Although peptide 3i lacks aromatic residues at positions R3 and R5, it instead has hydrophobic residues Leu and Val. Even though these residues should stabilize the hydrophobic core of a tetrameric β-sheet sandwich, the poor folding of the monomer may impede oligomerization.
To determine whether the poor folding of the monomer or the lack of aromatic interactions at positions R3 and R5 prevents the oligomerization of peptide 3i, we synthesized and studied peptides 3j–3m. Like peptide 3g, these peptides have Tyr residues at positions R2 and R6, which should favor folding of the monomer. Peptide 3j contains two hydrophobic residues (Ile and Val) at positions R3 and R5, while peptide 3m contains two polar residues (Ser and Thr) at these positions. Peptides 3k and 3l each contains one hydrophobic residue and one polar residue at positions R3 and R5: Peptide 3k contains the hydrophobic residue Ile at position R3 and the polar residue Thr at position R5, while peptide 3l contains the polar residue Ser at position R3 and the hydrophobic residue Val at position R5.
NMR studies show that the monomers of peptides 3j–3m fold slightly better than that of peptide 3a (Figure 12 and Figure 13), but that only peptides 3j and 3k form oligomers with distinct NMR resonances.48 Peptide 3j, with two hydrophobic residues at positions R3 and R5, oligomerizes at substantially lower concentrations than peptide 3k, with only one hydrophobic residue. Peptide 3l and peptide 3m each exhibits only one set of 1H NMR resonances, which broaden and shift slightly with increasing concentration and decreasing temperature.51 The failure of peptides 3l and 3m to form distinct oligomers may reflect their paucity of stabilizing hydrophobic interactions.
It is interesting that peptide 3l does not form a distinct oligomer, while peptide 3k does. Both peptides have one hydrophobic residue (Val or Ile) and one hydrophilic residue (Ser or Thr) at positions R3 and R5. The difference in oligomerization properties may reflect the greater hydrophobicity of the Ile and Thr residues in peptide 3k. The oligomerization properties of peptides 3a and 3h–3m suggest that macrocyclic peptides with folding comparable to or greater than that of peptide 3a can oligomerize if the residues at positions R3 and R5 can participate in hydrophobic interactions that stabilize the oligomer.
To further investigate the effect of the placement of hydrophobic residues on the folding and oligomerization of the macrocyclic β-sheets, we synthesized and studied peptide 3n. Peptide 3n is a sequence isomer of peptide 3f in which the Phe and Tyr residues at positions R3 and R5 have been swapped with the Thr and Ser residues at positions R1 and R7. 1H NMR studies in D2O reveal significant differences in the folding and oligomerization properties of peptides 3f and 3n. In contrast to the monomer of peptide 3f, which is well folded, peptide 3n is only partially folded. The average ΔδHα value of peptide 3n is about 0.39 ppm lower than that of peptide 3f, and the average ΔδδOrn is about 0.24 ppm lower (Figure 12 and Figure 13). Unlike peptide 3f, which exhibits distinct NMR resonances corresponding to the monomer and the tetramer even at low concentrations, peptide 3n exhibits only one set of NMR resonances, which broaden slightly with increasing concentration and decreasing temperature. These differences in the folding and oligomerization properties of peptides 3f and 3n further support that aromatic residues at positions R3 and R5 of the macrocyclic β-sheets promote folding of the monomer and that they assist oligomerization by creating a stabilizing hydrophobic core.
Collectively, these studies of variation at positions R1, R3, R5, and R7 suggest that aromatic residues at positions R3 and R5 improve folding of the monomer and that peptides with folding comparable to or greater than that of peptide 3a are able to form oligomers with distinct NMR resonances if the peptides can form a stabilizing hydrophobic core. Other factors, such as β-sheet propensities of the residues and the packing of side chains, might also be important in oligomerization but have not been investigated here.
Conclusion
Combination of a suitable heptapeptide strand, two Hao β-strand mimics, two δ-linked ornithine turn units, and one additional α-amino acid gives water-soluble macrocyclic peptides 3 that mimic the quaternary β-sheet structure of proteins. Many of these peptides self-associate in water through edge-to-edge and face-to-face intermolecular β-sheet interactions to form tetramers. Aromatic residues at positions R2, R3, R5, and R6 improve the folding of the monomers through aromatic interactions with the Hao aromatic rings. Hydrophobic residues (including aromatic residues) at positions R3 and R5 are essential to the formation of the oligomers through stabilizing hydrophobic interactions. The folding and self-association of these peptides is cooperative. In peptides with partially folded monomers, the formation of β-sheet secondary and tertiary structures is intimately linked to the formation of β-sheet quaternary structure. These partially folded monomers self-associate to form strongly folded oligomers. In peptides with better-folded monomers, the formation of oligomers occurs with greater facility.
The formation of edge-to-edge dimers does not appear to occur without further self-association (e.g., tetramer formation) through face-to-face interactions. The need for further self-association and the formation of a hydrophobic core in these structures likely reflects the frailty of hydrogen bonds alone in water. Indeed, β-sheets almost never occur in proteins without being buttressed in against additional structures.52
Macrocyclic β-sheet peptides 3 represent a new class of model system with which to study β-sheet formation in water. Unlike prior aqueous model systems, which have largely focused on β-sheet secondary and tertiary structures, these peptides provide a window into β-sheet quaternary structure. The studies described in this paper illustrate how this model system can be used to evaluate the role of hydrophobic interactions in stabilizing β-sheet quaternary structure. In future studies, we will explore the β-sheet quaternary interactions of these systems with proteins and peptide β-sheet aggregates.
Supplementary Material
Experimental procedures and NMR spectra, mass spectra, and HPLC traces for cyclic peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGMENT
J. S. Nowick and O. Khakshoor thank the NIH for grant support (GM 49076) and Prof. P. Legault and Dr. B. D. Nguyen for providing and implementing the PFG NMR sLED pulse sequence used to determine diffusion coefficients. B. Demeler thanks the NSF for grant support (DBI-9974819). The authors thank V. Schirf for technical assistance with the AUC experiments.
References and Notes
- 1.(a) Maitra S, Nowick JS. In: The Amide Linkage: Structural Significance in Chemistry, Biochemistry, and Materials Science. Greenberg A, Breneman CM, Liebman JF, editors. New York: Wiley; 2000. pp. 495–518. [Google Scholar]; (b) Remaut H, Waksman G. Trends Biochem. Sci. 2006;31:436–444. doi: 10.1016/j.tibs.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Navia MA, Fitzgerald PMD, McKeever BM, Le CT, Heimbach JC, Herber WK, Sigal IS, Darke PL, Springer JP. Nature. 1989;337:615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- 3.Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. Nature. 1995;375:554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- 4.(a) Ross CA, Poirier MA. Nat. Med. 2004;10 Suppl.:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]; (b) Tycko R. Curr. Opin. Struct. Biol. 2004;14:96–103. doi: 10.1016/j.sbi.2003.12.002. [DOI] [PubMed] [Google Scholar]; (c) Makin OS, Serpell LC. FEBS J. 2005;272:5950–5961. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]; (d) Nelson R, Eisenberg D. Curr. Opin. Struct. Biol. 2006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]; (e) Chiti F, Dobson CM. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]; (f) Wetzel R. Acc. Chem. Res. 2006;39:671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- 5.Sommers WS, Phillips SEV. Nature. 1992;359:387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- 6.Hill CP, Yee J, Selsted ME, Eisenberg D. Science. 1991;251:1481–1485. doi: 10.1126/science.2006422. [DOI] [PubMed] [Google Scholar]
- 7.(a) Smith CK, Regan L. Acc. Chem. Res. 1997;30:153–161. [Google Scholar]; (b) Gellman SH. Curr. Opin. Chem. Biol. 1998;2:717–725. doi: 10.1016/s1367-5931(98)80109-9. [DOI] [PubMed] [Google Scholar]; (c) Ramirez-Alvarado M, Kortemme T, Blanco FJ, Serrano L. Bioorg. Med. Chem. 1999;7:93–103. doi: 10.1016/s0968-0896(98)00215-6. [DOI] [PubMed] [Google Scholar]; (d) Searle MS. J. Chem. Soc., Perkin Trans. 2001;2:1011–1020. [Google Scholar]; (e) Venkatraman J, Shankaramma SC, Balaram P. Chem. Rev. 2001;101:3131–3152. doi: 10.1021/cr000053z. [DOI] [PubMed] [Google Scholar]; (f) Searle MS, Ciani B. Curr. Opin. Struct. Biol. 2004;14:458–464. doi: 10.1016/j.sbi.2004.06.001. [DOI] [PubMed] [Google Scholar]; (g) Hughes RM, Waters ML. Curr. Opin. Struct. Biol. 2006;16:514–524. doi: 10.1016/j.sbi.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 8.For examples of large peptides that fold into larger β-sheet structures, see: Pessi A, Bianchi E, Crameri A, Venturini S, Tramontano A, Sollazzo M. Nature. 1993;362:367–369. doi: 10.1038/362367a0. Quinn TP, Tweedy NB, Williams RW, Richardson JS, Richardson DC. Proc. Natl. Acad. Sci. USA. 1994;91:8747–8751. doi: 10.1073/pnas.91.19.8747. Yan YB, Erickson BW. Protein Sci. 1994;3:1069–1073. doi: 10.1002/pro.5560030709. Ilyina E, Roongta V, Mayo KH. Biochemistry. 1997;36:5245–5250. doi: 10.1021/bi963064o. Kortemme T, Ramirez-Alvarado M, Serrano L. Science. 1998;281:253–256. doi: 10.1126/science.281.5374.253. Schenck HL, Gellman SH. J. Am. Chem. Soc. 1998;120:4869–4870. De Alba E, Santoro J, Rico M, Jiménez MA. Protein Sci. 1999;8:854–865. doi: 10.1110/ps.8.4.854. Griffiths-Jones SR, Searle MS. J. Am. Chem. Soc. 2000;122:8350–8356. Carulla N, Woodward C, Barany G. Protein Sci. 2002;11:1539–1551. doi: 10.1110/ps.4440102. Venkatraman J, Gowda GAN, Balaram P. J. Am. Chem. Soc. 2002;124:4987–4994. doi: 10.1021/ja0174276. Neidigh JW, Fesinmeyer RM, Anderson N. Nat. Struct. Biol. 2002;9:425–430. doi: 10.1038/nsb798. Nanda V, Rosenblatt MM, Osyczka A, Kono H, Getahun Z, Dutton PL, Saven JG, DeGrado WF. J. Am. Chem. Soc. 2005;127:5804–5805. doi: 10.1021/ja050553f.
- 9.(a) Nowick JS, Smith EM, Pairish M. Chem. Soc. Rev. 1996:401–415. [Google Scholar]; (b) Nowick JS. Acc. Chem. Res. 1999;32:287–296. [Google Scholar]; (c) Loughlin WA, Tyndall JDA, Glenn MP, Fairlie DP. Chem. Rev. 2004;104:6085–6117. doi: 10.1021/cr040648k. [DOI] [PubMed] [Google Scholar]
- 10.For some recent examples, see: Matsumura S, Uemura S, Mihara H. Chem.—Eur. J. 2004;10:2789–2794. doi: 10.1002/chem.200305735. Ozbas B, Kretsinger J, Rajagopal K, Schneider JP, Pochan DJ. Macromolecules. 2004;37:7331–7337. Litvinchuk S, Bollot G, Mareda J, Som A, Ronan D, Shah MR, Perrottet P, Sakai N, Matile S. J. Am. Chem. Soc. 2004;126:10067–10075. doi: 10.1021/ja0481878. Deechongkit S, Powers ET, You S-L, Kelly JW. J. Am. Chem. Soc. 2005;127:8562–8570. doi: 10.1021/ja050558c. Davies RVP, Aggeli A, Beevers AJ, Boden N, Carrick LM, Fishwick CWG, McLeish TCB, Nyrkova I, Semenov AN. Supramol. Chem. 2006;18:435–443. Koga T, Higuchi M, Kinoshita T, Higashi N. Chem. —Eur. J. 2006;12:1360–1367. doi: 10.1002/chem.200500611.
- 11.For some recent reviews, see: Bong DT, Clark TD, Granja JR, Ghadiri MR. Angew. Chem., Int. Ed. Engl. 2001;40:988–1011. Zhang SG. Nat. Biotechnol. 2003;21:1171–1178. doi: 10.1038/nbt874. Rajagopal K, Schneider JP. Curr. Opin. Struct. Biol. 2004;14:480–486. doi: 10.1016/j.sbi.2004.06.006.
- 12.(a) Nowick JS, Tsai JH, Bui Q-CD, Maitra S. J. Am. Chem. Soc. 1999;121:8409–8410. [Google Scholar]; (b) Nowick JS, Chung DM, Maitra K, Maitra S, Stigers KD, Sun Y. J. Am. Chem. Soc. 2000;122:7654–7661. [Google Scholar]; (c) Nowick JS, Lam KS, Khasanova TV, Kemnitzer WE, Maitra S, Mee HT, Liu R. J. Am. Chem. Soc. 2002;124:4972–4973. doi: 10.1021/ja025699i. [DOI] [PubMed] [Google Scholar]; (d) Nowick JS, Chung DM. Angew. Chem., Int. Ed. Engl. 2003;42:1765–1768. doi: 10.1002/anie.200250750. [DOI] [PubMed] [Google Scholar]; (e) Chung DM, Nowick JS. J. Am. Chem. Soc. 2004;126:3062–3063. doi: 10.1021/ja031632z. [DOI] [PubMed] [Google Scholar]; (f) Chung DM, Dou Y, Baldi P, Nowick JS. J. Am. Chem. Soc. 2004;127:9998–9999. doi: 10.1021/ja052351p. [DOI] [PubMed] [Google Scholar]
- 13.(a) Ghadiri MR, Kobayashi K, Granja JR, Chadha RK, McRee DE. Angew. Chem. Int. Ed. Engl. 1995;34:93–95. [Google Scholar]; (b) Gong B, Yan Y, Zeng H, Skrzypczak-Jankunn E, Kim YW, Zhu J, Ickes H. J. Am. Chem. Soc. 1999;121:5607–5608. [Google Scholar]; (c) Phillips ST, Rezac M, Abel U, Kossenjans M, Bartlett PA. J. Am. Chem. Soc. 2002;124:58–66. doi: 10.1021/ja0168460. [DOI] [PubMed] [Google Scholar]
- 14.Mayo KH, Ilyina E. Protein Sci. 1998;7:358–368. doi: 10.1002/pro.5560070216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali MH, Peisach E, Allen KN, Imperiali B. Proc. Natl. Acad. Sci. USA. 2004;101:12183–12188. doi: 10.1073/pnas.0401245101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Dou Y, Baisnée P-F, Pollastri G, Pécout Y, Nowick J, Baldi P. Bioinformatics. 2004;20:2767–2777. doi: 10.1093/bioinformatics/bth326. [DOI] [PubMed] [Google Scholar]; (b) http://www.igb.uci.edu/servers/icbs/
- 17.Nowick JS, Brower JO. J. Am. Chem. Soc. 2003;125:876–877. doi: 10.1021/ja028938a. [DOI] [PubMed] [Google Scholar]
- 18.Woods RJ, Brower JO, Castellanos E, Hashemzadeh M, Khakshoor O, Russo WA, Nowick JS. J. Am. Chem. Soc. doi: 10.1021/ja0667965. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.For other examples of designed cyclic peptides that fold into β-sheet structures, see: Spath J, Stuart F, Jiang L, Robinson JA. Helv. Chim. Acta. 1998;81:1726–1738. Syud FA, Espinosa JF, Gellman SH. J. Am. Chem. Soc. 1999;121:11577–11578. Kiehna SE, Waters ML. Protein Sci. 2003;12:2657–2667. doi: 10.1110/ps.03215403.
- 20.Other cyclic peptides containing 54-membered rings (cyclo-octadecapeptides), including naturally occurring θ-defensin, have been previously synthesized and studied. For examples, see reference 19c and Ando S, Nishikawa H, Takiguchi H, Lee S, Sugihara G. Biochimica et Biophysica Acta, Biomembranes. 1993;1147:42–49. doi: 10.1016/0005-2736(93)90314-p. Tang Y-Q, Yuan J, Ösapay G, Ösapay K, Tran D, Miller CJ, Quellette AJ, Selsted ME. Science. 1999;286:498–502. doi: 10.1126/science.286.5439.498. Trabi M, Schirra HJ, Craik DJ. Biochemistry. 2001;40:4211–4221. doi: 10.1021/bi002028t. Tran D, Tran PA, Tang Y-Q, Yuan J, Cole T, Selsted ME. J. Biol. Chem. 2002;277:3079–3084. doi: 10.1074/jbc.M109117200.
- 21.Nauli S, Kuhlman B, Trong IL, Stenkamp RE, Teller D, Baker D. Protein Sci. 2002;11:2924–2931. doi: 10.1110/ps.0216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.PS-PEG-trityl chloride resin was prepared from PS-PEG-trityl alcohol resin (Novabiochem NovaSyn TGT alcohol resin) by treatment with acetyl chloride, as described in the Novabiochem catalog.
- 23.Oligomerization of peptide 3a decreases modestly upon increasing temperature; NMR concentration studies at 298 K show that about 50% of peptide 3a exists as oligomer in 5 mM D2O solution, while at 280 K, 50% of the peptide exists as oligomer in 2 mM D2O solution.
- 24.Wishart DS, Sykes BD. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 25.The monomer of peptide 3a shows slightly higher ΔδHα values (average of 0.05 ppm) than those of peptide 6 (Figure 3). This slight downfield shifting of the peptide 3a monomer suggests a somewhat greater populations of β-strand conformer.
- 26.NOEs among side chains that might arise from further self-association of the dimers cannot be distinguished from those resulting from folding and dimerization.
- 27.For an introduction to PFG NMR, see: Price WS. Concepts Magn. Reson. 1997;9:299–336. Price WS. Concepts Magn. Reson. 1998;10:197–237. Johnson CS. Progr. NMR Spectrosc. 1999;34:203–256.
- 28.(a) Altieri AS, Hinton DP, Byrd RA. J. Am. Chem. Soc. 1995;117:7566–7567. [Google Scholar]; (b) Yao S, Howlett GJ, Norton RS. J. Biomol. NMR. 2000;16:109–119. doi: 10.1023/a:1008382624724. [DOI] [PubMed] [Google Scholar]; (c) Cohen Y, Avram L, Frish L. Angew. Chem. Int. Ed. 2005;44:520–554. doi: 10.1002/anie.200300637. [DOI] [PubMed] [Google Scholar]
- 29.Exchange between the monomer and the oligomer occurs relatively slowly, on the time scale of the PFG NMR experiment, and has little effect on the diffusion coefficients observed for either species.
- 30.Ilyina E, Roongta V, Pan H, Woodward C, Mayo KH. Biochemistry. 1997;36:3383–3388. doi: 10.1021/bi9622229. [DOI] [PubMed] [Google Scholar]
- 31.(a) Longsworth LG. J. Phys. Chem. 1954;58:770–773. [Google Scholar]; (b) Barshtein G, Almagor A, Yedgar S, Gavish B. Phys. Rev. E. 1995;52:555–557. doi: 10.1103/physreve.52.555. [DOI] [PubMed] [Google Scholar]
- 32.Teller DC, Swanson E, DeHaen C. Methods Enzymol. 1979;61:103–124. doi: 10.1016/0076-6879(79)61010-8. [DOI] [PubMed] [Google Scholar]
- 33.Polson A. J. Phys. Colloid Chem. 1950;54:649–652. [Google Scholar]
- 34.(a) Ralston G. Introduction to Analytical Ultracentrifugation. Fullerton, CA: Beckman Instruments, Inc.; 1993. [Google Scholar]; (b) McRorie DK, Voelker PJ. Self-Associating Systems in the Analytical Ultracentrifugation. Palo Alto, CA: Beckman Instruments, Inc.; 1993. [Google Scholar]; (c) Howlett GJ, Minton AP, Rivas G. Curr. Opin. Chem. Biol. 2006;10:430–436. doi: 10.1016/j.cbpa.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 35.NMR studies establish that the oligomerization of peptide 3a increases slightly with added NaCl. For example, more than 50% of peptide 3a exists as oligomer in 1 mM D2O solution containing 100 mM NaCl at 298 K, while 50% of peptide 3a exists as oligomer in 5 mM D2O solution without NaCl at the same temperature. PFG NMR diffusion coefficient measurements of peptide 3a with 100 mM NaCl in D2O solution show that the diffusion coefficients for the monomer and oligomer are similar to those in D2O solution without NaCl. This result indicates that the oligomerization states do not change upon adding NaCl.
- 36.(a) Durchschlag H. In: Thermodynamic Data for Biochemistry and Biotechnology. Hinz H-J, editor. New York: Springer-Verlag; 1986. pp. 45–128. [Google Scholar]; (b) Durchschlag H, Zipper P. Progr. Colloid Polym. Sci. 1994;94:20–39. [Google Scholar]
- 37.The Hao subunit was calculated to have a MW of 235.12, a volume of 152.8 cm3/mol, and a partial specific volume of 0.65 cm3/g according to reference 36b.
- 38.(a) Demeler B. In: Modern Analytical Ultracentrifugation: Techniques and Methods. Scott DJ, Harding SE, Rowe AJ, editors. UK: Royal Society of Chemistry; 2005. pp. 210–229. [Google Scholar]; (b) http://www.ultrascan.uthscsa.edu.
- 39.Demeler B, Van Holde KE. Anal. Biochem. 2004;335:279–288. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 40.Finite element analysis of the data from the 39 µM experiment suggests that this peak corresponds to the monomer. For a description of finite element analysis, see: Cao W, Demeler B. Biophys J. 2005;89:1589–1602. doi: 10.1529/biophysj.105.061135. Demeler B, Saber H. Biophys. J. 1998;74:444–454. doi: 10.1016/S0006-3495(98)77802-6.
- 41.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Biophys. J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The UV-absorbance data from the 15 µM samples (g and h) are scaled to fringes and appear relatively shallow because they are plotted on the same scale as the data from the 1.5 and 9 mM experiments.
- 43.Foss TR, Kelker MS, Wiseman RL, Wilson IA, Kelly JW. J. Mol. Biol. 2005;347:841–854. doi: 10.1016/j.jmb.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 44.Hagihara Y, Oobatake M, Goto Y. Protein Sci. 1994;3:1418–1429. doi: 10.1002/pro.5560030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The ionization state of the Glu residue in peptides 3b and 3h is not certain, because the studies were done without buffer. Attempts to control the pH with phosphate buffer resulted in precipitation of these peptides.
- 46.PFG NMR diffusion measurements confirm that peptide 3f participates in a monomer-tetramer equilibrium. At 298 K, the diffusion coefficients of the monomer and oligomer in D2O were measured as 2.04 × 10−6 cm2/s and 1.16 × 10−6 cm2/s, respectively, which are about the same as those values for the monomer and tetramer of peptide 3a. The ratio of these values (0.57) is consistent with tetramer formation.
- 47.PFG NMR diffusion measurements confirm that peptide 3h participates in a monomer-tetramer equilibrium. At 280 K, the diffusion coefficient of the oligomer of a 10 mM D2O solution of the peptide was measured as 6.5 ×10−7 cm2/s, which is about the same as that of the tetramer of peptide 3a at the same temperature.
- 48.The diffusion coefficients of the oligomers of peptides 3j and 3k are about 0.80–0.85 times that of the tetramer of peptide 3a. The smaller diffusion coefficients of these peptides suggest that peptides 3j and 3k form higher oligomers, such as hexamers or octamers, rather than tetramers. ROESY experiments show that the oligomers of these peptides comprise edge-to-edge dimers. The higher oligomers may be thought of as small micelles in which the hydrophobic faces of the β-sheet dimers form a hydrophobic core.
- 49.We have observed similar improvements in the folding of the 42-membered-ring macrocyclic peptides 2 upon placing Phe across from the aromatic ring of Hao. For details, see reference 18.
- 50.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. p. 17. [Google Scholar]
- 51.Increasing the concentration of a D2O solution of peptide 3l from 1.2 mM to 10.0 mM results in an average downfield shifting of the α-proton resonances by ca. 0.05 ppm at 298 K. Increasing the concentration of a D2O solution of peptide 3m from 1.0 mM to 16.0 mM results in similar downfield shifting.
- 52.For a notable exception, see: Li H, Dunn JJ, Luft BJ, Lawson CL. Proc. Natl. Acad. Sci. USA. 1997;94:3584–3589. doi: 10.1073/pnas.94.8.3584. Koide S, Huang X, Link K, Koide A, Bu Z, Engelman DM. Nature. 2000;403:456–460. doi: 10.1038/35000255.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental procedures and NMR spectra, mass spectra, and HPLC traces for cyclic peptides. This material is available free of charge via the Internet at http://pubs.acs.org.