Abstract
Virus-immune CD8+ TCR repertoires specific for particular peptide-MHC class I complexes may be substantially shared between (public), or unique to, individuals (private). Since public TCRs can show reduced terminal deoxynucleotidyl transferase (TdT)-mediated N-region additions, we analysed how TdT shapes the heavily public (to DbNP366) and essentially private (to DbPA224) CTL repertoires generated following influenza A virus infection of C57Bl/6 (B6, H2b) mice. The DbNP366-specific CTL response was virtually clonal in TdT−/− B6 animals, with one of the three public clonotypes prominent in the wt response consistently dominating the TdT−/− set. Furthermore, this massive narrowing of TCR selection for DbNP366 reduced the magnitude of DbNP366-specific CTL response in the virus-infected lung. Conversely, the DbPA224-specific responses remained comparable in both magnitude and TCR diversity within individual TdT−/− and wt mice. However, the extent of TCR diversity across the total population was significantly reduced, with the consequence that the normally private wt DbPA224-specific repertoire was now substantially public across the TdT−/− mouse population. The overall conclusion is thus that the role of TdT in ensuring enhanced diversity and the selection of private TCR repertoires promotes optimal CD8+ T cell immunity, both within individuals and across the species as a whole.
Keywords: T Cells, Cytotoxic, T Cell Receptors, Repertoire Development, Viral
Introduction
Host response profiles tend to be highly consistent for virus-specific CD8+ T cell-mediated immunity, with the same few epitopes being targeted in individuals sharing MHC class I haplotypes. This reproducibility can extend to epitope-specific T cell responses with, in some cases, the same TCRs (usually defined at the level of identical CDR3β amino acid sequence) being used repeatedly in different individuals to constitute an essentially ‘public’ TCR repertoire, (1). Alternatively, part (or all) of an epitope-specific TCR response may be substantially “private” to a given individual. Overall, a TCR repertoire is defined as broadly public or private by the extent to which the response is shared or unique (2). This private versus public character looks to be determined by the particular peptide+MHC (pMHC) epitope, the extent and character of the available naïve TCRs and the stochastic nature of T cell antigen encounter and activation (3).
Diversity within the TCR repertoire is generated via several mechanisms. While different combinations of variable (V), diversity (D), and junctional (J) gene segments, along with the pairing of various TCR α and β (or γ and δ) chains, provide a measure of variability, much greater levels of diversity are generated at the junctions of the V, D, and J gene segments by template dependent (P additions) and template independent (N regions) nucleotide additions. The role of TdT as the DNA polymerase responsible for the template independent addition of nucleotides during both TCR and antibody gene rearrangement has long been established (4, 5), with studies in mice genetically deficient in this enzyme showing that it is normally responsible for establishing at least 90% of the naïve TCRαβ repertoire (6). Even so, despite the fact that the more limited TCR pool in TdT−/− animals results in a very substantial narrowing in epitope-specific TCR diversity, the responses to multiple epitopes in TdT−/− mice are generally comparable in magnitude to those elicited in the wt controls (7, 8).
The present analysis focuses on the role of TdT in determining the private or public nature of virus-specific CD8+ T cell TCRβ repertoires. Prior analysis with a number of epitope-specific T cell responses has shown that public TCR clonotypes have a substantially reduced number of N-region additions (7, 9), a finding that has in turn contributed to the idea that such TCRs tend to be both public and dominant within individuals because they are more readily generated by the recombination machinery (9). In this study we analyse the simultaneous establishment of distinctive CD8+ T cell repertoires, directed at H2Db complexed with peptides from the influenza A virus nucleoprotein (DbNP366-374) and acid polymerase (DbPA224-233), in virus infected TdT+/+ (wt) and TdT−/− C57BL6 mice. While both NP366 and PA224 bind to H2Db, they induce different TCRVβ biases, each with unique preferences for CDR3β length and Jβ usage (10, 11). Importantly, detailed CDR3β sequence analysis has established that while the wt DbNP366-specific repertoire tends to be restricted in clonotype diversity and is heavily public, the wt DbPA224-specific population is much more “individualized”, or private. This has allowed us to probe how TdT determines TCR repertoire diversity, the extent of sharing (public) or uniqueness (private) and response magnitude for parallel, in vivo viral epitope specific CD8+ T cell responses.
Materials and Methods
Mice and virus infections
Female C56Bl/6J (H-2b) mice were purchased from The Jackson Laboratory and TdT−/− mice (4) were a kind gift from Dr Ann Feeney from The Scripps Research Institute. Naïve mice were anaesthetised by i.p. injection of 2,2,2-tribromoethanol (Avertin) (250 mg/kg) and primed i.n. with 106 EID50 of the HKx31 (H3N2) influenza A virus (12) in 30 μl of PBS. “Memory” mice were primed by i.p. injection with 108 EID50 of the serologically distinct PR8 (H1N1) influenza A virus that shares NP and PA proteins of HKx31 (13, 14), then challenged i.n. with HKx31 virus at least 4 weeks later to generate a secondary response. Virus stocks were grown in the allantoic cavity of 10-day (d) embryonated hen’s eggs and quantified as EID50. All animal work was performed in compliance with the guidelines set out by the St Jude Research Children’s Hospital Animal Experimental Ethics Committee.
Tissue sampling and cell preparation
Spleens and bronchoalveolar lavage (BAL) samples were recovered from mice at the acute (d10) and memory (d33) phases of primary infection or acute (d8) time-point of the secondary response. BAL samples were incubated on plastic petri-dishes for 1hr at 37°C to remove macrophages. Spleens were disrupted and enriched for CD8+ T cells by panning on goat anti-mouse IgG and IgM antibody coated plates (Jackson ImmunoResearch Labs, West Grove, PA, USA) for 1hr at 37°C. Cells were washed and resuspended either in FACS buffer (1%BSA/0.02%NaN3 in PBS) for phenotypic analysis or sort buffer (0.1%BSA in PBS) for single-cell sorting.
Tetramer staining and tetramer dissociation analysis
CD8+ T cell-enriched lymphocytes from spleen and BAL were stained with DbNP366 or DbPA224 tetramers conjugated to Streptavidin-PE (Molecular Probes, Eugene, OR, USA) for 60mins at room temperature. Cells were washed twice in FACS buffer (10%BSA/0.02%NaN3 in PBS) and stained with anti-CD8α-FITC (BD Biosciences Pharmingen) for 30mins on ice, washed twice and analysed by flow cytometry. For investigation of TCR avidity, the relative off-rates of TCR binding were determined by tetramer dissociation (15–17). Briefly, cells were stained with tetramer, then incubated in the presence of anti-H2Db antibody (28-14-8, BD Biosciences Pharmingen) at 5μg/ml at 37°C to prevent tetramer rebinding. At designated times, cells were removed into FACS buffer and placed on ice, stained with anti-CD8α-FITC, and analysed by flow cytometry. Loss of tetramer+CD8+ T cells at particular time-points was calculated relative to tetramer staining at t=0mins.
Peptide stimulation and intracellular cytokine staining
Enriched T cell populations from spleen and BAL samples were stimulated with the NP366 (ASNENMETM) or PA224 (SSLENFRAYV) peptides (Hartwell Center, St Jude Children’s Research Hospital) for 5hrs in 200μl complete-RPMI medium containing 1μg/ml Golgi-Plug (BD Biosciences Pharmingen) and 10U/ml recombinant human IL-2 (Roche, Germany). Cells were washed in FACS buffer and stained with anti-CD8α-PerCP-Cy5.5 (BD Biosciences Pharmingen) for 30mins on ice. After two washes, cells were fixed and permeabilized using BD Cytofix/Cytoperm kit (BD Biosciences Pharmingen) according to manufacturer’s instructions. Subsequently, cells were stained with anti-IFN-γ-FITC, anti-TNF-α-APC and anti-IL-2-PE (BD Biosciences Pharmingen) for 30mins on ice. After two washes, cells were analysed by flow cytometry using a FACSCalibur flow cytometer (BD Immunocytometry Sustems, San Jose, CA).
Vβ staining of DbNP366+CD8+ and DbPA224+CD8+ T cells
Lymphocytes from spleens of primary or secondary mice were stained with either the DbNP366-PE or DbPA224-PE tetramers for 1hr at room temperature. Cells were then washed twice, stained with anti-CD8α-APC, and a panel of anti-Vβ mAbs conjugated to FITC, for 30mins at 4°C (all from BD Pharmingen, San Diego, CA, USA). After two washes, lymphocytes were analyzed by flow cytometry.
Single cell RT-PCR and sequencing
CD8+ T cell-enriched lymphocyte populations were stained either with DbNP366-PE or DbPA224-PE for 60 min at room temperature, followed by two washes in sort buffer. Cells were then stained with anti-CD8α-APC, and anti-Vβ8.3-FITC or anti-Vβ7-FITC mAbs for DbNP366+CD8+ or DbPA224+CD8+ T cells respectively. Individual DbNP336+Vβ8.3+CD8+ or DbPA224+Vβ7+CD8+ T cells were sorted, using a MoFlo sorter (Cytomation, Fort Collins, CO, USA), into wells of a 96-well PCR plate (Eppendorf, Hamburg, Germany). Negative controls were interspersed between the samples (1 in 10), and 80 cells were sorted per plate. cDNA synthesis was performed in 5ql of cDNA reaction mix (10, 11) for 90mins at 37°C, followed by 5 min at 95°C. The Vβ8.3 and Vβ7 cDNA was then amplified by nested PCR, and the purified PCR products sequenced (10, 11).
Repertoire Analysis Statistics
We focused on a set of statistics to describe clonotype diversity within and between individuals and clonotype sharing between individuals. The Simpson’s Diversity Index (D) (18, 19) is calculated as, where ni is the number of individuals in the ith species and N is the total number of individuals in the whole population. This is a measure of diversity, and it is expressed as 1-D so that a higher number corresponds to a higher level of diversity.
To measure clonotype sharing, we have determined the proportion of sequences that are found in a specific percentage of mice that have been examined. We developed it and refer to it as PTICq (Proportion of TCRs in Common), with the q referring to the percentage of mice the sequences must be found in. For example, the PTIC0.4 would be calculated as, for all sequences “n” where “n” is present in at least 40% (q) of the mice. Essentially, PTICq calculates the proportion of clones that are shared, based on a given percentage of sharing, q.
Student’s t-tests were used to determine significance for all individual statistical comparisons. For population-level comparisons of Simpson’s Diversity, we used EstimateS v 7.51 (20). The statistics presented are generated by random sampling with replacement from our database of wt primary and secondary DbNP366- and DbPA224-specific sequences, or the TdT sequences determined in this study. The statistics represent the average Simpson’s Diversity Index of sequences pooled from four mice generated by 10000 randomizations, in order to compare our wt data directly with the four mice in each TdT−/− group.
Results
Existing TCR Vβ preferences are exacerbated in TdT−/− mice
A broad investigation of DbNP366- and DbPA224-specific CD8+ T cell repertoires in TdT−/− mice was initiated to analyse profiles of TCR Vβ usage in wt and TdT−/− mice infected i.n with the HKx31 influenza virus. Previous studies identified substantial yet distinct Vβ biases in the DbNP366- and DbPA224-specific T cell responses, with Vβ8.3+ T cells constituting around 30–50% of the DbNP366-specific response (21, 22), and Vβ7+ cells comprising 50–65% of the DbPA224-specific population (11, 23). Interestingly, for both the DbNP366- and DbPA224-specific sets, there was a significant increase (to 77% and 81%, respectively) in the proportion of CD8+ T cells expressing the preferred Vβ8.3+ or Vβ7+ TCRs in TdT−/− mice (Fig. 1). Thus, a clear narrowing in TCRβ diversity was already evident at this superficial level of analysis.
Figure 1. Vβ bias within DbNP366+CD8+ and DbPA224+CD8+ T cells in TdT−/− mice.
Vβ usage within DbNP366+CD8+ and DbPA224+CD8+ T cell populations was compared for B6 and TdT−/− mice at d10 following primary influenza virus infection. Splenocytes were stained with DbNP366 or DbPA224 tetramers, and anti-CD8α and anti-Vβ mAbs. Tetramer+ CD8+ cells were analysed for the spectrum of Vβ usage. Data from 4 to 5 mice per group are expressed as a pie chart with the mean±SD of the dominant Vβ shown. * statistical comparisons are made for the dominant Vβ usage between wt and TdT−/− groups.
CDR3β repertoire usage in TdT−/− mice
Single cell CDR3β sequencing was then used to analyse sorted CD8+Vβ8.3+DbNP366- and CD8+Vβ7+DbPA224-specific cells from TdT−/− mice, for comparison with the extensive database of wt DbNP366 and DbPA224 sequences that have been generated using the same methodology (10, 11, 24). The results obtained following both primary (Tables I & II) and secondary (not shown) challenge were broadly comparable. Initial analysis of the DbNP366- and DbPA224-specific CDR3β profiles from TdT−/−animals showed that the characteristic modal CDR3β lengths (9 aa and 6 aa respectively) were maintained, as were the respective preferences for Jβ2S2 and Jβ2S6 (however the contribution of Jβ1S4 was increased) (Tables I & II) (2, 10, 11). Beyond that, though, there were major differences between wt and TdT−/− mice for both the DbNP366- and DbPA224-specific repertoires.
Table I.
TCRβ repertoire of DbNP366+Vβ8.3+CD8+ T cells in TdT−/− mice following primary influenza A virus infection.
| CDR3βregiona | Jβ | aa | Frequency | |||
|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | |||
| SGGANTGQL | 2S2 | 9 | 44 | 43 | 59 | 53 |
| SGGGNTGQL | 2S2 | 9 | 1 | 3 | ||
|
| ||||||
| Number of sequences | 45 | 43 | 62 | 53 | ||
mRNA from individual sorted cells isolated from TdT−/− mice at the acute phase following primary infection was reverse transcribed followed by two rounds of nested Vβ8.3-specific PCR amplification. PCR products were purified and sequenced using the internal Vβ8.3 oligonucleotide primer.
Table II.
TCRβ repertoire of Db PA224+Vβ7+CD8+ T cells in TdT−/− mice following primary influenza A virus infection.
| CDR3β regiona | Jβ | aa | ||||
|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | |||
| SLGGEQb | 2S6 | 6 | 10 | 15 | 4 | 1 |
| SLGAEQ | 2S1 | 6 | 2 | 2 | 3 | 2 |
| SQGERL | 1S4 | 6 | 13 | 10 | 8 | 5 |
| SLGERL | 1S4 | 6 | 6 | 2 | 8 | 8 |
| TGGERL | 1S4 | 6 | 5 | 1 | 6 | 1 |
| SWGGEQ | 2S6 | 6 | 2 | 1 | 1 | 2 |
| SLGAEV | 1S1 | 6 | 1 | 2 | 2 | |
| SWGDTL | 2S4 | 6 | 1 | 1 | 1 | |
| SGGAEQ | 2S6 | 6 | 2 | 3 | 3 | |
| SWGDTQ | 2S5 | 6 | 1 | 3 | 1 | |
| SSGERL | 1S4 | 6 | 3 | 1 | 1 | |
| TGGAEQ | 2S1 | 6 | 2 | 2 | 1 | |
| SWGAEQ | 2S1 | 6 | 1 | 2 | 1 | |
| SWGAEQ | 2S6 | 6 | 1 | 2 | ||
| SGGAEV | 1S1 | 6 | 2 | 1 | ||
| TGGTGQL | 2S2 | 7 | 1 | 4 | ||
| TNTGQL | 2S2 | 6 | 1 | 1 | ||
| SQGAEV | 1S1 | 6 | 1 | 7 | ||
| SGGQAP | 1S5 | 6 | 2 | |||
| SLGDEQ | 2S6 | 6 | 2 | |||
| SFGGEQ | 2S6 | 6 | 2 | |||
| TGGDEQ | 2S6 | 6 | 1 | |||
| SLGGAV | 1S1 | 6 | 1 | |||
| SSAETL | 2S3 | 6 | 1 | |||
| STGGEV | 1S1 | 6 | 1 | |||
| TAETL | 2S3 | 5 | 1 | |||
| SLGREQ | 2S6 | 6 | 2 | |||
| SRGERL | 1S4 | 6 | 1 | |||
| SSGGEQ | 2S6 | 6 | 1 | |||
| SSGTAP | 1S5 | 6 | 1 | |||
| SSYEQ | 2S6 | 5 | 2 | |||
| SWGDEQ | 2S6 | 6 | 1 | |||
| TGGAAP | 1S5 | 6 | 1 | |||
| TGGAEV | 1S1 | 6 | 1 | |||
| STGERL | 1S4 | 6 | 1 | |||
| SSGAEV | 1S1 | 6 | 1 | |||
| SLDRAEV | 1S1 | 7 | 2 | |||
| SLGGAQ | 2S6 | 6 | 1 | |||
| STGGEQ | 2S6 | 6 | 1 | |||
|
| ||||||
| Number of sequences | 53 | 52 | 53 | 43 | ||
mRNA from individual sorted cells isolated from TdT−/− mice at the acute phase following primary infection was reverse transcribed followed by two rounds of nested Vβ7-specific PCR amplification. PCR products were purified and sequenced using the internal Vβ7 oligonucleotide primer.
Shown in bold are public sequences found in all 8 mice analysed (4 × 1°, 4 × 2°).
Compared to the wt DbNP366-specific repertoire, which normally utilizes 5–10 clonotypes per individual, the TdT−/− DbNP366-specific set was virtually clonal, with 95–100% of the Vβ8.3+ response (which now constituted 77% of the DbNP366-specific repertoire) utilizing a single CDR3β clonotype (SGGANTGQL). However, despite the extreme restriction at the amino acid level, there was some diversity in the nucleotide usage with a total of 4 different nucleotide sequences encoding the SGGANTGQL clonotype, and 2 encoding the SGGGNTGQL sequence (Table III). Though the SGGANTGQL clonotype is often prominent in wt DbNP366-specific responses (10), it is one of several such clonotypes, the remainder of which were absent from the 8 TdT−/− animals tested. Interestingly, the only other CDR3β observed in the TdT−/− mice (SGGGNTGQL) can be both dominant and public in wt responses, but it was never a major component of the TdT−/− animals (Table I and data not shown).
Table III.
Nucleotide sequences encoding DbNP366-specific CDR3β aa clonotypes in TdT−/− mice.
| Frequency
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| N additions | Primary, d10
|
Secondary, d8
|
|||||||
| DbNP366-specific CDR3βa | M1 | M2 | M3 | M4 | M1 | M2 | M3 | M4 | |
| SGGANTGQL | 44a | 43 | 59 | 53 | 41 | 122 | 32 | 38 | |
| agtgggggggcaaacaccgggcagctc | 0 | 44 | 39 | 58 | 52 | 30 | 115 | 29 | 36 |
| agtgggggggcgaacaccgggcagctc | 0 | - | 4 | 1 | 1 | 1 | 1 | 2 | - |
| agtgggggcgcaaacaccgggcagctc | 0 | - | - | - | - | 10 | 4 | 1 | 2 |
| agtgggggagcaaacaccgggcagctc | 1 | - | - | - | - | - | 2 | - | - |
| SGGGNTGQL | 1 | - | 3 | - | 1 | - | - | - | |
| agtggggggggaaacaccgggcagctc | 0 | 1 | - | - | - | 1 | - | - | - |
| agtggggggggcaacaccgggcagctc | 0 | - | - | 3 | - | - | - | - | - |
|
| |||||||||
| Number of sequences | 45 | 43 | 62 | 53 | 42 | 122 | 32 | 38 | |
Numbers shown in bold are the frequencies of amino acid clonotypes in each mouse, with the numbers in italics depicting the frequency of nucleotide sequences encoding those amino acid clonotypes.
In an attempt to explain this observation, detailed analysis was performed on the number of different ways each of these normally public amino acid sequences can potentially be generated in the presence (determined from a simulation of a random V(D)J recombination process in which one million in-frame TCR sequences were generated (9)) or absence of N-region addition. If both the number of different nucleotide sequences, as well as the number of ways each nucleotide sequence can be generated (i.e. recombination mechanisms, referring to the fact that certain nucleotides in a CDR3β sequence have the potential to be encoded by more than one gene segment or by N-region addition (9)) are taken into account, SGGANTGQL, SGGGNTGQL, and SGGSNTGQL can all be independently generated in a vast number of different ways in wt mice (130, 80, and 60, respectively) (Table IV). This fact likely contributes to the dominance and public nature of these clonotypes in the wt DbNP366-specific response (9). In the absence of N-region addition, however, the SGGANTGQL clonotype, which is prevalent in the TdT−/− responses, can be encoded by four different nucleotide sequences, three of which can also be made by several different possible recombination mechanisms. The SGGGNTGQL clonotype, which is less prevalent in the responses in the TdT−/− mice, can be encoded by two germline-encoded nucleotide sequences but there is only one way that each of these can be made (Table IV). The public SGGSNTGQL clonotype that is observed in wt mice, but not the TdT−/− mice, cannot be made without at least one n-addition and can only be made two ways(Table IV). Thus, it appears that the dominance of SGGANTGQL, and the infrequency or absence, in TdT−/− mice, of the other two typically public DbNP366-specific clonotypes, likely arises as a consequence of a shift in the precursor numbers of these clonotypes due to their differential dependence on N-region addition.
Table IV.
A comparison of the production mechanisms of public DbNP366-specific CDR3β amino acid sequences with and without N-addition.
| CDR3βregion | Minimum no. of required n- additions | No. of recombination mechanisms
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Without N- additions | With N- additions | ||||||||||
| S | G | G | A | N | T | G | Q | L | 40
|
130 | |
| agt | ggg | ggg | gca | aac | acc | ggg | cag | ctc | 0 | 14 | |
| agt | ggg | ggc | gca | aac | acc | ggg | cag | ctc | 0 | 12 | |
| agt | ggg | ggt | gca | aac | acc | ggg | cag | ctc | 0 | 11 | |
| agt | ggg | ggg | gcg | aac | acc | ggg | cag | ctc | 0 | 3 | |
| S | G | G | G | N | T | G | Q | L | 2
|
80 | |
| agt | ggg | ggg | gga | aac | acc | ggg | cag | ctc | 0 | 1 | |
| agt | ggg | ggg | ggc | aac | acc | ggg | cag | ctc | 0 | 1 | |
| S | G | G | S | N | T | G | Q | L | 0
|
60 | |
| agt | ggg | ggg | tca | aac | acc | ggg | cag | ctc | 1 | ||
| agt | ggg | ggc | tca | aac | acc | ggg | cag | ctc | 1 | ||
|
| |||||||||||
| Germline genes: | |||||||||||
| Vβ 8.3: agt gat g, Jβ2S2: t gca aac acc ggg cag ctc | |||||||||||
| Dβ 1: gggacagggggcg, D β2: gggactgggggggcg | |||||||||||
Shown are the nucleotide sequences that encode commonly public DbNP366-specific CDR3β amino acid sequences and require a minimal number of N-additions to be produced. One possible V(D)J recombination mechanism is shown for each nucleotide sequence, with the nucleotides attributed by the germline Vβ, Dβ and Jβ genes shown in blue, red, and green, respectively. N-additions are shown in black and p-additions are shown in bold text. The number of possible recombination mechanisms that can produce these TCR sequences with no N-additions are shown. These different recombination mechanisms involve different splicings of the germline TCR genes and different p-additions (see Ref. 9 for more detail on different recombination mechanisms). Also shown is the number of different recombination mechanisms allowing for N-additions, determined from a simulation of a random V(D)J recombination process in which one million in-frame TCR sequences were generated (9).
While the CDR3β aa motifs associated with DbPA224-specific recognition were obviously maintained in the TdT−/− repertoire, and several of these clonotypes could be identified in the wt DbPA224-specific repertoires (11), some previously unidentified or rare clonotypes dominated. Of the five public clonotypes (Table II, shown in bold) observed in the DbPA224-specific repertoire in TdT−/− animals (note that SWGGEQ was not observed in all mice following secondary infection), only three have been seen in wt DbPA224-specific repertoires across multiple individuals (SLGERL 7/29 mice, SLGGEQ 9/29 mice, SLGAEQ 9/29 mice). This suggests that the restriction imposed by a lack of TdT has forced the appearance and/or dominance of typically rare clonotypes.
CDR3β clonotype diversity and sharing in TdT−/− mice
The following analyses (Fig. 2) exploit two distinct approaches for assessing TCRβ diversity. The first reflects diversity at a population level across all the mice that were analyzed, while the second measures TCRβ diversity within those individuals. The “overall” analysis of clonotype diversity at a population level showed that both the DbNP366- and DbPA224-specific CTL repertoires were significantly more restricted in TdT−/− compared to wt animals following both primary and secondary infection (Fig. 2A). Interestingly, however, while the restriction in population diversity was also reflected within individuals for the DbNP366 repertoire (Fig. 2B, left panel), the spectrum of “within mouse” diversity was unchanged for the DbPA224-specific repertoire between wt and TdT−/− animals (Fig. 2B, right panel), reflecting that approximately 15–20 clonotypes were commonly observed in both cases (2, 11).
Figure 2. CDR3β clonotypic diversity in TdT−/− mice.
Individual CD8+Vβ8.3+DbNP366+ or CD8+Vβ7+DbPA224+ cells were sorted from wt or TdT−/−splenocytes harvested at acute primary (d10) or secondary (d8) timepoints after infection. mRNA was reverse transcribed followed by nested Vβ8.3 or Vβ7 specific PCR. PCR products were purifed and sequenced and individual clonotypes defined according to CDR3β sequence. Clonotypic diversity across all sequences (A) and within individuals (B) was measured using Simpson’s Diversity Index and statistical analyses were performed using Student’s t-test. *p<0.02, **p<0.0005.
In order for DbPA224-specific overall diversity to be narrowed without a corresponding restriction in individual diversity, the degree of clonotype sharing between individuals must be increased (Fig. 3). In fact, the proportion of an individual’s response which was shared by >40% of mice in the group (referred to as Proportion of TCRs in Common, or PTIC in Fig. 3) was significantly enhanced for both the DbNP366- and DbPA224-specific repertoires in TdT−/− mice, effectively ensuring the public nature of both responses. The increase for the DbNP366 repertoire was primarily due to the fact that the repertoire was now clonal, while the greater sharing in the TdT−/− DbPA224-specific repertoire was highlighted by the fact that 5 of 39 CDR3β sequences were observed in all 8 of the mice analysed (4 primary, 4 secondary) (Table II). Such use of public clonotypes is never observed for the wt DbPA224-specific response (11), with no single clonotype being observed in all mice analysed. Thus, while TdT appeared to be dispensable for the generation of DbPA224-specific repertoire diversity within any given individual, it was critical for maintaining TCR diversity within the population at large.
Figure 3. Degree of CDR3β clonotype sharing in TdT−/− mice.
For CDR3β sequencing details, see legend to Figure 2. The proportion of TCRs in common (PTIC) shows the proportion of the clonotypic response from individual mice which are shared by at least 40% of mice sampled. Statistical analyses were performed using Student’s t-test. *p<0.02, **p<0.0005. *p<0.001, **p<0.0001.
TdT−/− DbNP366-specific CTL magnitude is selectively reduced at the site of infection
The DbNP366- and DbPA224-specific CD8+ T cell responses following influenza virus infection of B6 mice have been well characterized (23, 13, 15, 16). These epitopes elicit primary responses of similar magnitude but, following secondary infection, the DbNP366-specific set dominates by a factor of 5–10-fold. Given the severely restricted TCR clonotype usage in the TdT−/− DbNP366-specific repertoire compared to the still relatively diverse Tdt−/− DbPA224 response, it was of particular interest to determine whether there was any obvious difference in the magnitude and/or functionality of these populations. Freshly isolated spleen and BAL cells were analysed by tetramer staining following primary and secondary infection. No difference in the magnitude of either epitope-specific response was observed for wt and TdT−/−spleen populations (Fig. 4). However, the extent of DbNP366-specific CTL localization to the virus-infected lung was significantly (p<0.05) diminished in the TdT−/− mice following both primary and secondary challenge (Fig. 4). This effect was not observed for the more diverse DbPA224-specific response, indicating that repertoire limitation can diminish effector T cell magnitude in a site of pathogen-induced pathology. This effect may not just be a consequence of restrictions within the TCRβ repertoire but may also reflect limitations within the TCRα compartment.
Figure 4. Magnitude of epitope-specific CD8+ T cells in TdT−/− mice.
Naïve mice were either infected i.n. (1°, d10) or i.p (1°, d34), while PR8-immune B6 mice were infected i.n. (2°, d8) (see Materials and Methods), to analyze various phases of the immune response. Enriched splenocytes and BAL cells were stained with DbNP366-PE or DbPA224-PE tetramer, followed by anti-CD8α-FITC. Shown are the total numbers of CD8+ tetramer+ cells, calculated from the cell counts per organ and the % cells staining. * p< 0.05 using a two-tailed Student’s t-test, comparing wt and TdT−/− responses.
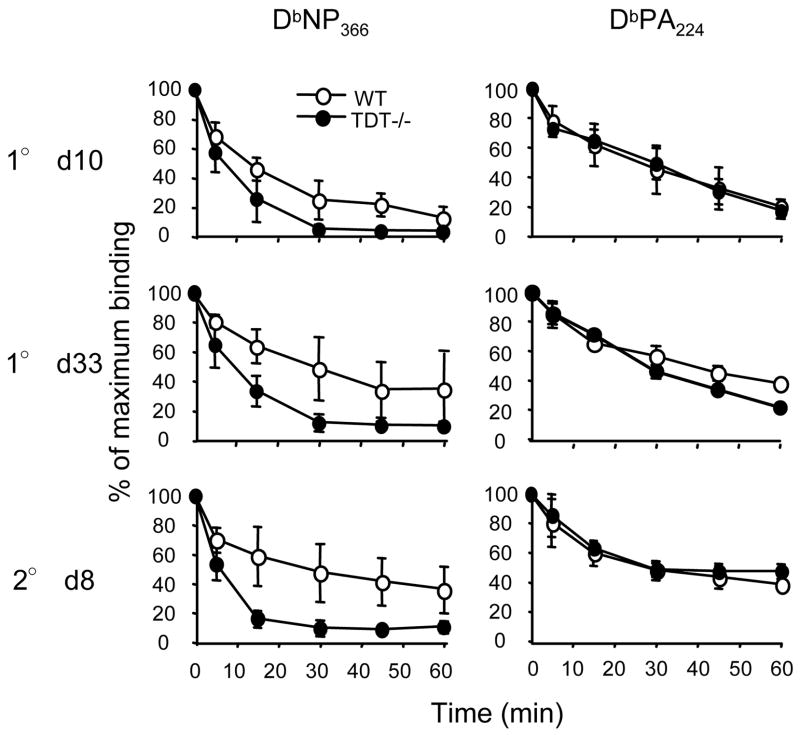
DbNP366-specific, but not DbPA224-specific, CTL exhibit reduced avidity in TdT−/− mice
Did these differential effects of TdT deficiency on the diversity of the DbNP366-and DbPA224-specific TCRβ repertoires in any way modify TCR avidity for the pMHCI complex? The measurement of tetramer dissociation kinetics provides a reliable and highly relevant measure of TCR/pMHCI avidity (15–17). Interestingly, irrespective of the stage of the immune response to influenza virus infection analysed (primary acute, primary memory, and secondary acute), the DbNP366-specific CTL population from TdT−/− mice showed a substantially enhanced TCR off-rate when compared to DbNP366-specific cells from wt mice (Fig. 5). On the other hand, the relative off-rates for the DbPA224-specific populations were equivalent for the two groups of mice (Fig. 5). Evidently the single clonotype (found in both wt and TdT−/− mice) that dominates the CTL response to DbNP366 in the deficient animals is of lower TCR avidity than the wt population overall. However, we did not determine whether this is solely a consequence of the SGGANTGQL CDR3β profile, as there could also be differential TCRα utilization. Furthermore, though we found evidence of reduced TCR avidity for the TdT−/− DbNP366-specific set, this did not translate into different profiles of induced cytokine production following short term in vitro stimulation with peptide (data not shown).
Figure 5. Avidity of influenza-specific CD8+ T cells in TdT−/− mice.
The kinetics of tetramer dissociation for splenic CD8+ T cells were analysed directly ex vivo on d10 after primary i.n. infection with the HKx31virus (1° d10) (n=5), on d33 after priming i.p. with PR8 to establish memory (1° d33) (n=3), or on d8 after secondary i.n. challenge with HKx31 (2° d8) (n=3). Enriched CD8+ T cells were stained with the DbNP366-PE or DbPA224-PE tetramer for 1h at room temperature. Cells were washed and incubated for designated times at 37°C in the presence of mAb to H-2Db, before co-staining with anti-CD8α-FITC. Shown are CD8+ tetramer+ cells expressed as a percentage of the maximum binding observed at time 0.
Discussion
As it has been shown previously that public T cell clonotypes generally contain fewer TdT-mediated N-region additions than the population average (7, 9), it might be thought that the absence of TdT would be more likely to perturb a private CTL repertoire (DbPA224) than a broadly public one (DbNP366). In fact the substantially public TCRVβ8.3 DbNP366-specific response was greatly disrupted, promoting the emergence of only two clonotypes (identified by CDR3β sequence) within or between individual mice. Furthermore, the one TCR (SGGANTGQL) accounted for 95–100% of these Vβ8.3+ T cells, while other public TCRS that are prevalent in wt animals were not found at all in the TdT−/− mice. This extreme skewing of the selected DbNP366-specific TCR repertoire to SGGANTGQL has been described previously following immunization of H2b TdT−/− mice with plasmid DNA encoding the influenza nucleoprotein (7), though such priming was not shown to elicit the minority SGGGNTGQL Vβ8.3 clonotype or the further 20% or so of tetramer+ T cells that did not express a Vβ8.3 TCR but were elicited here by active infection. Overall, these findings demonstrate that, while a lack of dependence on N-region addition could potentially increase the likelihood of a clonotype being public, it cannot account for all public clonotypes, or even the majority of TCRVβ8.3 DbNP366-specific public TCRs.
It has been suggested that the emergence of particular TCRβ clonotypes as dominant and public in an epitope-specific T cell response is linked to structural qualities which confer optimal recognition of the specific pMHC (25, 26). This does not appear to be the case for the epitope-specific responses studied here. The lack of any “public advantage” is exemplified with the SGGGNTGQL clonotype, which can be generated in the absence of N-region addition, but is a minor component of the response in TdT−/−mice. Furthermore, the extreme selection of the low avidity SGGANTGQL clonotype in the TdT−/−DbNP366-specific repertoire also argues against this being a superior ‘fit’ for DbNP366. It also suggests that avidity does not play a substantial role in determining clonotype abundance following influenza virus infection. This seems to contradict studies in which clonal dominance for EBV and CMV viral epitopes has been correlated directly with TCR/pMHCI avidity (27, 28). However, unlike the influenza A virus, EBV and CMV both persist so that the continued, or sporadic, emergence of antigen may well facilitate the progressive enrichment of high avidity T cells. Conversely, increasing the precursor frequency for low avidity CTLs has been shown to abrogate the preferential expansion of high avidity T cells when both populations are present at the same frequency (29). Perhaps higher precursor frequency of the low avidity SGGANTGQL in the naïve pool of the TdT−/− mice may explain the later dominance in the immune repertoire. This is certainly supported by the finding that the number of recombination mechanisms which can be used to generate a particular nucleotide sequence correlated with the prevalence of a particular amino acid clonotype in the immune response in both wt (9) and TdT−/− mice. Together, it seems likely that whether a particular TCR emerges as public in any given set of responses is simply a matter of its capacity to achieve a sufficient “fit” with the pMHC complex, and its prevalence in the naïve repertoire.
The observation that the TdT−/− DbPA224-specific repertoire diversity was significantly decreased overall, while remaining unchanged within individuals was particularly intriguing. Apparently the extent of diversity within the naive DbPA224-specific CTL pool confers an inherent plasticity on the responding repertoire. That is, there is sufficient naïve DbPA224-specific TCR diversity such that elimination of clonotypes requiring N-region addition does not significantly restrict the response spectrum within individuals. However, these clonotypes must also be repeated at much higher prevalence in the naïve repertoires of different TdT −/− mice, ensuring diminished diversity at the host population level.
The fact that the varied and private TCR repertoire normally induced by DbPA224 assumed a substantially public character in the TdT−/− mice supports the view that the generation of essentially private TCR repertoires is heavily dependent on TdT-mediated N-region additions. The homogenisation of the response between mice that is a consequence of the lack of N-region diversity could potentially limit the capacity to control virus escape mutants at the population level. Certainly, there is evidence that such MHC allele-restricted virus selection can occur for large populations (30, 31), while recent studies suggest that limited repertoire diversity facilitates virus escape within individuals (32, 33).
Furthermore, the relative importance of private versus public TCR repertoires has been directly demonstrated for “heterologous immunity”, defined as the reactivation of memory T cells generated by an earlier infection in response to subsequent exposure to a seemingly unrelated virus (34). Adoptive transfer of LCMV-immune splenocytes from one individual into multiple, naive recipients that were then infected with vaccinia virus elegantly demonstrated that the extent of cross-reactivity to a newly encountered vaccinia epitope was a function of private, rather than public, LCMV-specific TCR specificities (35, 36). Since heterologous immune responses are likely to be generally beneficial (36–38), the prevalence of TCR repertoires that are private for individuals has the potential to increase the diversity of response profiles for any given population, an effect that presumably enhances overall fitness. Taken together with the possibility (discussed above) that the extent of epitope-specific TCR repertoire diversity both within and between individuals is likely to minimise the emergence of viral escape mutants (32, 33), it seems likely that the TdT mechanism that facilitates the emergence of private TCR repertoires is advantageous in the evolutionary sense.
Acknowledgments
The authors would like to thank Dr Ann Feeney for providing the TdT−/− mice, Cory Reynolds for development of data analysis tools and technical assistance, and Elvia Olivas and Melissa Morris for technical assistance. This work was supported by Australian National Health and Medical Research Council (NHMRC) Project Grants AI454595 (to P.C.D.); AI454312 (to K.K.), and AI350395 (to N.L.G.), and an NHMRC Burnet Award, Science Technology Innovation funds from the Government of Victoria, Australia (AI29579), and an NIH grant AI70251 (to P.C.D.). K.K. is the recipient of a University of Melbourne Early Career Researcher grant. K.K. and N.L.G. are NHMRC RD Wright Fellows and S.J.T. is a Pfizer Australia Research Fellow.
Footnotes
This work was supported by Australian NHMRC Project Grants AI454595 (to P.C.D.); AI454312 (to K.K.), and AI350395 (to N.L.G.), the Australian Research Council, an NHMRC Burnet Award, Science Technology Innovation funds from the Government of Victoria, Australia (AI29579), and an NIH grant AI70251 (to P.C.D.). K.K. is the recipient of a University of Melbourne Early Career Researcher grant. K.K. and N.L.G. are NHMRC RD Wright Fellows, S.J.T. is a Pfizer Australia Research Fellow, and M.P.D is a Sylvia and Charles Viertel Senior Medical Research Fellow.
Abbreviations used in this paper: peptide+ MHC class I glycoprotein, pMHC; DbNP366, nucleoprotein amino acid residues 366-374+MHC class I H-2Db; DbPA224, acid polymerase amino acid residues 224-233+H-2Db; NA, neuraminidase; B6, C57BL/6J mice; PR8, A/Puerto Rico/8/34 influenza A virus; HKx31, A/Hong Kong x31 virus; BAL, bronchoalveolar lavage.
References
- 1.Cibotti R, Cabaniols JP, Pannetier C, Delarbre C, Vergnon I, Kanellopoulos JM, Kourilsky P. Public and private V beta T cell receptor repertoires against hen egg white lysozyme (HEL) in nontransgenic versus HEL transgenic mice. J Exp Med. 1994;180:861–872. doi: 10.1084/jem.180.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner SJ, Kedzierska K, La Gruta NL, Webby R, Doherty PC. Characterization of CD8+ T cell repertoire diversity and persistence in the influenza A virus model of localized, transient infection. Seminars in immunology. 2004;16:179–184. doi: 10.1016/j.smim.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Welsh RM. Private specificities of heterologous immunity. Current opinion in immunology. 2006;18:331–337. doi: 10.1016/j.coi.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 5.Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 6.Cabaniols JP, Fazilleau N, Casrouge A, Kourilsky P, Kanellopoulos JM. Most alpha/beta T cell receptor diversity is due to terminal deoxynucleotidyl transferase. J Exp Med. 2001;194:1385–1390. doi: 10.1084/jem.194.9.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fazilleau N, Cabaniols JP, Lemaitre F, Motta I, Kourilsky P, Kanellopoulos JM. Valpha and Vbeta public repertoires are highly conserved in terminal deoxynucleotidyl transferase-deficient mice. J Immunol. 2005;174:345–355. doi: 10.4049/jimmunol.174.1.345. [DOI] [PubMed] [Google Scholar]
- 8.Gilfillan S, Bachmann M, Trembleau S, Adorini L, Kalinke U, Zinkernagel R, Benoist C, Mathis D. Efficient immune responses in mice lacking N-region diversity. European journal of immunology. 1995;25:3115–3122. doi: 10.1002/eji.1830251119. [DOI] [PubMed] [Google Scholar]
- 9.Venturi V, Kedzierska K, Price DA, Doherty PC, Douek DC, Turner SJ, Davenport MP. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc Natl Acad Sci U S A. 2006;103:18691–18696. doi: 10.1073/pnas.0608907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedzierska K, Turner SJ, Doherty PC. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc Natl Acad Sci U S A. 2004;101:4942–4947. doi: 10.1073/pnas.0401279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner SJ, Diaz G, Cross R, Doherty PC. Analysis of clonotype distribution and persistence for an influenza virus-specific CD8+ T cell response. Immunity. 2003;18:549–559. doi: 10.1016/s1074-7613(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 12.Kilbourne ED. Future influenza vaccines and the use of genetic recombinants. Bulletin of the World Health Organization. 1969;41:643–645. [PMC free article] [PubMed] [Google Scholar]
- 13.Flynn KJ, Belz GT, Altman JD, Ahmed R, Woodland DL, Doherty PC. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 14.Allan W, Tabi Z, Cleary A, Doherty PC. Cellular events in the lymph node and lung of mice with influenza. Consequences of depleting CD4+ T cells. J Immunol. 1990;144:3980–3986. [PubMed] [Google Scholar]
- 15.La Gruta NL, Kedzierska K, Pang K, Webby R, Davenport M, Chen W, Turner SJ, Doherty PC. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. Proc Natl Acad Sci U S A. 2006;103:994–999. doi: 10.1073/pnas.0510429103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Gruta NL, Turner SJ, Doherty PC. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J Immunol. 2004;172:5553–5560. doi: 10.4049/jimmunol.172.9.5553. [DOI] [PubMed] [Google Scholar]
- 17.Kalergis AM, Boucheron N, Doucey MA, Palmieri E, Goyarts EC, Vegh Z, Luescher IF, Nathenson SG. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nature immunology. 2001;2:229–234. doi: 10.1038/85286. [DOI] [PubMed] [Google Scholar]
- 18.Magurran AE. Measuring Biological Diversity. Blackwell; Oxford: 2004. [DOI] [PubMed] [Google Scholar]
- 19.Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods. 2007;321:182–95. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 20.Colwell RK. EstimateS: Statistical Estimation of Species Richness and Shared Species from Samples. S. University of Connecticut; CT: 2006. [Google Scholar]
- 21.Deckhut AM, Allan W, McMickle A, Eichelberger M, Blackman MA, Doherty PC, Woodland DL. Prominent usage of V beta 8.3 T cells in the H-2Db-restricted response to an influenza A virus nucleoprotein epitope. J Immunol. 1993;151:2658–2666. [PubMed] [Google Scholar]
- 22.Kedzierska K, La Gruta NL, Stambas J, Turner SJ, Doherty PC. Tracking phenotypically and functionally distinct T cell subsets via T cell repertoire diversity. Molecular immunology. 2008;45:607–618. doi: 10.1016/j.molimm.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belz GT, Xie W, Altman JD, Doherty PC. A previously unrecognized H-2D(b)-restricted peptide prominent in the primary influenza A virus-specific CD8(+) T-cell response is much less apparent following secondary challenge. J Virol. 2000;74:3486–3493. doi: 10.1128/jvi.74.8.3486-3493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong W, Reinherz EL. In vivo selection of a TCR Vbeta repertoire directed against an immunodominant influenza virus CTL epitope. Int Immunol. 2004;16:1549–1559. doi: 10.1093/intimm/dxh156. [DOI] [PubMed] [Google Scholar]
- 25.Kjer-Nielsen L, Clements CS, Purcell AW, Brooks AG, Whisstock JC, Burrows SR, McCluskey J, Rossjohn J. A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity. 2003;18:53–64. doi: 10.1016/s1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- 26.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nature immunology. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 27.Callan MF, Steven N, Krausa P, Wilson JD, Moss PA, Gillespie GM, Bell JI, Rickinson AB, McMichael AJ. Large clonal expansions of CD8+ T cells in acute infectious mononucleosis. Nature medicine. 1996;2:906–911. doi: 10.1038/nm0896-906. [DOI] [PubMed] [Google Scholar]
- 28.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, Sewell AK, Connors M, Douek DC. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rechtsteiner G, Warger T, Hofmann M, Rammensee HG, Schild H, Radsak MP. Precursor frequency can compensate for lower TCR expression in T cell competition during priming in vivo. European journal of immunology. 2006;36:2613–2623. doi: 10.1002/eji.200636331. [DOI] [PubMed] [Google Scholar]
- 30.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science (New York, NY) 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 31.Neumann-Haefelin C, McKiernan S, Ward S, Viazov S, Spangenberg HC, Killinger T, Baumert TF, Nazarova N, Sheridan I, Pybus O, von Weizsacker F, Roggendorf M, Kelleher D, Klenerman P, Blum HE, Thimme R. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology (Baltimore, Md) 2006;43:563–572. doi: 10.1002/hep.21049. [DOI] [PubMed] [Google Scholar]
- 32.Cornberg M, Chen AT, Wilkinson LA, Brehm MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM, Selin LK. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. The Journal of clinical investigation. 2006;116:1443–1456. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, Letvin NL, Franchini G, Wolinsky SM, Koup RA, Douek DC. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Rehermann B, Shin EC. Private aspects of heterologous immunity. J Exp Med. 2005;201:667–670. doi: 10.1084/jem.20050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SK, Cornberg M, Wang XZ, Chen HD, Selin LK, Welsh RM. Private specificities of CD8 T cell responses control patterns of heterologous immunity. J Exp Med. 2005;201:523–533. doi: 10.1084/jem.20041337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urbani S, Amadei B, Fisicaro P, Pilli M, Missale G, Bertoletti A, Ferrari C. Heterologous T cell immunity in severe hepatitis C virus infection. J Exp Med. 2005;201:675–680. doi: 10.1084/jem.20041058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen HD, Fraire AE, Joris I, Brehm MA, Welsh RM, Selin LK. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nature immunology. 2001;2:1067–1076. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- 38.Selin LK, Varga SM, Wong IC, Welsh RM. Protective heterologous antiviral immunity and enhanced immunopathogenesis mediated by memory T cell populations. J Exp Med. 1998;188:1705–1715. doi: 10.1084/jem.188.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]