Abstract
U-insertion/deletion RNA editing in the single mitochondrion of ancient kinetoplastids is a unique mRNA maturation process needed for translation. Multi-subunit editing complexes recognize many pre-mRNA sites and modify them via cycles of three catalytic steps: guide-RNA (gRNA) mediated cleavage, insertion or deletion of uridylates at the 3’ terminus of the upstream cleaved piece, and ligation of the two mRNA pieces. While catalytic and many structural protein subunits of these complexes have been identified, the mechanisms and basic determinants of substrate recognition are still poorly understood. The current study defined relatively simple single- and double-stranded determinants for association and gRNA-directed cleavage. To this end, we used an electrophoretic mobility shift assay to directly score the association of purified editing complexes with RNA ligands, in parallel with U.V. photo-crosslinking and functional studies. The cleaved strand required a minimal 5’ overhang of 12-nt and a ~15-bp duplex with gRNA to direct the cleavage site. A second protruding element in either the cleaved or the guide strand was required unless longer duplexes were used. Importantly, the single-stranded RNA requirement for association can be upstream or downstream of the duplex, and the binding and cleavage activities of purified editing complexes could be uncoupled. The current observations together with our previous reports (Cifuentes-Rojas et al., 2005 and 2006) show that association, cleavage and full-round editing by purified editing complexes have distinct determinants that increase in complexity as these editing stages progress. Finally, we found that the endonuclease KREN1 in purified complexes photo-crosslinks with a targeted editing site. A model is proposed whereby one or more RNase III-type endonucleases in editing complexes mediate the initial binding and scrutiny of potential ligands, and subsequent catalytic selectivity triggers either insertion or deletion editing enzymes.
Introduction
The majority of primary mRNA transcripts in the single mitochondrion of kinetoplastids, including species of Trypanosoma and Leishmania, are plagued with frameshifts and stop codons. Protein-encoding sequences are produced via an extraordinary maturation process involving specific insertion and deletion of uridylates at often hundreds of editing sites (ESs) in a single transcript. This process is catalyzed by megadalton multi-subunit assemblies known as L-complexes, 20S editosomes, or editing complexes that contain between 16 and 20 known subunits and target ESs specified by the partial complementarity of pre-edited mRNA (pre-mRNA) and guide RNAs (gRNAs). For recent reviews see 1; 2.
RNA editing has been recreated in vitro at single model ESs in either natural-like 3; 4 or completely artificial 5 substrates. Early mechanistic studies indicated that all steps of deletion and insertion editing were catalyzed by distinct enzymatic activities 6; 7; 8; 9; 10. More recently it was shown that a deletion cycle involves the consecutive action of endonuclease KREN1, 3’ exo-uridylylase KREX1 and/or KREX2, and ligase KREL1 9; 11; 12; 13; 14; 15. Similarly, an insertion cycle involves endonuclease REN2 or REN3, terminal uridylyl transferase KRET2, and preferentially, ligase KREL2 9; 14; 16; 17; 18. Yet, KREL1 may be used in absence of KREL2 in vitro and in vivo 9; 14; 19; 20. Potentially, KREN1 and KREX enzymes could also help proofread misedited insertion ESs bearing extra Us; i.e., misedited insertion sites could be targeted and repaired by deletion editing6. Additional observations also suggest that deletion and insertion activities may occur at individual ESs in vivo. Namely, RNAi of KREN1 dowregulates editing of CYb and COII pre-mRNAs in vivo, which only contain insertion ESs11. Also, RET2 was shown to add Us at deletion sites in vitro21.
Pre-mRNA/gRNA hybrids are proposed to form two helical regions flanking an internal loop. The downstream (relative to the scissile bond) “anchor” duplex directs endonuclease cleavage immediately 5’ to it, whereas the upstream duplex is thought to tether the cleaved 5’ piece during U-specific processing and re-ligation. The mechanisms of substrate recognition in assembled editing complexes are currently been addressed (for a recent review see 22). Previous studies in our laboratory using purified native complexes have shown that secondary structure rather than sequence-specific features are primarily required for full-round insertion editing 5; 23. In a completely artificial 43-nt pre-mRNA/gRNA model substrate with single-helical turns flanking the central loop, simple features of this loop were manipulated to interconvert sites between insertion and deletion editing. Important insights on the specificity of substrate association with purified editing complexes were obtained in competition studies using parallel U.V. photo-crosslinking and full-round catalytic editing assays. Such studies, using a single photo-reactive 4-thioU and a 32P atom at targeted ESs, showed a preferential association of complexes with deletion and insertion substrates, particularly with the most efficient model substrate currently available for full-round editing (A6 pre-mRNA/D33 gRNA hybrid) 5; 8. The native complexes also exhibited a level of non-specific binding to unrelated transcripts. Interestingly, ribose 2’ H substitutions on the downstream helix and gRNA-side of the central loop significantly inhibited both pre-mRNA cleavage and photo-crosslinking activities at a targeted ES. Furthermore, a single 2’ H substitution adjoining the scissile bond obliterated the endonucleolytic activity but had no effect on photo-crosslinking, suggesting that the ribose 2’ hydroxyl at this position is relevant for catalysis not association of editing complexes 5.
One of the photo-crosslinking subunits in assembled editing complexes was proposed to be KREPA2 (MP63)24, which as several other subunits, contains conserved domains that predict interaction with nucleic acids 25; 26. Studies of purified recombinant proteins established that KREPA3 (MP42), KREPA4 (MP24) and KREPA6 (MP18) exhibit RNA-binding activity 27; 28; 29, but their precise function in assembled editing complexes remains to be determined. KREPA4 and KREPA6 exhibited preferential binding to poly(U) homopolymers, suggesting a role in the recognition of the natural 3’ poly(U) extension of gRNAs. These recombinant proteins also showed a general low-affinity binding for RNA.
While previous photo-crosslinking analyses provided insights on the specificity of the editing enzyme/substrate association, absence of crosslinking with certain mutant substrates could not be interpreted with certainty. Furthermore, whether purified editing complexes form transient or stable ribonucleoprotein complexes (RNPs) with cognate substrates is unknown. In the current study we used an electrophoretic mobility shift assay (EMSA) to directly examine, for the first time, RNPs formed by purified editing complexes. We applied EMSA, photo-crosslinking and endonuclease analyses to define substrate determinants for association and endonuclease cleavage, the first catalytic step of RNA editing. Both single-stranded (ssRNA) and double-stranded (dsRNA) RNA were required for these two stages of editing, but ssRNA for association can be satisfied in different ways, whether or not endonuclease cleavage activity is observed. Importantly, the determinants for association and cleavage can be uncoupled, and the determinants for endonuclease cleavage are more complex than for association but less intricate than for full-round editing.
Finally, we compared preparations of native and affinity-purified editing complexes in association and catalytic assays, and established that one subunit that photo-crosslinks at a targeted ES is the essential endonuclease KREN1. The subunit KREPA2 (MP63) was also confirmed to photo-crosslink. A model is proposed whereby recognition of basic determinants including those defined here, leads to a preferential association of editing complexes with potential substrates. Such initial interactions may precede subsequent specialized contacts that trigger catalysis by either deletion or insertion editing.
Results
Our previous RNA-protein photo-crosslinking studies showed that purified native editing complexes preferentially associate with a model A6 substrate for full-round editing (Fig. 1A) via recognition of secondary structure not sequence-specific features 5; 24. However, absence of crosslinking due to certain substrate modifications or reaction conditions leaves uncertainties about the editing enzyme/substrate association.
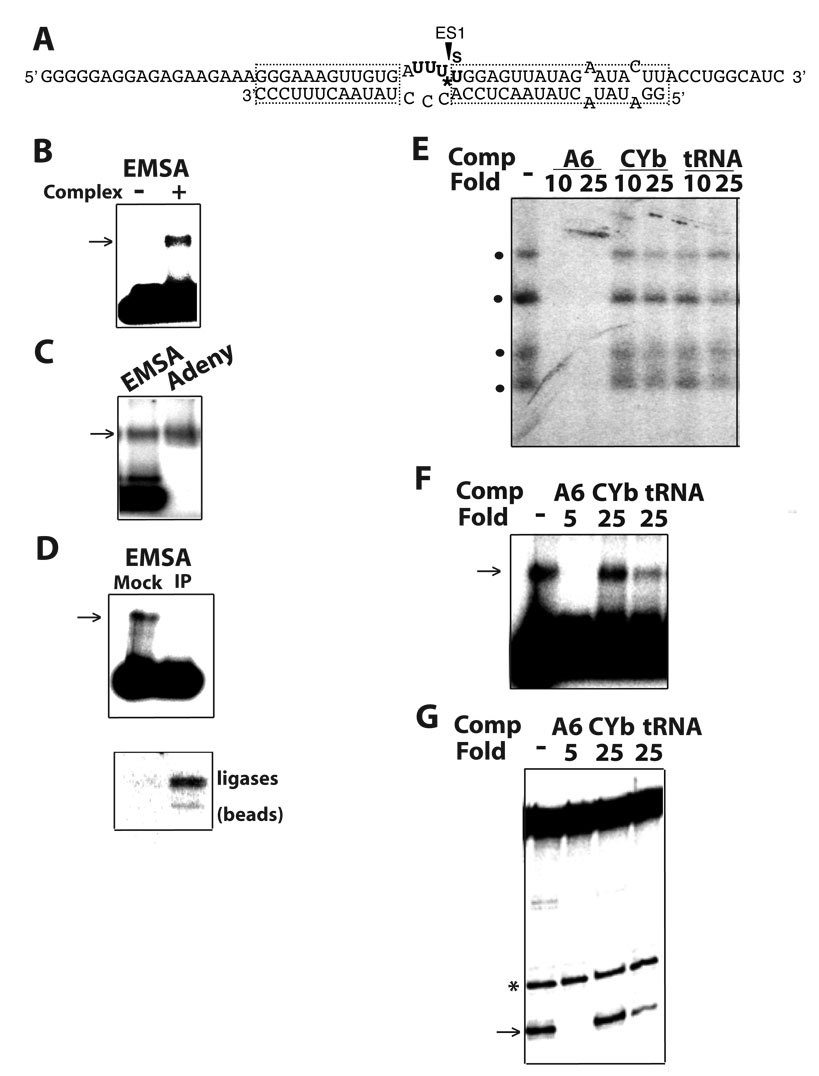
Fig. 1.
The association of purified editing complexes with substrates can be directly scored by EMSA, in parallel with U.V. photo-crosslinking and pre-mRNA cleavage assays. (A) Scheme of an ES1-32P labeled (*) and thio-labeled (s) model A6 substrate for EMSA, U.V. photo-crosslinking and full-round U-deletion editing. (B) EMSA in a native agarose gel showing a shifted band (arrow) only in presence of editing complexes; (C) Co-migration of the shifted substrate with editing complexes that were radiolabeled by auto-adenynylation (Adeny); (D) Specific depletion of the shifted product by co-immunoprecipitation (IP; upper) and recovery of self-adenylytable ligase subunits in the beads (lower). A mock reaction was devoid of antibodies. (E) Preferential association of editing complexes with a (A6) substrate for full-round editing in competition studies using U.V. photo-crosslinking (dots indicate four major crosslinks) or parallel assays of (F) EMSA and (G) endonuclease cleavage (arrow). A spurious cut (*) serves as loading control. In the EMSA, most substrate remained unbound. The fold excess of unlabeled homologous A6 (5 or 10-fold) and heterologous CYb and tRNA competitors is indicated. No competitor is (−).
To directly score substrate binding by editing complexes, we established an electrophoretic mobility shift assay (EMSA). A standard reaction mixture for full-round editing or photocrosslinking studies, using purified editing complexes and an ES1-labeled substrate (Fig. 1A)24, was briefly incubated and loaded onto a native agarose gel. A fraction of radiolabeled substrate exhibited delayed electrophoretic mobility only in the presence of editing complexes (Fig. 1B). This shifted product comigrated with complexes that were radiolabeled by adenylylation of ligase subunits (Fig. 1C)30 and was specifically immunodepleted by monoclonal antibodies to editing subunits (Fig. 1D, upper). As expected, adenylylatable editing ligases were enriched in the antibody-conjugated IgG beads but not in beads without antibodies (lower).
To further confirm that these ribonucleoprotein assemblies (RNPs) include editing complexes, we examined their substrate specificity using competition analysis as those performed in photo-crosslinking and full-round editing studies 24. Importantly, the competition profiles in photo-crosslinking (that we reported 24) and EMSA assays were equivalent, i.e., the homologous A6 competitor was strongly inhibitory at 5–10 fold excess whereas tRNA and CYb were significantly less inhibitory at 25-fold excess (Figs. 1E and 1F, respectively; and data not shown). Moreover, a similar competition pattern was observed in assays of gRNA-directed endonuclease cleavage, the first enzymatic step of a full-round editing cycle (Fig. 1G). Together, these data indicate that the EMSA directly scores the editing enzyme/substrate association and specificity of editing complexes. The data using EMSA also mirrors the observations in parallel studies of RNA-protein photo-crosslinking and editing enzymatic activities. Furthermore, all these activities of editing complexes can be examined using common substrates and reaction conditions.
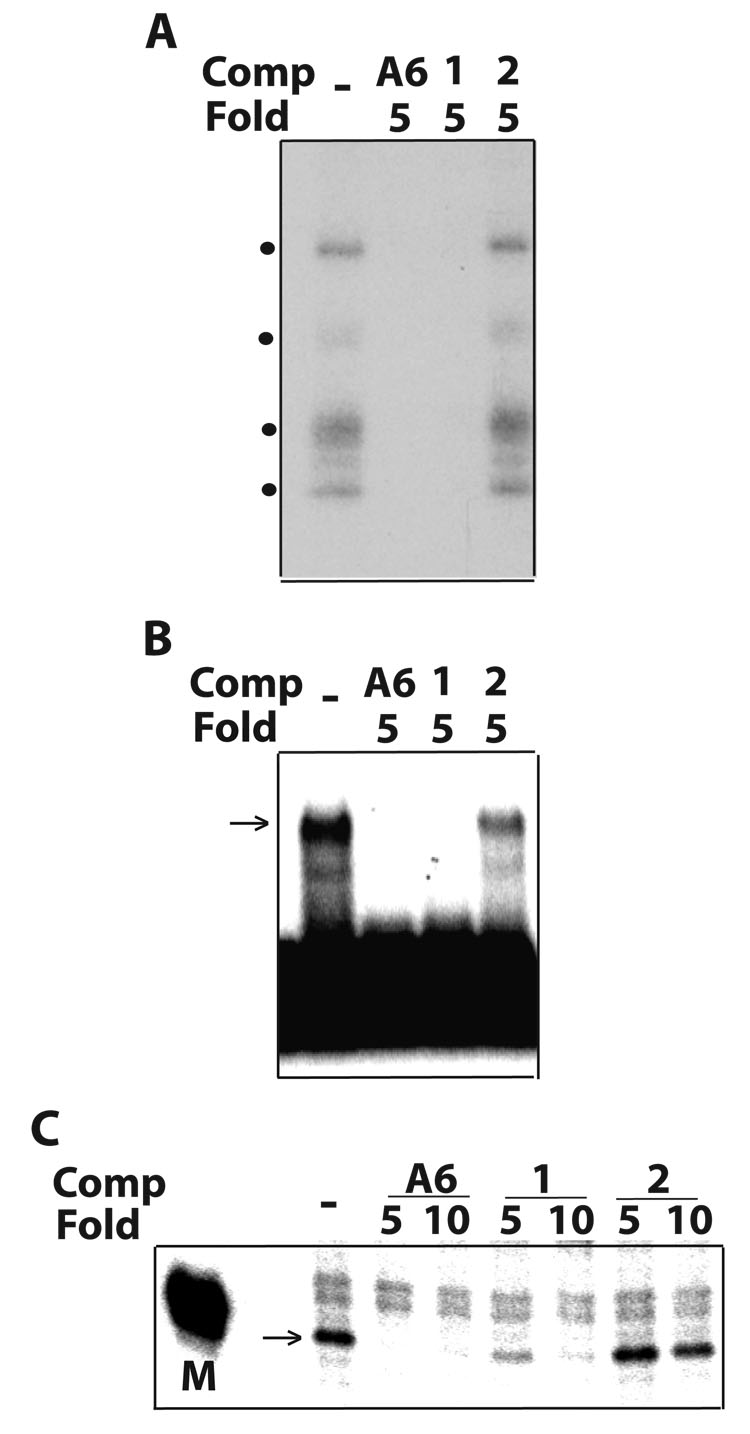
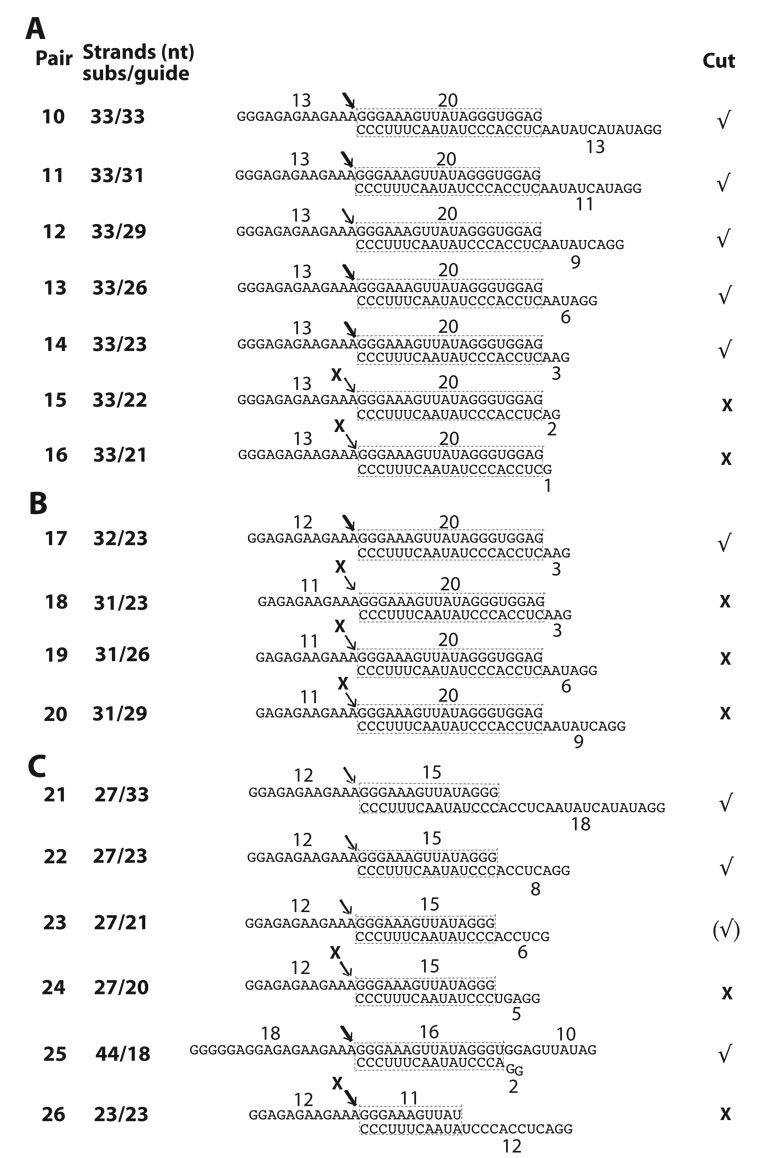
Based on these observations, we sought to define substrate determinants for association and guide-directed cleavage by editing complexes. We performed competition analyses, as in Figs. 1E–G, to examine the effects of unlabeled derivatives of the homologous (A6) competitor (diagramed in Fig. 2A). Our standard editing mixtures include gRNA at ~120-fold excess over radiolabeled A6 pre-mRNA to ensure quantitative annealing 5. In order to form competitor duplexes, the abundant free gRNA (“guide strand”) in the standard mixture was allowed to pre-anneal with each pre-mRNA derivative (“substrate strand”) added at a small, 5–10 fold, excess over radiolabeled pre-mRNA (Fig. 2B). All constructs in Fig. 2A used the same guide strand, and quantitative annealing was confirmed in native gels 5 (see methods). Such analysis in binding and catalytic assays performed in parallel is illustrated in Fig. 3. In this example, both the homologous A6 pair and derivative Pair-1, whose guide strand is fully based paired (i.e., it forms a continuous 33-bp duplex), were strong competitors in photo-crosslinking, EMSA and cleavage assays (Figs. 3A–C, respectively). However, a second derivative that conserves the 33-bp duplex but lacks overhangs (Pair-2) was a poor competitor in all assays. These data suggest that editing complexes associate with Pair-1 but not Pair-2. Thus the presence or absence of the central loop region in the parental A6 construct does not significantly affect the binding efficiency of editing complexes, although ssRNA seems required for association.
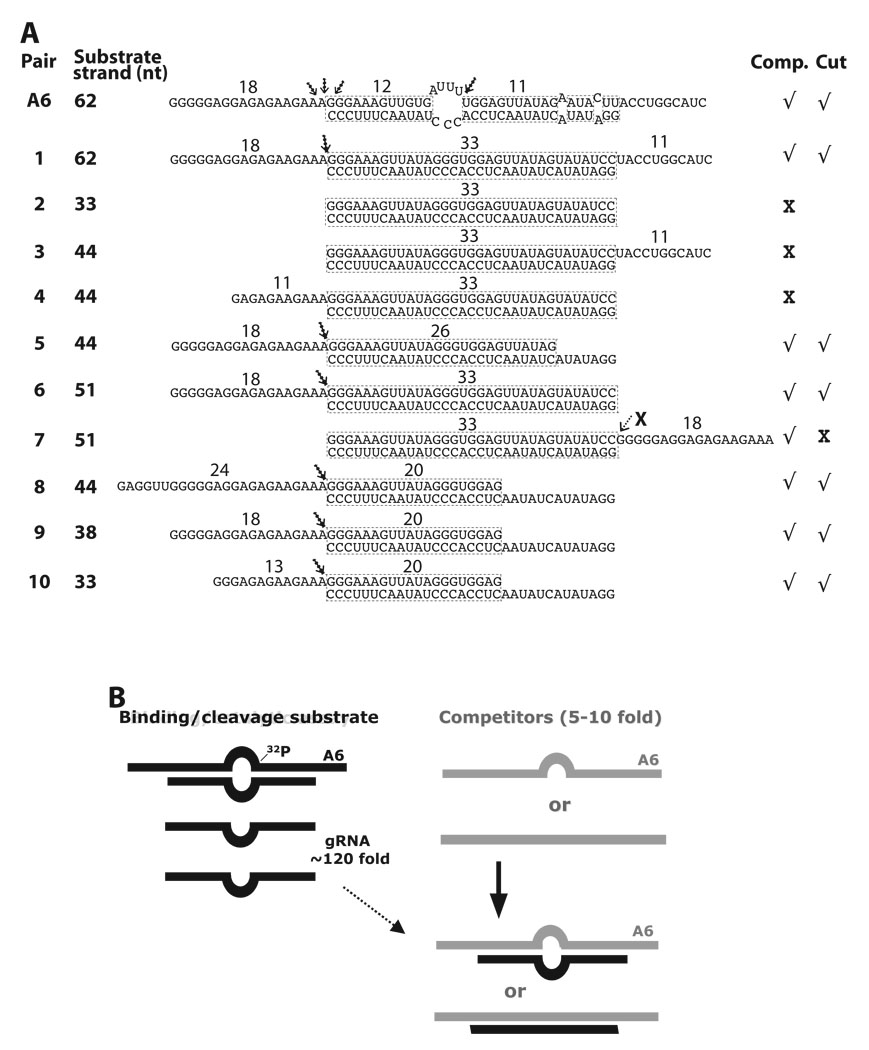
Fig. 2.
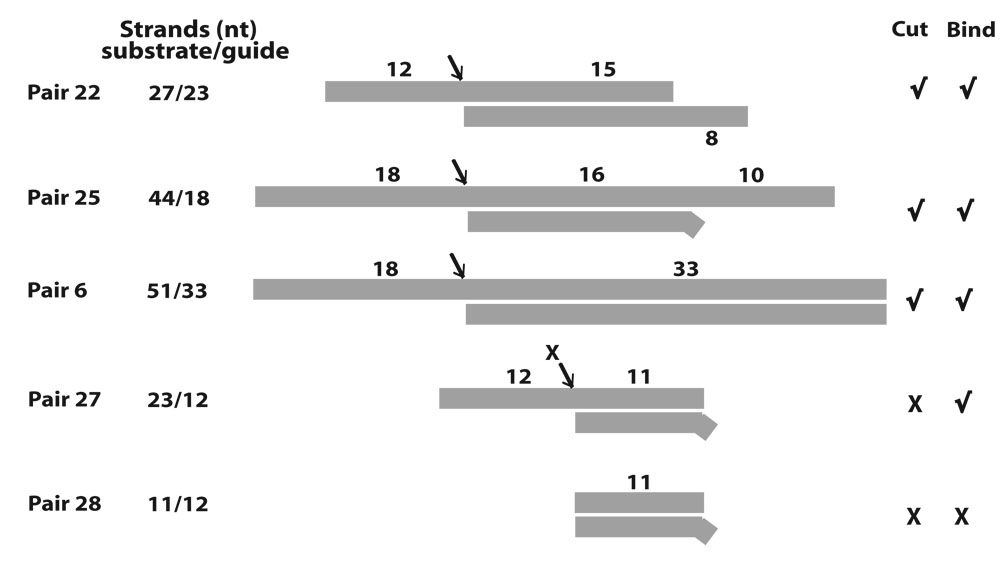
Constructs tested and scheme for competition assays. (A) Homologous A6 and derivative competitors (top “substrate” strand) paired with gRNA.D33 (lower “guide” strand). The assigned number of each competitor RNA pair and size (nt) of the substrate strand are indicated, as well as the size of the predicted helix and overhangs. The demonstrated cleavage sites are noted with an arrow. Evident (√) or weak-to-undetected (X) competition and cleavage activity for each construct is indicated at right. Some pairs were not tested for cleavage activity. Pair-7 was tested for cleavage although a negative result was expected (see text). (B) Cartoon of model RNA construct in standard functional (left) and modified competition assays (right). 32P labeled A6 pre-mRNA is usually annealed with complementary gRNA at ~120 fold excess in our standard editing assays. Unlabeled A6 substrate strand or variants (light strand) at 5–10 fold excess, over radiolabeled A6, anneal with free gRNA (both as dark strands) forming competitor pairs (light/dark hybrids).
Fig. 3.
Parallel competitions in (A) U.V. photo-crosslinking, (B) EMSA and (C) RNA cleavage assays with purified editing complexes, as in Fig. 1. A6 and variant competitors (Comp) diagrammed in Fig. 2A were examined at the indicated fold excess. Our cleavage assays typically included a size marker (M) such as the 32P kinased donor fragment used to prepare the parental A6 substrate (Fig. 1A), or control lanes with and without gRNA.
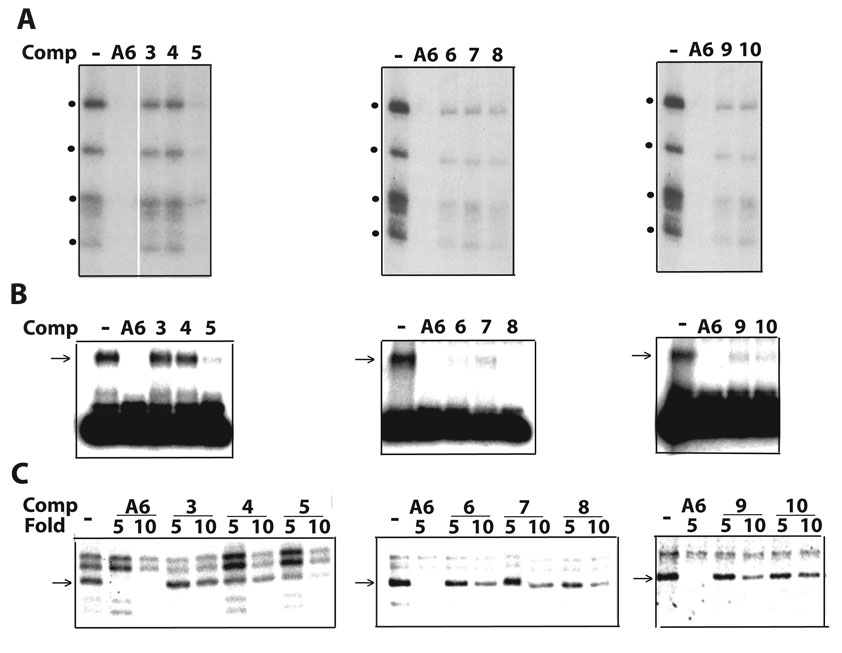
To dissect RNA requirements for association with editing complexes that distinguish Pair-1 from Pair-2, we designed competitors based on Pair-2 that contain upstream and/or substrate-strand downstream overhangs of various lengths (Fig. 4; diagrammed in Fig. 2A). While 13-nt, 18-nt and 24-nt extensions favored association of editing complexes (Pairs 5–10), 11-nt extensions at either side of the duplex (Pairs 3–4) did not. Furthermore, constructs with shorter duplexes, 26-bp (Pair-5) and 20-bp long (Pairs 8–10), were also effective competitors. Most of these constructs used a 44-nt substrate strand, however, Pair-10 with a 33-nt substrate strand was also a significant competitor. Some competitions are more evident in crosslinking and EMSA than in cleavage studies (Figs. 4A–C). This difference may reflect different dynamics in the assays; that is, the former two score RNP complexes that either are present at the time of crosslinking or that withstand gel electrophoresis, respectively, whereas the latter scores accumulation of cleaved product over time, regardless of the relative stability of RNPs. Together, the competition studies in Fig. 1–Fig. 4 suggest that association with editing complexes requires recognition of a relatively simple structure bearing discrete ssRNA and dsRNA determinants.
Fig. 4.
A–C. Parallel competitions as in Fig. 3. The homologous A6 and derived competitors are diagrammed in Fig. 2A.
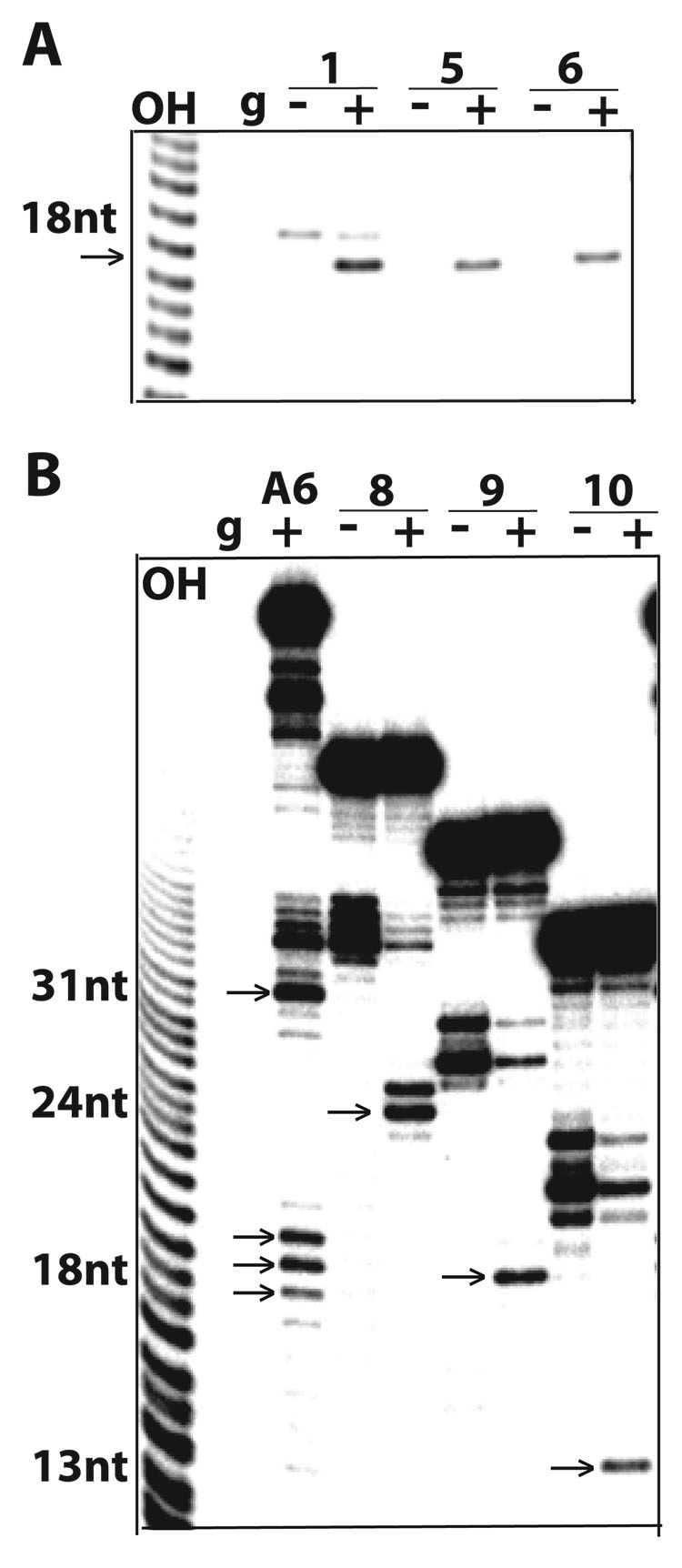
Several constructs examined so far were effective competitors, indicating that are bound by editing complexes, but it was unclear whether they were also active in enzymatic assays. To directly address this, we tested these constructs for specific gRNA-directed cleavage by editing complexes (Fig. 5). Since the guide strand in these pairs fully complements the substrate strand we assayed for potential guide-directed cleavage at the phosphodiester bond just 5’ of the duplex3. We have reported that this particular bond is cleaved just 5’ of the upstream duplex in the parental construct (Fig. 2A, top construct; and ahead in Fig. 5B) 8. Pairs 1, 5 and 6 generated a predicted 18-nt cleaved product (Fig. 5A) that corresponds to the 5’-end labeled overhang. This cleavage occurred only in presence of the guide strand. Furthermore, Pairs 8–10 which form a shorter 20-bp duplex were also cleaved with comparable efficiencies to the parental A6 construct (Fig. 5B). The expected 24-nt, 18-nt and 13-nt cleavage products, respectively, were gRNA dependent. In the parental A6 construct, gRNA-directed cleavages occur 5’ of both downstream (ES1) and upstream duplexes: the 5’ end-labeled substrate strand accumulates a 31-nt product, as a result of consecutive cleavage and removal of 3Us by U-specific exonuclease activity at ES1 13; also, multiple cuts 5’ of the upstream duplex are observed probably due to misannealing of this helix. Spurious fragments of the substrate strand often accumulate due to breakage or RNase contamination that preferentially target Us in absence of guide strand, and are more evident with 5’ labeled substrates.
Fig. 5.
5A–B. Direct cleavage assays of 5’end-labeled substrate-strand transcripts paired with the parental gRNA.D33. The homologous A6 and derived competitors are diagrammed in Fig. 2A. Lanes with “+” and without “−“ gRNA (g) are shown. Specific cleavage only occurs in the presence of gRNA (marked by an arrow). Spurious fragmentation of these transcripts occurs without gRNA but is inhibited by annealing of gRNA. Partial alkaline RNA hydrolysis “OH” was used as sizing ladder. Guide-directed cleavage of the A6 construct is directed by the downstream duplex (ES1) and by the upstream duplex. The latter occurs at three adjacent positions (~18-nt products) possibly due to alternative pairing. The short upstream duplex may be stabilized by co-axially stacking with the downstream duplex 8.
Among constructs found to associate with editing complexes, Pair-7 was not subject to guide-directed endonuclease cleavage as its substrate strand lacks a 5’ overhang and its 3’ ssRNA extension does not undergo cleavage (Fig. 2A; data not shown). The 18-nt protrusion of Pair-7 rescued the inactive Pairs 2–4 in crosslinking (Fig. 2A) and EMSA (data not shown) assays. In summary, all efficient competitors in EMSA and photo-crossliking assays were also functional for endonuclease cleavage, except for Pair-7. While both association and endonuclease cleavage activities of editing complexes have ssRNA and dsRNA requirements, these could be present in a way that promotes association but not cleavage. Thus, association and catalysis by editing complexes can be uncoupled.
We decided to further analyze derivatives of Pair-10, the shortest construct tested that supported editing complex association and specific endonuclease cleavage activity. This symmetric construct with 13-nt overhangs flanking a 20-bp duplex was ideal to dissect determinants involved in selection of the substrate strand. That is, how are the substrate and guide strands distinguished in a duplex? We tested Pair-10 derivatives (Pairs 11–16) bearing progressively shortened 5’ overhangs in the guide strand (Figs. 6A and ahead in 6D). In these reduced structures, a 3-nt 5’ overhang in the guide strand promoted efficient cleavage of the substrate strand, but 1-nt and 2-nt extensions were strongly inhibitory (Pairs 15–16). Also, the latter constructs were not rescued by longer (18-nt) 5’ overhangs in the substrate strand (not shown). This suggests that 5’ overhangs in the substrate and guide strands are not compensatory.
Fig. 6.
Diagram of minimized substrates for endonuclease cleavage by purified editing complexes. (A–C) A6 and derivative competitors (substrate strand) paired with parental gRNA D33 or shorter versions (guide strand). The size of both strands in each pair is indicated. All other labeling is as in Fig. 2A. Detected (√) or undetected (X) cleavage activity is indicated for each construct. Cleavage activity on Pair-23 was relatively weak. (D–G) Cleavage assays using 3’ end-labeled substrate strand derivatives.
Analysis of constructs bearing shorter 5’ extensions in the substrate strand (Pairs 17–20; Fig. 6B) showed that 12-nt are minimally required for endonuclease cleavage activity (Figs. 6D–E; and data not shown). Constructs with 11-nt 5’ overhangs in the substrate strand were inactive and not rescued by the presence of longer guide-strand overhangs (e.g., Pairs 19–20).
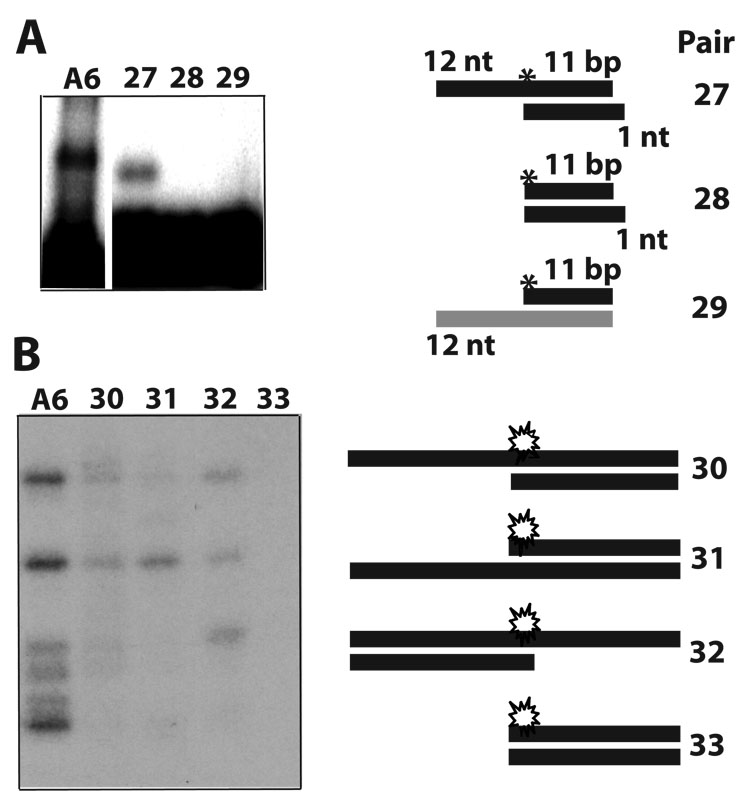
To determine whether constructs with duplexes shorter than 20-bp are functional we examined Pairs-21–26 (Figs. 6C and 6F). Efficient endonuclease cleavage was supported by Pair-21, which forms a 15-bp duplex, but progressive truncations of the guide-strand 5’ overhang were increasingly inhibitory (Pairs 22–24). Pair-21 also showed that the substrate strand can be shorter that the guide strand, and that a ~27-nt substrate strand bearing a 12-nt 5’ overhang supports efficient endonuclease cleavage. In the above constructs the substrate-strand 5’ extension appears to be separately recognized, as inactivating truncations of this element were not compensated by a longer duplex or extended guide-strand 5’ ssRNA. In contrast, the guide-strand 5’ overhang could be replaced by using either an extended double-stranded terminus (e.g., Pair-6; Fig. 2A), or a 3’ overhang of the substrate-strand (Pair-25; Fig. 6C). The latter pair also showed that an 18-nt guide strand, largely annealed with the substrate strand, directs efficient endonuclease cleavage activity. Seiwert et al. reported that an 18-nt guide strand directs endonuclease cleavage of a complementary 73-nt A6 mRNA 3. Pair-25 and Pair-5, both of which generate the same cleaved product, were nearly as efficient as the parental A6 construct (Fig. 6G; see also Fig. 5A). Finally, we found that an 11-bp duplex in Pair-26 failed to direct detectable cleavage of the substrate strand (not shown). Such 11-bp duplex seems relatively stable (−18.4 kcal/mol) and we confirmed efficient annealing with the substrate strand in native gels 5. Although this simple pair is not cleaved, it binds editing complexes in an EMSA (see the site-specific labeled Pair-27 in Fig 7A). Importantly, the ssRNA overhang was essential for binding, and substitution of a paired strand with DNA was inhibitory (Pair-28 and Pair-29, respectively). We examined additional constructs for association, whether or not they are cleaved, (Fig. 7B). In this case, we prepared derivatives of the thiolated parental A6 (diagrammed in Fig. 1) and tested their ability to photo-crosslink with editing complexes. For example, Pair-30 photo-crosslinks and is also cleaved (Fig. 7B; and data not shown). Other derivatives with an ssRNA overhang that crosslinked are not cleaved, whereas a blunt helix did not exhibit detectable crosslinking (Pairs 31–33, respectively). The parental A6 substrate generates more robust signals in association assays that most derivatives tested in our study.
Fig. 7.
Additional RNA pairs that associate with purified editing complexes but are not cleaved. (A) EMSA of Pair-27 (derived from Pair-26) that forms an RNP but is not cleaved. This RNP exhibits a faster electrophoretic mobility than with the parental A6 but the reason of this is unclear. Duplexes without the 12-nt overhang or bearing a DNA strand failed to form an RNP (Pairs 28 and 29, respectively; (B) U.V. photo-crosslinking assays of the A6 parental construct in Fig. 1 and derivatives with or without an ssRNA overhang (Pairs 30–33). The site-specific 32P label in (A) and the 32P and thio labels in (B) are depicted by an asterisk and a star, respectively.
In summary, the construct series in Fig. 6 showed that purified editing complexes only cleave substrate strands bearing a minimal 5’ overhang of 12-nt. The minimal duplex directing specific cleavage was not determined to the nucleotide but it could be ~15-bp long, if not smaller. In addition to these two features, cleavage activity required the presence of (a) either a substrate 3’-overhang or a guide 5’-overhang when using a 15-bp duplex, or (b) a larger duplex without additional ssRNA. Fig. 7 confirmed that association and cleavage can be uncoupled although an ssRNA overhang is essential for both these two stages of editing. Importantly, association exhibits simpler determinants than cleavage.
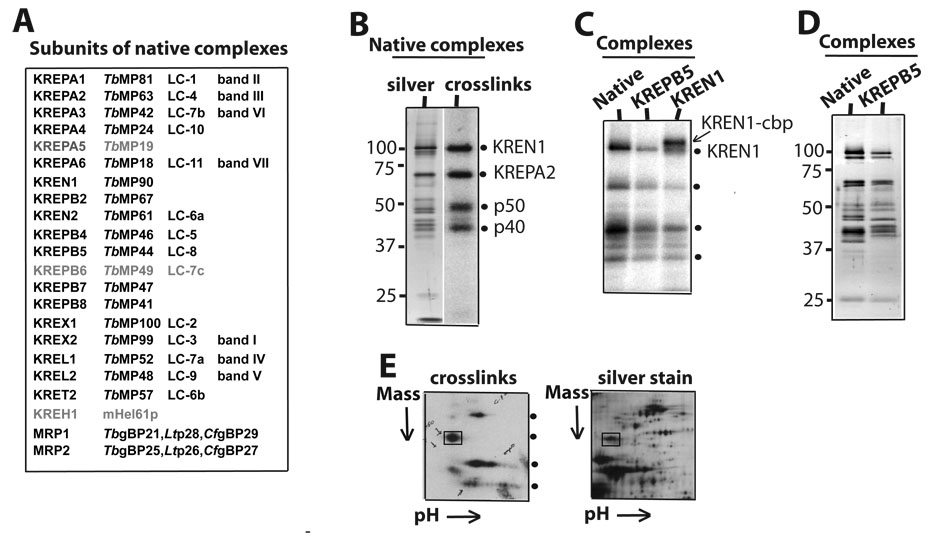
It is feasible that some if not all determinants defined in the current study may be recognized by one or more RNA-binding subunits of editing complexes, including RNase III-type, OB-fold and zinc-finger domain bearing subunits. At least three RNase III-type endonucleases identified in editing complexes are thought to catalyze pre-mRNA cleavage in insertion and deletion editing 11; 12; 16; 18. However, the composition of the native editing complexes used here, including the presence of reported endonucleases, was unclear. A mass spectrometric analysis of this protein preparation revealed nearly all reported subunits of affinity-purified ~20S editing complexes in T. brucei and L. tarentolae 11; 12; 16, in addition to subunits of the MRP complex which are thought to transiently associate with ~20S editing complexes via an RNA linker (Fig. 8A)31. Three other proposed editing subunits, KREPA5, KREPA6 and KREH1, were not detected likely because they were either substoichiometric, insufficiently ionized in our preparation or absent. However, KREPA6 was recently reported to be essential 29 and was most likely undetected in our samples.
Fig. 8.
Composition of native editing complexes and identification of two photo-crosslinking subunits: RNase III-type endonuclease KREN1 and structural KREPA2 (MP63). (A) Listing of all subunits detected by mass spectrometry. Alternative nomenclatures used in the literature are indicated. Three subunits were not detected (faded). (B) Native editing complexes stained with silver (lane 1) or exposed onto an X-ray film after U.V. photo-crosslinking (lane 2). The crosslinks (dots) by KREN1 and KREPA2 and two more subunits to be identified (p50 and p40) are indicated. (C) Crosslinks by native (lane 1) or affinity-purified KREPB5 (MP44) (lane 2) and KREN1 (lane 3) complexes. Both, cbp-tagged (up-shift) and endogenous KREN1 are indicted. (D) Silver staining of native and affinity-purified KREPB5 complexes. This panel was prepared using complexes purified during the current study (see Material and Methods section). Preliminary studies using aliquots from KREN1 and KREN2 complexes characterized in a previous study 32 showed that only the former generate the 100 kDa crosslink (see text). (E) 2D gel of partially purified complexes after photo-crosslinking (left) or silver staining (right). Crosslinked KREPA2 (boxed) was excised from the gel and identified by mass spectrometry.
Since our previous photo-croslinking studies indicated that at least four subunits of purified ~20S native complexes make intimate contact with model editing sites (Fig. 8B) 5; 24 we attempted the identification of a crosslinking subunit that migrates at about 100 kDa, where the endonuclease KREN1 was expected. To this end, we made a TAP-KREN1 construct and expressed it in T. brucei procyclic cells (see Materials and Methods section) based on a reported protocol that generated the same cell line 32. Tagged-editing complexes were purified through IgG and calmodulin-binding peptide (CBP) coupled resins and then examined by photo-crosslinking. We found that cbp-KREN1 complexes produced a shift of the ~100 kDa crosslink due to the mass added by the tag (~5kDa; Fig. 8C). These complexes also exhibited the crosslink by endogenous KREN1 and the other major crosslinks observed in native complexes. As far as we know this is the first evidence that at least two copies of KREN1 are present in editing complexes. Previous characterization of KREL1, KREN2 and KREN3 (KREPB2) affinity-purified complexes showed that endogenous and ectopic copies of these subunits were also present 14; 31; 32. Importantly, the shifted crosslink is specific of our tagged-KREN1 cell line, and not associated with the cell culture or protein purification conditions, as affinity-purified complexes using a different tagged subunit (TAP-KREPB5; i.e., MP44) exhibited the same crosslinking pattern of native complexes (Fig. 8C), as well as a similar silver staining pattern (Fig. 8D) and full-round insertion and deletion editing activity (not shown). Consistent with the identification of KREN1 in the current study, our preliminary crosslinking analysis using aliquots of KREN1 and KREN2 complexes purified and characterized in another study 32 showed that the former but not the latter forms the 100 kDa crosslink (data not shown). The presence of these KREN proteins was mutually exclusive in the reported purified complexes 32.
Our previous 1D-analyses suggested that the crosslink at ~60 kDa was KREPA2 24. We confirmed this identification by performing a 2D-gel analysis of a partially purified protein preparation exhibiting significant crosslinking activity by editing complexes (Fig. 8D, lower panel). The ~60 kDa crosslink was resolved in a discrete region of the gel, and mass spectrometric analysis of the excised region only contained KREPA2. The crosslinking subunits at about 50 and 40 kDa were more disperse and mass spectrometric analyses of these gel regions were unsuccessful. Thus, they remain to be identified.
Overall, the native editing complexes used in the current studies contain most subunits previously observed in purifications by other labs including the RNase III-type endonuclease KREN1, which we showed directly photo-croslinks with model editing sites. This subunit may be involved in the editing complex recognition of the substrate determinants defined here for association and endonuclease cleavage, but additional work is needed to explore this possibility.
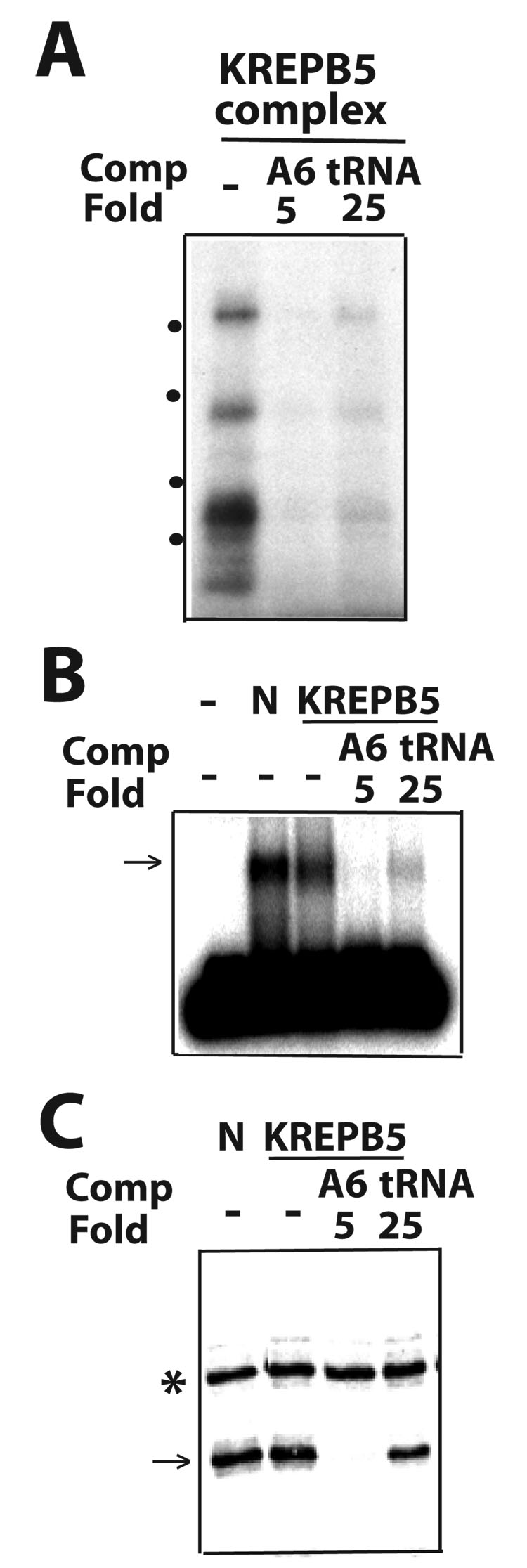
Finally, we compared the substrate specificity of native editing complexes and KREPB5 affinity-purified complexes in parallel EMSA, photo-crosslinking and endonuclease cleavage assays (Fig. 8A–C). Native and tagged-KREPB5 editing complexes exhibited similar substrate specificity, in presence of the homologous A6 (5-fold excess) and tRNA (25-fold), as positive control and relatively poor competitors, respectively. Thus, the approaches adopted in these studies should be useful in further comparisons of native and affinity-purified editing RNPs that exhibit different protein and functional composition.
Discussion
The goal of the current work was to define substrate requirements for association of purified editing complexes and gRNA-directed cleavage, the first catalytic step of an editing cycle. To this end, we used an EMSA, for the first time, in parallel with U.V. photo-crosslinking and gRNA-directed cleavage assays. Importantly, these assays were performed under comparable reaction conditions and the data obtained was complementary. The RNP assemblies detected by EMSA contained adenylylatable ligases and co-immunoprecipitated with known editing subunits (Figs. 1B–D), and their substrate specificity was conserved in the association and catalytic assays (Figs. 1E–G). Our combined EMSA, photo-crosslinking and enzymatic studies defined ssRNA and dsRNA determinants for association and cleavage, summarized in Fig. 10. Three main combinations of ssRNA and dsRNA determinants supported endonuclease cleavage are represented by the following pairs: Pair-22 (27-nt substrate and 23-nt guide strands) exhibits minimal 5’ overhangs and ~15-bp duplex for cleavage. In this context, a 12-nt 5’ overhang in the substrate strand was minimally required, whereas truncations of the 8-nt 5’ overhang in the guide strand were gradually inhibitory. The size of one overhang did not compensate for the size of the other, and thus appear to involve separate recognitions. Pair-25, a long substrate-strand annealed to a minimal guide-strand of 18-nt (16-nt in a duplex) supports efficient cleavage. This confirms the observation by Seiwert et al., that an 18-nt guide strand directed endonuclease cleavage of a complementary 73-nt A6 mRNA 3. Thus, a substrate 3’-overhang can substitute for a guide 5’-overhang. Pair-6, a long duplex overrides a requirement for ssRNA rightward of the duplex. Thus neither these overhangs are essential but an ssRNA extension, abutting a short duplex, may suffice. In this type of construct, the size of the substrate 5’-overhang was also tested. 12-nt or more supported cleavage (e.g., Pair-6, and data not shown) but 11-nt was inactivating (i.e., Pair-4; data not shown). Additional pairs were bound but not cleaved by editing complexes, showing that these two aspects of editing can be uncoupled. Pair-27 is the simplest construct of this kind. Competition studies or straight association assays by crosslinking or EMSA showed that pairs bearing blunt-ended helices or insufficient ssRNA cannot associate with editing complexes. Pair-2 and Pair-28 reproducibly failed to form detectable RNPs and Pair-3 was significantly less effective than the parental A6 substrate (data not shown). Some constructs that bind but are not cleaved were examined by photo-croslinking or EMSA using 5’-end labeled rather than more sensitive site-specific labeled RNAs (Fig. 7B; and data not shown).
Fig. 10.
Summary of defined ssRNA and dsRNA determinants for endonuclease cleavage and association by purified editing complexes. Important variations were observed depending on the secondary structure context. Three main types of cleaved constructs are illustrated by Pair-22: It bears minimal substrate 5’ and guide 3’ overhangs. In this context, further shortening of either element was strongly inhibitory and not rescued by lengthening of the other. Pair-25: Its long substrate-strand allowed reducing the guide-strand to 18-nt. Thus, a substrate 3’ overhang can substitute for a guide 5’ overhang. Pair-6: Its long duplex can substitute for either substrate 3’ or guide 5’ overhangs. Thus, neither these overhangs are essential but one may suffice in cleaved constructs. Importantly, association can occur without cleavage although it also requires an essential overhang either upstream or downstream of the helix. This is illustrated by Pair-27 and Pair-28. Detected (√) or undetected (X) binding (bind) and cleavage (cut) are indicated.
Together, these constructs as well as others examined indicated that an appropriate combination of dsRNA and ssRNA determinants, rather than overall size of the bi-molecular structure, is required for both association and endonuclease cleavage by purified editing complexes. The ssRNA requirement (12-nt) 5’ of the scissile bond and the dsRNA/ssRNA combinations 3’ of it seem to involve separate recognitions. The smallest helix tested that directed endonuclease cleavage was 15-bp long (~1.5 helices) but shorter versions similar to Pair-26 may be feasible (Fig. 6C). Although the shortest functional guide strand tested was 18-nt long, a functional guide strand may be longer than the substrate strand (e.g., Pair-21).
Importantly, the requirements for association and for catalysis can be uncoupled. This was shown by Pair-7 (Fig. 2), Pair-27 and A6 thio-lated derivatives (Fig. 7) that bind editing complexes but are not cleaved. In Pair-7, the substrate-strand forms a 3’ overhang but not a 5’ overhang. Its substrate-strand 3’ ssRNA stimulates association (compare with the inactive Pair-2) but, as expected, is not cleaved since editing endonucleases specifically target the phosphodiester bond immediately 5’ of the guiding “anchor” duplex 3; 6. On the other hand Pair-27 bears the critical 12-nt 5’ overhang but either insufficient duplex or overall length for cleavage. Furthermore, while a substrate 5’ 12-nt overhang is minimally required for cleavage, whether all residues need be unpaired or some may partially complement apposing guide-strand residues was not determined. In full-round editing substrates, single-strandedness of residues near the downstream “anchor” duplex is strongly stimulatory. More distal residues can engage in formation of a proposed upstream “tether” duplex in deletion or insertion in vitro 8; 33. Furthermore, the presence and/or the nature of gRNA residues in the internal loop may stimulate full-round editing. Consistent with this idea, the lack or inappropriate number of such residues inhibited full-round deletion and insertion editing 8; 33 (and unpublished data), and 2’ deoxy substitutions on the gRNA-side of the internal loop inhibited both photo-crosslinking and cleavage at the scissile bond 5.
Previously, our lab defined a minimal 43-nt pre-mRNA/gRNA hybrid for efficient full-round editing, which formed 10-bp helices flanking the ES. These nearby helices may be stabilized by coaxial stacking interactions, resembling a continuous helix. The smaller hybrid for endonuclease cleavage activity (including a ~27-nt substrate strand) and even simpler structure for binding imply that editing complexes require more extensive RNA contacts for the complete editing reaction, than for the intermediate cleavage step and the initial association step. Consistent with this concept, the artificially enhanced A6 parental substrate 8 is more efficient in all EMSA, photo-crosslinking and cleavage assays than most simpler derivatives tested here.
The fact that only one shifted product is reproducibly detected in the EMSA of the constructs examined suggests binding by a single editing complex, whether dimeric or of higher-order composition consistent with the co-purification of endogenous and ectopically expressed editing subunits, i.e., KREN1 in the current study (Fig. 8C) and KREN2, KREN3 (KREPB2) and KREL1 in previous studies 14; 31; 32. A mass spectrometric analysis revealed that the native complexes used in this study contain most known subunits of catalytic ~20S editing complexes, as expected from similar biochemical purifications 34. In addition, we found subunits of the MRP subcomplex as it was reported in purified L-complexes 25; 31, suggesting that at least some purified assemblies represent holoenzyme rather than core complexes.
Several observations lead us to suggest that some if not all determinants defined in this study may be recognized by one or more RNase III type proteins. Namely, (a) the shortest duplex tested that directed efficient endonuclease cleavage activity spanned ~1.5 turns. This is also the size of the smallest substrate identified that binds bacterial RNase III 35; (b) the critical role of 5’ and 3’ overhangs for cleavage at ssRNA-dsRNA junctions by the RNase III family member Drosha 36, and (c) the fact that KREN1 photo-crosslinks with a site for full-round editing (Fig. 8). This photo-crosslink was defined at a deletion site (Fig. 8C) but most likely also corresponds to a co-migrating crosslink at insertion sites 5. KREN1 endonuclease was proposed to specifically cleave deletion sites 11, however since association and cleavage are uncoupled we propose a model whereby KREN1 and related RNA-binding subunits may help scrutinize potential ligand determinants in the earliest checkpoint of RNA editing. Subsequent to the binding step, catalytic selectivity based on additional specific substrate recognitions may activate either the deletion or insertion enzymes, including proofreading of misedited insertion sites. A role of REN1 in an early checkpoint of ligand binding may explain why KREN1 downregulation inhibits editing of CYb and COII pre-mRNAs in vivo, which only have insertion sites. It is known that bacterial RNase III can undertake a modulatory role as a general dsRNA-binding protein regardless of its catalytic action 37. Importantly, the crosslinking activity of KREN1, KREPA2 (MP63) and at least two other major crosslinking subunits is conserved in both native and tap-tagged affinity-purified complexes. Such conservation further suggests that the interactions are relevant, and independent of purification protocols and cell lines used. The conserved OB-fold and zinc fingers of KREPA2 may also be involved in recognition of single-stranded determinants defined here.
Finally, while the current study shows that RNPs formed by purified editing complexes can be directly visualized, it is currently unclear if the fraction of substrate that remains unbound in association assays reflects the concentration and/or affinity of either total complexes or functional complexes. Also, not all RNPs formed in solution may be stable enough to withstand the forces of gel electrophoresis. These and related questions will be addressed in separate studies.
Methods
Synthesis and labeling of RNA
The ES1-radiolabeled A6 mRNA substrate was prepared by splint ligation as described 24. All other RNAs were synthesized in vitro by the Uhlenbeck single-stranded enzymatic method 38 and gel purified.
For the preparation of 5’-end labeled substrates, gel-purified RNA was dephosphorylated by treatment by alkaline phosphatase at 37°C for one hour, followed by addition of SDS, EDTA and proteinase K to a final concentration of 1.5%, 5 mM, and 40 µg/mL respectively and additional incubation at 50°C for 30 minutes. RNA was purified by phenol/chloroform extraction and precipitated with ethanol. 5 pmols of dephosphorylated RNA were incubated at 37°C for 30 minutes with [γ-32P] ATP (1:2 ratio of 5’-ends to ATP) and T4 polynucleotide kinase and gel-purified. For 3’-end labeling, 5 pmoles of gel-purified RNA were incubated at 4°C for 12 hours with an equimolar amount of [5’-32P] Cytidine 3’, 5’-Bis (Phosphate) and 15 units of T4 RNA ligase in RNA ligase buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 10 µg/mL BSA, 50 µM ATP, 10 mM DTT, 2U/µL anti-RNase (Ambion),10% DMSO) and gel-purified.
Cloning, cell culture and transfection
ORFs were amplified from T. brucei genomic DNA, kindly provided by Larry Simpson. The primers for KREN1 were designed as reported 32. For KREPB5, the primers were: forward CCC aagctt ATG AGA CGG GCT GTG GTA CTC CGT AC; and reverse CGC ggatcc CCG CCC TCC CAG TGC CAG CGC AAC TA (Hind III and Bam HI sites are in small case letters, respectively). The amplified products using Pfu DNA polymerase were treated with HindIII, BamHI and ligated to the pLEW79TAP expression vector, kindly provided by Achim Schnaufer 32. Constructs were linearized with NotI and used to transfect T. brucei strain 29.13 as described 39. Selection of transfectants was applied with 2.5 µg/mL phleomycin. KREN1 and KREPB5 expression was induced with 100 ng/mL and 1 µg/mL tetracycline, respectively, and confirmed by immunoblotting with the PAP reagent (Sigma).
Purification and protein composition determination of Editing Complexes
Chromatographic purification of RNA editing complexes
Mitochondrial extracts were prepared from procyclic T. brucei strain TREU667 as described 40; 41. Editing complexes were purified from mitochondrial extracts by consecutive anion exchange and DNA-affinity chromatography as described 41; 42.
Tandem affinity purification of RNA editing complexes
Four liters of culture at a density of ~2.0×107 cells/mL were pelleted and lysed in 25mL of 10 mM Tris-HCl, pH 8.0, 150 mM KCl, 0.1% NP-40, 1% Triton-X-100 and one tablet of EDTA-free complete protease inhibitors (Roche) for 30 minutes on ice. Lysis was confirmed by microscopy. Lysates were spun at 6000 X g for 15 minutes and the clarified extract purified by sequential IgG and Calmodulin affinity chromatography as described 43.
Mass spectrometric analysis of native RNA editing complexes
Proteins in gel bands and complex mixtures were identified by LC-MS/MS analysis as described 34.
Photocross-linking, RNA cleavage, Electrophoretic Mobility Shift, Competitions and Adenylylation assays
All assays are variations of the standard editing assay in our lab which consists of a mixture of a pre-annealed mixture of 10 fmols 32P-labeled RNA and 1.25 pmoles unlabeled gRNA, completed to 20 µL with MRB [25 mM Tris-HCl, pH 8, 10 mM Mg(OAc)2, 10 mM KCl, 1 mM EDTA, pH 8, 50 µg/mL hexokinase and 5% glycerol] and, if applicable, competitor RNA at the indicated molar excess relative to the 32P-labeled substrate. The mixture was pre-equilibrated for 10 minutes at 26°C and 2 µL of peak editing or TAP fraction was added. Prior to the assays, quantitative annealing of the RNA pairs tested was confirmed in native gels [as in 5]. The sample was incubated at 26°C for 10 minutes then treated in an assay-specific manner. For cross-linking, samples were irradiated for 10 minutes under a 365 nm UV lamp, treated with RNase A and RNase T1 (50 µg/mL and 125 units/mL final concentrations respectively) at 37°C for 15 minutes, supplemented with SDS loading dye and loaded onto an SDS-polyacrylamide gel. For mRNA cleavage, purified editing complexes were pre-treated with 10 mM tetrapotassium pyrophosphate, pH 8, in MRB, for 5 minutes on ice to inhibit ligase activity 9; after incubation the mixture was deproteinized and RNAs were resolved on denaturing polyacrylamide gels. For electrophoretic mobility shift assays, the reaction mixture was loaded directly (no loading dye) onto a 1.5% agarose gel in 0.5X TBE (45 mM Tris-borate and 1 mM EDTA) and run for 2 hours at ~5 V/cm at 4°C. Following electrophoresis, the agarose gel was dried under vacuum. EMSA with site-specific labeled transcripts were significantly more sensitive and reproducible than with end-labeled substrates, since the splint-ligation method used to generate the former (see above) exclusively incorporates phosphorylated fragments. Only the parental A6 substrate and Pair-27 were site-specific labeled using synthetic donor fragments [e.g., as in Fig. 1;24], although 5’-end labeled A6 parental and other constructs were also compared side-by-side in shift assays. Immunodepletions were carried out as described for the immunoprecipitation of RNA cross-linking proteins 24 using a monoclonal antibody against KREPA2 immobilized on goat anti-mouse IgG resin (Dynal). Adenylylation assays were performed as described 30. All assays can be scaled-up linearly to enhance signal. The data were reproducible in at least two independent experiments. Each experiment included repeat assays, and those show are representative. Data were visualized by phosphorimaging and/or autoradiography.
Fig. 9.
Association and endonuclease cleavage activity of affinity-purified editing complexes. Parallel (A) photo-crosslinking, (B) EMSA and (C) cleavage assays, respectively. All labeling is as in Fig. 1. KREPB5-tagged complexes were directly compared to native “N” complexes.
Acknowledgments
We want to thank Dr. C.T. Ranjith Kumar, and in the Cruz-Reyes lab to Bhaskara Reddy Madina and Ambrish Kumar for their comments on the manuscript and helpful discussions. We also thank Drs. Achim Schnaufer, Ruslan Aphasizhev and Juan D. Alfonzo for their generous advise and protocols for tandem-affinity purification and transfection of trypanosomes. pLEW79TAP was a gift from Achim Schnaufer. We are grateful to Dr. Larry Simpson for providing T. brucei genomic DNA and Dr. Andrew McMillan for his advise on the EMSA. This work was supported by grants from the NIH (GM067130) to JC-R, and (GM42188) to KS. The mass spectrometric studies were performed at the SBRI, Seattle.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cruz-Reyes J, Hernandez A. Protein-protein and RNA-protein interactions in U-insertion/deletion RNA editing complexes. In: Smith HC, editor. RNA and DNA editing. New Jersey: John Wiley & Sons, Inc.; 2008. pp. 71–98. [Google Scholar]
- 2.Carnes J, Stuart KD. Uridine insertion/deletion editing activities. Methods Enzymol. 2007;424:25–54. doi: 10.1016/S0076-6879(07)24002-9. [DOI] [PubMed] [Google Scholar]
- 3.Seiwert SD, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 4.Kable ML, Seiwert SD, Heidmann S, Stuart K. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science. 1996;273:1182–1183. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- 5.Cifuentes-Rojas C, Pavia P, Hernandez A, Osterwisch D, Puerta C, Cruz-Reyes J. Substrate determinants for RNA editing and editing complex interactions at a site for full-round U insertion. J Biol Chem. 2006;282:4265–4276. doi: 10.1074/jbc.M605554200. [DOI] [PubMed] [Google Scholar]
- 6.Cruz-Reyes J, Sollner-Webb B. Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc Natl Acad Sci U S A. 1996;93:8901–8906. doi: 10.1073/pnas.93.17.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz-Reyes J, Rusche LN, Piller KJ, Sollner-Webb B. T. brucei RNA editing: adenosine nucleotides inversely affect U-deletion and U-insertion reactions at mRNA cleavage. Mol Cell. 1998;1:401–409. doi: 10.1016/s1097-2765(00)80040-4. [DOI] [PubMed] [Google Scholar]
- 8.Cruz-Reyes J, Zhelonkina A, Rusche L, Sollner-Webb B. Trypanosome RNA editing: simple guide RNA features enhance U deletion 100-fold. Mol Cell Biol. 2001;21:884–892. doi: 10.1128/MCB.21.3.884-892.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz-Reyes J, Zhelonkina AG, Huang CE, Sollner-Webb B. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol Cell Biol. 2002;22:4652–4660. doi: 10.1128/MCB.22.13.4652-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang CE, Cruz-Reyes J, Zhelonkina AG, O'Hearn S, Wirtz E, Sollner-Webb B. Roles for ligases in the RNA editing complex of Trypanosoma brucei: band IV is needed for U-deletion and RNA repair. Embo J. 2001;20:4694–4703. doi: 10.1093/emboj/20.17.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K. A deletion site editing endonuclease in Trypanosoma brucei. Mol Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 12.Kang X, Gao G, Rogers K, Falick AM, Zhou S, Simpson L. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc Natl Acad Sci U S A. 2006;103:13944–13949. doi: 10.1073/pnas.0604476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang X, Rogers K, Gao G, Falick AM, Zhou S, Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc Natl Acad Sci U S A. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 15.Rogers K, Gao G, Simpson L. Uridylate-specific 3′ 5′-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J Biol Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- 16.Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernst NL, Panicucci B, Igo RP, Jr, Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 18.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA Editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2007 doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao G, Simpson L. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J Biol Chem. 2003;278:27570–27574. doi: 10.1074/jbc.M303317200. [DOI] [PubMed] [Google Scholar]
- 20.O'Hearn SF, Huang CE, Hemann M, Zhelonkina A, Sollner-Webb B. Trypanosoma brucei RNA editing complex: band II is structurally critical and maintains band V ligase, which is nonessential. Mol Cell Biol. 2003;23:7909–7919. doi: 10.1128/MCB.23.21.7909-7919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhelonkina AG, O'Hearn SF, Law JA, Cruz-Reyes J, Huang CE, Alatortsev VS, Sollner-Webb B. T. brucei RNA editing: action of the U-insertional TUTase within a U-deletion cycle. Rna. 2006;12:476–487. doi: 10.1261/rna.2243206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruz-Reyes J. RNA-protein interactions in assembled editing complexes in trypanosomes. Methods Enzymol. 2007;424:107–125. doi: 10.1016/S0076-6879(07)24005-4. [DOI] [PubMed] [Google Scholar]
- 23.Cifuentes-Rojas C, Halbig K, Sacharidou A, De Nova-Ocampo M, Cruz-Reyes J. Minimal pre-mRNA substrates with natural and converted sites for full-round U insertion and U deletion RNA editing in trypanosomes. Nucleic Acids Res. 2005;33:6610–6620. doi: 10.1093/nar/gki943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacharidou A, Cifuentes-Rojas C, Halbig K, Hernandez A, Dangott LJ, De Nova-Ocampo M, Cruz-Reyes J. RNA editing complex interactions with a site for full-round U deletion in Trypanosoma brucei. Rna. 2006;12:1219–1228. doi: 10.1261/rna.2295706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. Rna. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Brecht M, Niemann M, Schluter E, Muller UF, Stuart K, Goringer HU. TbMP42, a protein component of the RNA editing complex in African trypanosomes, has endo-exoribonuclease activity. Mol Cell. 2005;17:621–630. doi: 10.1016/j.molcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 28.Salavati R, Ernst NL, O'Rear J, Gilliam T, Tarun S, Jr, Stuart K. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. Rna. 2006;12:819–831. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarun SZ, Jr, Schnaufer A, Ernst NL, Proff R, Deng J, Hol W, Stuart K. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. Rna. 2007 doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabatini R, Hajduk SL. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J Biol Chem. 1995;270:7233–7240. doi: 10.1074/jbc.270.13.7233. [DOI] [PubMed] [Google Scholar]
- 31.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. Embo J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. Compositionally and functionally distinct editosomes in Trypanosoma brucei. Rna. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igo RP, Jr, Lawson SD, Stuart K. RNA sequence and base pairing effects on insertion editing in Trypanosoma brucei. Mol Cell Biol. 2002;22:1567–1576. doi: 10.1128/mcb.22.5.1567-1576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panigrahi AK, Schnaufer A, Stuart KD. Isolation and compositional analysis of trypanosomatid editosomes. Methods Enzymol. 2007;424:3–24. doi: 10.1016/S0076-6879(07)24001-7. [DOI] [PubMed] [Google Scholar]
- 35.Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 36.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 37.Calin-Jageman I, Nicholson AW. RNA structure-dependent uncoupling of substrate recognition and cleavage by Escherichia coli ribonuclease III. Nucleic Acids Res. 2003;31:2381–2392. doi: 10.1093/nar/gkg329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 40.Harris ME, Hajduk SL. Kinetoplastid RNA editing: in vitro formation of cytochrome b gRNA-mRNA chimeras from synthetic substrate RNAs. Cell. 1992;68:1091–1099. doi: 10.1016/0092-8674(92)90080-v. [DOI] [PubMed] [Google Scholar]
- 41.Sollner-Webb B, Rusche LN, Cruz-Reyes J. Ribonuclease activities of trypanosome RNA editing complex directed to cleave specifically at a chosen site. Methods Enzymol. 2001;341:154–174. doi: 10.1016/s0076-6879(01)41151-7. [DOI] [PubMed] [Google Scholar]
- 42.Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. Embo J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]