Abstract
The link between human herpesvirus 8 (KSHV) and Kaposi’s Sarcoma (KS) has been proven, but many important aspects including risk factors, genetic predisposition to tumor development, transmission of KHSV, and the pathogenic potential of different genotypes remain to be elucidated.
Possible associations between clinical parameters and antibody levels, viral load fluctuations, and viral genotype were analyzed by quantitative real-time PCR, an in-house developed IFA assay, and sequence analysis of ORF K1-VR1 in blood, serum and saliva of 38 subjects with classic KS (cKS). KHSV lytic antibodies were significantly increased in stage IV compared to stage I and II patients (p= 0.006 and p=0.041, respectively). KHSV blood, serum, and saliva viral load was comparable in all stages. The highest viral loads were detected in saliva, and they decreased in stage III-IV compared to stage I-II patients. Higher concentrations of lytic antibodies and higher viral loads were observed in fast progressing cKS patients, in whom KHSV detection from blood was also more frequent. Type A KSHV strain was almost exclusively present in fast progressors (12/17 cases), while C type was mainly present in slow progressing patients (6/7 cases). Finally, detection of type A KHSV strain associated with higher blood viral loads.
KHSV lytic antibody levels and viral load can be used to monitor clinical evolution of cKS. Infection supported by KHSV A subtype is associated with faster progressing disease. Careful monitoring and aggressive therapeutic protocols should be considered in patients with KHSV A-supported infection.
Keywords: classic Kaposi’s sarcoma, HHV8, HHV8 subtypes, tumor progression
INTRODUCTION
Kaposi’s Sarcoma-Associated Herpesvirus (KHSV), also known as Human Herpesvirus 8 (HHV-8), is a more recent member of the Herpesviridae family (Gammaherpesvirinae subfamily, Rhadinovirus genus) etiologically associated with all forms (classic, endemic African, AIDS-associated, iatrogen) of Kaposi’s sarcoma (KS) (Chang et al., 1994). Classic KS (cKS), the topic of this analysis, is a rare vascular tumor mainly observed in elderly Mediterranean individuals. The incidence rate of this tumor varies widely in different geographic areas; in Italy the estimated incidence rate is 0.4/100000 in women and 1/100000 in men (Dal Maso et al., 2005); this rate is higher compared to what is observed in other European areas. cKS is usually characterized by a slow progression that, nevertheless, can be abruptly interrupted by relapses, development of new lesions, and complications, such as lymphedema, ulcerations, superinfections, and, possibly, involvement of internal organs (gastrointestinal tract, lymph nodes, lungs).
KHSV infection is necessary, together with other co-factors not yet defined clearly (immune dysfunction, sex, genetic background) (Guerini et al., 2006), for the development of cKS. Viral transmission occurs by sexual and non-sexual routes, and KHSV seroprevalence varies in European general populations with geographical localization (Calabrò et al., 1998; Gao et al., 1996)
Sequence analysis of the highly variable ORF K1 region has allowed the identification of four main KHSV subtypes (A, B, C, D,)(Zong et al.,1999) that are differently distributed in the world: KSHV subtype A and C predominate in Europe and USA, while B subtype predominates in Africa, D is present in the Pacific islands; other recently identified subtypes are E, clusters in ancient populations, like Brazilian Amerindians (Biggar et al., 2000; Kazanji et al., 2006), Z detected in Zambian children (Kasolo et al., 1998) and F identified in Ugandan Bantu tribe (Kajumbula et al., 2006). It is still unclear whether different genotypes are associated with diverse rates of disease progression; analogously, the role of viral load, antibody titers and of other possible surrogate markers on the outcome of KS is still under discussion.
In an attempt to clarify the relative importance of virologic parameters in disesase progression we compared a number of these parameters in a group of Italian patients affected by classic KS (cKS).
MATERIALS AND METHODS
Patients
38 histologically-confirmed cKS patients (30 males, 8 female; median age 69 years; range 44–90) were enrolled in the study. All prior therapies for KS were completed at least two months before enrolment in the study. Patients were clinically classified in four stages defined as reported in Table I (Brambilla et al., 2003), considering the type and localization of skin lesions as well as the presence of complications and visceral involvement. Moreover, all patients were classified as fast or slow progressors on the basis of clinical parameters (Table II). After signing informed consent, peripheral blood, plasma and whole saliva samples were collected for each patients and stored at −20°C. All patients were HIV-1 seronegative.
Table II.
Demographic and clinical characterization of the patients with classic Kaposi Sarcoma (cKS) enrolled in the study.
| Patient number | Mean age | range | M/F | |
|---|---|---|---|---|
| STAGE I | 10 | 62 | 44–88 | 5/5 |
| STAGE II | 10 | 71 | 56–88 | 9/1 |
| STAGE III | 12 | 70 | 52–90 | 10/2 |
| STAGE IV | 6 | 73 | 61–80 | 6/0 |
| slow evolution | 9 | 64 | 44–88 | 6/3 |
| fast evolution | 29 | 70 | 52–90 | 24/5 |
| Total cKS | 38 | 69 | 44–90 | 30/8 |
Serologic assays
Antibodies against latent and lytic HHV8 antigens were detected using an immunofluorescence assay (IFA) based on the body-cavity B cell lymphoma (BCBL-1) cell line (Renne et al., 1996), as described elsewhere (Perna et al., 2000). Antibody titrations were done by twofold serial dilutions, starting from 1:100. Antibodies against latent antigens showed a specific granular punctate nuclear fluorescence in 80% of spotted cells; lytic antibodies showed cytoplasmic expression in at least 20–30% of cells for each microscopic field. Monoclonal antibodies to ORF73 (LNA-1), to ORF 59 and ORF K8.1 lytic proteins (Advanced Biotechnologies Inc., Maryland, USA) were utilized as controls of the specific patterns of fluorescence.
DNA extraction
DNA extraction from buffy-coat samples was performed by phenol-chloroform extraction, using standard procedures. The collected DNA, measured with a spectrophotometer, was stored at −20°C. Nucleic acid extraction was performed from 150 μl serum using Nucleospin virus, (Macharey-Nagel, Duren, Germany), as previously described (Boldorini et al., 2003), and from saliva samples from 2 ml using Nucleospin Blood Kit (Macherey–Nagel, Duren, Germany), according to the manufacturer’s instructions. These kits utilized silica columns which have a high binding capacity for nucleic acids in presence of guanidine thiocyanate. All procedures were carried out under stringent conditions to avoid contamination: each sample was processed individually; DNA extractions were performed one at time, in a clean, dust-free area with sterile and disposable materials, with a change of gloves for each sample.
Real-time PCR
KSHV DNA was determined by quantitative real-time PCR using an ABI PRISM 7000 SDS (Applied Biosystems, Foster City, CA). The primers and the probe were designed using the Primer express software (Applied Biosystems, Foster City, CA) (Table III) to obtain a 74 bp amplicon on ORF26 region of the viral genome, coding for KHSV minor capsid protein. PCR amplification was performed in 25 μl reaction mixtures using Taqman Universal Master Mix (Applied Biosystems), 0.9 mM each primers, 0.2 mM Taqman probe and 250 ng of blood DNA or 10 μl of DNA extracted from serum or saliva were used as template. Each sample was analyzed in triplicate: samples that did not yield three positive reactions were repeated in triplicate and viral load results were given by the mean of the three positive reactions. Each run contained a negative control, constituted by the reaction mixture without the DNA template. Viral load was analysed by interpolation of a standard curve obtained from serial dilution of KHSV quantitated purified viral DNA (Advanced Biotechnologies Inc., Columbia, MD). The assay was linear from 101 to 107 copies per reaction (dynamic range). The interassay variability, evaluated by repeating the quantitation of the same positive specimen for 8 times, was low, with the coefficient of variation less than 20% for viral load ranging from 130 copies/ml (2.11 log10) to 1.3 × 105 copies/ml (5.11 log10). The specificity was tested using as template DNA of EBV, (whose genome is very similar to KHSV genome) wich was not amplified in our assay. The lowest detection limit was 10 copies/reaction, which is equivalent to 40 copies/μg of DNA extracted from blood, 630 copies/ml of serum and 4000 copies/ml of saliva.
Table III.
Primers and probes utilized for real-time PCR.
| primers | nt.* | Sequence 5′-3′ |
|---|---|---|
| ORF26 F | 47311–47331 | 5′-CTCGAATCCAACGGATTTGAC-3′ |
| ORF26R | 47384–47366 | 5′-TGCTGCAGAATAGCGTGCC-3′ |
| PROBE | 47342–47360 | FAM-CCATGGTCGTGCCGCAGCA-TAMRA |
(nt. position referred to accession number: U75698)
KHSV strains genotyping
Genotype characterization of KHSV strains amplified from different KS patients was based on sequencing of the viral region ORFK1 coding for a transmembrane protein and containing two hypervariable regions (VR1 and VR2). Amplification was performed by nested PCR protocol that amplified one fragment of the ORF-K1 variable loop region, VR1 (spanning from nt. 23 to nt. 445) as previously described (Nascimento et al., 2005). Procedures were carried out under stringent conditions to avoid contamination; multiple negative and positive samples were included in each PCR.
To perform nucleotide sequence analysis, 10ng of PCR products were added to a mixture containing 4μl of Ready Reaction Premix 2.5X, 2μl of BigDye Sequencing (Applied Biosystems) 5X buffer, 3.2 pmol of either forward or reverse primer, and water up to a final volume of 20μl. The cycle sequencing was performed using the GeneAmp PCR System 9700 (Applied Biosystems), according to the following protocol: initial denaturation at 96°C for 1’, then 25 cycles with a first step at 96°C for 10’’, a second step at 50°C for 5’’ and the last rapid thermal ramp to 60°C for 4’. In order to purify the extension products, Centrisep columns (Princetons Separation, Inc, Adelphia) were used, following the manufacturer’s protocol. Finally, 5 μl of the purified product underwent electrophoresis on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). Sequence homology searches were performed using BLAST at NCBI (USA). The genotype of each samples was determined by comparing its sequence with those of KHSV prototypes (Zong et al., 1999) obtained from GeneBank. Multiple sequence alignment were performed using CLUSTALW 1.7 program (Thompson et al., 1994). Phylogenetic tree was obtained by neighbour-joining (NJ) method, calculating bootstrap values (100 replica samplings) using ClustalX 2.0 (Thompson et al, 1997) List of references sequences: AF133038 (A1); AF130305(A2) ; U86667(A3); AF133039(A4); AF178823 (A5); AF133040 (B1); AF130259(B2); AF133041(C1); AF133042 (C3) ; AF133043 (D1) AF133044 (D2); AF220292 (E).
Statistical analysis
As data were non distributed normally (Shapiro–Wilk’s test), they are reported as median and IQR (Inter Quartile Range). Comparisons between groups were performed using the Kruskal-Wallis ANOVA, and the Mann-Whitney U test for posthoc pair-wise comparisons with Bonferroni correction for multiple tests. Differences in frequencies were evaluated by means of Chi-square statistic or Fisher exact test, as appropriate. All tests were two–sided. Analyses were performed with Statistica for Windows software (StatSoft, Inc. 2004, Tulsa, OK, US.).
RESULTS
Antibody titers, KHSV DNA, and KHSV viral load in cKS patients
A total of thirty-eight cKS patients were enrolled in the study: 10 subjects were classified as KS stage I, 10 as stage II, 12 as stage III and 6 as stage IV. In terms of disease evolution, 29 patients had fast and 9 slow progressing cKS.
Specific antibodies to KHSV latent (anti-LANA) and lytic antigens (anti-lytic) were detected in 100% of patients. LANA-specific antibody titers (median reciprocal of titres: 3.54 log10) were higher than those towards lytic antigens (median titer:3.39 log10)(Table IV). Median levels of LANA antibodies did not differ among the four stages (stage I: 3.8, stage II: 3, stage III 3.65, stage IV 3.74), whereas lytic antibodies increased significantly in stage III (3.39) and IV (4.04) compared to stage I and II patients (I vs. IV :p= 0.006; II vs. IV :p=0.041). KHSV DNA was detected by real-time PCR in 66% of peripheral blood, in 42% of serum and in 58% of saliva samples. Whereas KHSV DNA was more frequently detected in the blood or serum of patients with more severe disease, salivary KHSV DNA was more frequently observed in patients belonging to stage I (70%) than in patients with other stages (60% on stage II and to 50% in III and IV stage). None of these differences reach statistical significance.
Table IV.
Antibody levels, detection KHSV DNA and viral load in cKS patients grouped according to clinical stage and development of disease.
| Antibodies a | KHSV DNA (%)b | viral load HHV8 (median) c | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n° | Anti latent Ag | anti lytic Ag | blood | Serum | Saliva | Blood | serum | saliva | ||
| Clinical stage | I | 10 | 3.80 (3.3–4) | 3.09* (2.7–3.6) | 40 (4/10) | 10 (1/10) | 70 (7/10) | 1.54 (1.54–3.22) | 3.04 (3.04–3.04) | 6.97 (3.54–7.79) |
| II | 10 | 3.00 (2.7–4) | 3.00# (2.3–3) | 80 (8/10) | 60 (6/10) | 60 (6/10) | 2.8 (2.4–3.38) | 3.55 (3.0 – 4.15) | 5.11 (3.54 – 5.91) | |
| III | 12 | 3.65 (3.0–4.15) | 3.39 (3.15–3.74) | 67 (8/12) | 50 (6/12) | 50 (6/12) | 2.75 (1.54–3.41) | 3.22 (3.04 –3.62) | 3.54 (3.54–6.96) | |
| IV | 6 | 3.74 (3.3–4) | 4.04*# (3.7–4.3) | 83 (5/6) | 50 (3/6) | 50 (3/6) | 2.87 (2.19–3.2) | 3.45 (3.04–4.54) | 4.08 (3.54–5.26) | |
|
| ||||||||||
| Clinical Evolution | slow | 9 | 3.60 (3.3–4) | 3.00 (2.7–3.48) | 44 (4/9) | 11 (1/9) | 67 (6/9) | 1.54 (1.54–2.57) | 3.04 (3.04–3.04) | 5.83 (3.54–7.79) |
| fast | 29 | 3.48 (3–4) | 3.48 (3–4) | 72 (21/29) | 52 (15/29) | 55 (16/29) | 2.9 (1.54–3.36) | 3.18 (3.04–4.15) | 4.63 (3.54–6.96) | |
|
| ||||||||||
| Total | 38 | 3.54 (3–4) | 3.39 (2.95–4) | 66 (25/38) | 42 (16/38) | 58 (22/38) | 2.54 (1.54–3.34) | 3.04 (3.04–3.92) | 4.94 (3.54–6.97) | |
The median log of reciprocal titre and interquartile range (IQR) are given.
Percentage and absolute number are shown
Viral load is expressed as median log copies/μg in blood and in median log copies/ml in serum and saliva samples. In bracket IQR
p = 0.006
p = 0.041
Viral load was lower in blood (median 2.54 log10 copies/μg DNA) and in serum samples (median value 3.04 log10copies/ml) compared to the results obtained in saliva (median value 4.94 log10 copies/ml). An increasing trend in KHSV load was observed in blood and in serum samples of later stages patients (median values in stage I, II, II, IV: blood= 1.54, 2.8, 2.75, 2.87; serum= 3.04, 3.55, 3.22, 3.45). Also in this case results obtained in saliva were different. Thus, KHSV salivary load decreased from stage I (6.97 log10 copies/ml) to stage IV (4.08 log10 copies/ml). Again, these differences did not reach statistical significance.
Association between viro-immunologic and clinical parameters in cKS patients
Higher median value of lythic antigensspecific antibodies were detected in serum of fast (median 3.48 log10) compared to slow (median 3 log 10) progressors; KHSV DNA detection was also more frequent in blood (72% vs. 44%; p=0.22) and serum (52% vs. 11%; p=0.05) of these patients.
Finally, higher viral loads were present in blood (median values: 2.9 vs. 1.54 log10copies/μg) and serum (median: 3.18 vs 3.04 log10copies/ml; p=0.06) of patients with faster disease progression. Interestingly, though, KHSV viral load was decreased in saliva of faster-compared to slower-progressing patients (median: 5.83 vs. 4.63 log10copies/ml).
Association of clinical parameters with KHSV genotype
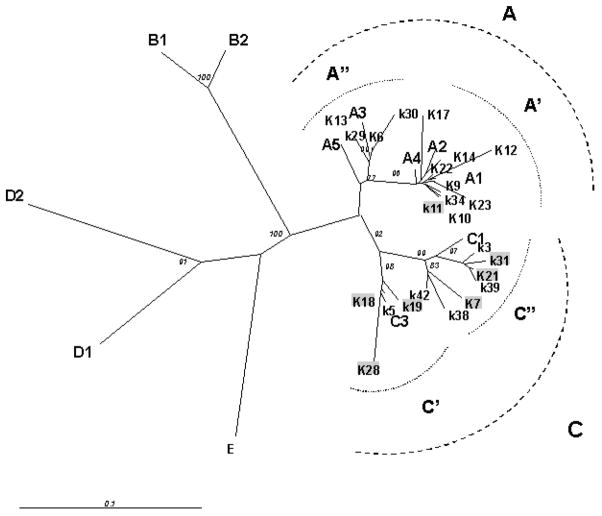
VR-1 PCR products was obtained from 24 /38 cKS subjects . Molecular characterization was performed by direct sequence analysis of 24 amplicons. Phylogenetic tree obtained from these sequences by NJ method is shown in Figure 1. KHSV subtypes identification, based on the classification of Cook et al (1999) and Zong et al (2002), are reported in Table V.
Figure 1.
Radial unrooted phylogenetic tree of 24 partial VR1 DNA sequences (348 bp) obtained from cKS italian patients. Neighbour joining (NJ) analysis with 100 bootstrap replicas was carried out including the following KHSV DNA sequences deposited in Genebank : A1: AF133038 A2:AF130305 A3: U86667 A4: AF133039 A5: AF178823; B1:AF133040; B2: AF130259 C1: AF133041 C3:AF133042; D1: AF133043 D2: AF133044 ;E:AF220292). Bootstrap values ≥ 70 are shown for support. Sequences highlighted in gray correspond to strain obtained from patients with slow evolution of cKS. 254×190mm (96 × 96 DPI)
Table V.
Molecular typing of 24 KHSV strains obtained from cKS italian patients according to clinical evolution of disease.
| Patient ID | sex | clinical stage | clinical evolution | KHSV (K1) subtype |
|---|---|---|---|---|
| K6 | F | I | fast | A3 |
| K9 | M | III | fast | A1 |
| K10 | M | III | fast | A′ |
| K12 | M | II | fast | A1 |
| K13 | M | IV | fast | A″ |
| K14 | M | II | fast | A1 |
| K17 | F | III | fast | A2 |
| K22 | M | II | fast | A′ |
| K23 | M | II | fast | A′ |
| K29 | M | III | fast | A3 |
| K30 | M | I | fast | A3 |
| K34 | M | IV | fast | A1 |
| K3 | M | III | fast | C1 |
| K5 | M | IV | fast | C3 |
| K38 | M | III | fast | C1 |
| K40 | M | III | fast | C1 |
| K42 | F | III | fast | C1 |
| K11 | M | I | slow | A1 |
| K7 | M | I | slow | C1 |
| K18 | M | II | slow | C3 |
| K19 | F | I | slow | C3 |
| K21 | M | I | slow | C1 |
| K28 | F | I | slow | C3 |
| K31 | M | II | slow | C1 |
Seventeen strains were analysed in faster-progressing patients: in 12/17 cases (71%) KHSV A subtype was observed, whereas subtype C was present only in 5/17 (29%) patients. In contrast with these results, subtype A KHSV was observed in only 1/7 (14%) patients with slower disease evolution, in whom subtype C was highly prevalent (6/7; 86%). These differences were statistically significant (Fisher exact test: p= 0.023). Moreover, KSHV blood viral load was significantly higher in patients infected with an A (median 3.39 log10 copies/ml, IQR 3.16–3.82) compared to those infected with a C subtype (median 2.5 log10 copies/ml, IQR 1.3–3.19) (Mann- Whitney test, p= 0.0054). These results are presented in Figure 2.
Figure 2.
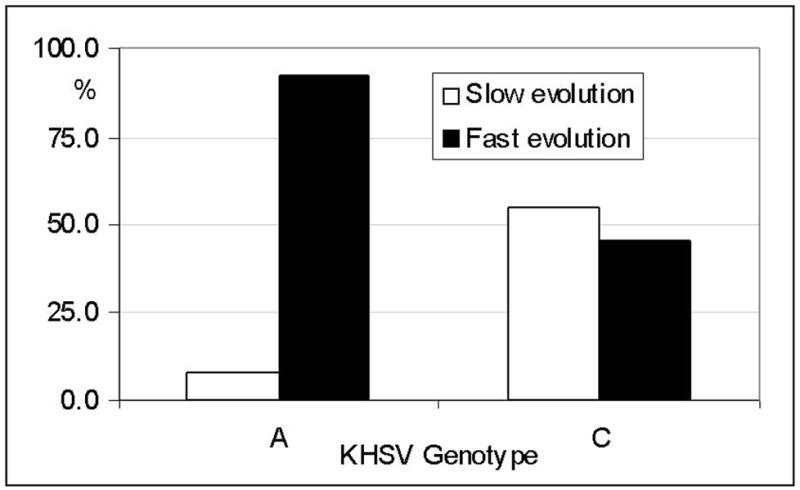
KHSV A and C subtypes frequency (%) in cKS patients with slow and fast evolution. 254×190mm (96 × 96 DPI)
DISCUSSION
Different studies have assessed the association of KHSV viral parameters and KS progression in HIV-infected patients (Campbell et al., 2000; Pellet et al., 2001), but few reports are available in patients with classical KS.
In this study immuno-virological analyses were performed in 38 Italian patients with a diagnosis of classical Kaposi’s sarcoma in order to search possible link with clinical stage and type of evolution of sarcoma.
KHSV was detected in blood samples of 66% of patients, a result that confirms previous data (Boneschi et al., 2001; Pellet et al., 2006). Additional results showed that KHSV viral loads increase in blood and serum samples of patients with more severe disease; detection of KHSV DNA in blood and serum samples was also more frequent in these patients. No clearcut, statistically significant association were nevertheless observed between rate of KHSV detection, KHSV viral load, and clinical stage. These results might be secondary to the low KHSV copy numbers present in patients with KS or, more simply, to the small number of patients for each clinical stage and the large dispersion of values around the median. These data confirm a previous report indicating that associations between KHSV viral load and disease progression was only possible in HIV coinfected patients (Engels et al., 2003) : a scenario of profound immunologic damage that facilitates KHSV viral replication. When these parameters were analyzed based on rate of disease progression a clearer picture nevertheless emerged. Thus, both KHSV viral load and rate of virus detection from blood differed in patients with fast disease progression.
Finally, serum titers of KHSV lytic antigens-specific antibodies were significantly increased both in later stage and in fast progressing patients; these data suggest that lytic viral replication may occur, probably in other tissue other than blood cells, (where any viral load increase was observed) leading to increase of lytic antibody, underlining a possible pivotal role for viral replication in the natural history of cKS.
The highest KHSV viral loads were observed in saliva samples; this observation strongly suggests that oral shedding is an important source of transmission of KHSV, at least in the classic KS form of disease, and will be interesting to study virus diffusion in families of cKS patients KHSV detection and KHSV viral loads were higher in saliva of patients in the initial phase of disease; this trend was specular to what observed in blood and plasma: in these body fluids KHSV loads augmented in the later stages of disease. These results indicate the KHSV does compartmentalize, and support previous observations indicating that KHSV preferentially replicates in the oro-pharingeal tissues (Pauk et al., 2000; Lampinen et al., 2000; Marcelin et al., 2004; Vieira et al., 1997). This biologic behaviour is seen in other human herpes viruses as well, and in particular, seems to be shared with the closely related Epstein-Barr virus (EBV) (Ikuta et al., 2000). In this regard it interesting to observe that the increase of serum lytic antibodies seen in later stages disease correlated with a decreased salivary KHSV concentration: it is possible to hypothesize that the high level of lytic antibodies observed in the later disease stages could reduce the oro-pharingeals lytic KHSV replication.
KHSV A and C subtypes predominate in Italy (Cook et al., 1999; Zong et al., 2002). Interestingly, the dominant KHSV genotype was different in fast compared to slow progressing patients; subtype A was significantly more frequently isolated in patients with fast progression; subtype C prevailed in individuals with slow progression. Moreover, KHSV viral load was significantly increased in patients in whom subtype A was isolated. KHSV A genotype has been associated with higher transforming potential and higher aggressiveness (Luppi et al., 1997; Boralevi et al., 1998; Gazouli et al., 2004); possible correlations between genotype and KHSV-related pathologies with a different degree of aggressiveness have nevertheless not been confirmed by all authors (Cook et al., 1999; Lacoste et al., 2000; Kadirova et al., 2003). Our results support the claim that infection with KHSV A subtype is indeed associated with worst clinical parameters, including faster disease progression and higher KHSV viral loads.
The sequencing of whole the K1 gene will be an important improvemement of this study, to evidence intratypic or intertypic recombinant events. Although the limited number of patients included in sequence analysis will require confirmation by wider studies, our results indicate the possibility that KHSV strain variability could be associated with different clinical evolution of cKS. Careful monitoring and aggressive therapeutic protocols should be considered in patients with KHSV A genotype-supported infection.
Table 1.
Classical Kaposi’s sarcoma staging (Brambilla et al 2003).
| Stage | Skin lesions | Behaviour | Evolution | Complications* |
|---|---|---|---|---|
| I) Macular/ nodular | Maculae and/or nodules | Non aggressive | Slow
Fast |
Lymphoedema
Lymphorrea Hemorrhage Ulceration Pain Functional impairment |
| II) Infiltrative (+/− v) | Plaques | Locally aggressive | Slow
Fast |
|
| III) Florid (+/− v) | Angiomatous nodules and plaques | Locally aggressive | Slow
Fast |
|
| IV) Generalized (+/−) | Angiomatous nodules and plaques | Disseminated aggressive | Slow
Fast |
v = visceral involvement (pharyngo-oral cavity, gastroenteric tract, lymph nodes, bone marrow, lungs)
Fast = increase in total number of nodules/plaques or in total area of plaques in the three months following an examination
All of them prevalent in stage III and IV, lymphedema and lymphorrea often observed in stage II, lymphedema, ulceration and hemorrage sometimes present in stage I.
Acknowledgments
This study was supported by grants from the Istituto Superiore di Sanita’ “Programma Nazionale di Ricerca sull’ AIDS”; NIH grant n. MH072528; the EMPRO and AVIP EC WP6 Projects; the Japan Health Science Foundation; 2007 Ricerca Finalizzata (Italian Ministry of Health); 2007 Ricerca Corrente (Italian Ministry of Health).
References
- Biggar RJ, Whitby D, Marshall V, Linhares AC, Black F. Human herpesvirus 8 in Brazilian Amerindians: a hyperendemic population with a new subtype. J Infect Dis. 2000;181(5):1562–8. doi: 10.1086/315456. [DOI] [PubMed] [Google Scholar]
- Boldorini R, Pagani E, Car PG, Omodeo-Zorini E, Borghi E, Tarantini L, Bellotti C, Ferrante P, Monga G. Molecular characterization of JCV virus strains detected in human brain tumors. Pathology. 2003;35:248–253. doi: 10.1080/0031302031000123245. [DOI] [PubMed] [Google Scholar]
- Boneschi V, Brambilla L, Berti E, Ferrucci S, Corbellino M, Parravicini C, Fossati S. Human herpesvirus 8 DNA in the skin and blood of patients with Mediterranean Kaposi’s sarcoma: clinical correlations. Dermatology. 2001;203(1):19–23. doi: 10.1159/000051697. [DOI] [PubMed] [Google Scholar]
- Boralevi F, Masquelier B, Denayrolles M, Dupon M, Pellegrin JL, Ragnaud JM, Fleury HJ. Study of human herpesvirus 8 (HHV-8) variants from Kaposi’s sarcoma in France: is HHV-8 subtype A responsible for more agressive tumors? J Infect Dis. 1998;178(5):1546–7. doi: 10.1086/314471. [DOI] [PubMed] [Google Scholar]
- Brambilla L, Boneschi V, Taglioni M, Ferrucci S. Staging of classic Kaposi’s sarcoma: a useful tool for therapeutic choices. Eur J Dermatol. 2003;13(1):83–6. [PubMed] [Google Scholar]
- Calabrò ML, Sheldon J, Favero A, Simpson GR, Fiore JR, Gomes E, Angarano G, Chieco-Bianchi L, Schulz TF. Seroprevalence of Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 in several regions of Italy. J Hum Virol. 1998;1(3):207–13. [PubMed] [Google Scholar]
- Campbell TB, Borok M, Gwanzura L, MaWhinney S, White IE, Ndemera B, Gudza I, Fitzpatrick L, Schooley RT. Relationship of human herpesvirus 8 peripheral blood virus load and Kaposi’s sarcoma clinical stage. AIDS. 2000;14(14):2109–16. doi: 10.1097/00002030-200009290-00006. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–69. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Cook PM, Whitby D, Calabro ML, Luppi M, Kakoola DN, Hjalgrim H, Ariyoshi K, Ensoli B, Davison AJ, Schulz TF. Variability and evolution of Kaposi’s sarcoma-associated herpesvirus in Europe and Africa. International Collaborative Group. AIDS. 1999;13(10):1165–76. doi: 10.1097/00002030-199907090-00004. [DOI] [PubMed] [Google Scholar]
- Dal Maso L, Polesel J, Ascoli V, Zambon P, Budroni M, Ferretti S, Tumino R, Tagliabue G, Patriarca S, Federico M, Vercelli M, Giacomin A, Vicario G, Bellù F, Falcini F, Crocetti E, De Lisi V, Vitarelli S, Piffer S, Stracci F, Serraino D, Rezza G, Franceschi S. Classic Kaposi’s sarcoma in Italy, 1985–1998. Br J Cancer. 2005;92:188–93. doi: 10.1038/sj.bjc.6602265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels EA, Biggar RJ, Marshall VA, Walters MA, Gamache CJ, Whitby D, Goedert JJ. Detection and quantification of Kaposi’s sarcoma-associated herpesvirus to predict AIDS-associated Kaposi’s sarcoma. AIDS. 2003;17(12):1847–51. doi: 10.1097/00002030-200308150-00015. [DOI] [PubMed] [Google Scholar]
- Gao SJ, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo CR, Saah A, Phair J, Detels R, Chang Y, Moore PS. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi’s sarcoma. Nat Med. 1996;2(8):925–8. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- Gazouli M, Papaconstantinou I, Zavos G, Metaxa-Mariatou V, Nasioulas G, Boletis J, Arapadoni-Dadioti P, Giaslakiotis K, Zografidis A, Kostakis A. Human herpesvirus type 8 genotypes in iatrogenic, classic and AIDS-associated Kaposi’s sarcoma from Greece. Anticancer Res. 2004;24(3a):1597–602. [PubMed] [Google Scholar]
- Guerini FR, Agliardi C, Mancuso R, Brambilla L, Biffi R, Ferrucci S, Zanetta L, Zanzottera M, Bramati M, Boneschi V, Ferrante P. Association of HLA-DRB1 and –DQB1 with Classic Kaposi’s Sarcoma in Mainland Italy. Cancer Genomic & Proteomics. 2006;3(3):167-G. [PubMed] [Google Scholar]
- Ikuta K, Satoh Y, Hoshikawa Y, Sairenji T. Detection of Epstein-Barr virus in salivas and throat washings in healthy adults and children. Microbes Infect. 2000;2(2):115–20. doi: 10.1016/s1286-4579(00)00277-x. [DOI] [PubMed] [Google Scholar]
- Kadyrova E, Lacoste V, Duprez R, Pozharissky K, Molochkov V, Huerre M, Gurtsevitch V, Gessain A. Molecular epidemiology of Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 strains from Russian patients with classic, posttransplant, and AIDS-associated Kaposi’s sarcoma. J Med Virol. 2003;71(4):548–56. doi: 10.1002/jmv.10530. [DOI] [PubMed] [Google Scholar]
- Kajumbula H, Wallace RG, Zong JC, Hokello J, Sussman N, Simms S, Rockwell RF, Pozos R, Hayward GS, Boto W. Ugandan Kaposi’s sarcoma-associated herpesvirus phylogeny: evidence for cross-ethnic transmission of viral subtypes. Intervirology. 2006;49(3):133–43. doi: 10.1159/000089374. [DOI] [PubMed] [Google Scholar]
- Kasolo FC, Monze M, Obel N, Anderson RA, French C, Gompels UA. Sequence analyses of human herpesvirus-8 strains from both African human immunodeficiency virus-negative and -positive childhood endemic Kaposi’s sarcoma show a close relationship with strains identified in febrile children and high variation in the K1 glycoprotein. J Gen Virol. 1998;79:3055–65. doi: 10.1099/0022-1317-79-12-3055. [DOI] [PubMed] [Google Scholar]
- Kazanji M, Dussart P, Duprez R, Tortevoye P, Pouliquen JF, Vandekerkhove J, Couppié P, Morvan J, Talarmin A, Gessain A. Serological and molecular evidence that human herpesvirus 8 is endemic among Amerindians in French Guiana. J Infect Dis. 2005;192(9):1525–9. doi: 10.1086/491744. [DOI] [PubMed] [Google Scholar]
- Lacoste V, Judde JG, Brière J, Tulliez M, Garin B, Kassa-Kelembho E, Morvan J, Couppié P, Clyti E, Forteza Vila J, Rio B, Delmer A, Mauclère P, Gessain A. Molecular epidemiology of human herpesvirus 8 in africa: both B and A5 K1 genotypes, as well as the M and P genotypes of K14.1/K15 loci, are frequent and widespread. Virology. 2000;278(1):60–74. doi: 10.1006/viro.2000.0629. [DOI] [PubMed] [Google Scholar]
- Lampinen TM, Kulasingam S, Min J, Borok M, Gwanzura L, Lamb J, Mahomed K, Woelk GB, Strand KB, Bosch ML, Edelman DC, Constantine NT, Katzenstein D, Williams MA. Detection of Kaposi’s sarcoma-associated herpesvirus in oral and genital secretions of Zimbabwean women. J Infect Dis. 2000;181(5):1785–90. doi: 10.1086/315426. [DOI] [PubMed] [Google Scholar]
- Luppi M, Barozzi P, Marasca R, Ferrari MG, Torelli G. Human herpesvirus 8 strain variability in clinical conditions other than Kaposi’s sarcoma. J Virol. 1997;71(10):8082–3. doi: 10.1128/jvi.71.10.8082-8083.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelin AG, Gorin I, Morand P, Ait-Arkoub Z, Deleuze J, Morini JP, Calvez V, Dupin N. Quantification of Kaposi’s sarcoma-associated herpesvirus in blood, oral mucosa, and saliva in patients with Kaposi’s sarcoma. AIDS Res Hum Retroviruses. 2004;20(7):704–8. doi: 10.1089/0889222041524689. [DOI] [PubMed] [Google Scholar]
- Nascimento MC, Wilder N, Pannuti CS, Weiss HA, Mayaud P. Molecular characterization of Kaposi’s sarcoma associated herpesvirus (KSHV) from patients with AIDS-associated Kaposi’s sarcoma in Sao Paulo, Brazil. J Clin Virol. 2005;33(1):52–9. doi: 10.1016/j.jcv.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Pauk J, Huang ML, Brodie SJ, Wald A, Koelle DM, Schacker T, Celum C, Selke S, Corey L. Mucosal shedding of human herpesvirus 8 in men. N Engl J Med. 2000;343(19):1369–77. doi: 10.1056/NEJM200011093431904. [DOI] [PubMed] [Google Scholar]
- Pellet C, Chevret S, Blum L, Gauville C, Hurault M, Blanchard G, Agbalika F, Lascoux C, Ponscarme D, Morel P, Calvo F, Lebbe C. Virologic and immunologic parameters that predict clinical response of AIDS-associated Kaposi’s sarcoma to highly active antiretroviral therapy. J Invest Dermatol. 2001;117(4):858–63. doi: 10.1046/j.0022-202x.2001.01465.x. Erratum in: J Invest Dermatol 2002 Apr;118(4):741. [DOI] [PubMed] [Google Scholar]
- Pellet C, Kerob D, Dupuy A, Carmagnat MV, Mourah S, Podgorniak MP, Toledano C, Morel P, Verola O, Dosquet C, Hamel Y, Calvo F, Rabian C, Lebbe C. Kaposi’s sarcoma-associated herpesvirus viremia is associated with the progression of classic and endemic Kaposi’s sarcoma. J Invest Dermatol. 2006;126(3):621–7. doi: 10.1038/sj.jid.5700083. [DOI] [PubMed] [Google Scholar]
- Perna AM, Bonura F, Vitale F, Viviano E, Di Benedetto MA, Ajello F, Villafrate MR, Prestileo T, Mancuso S, Goedert JJ, Romano N. Antibodies to human herpes virus type 8 (HHV8) in general population and in individuals at risk for sexually transmitted diseases in Western Sicily. Int J Epidemiol. 2000;29(1):175–9. doi: 10.1093/ije/29.1.175. [DOI] [PubMed] [Google Scholar]
- Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–46. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, Huang ML, Koelle DM, Corey L. Transmissible Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of men with a history of Kaposi’s sarcoma. J Virol. 1997;71(9):7083–7. doi: 10.1128/jvi.71.9.7083-7087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong JC, Ciufo DM, Alcendor DJ, Wan X, Nicholas J, Browning PJ, Rady PL, Tyring SK, Orenstein JM, Rabkin CS, Su IJ, Powell KF, Croxson M, Foreman KE, Nickoloff BJ, Alkan S, Hayward GS. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi’s sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73(5):4156–70. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong J, Ciufo DM, Viscidi R, Alagiozoglou L, Tyring S, Rady P, Orenstein J, Boto W, Kalumbuja H, Romano N, Melbye M, Kang GH, Boshoff C, Hayward GS. Genotypic analysis at multiple loci across Kaposi’s sarcoma herpesvirus (KSHV) DNA molecules: clustering patterns, novel variants and chimerism. J Clin Virol. 2002;23(3):119–48. doi: 10.1016/s1386-6532(01)00205-0. [DOI] [PubMed] [Google Scholar]