Abstract
Vaccine development for possible influenza pandemics has been challenging. Conventional vaccines such as inactivated and live attenuated virus preparations are limited in terms of production speed and capacity. DNA vaccination has emerged as a potential alternative to conventional vaccines against influenza pandemics. In this study, we use a novel, cell-free DNA manufacturing process (synDNA™) to produce prototype linear DNA vaccines against the influenza virus type A/H5N1. This synDNA™ process does not require bacterial fermentation, so it avoids the use of antibiotic resistance genes and other nucleic acid sequences unrelated to the antigen gene expression in the actual therapeutic DNA construct. The efficacy of various vaccines expressing the hemagglutinin and neuraminidase proteins (H5N1 synDNA™), hemagglutinin alone (H5 synDNA™) or neuraminidase alone (N1 synDNA™) was evaluated in mice. Two of the constructs (H5 synDNA™ and H5N1 synDNA™) induced a robust protective immune response with up to 93% of treated mice surviving a lethal challenge of a virulent influenza A/Vietnam/1203/04 H5N1 isolate. In combination with a potent biological activity and simplified production footprint, these characteristics make DNA vaccines prepared with our synDNA™ process highly suitable as alternatives to other vaccine preparations.
Keywords: DNA vaccine, synDNA™ vaccine, cell-free DNA vaccine production, influenza virus type A/H5N1, immune response
Introduction
The emergence of highly pathogenic avian influenza virus type A/H5N1 in domestic poultry and the increasing number of cases of transmission of H5N1 viruses direct to humans indicate a significant threat to public health because of the potential for pandemic spread of these viruses. Since 1997, H5N1 viruses have killed millions of domestic fowl in Asia and over 300 human cases of infection were reported to WHO, 60% of which, were fatal.1
Currently, therapeutic and preventive options for H5N1 viral infections are limited. Antiviral drugs such as adamantanes (amantadine and rimantadine) and neuraminidase inhibitors (peramivir, oseltamivir and zanamivir) have been evaluated in preclinical tests as well as in the treatment of H5N1 infections.2,3 However, no conclusions have been made regarding their clinical efficacy. In addition, drug resistance in patients has already been reported.4 Combinational chemotherapy with multiple drugs that target different viral proteins might be more effective in combating drug resistant H5N1 strains5 and needs to be further explored.
Vaccination is still the preferred strategy for the prevention and control of pandemic influenza. Inactivated and live attenuated virus vaccines that are being developed for pandemic H5N1 virus are based on existing technologies licensed for production of seasonal influenza vaccines. In 2007, the first H5N1 vaccine, an inactivated virus, developed by Sanofi-Aventis, was approved by the U.S. Food and Drug Administration (FDA) although its effectiveness is limited.6 Other approaches that are also being explored include: (1) a recombinant HA subunit; (2) viral vectors expressing HA protein; and (3) plasmid DNA (pDNA) expressing H5N1 genes. These studies have shown some promises in preclinical as well as clinical studies.7
The immunogenic potential of DNA was first described in 1990 and since then significant progress has been made in developing DNA vaccines against various pathogens.8 DNA vaccination involving the administration of pDNA encoding one or more of antigenic proteins is an attractive alternative to other approaches. The main strength of the utilization of DNA vaccines for protection against pandemic influenza is that once developed against a particular target strain, new constructs representing the emerging reassortant variants could be created rapidly. To date, although there are no marketed pDNA vaccine products for humans, three pDNA vaccines have been approved for use against (1) West Nile virus in horses,9 (2) infectious haematopoietic necrosis virus (IHNV) in salmon,9 and (3) melanoma in dogs.10
Production of pDNA relies upon bacterial fermentation and requires rigorous purification processes to remove unwanted contaminants derived from bacterial cells (including endotoxins) and culture medium.11 Such impurities not only minimize the efficiency of DNA vaccines, they can also lead to dose-related toxicity.12 pDNA vaccines contain two critical moieties. The first component constitutes the plasmid backbone, required for maintenance and replication of pDNA in the host bacteria. The second component consists of a eukaryotic expression cassette containing the heterologous gene(s) to be expressed. In most instances, the pDNA backbone sequence represents a significant inactive portion of the therapeutic dose. In essence, this backbone is not required for proper gene expression in mammalian cells and may even be detrimental. Indeed, although immunogenic,13,14 CpG dinucleotides presented in the backbone sequences when prepared from bacteria have been shown to participate in gene silencing.15 In addition, other DNA elements found within the backbone sequences can also downregulate gene expression.16 Moreover, antibiotic resistance gene(s) and replication origin sequences could later lead to adverse effects in the host. These issues have spearheaded efforts to develop processes that produce therapeutic DNA devoid of any unnecessary backbone sequence.17–20 In addition, current manufacturing processes still require bacterial fermentation to produce therapeutic pDNA and do not address the issue of potential bacterial contamination in the DNA production. To avoid potential bacterial contaminants and to remove the pDNA backbone sequence, we have developed a novel, cell-free DNA synthesis process (synDNA™) that is suited for scale-up production of DNA vaccines or therapeutic DNA products.
Inbred mice (female Balb/C strain) represent a widely accepted animal model for the study of influenza A. To develop an effective vaccine for pandemic influenza, we utilized the influenza A/H5N1 isolate from a fatal human case in 2004 (A/Vietnam/1203/04). Several studies suggest this isolate is highly virulent in humans and reports suggest that rapid infection of and replication in the central nervous system may account for this difference.21–28 Thus, due to the highly virulent and rapid systemic dissemination of this highly pathogenic avian H5N1 influenza, particularly with CNS involvement, we have further examined the pathogenesis of this isolate in the mouse model. In six-week-old mice, intranasal infection with this viral isolate results in dose-dependent disease development and lethality (LD50 between 1 × 102 and 1 × 103 TCID50), with signs of encephalitis occurring between days 4 and 10.3 Our results indicate that doses >6 × 102 TCID50 typically cause lethal infection in > 75% of mice,3 suggesting that mice represent a useful model for preclinical studies of vaccine efficacy for pandemic influenza.
Here we present the results of using three synDNA™ constructs as vaccines against influenza A/H5N1 in a mouse model. We show that both H5N1 and H5 synDNA™ vaccines are able to confer protection against a lethal virus challenge with influenza A/Vietnam/1203/04 and are comparable to plasmid based DNA vaccines.
Results
In vitro expression of HA and NA genes
Linear synDNA™ vaccine constructs encoding the mammalian codon optimized H5 or N1 genes from the influenza A/H5N1 (Vietnam/1203/04) isolate were prepared as described in the Material and Methods. In one instance, a tandem H5N1 construct was designed consisting of an H5 expression cassette followed directly by an N1 expression cassette. An expression cassette expressing the firefly luciferase gene was used as a synDNA™ control (Fig. 1). Human A549 cells were transfected with synDNA™ vaccine constructs by electroporation to confirm the in vitro expression of the gene(s) of interest. Western blotting using polyclonal serum against influenza A/H5N1 (Vietnam/1203/04) whole virions or commercially available anti-NA antibodies were used to detect the presence of either H5 or N1 polypeptides. Figure 2 shows the results of the immunoblot of crude lysates from transfected cultured human cells at 16 hr post-transfection. All linear synDNA™ vaccine constructs expressed the intended proteins.
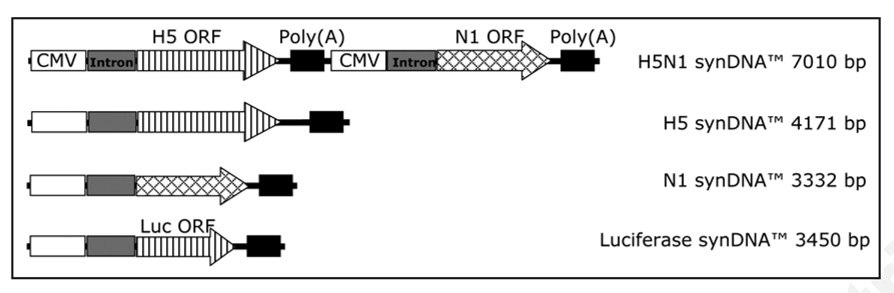
Figure 1.
Schematic representation of linear synDNA™ constructs. Three constructs were prepared expressing the H5 and/or N1 proteins from Influenza A/H5N1 (Vietnam/1203/04). All construct were synthesized using the cell-free synthesis process (synDNA™). As a control, a luciferase expressing construct was synthesized (Luciferase synDNA™) hereafter referred to as control synDNA™.
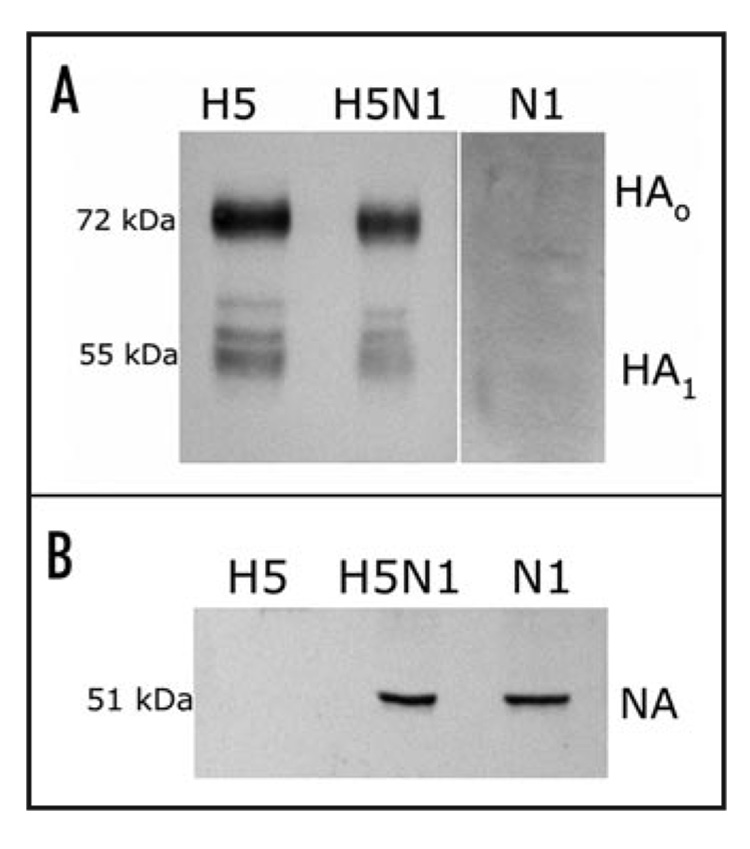
Figure 2.
Analysis of gene expression from synDNA™ constructs in cultured human cells. A549 cells transfected by electroporation were lysed and crude lysates were used in immunobloting experiments, as described in the Materials and Methods. (A) Proteins were blotted with goat anti-H5N1 polyclonal serum. H5 protein can be detected as full-length HA (HA0; 72 kDa) and as a proteolytically cleaved component, HA1 (55 kDa); (B) Transferred proteins were blotted with affinity purified rabbit anti-NA antibody raised against the 15 amino acids in the C-terminal region of the NA protein (Biodesign Int.). A ~51 kDa protein was detected in crude lysates from H5N1 or N1 synDNA™ transfected cells but not H5 synDNA™ alone.
Short term protection against lethal virus challenge
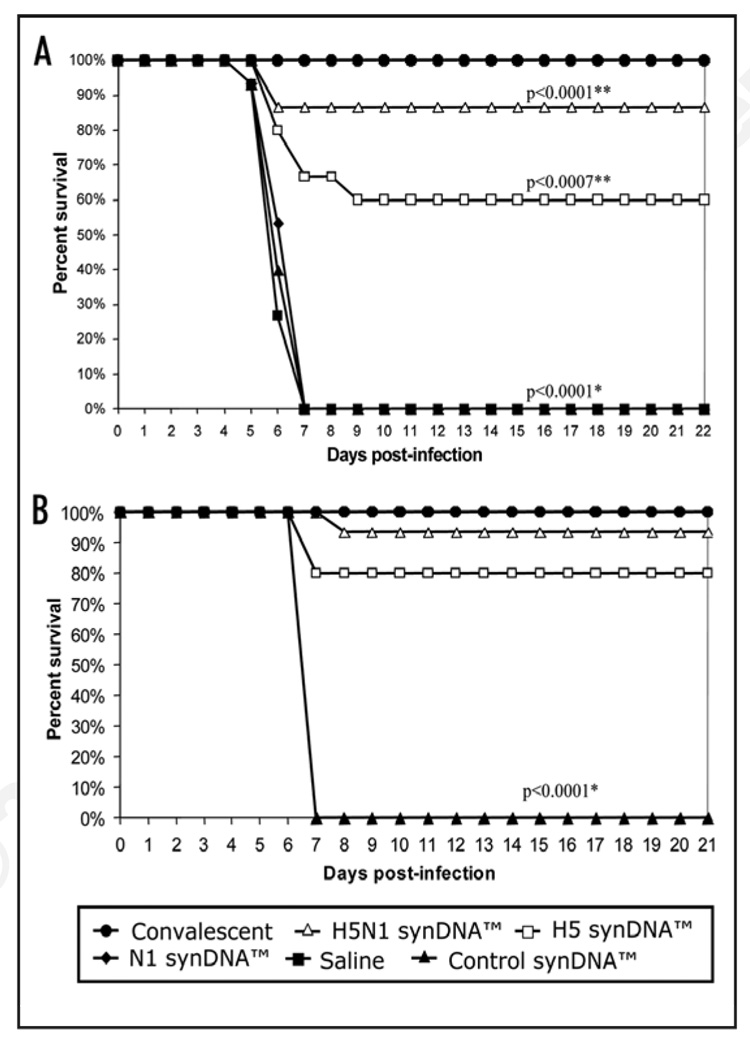
In order to test the ability of synDNA™ vaccines to trigger a protective immune response in mice, each construct was injected intramuscularly as described in the Materials and Methods, i.e., 1 injection with 2 boosters at 2 week intervals. Two weeks following immunization with vaccine constructs or with the control synDNA™ vector, all mice were challenged with a lethal dose of influenza A/H5N1 (Vietnam/1203/04) virus and monitored over time. The target challenge dose utilized in this study was selected based upon extensive prior studies that included a wide range of intranasal doses in multiple. For mice inoculated with a H5N1 dose of >2 × 103 TCID50 per mouse, 100% developed paralysis or died between day 7 and 10 post-infection. Therefore, delivered H5N1 doses greater than this cutoff, based upon back-titration of the viral inoculum (day 0), are considered to be a “high dose” (lethal) challenge.3 As shown in Figure 3A, 87% of the H5N1 synDNA™ vaccine-immunized mice survived lethal challenge compared to 60% survival rate in the H5 synDNA™ vaccine-immunized group and a statistically significant improvement in survival in comparison with the placebo (p < 0.0001 and p = 0.007, respectively). Interestingly, we did not detect any protective response in mice immunized with N1 synDNA™ vaccine alone (Fig. 3) although the N1 protein expression was detected in transfected A549 cells (Fig. 2). In order to confirm these results, the vaccine trial was repeated with a larger group of animals using syn-DNA™ vaccines prepared independently from the previous vaccine trial. Figure 3B shows that 14 out of 15 mice (93%) immunized with the H5N1 synDNA™ survived lethal H5N1 challenge. Similar to the first vaccine trial, the H5 synDNA™ appeared to have been slightly less effective than the H5N1 synDNA™ in triggering a protective immune response in mice, however the difference was not statistically significant.
Figure 3.
Survival of synDNA™ vaccinated mice following lethal challenge with influenza A/H5N1. (A) Vaccine trial 1. Four-week-old female Balb/C mice (N = 15 per group) were vaccinated via intramuscular (i.m.) route with the indicated synDNA™ vaccine (50 µg per mouse) or, as controls, with vehicle saline or control synDNA™, on day -42, -28 and -14 relative to H5N1 challenge. Two weeks after the last injection (day 0), mice were challenged via intranasal (i.n.) route with a lethal dose of virulent H5N1 virus (6.8 × 104 TCID50 per mouse). As positive control for protection, Balb/C mice that survived a prior i.n. infection with a low dose of H5N1 were infected in parallel with synDNA™ vaccinated mice on day 0. Animals were monitored daily up to 21 days; (B) Vaccine trial 2. A second survival trial was performed with identical dose and immunization schedule used in trial 1. Four-week-old mice (N = 19 per group) were vaccinated via i.m. route with the indicated synDNA™ (50 µg per mouse) or, as control, luciferase synDNA™, on day -42, -28 and -14 relative to H5N1 challenge. Two weeks after the last injection (day 0), mice were challenged with a lethal dose of virulent H5N1 virus (5.3 × 103 TCID50 per mouse). Four mice from each group were euthanized at day 5 post-challenge for viral load analysis (see text). For both trials, statistical analysis of survival comparing survival time for all groups over the post-challenge monitoring period was performed using logrank test at a significant level of α < 0.05 (see Materials and Methods). For pairwise comparison of the survival of the H5-, N1- or H5N1 SynDNA™ vaccinated versus the control (saline or SynDNA™ group, 0/15 survivors), Fisher’s Exact Test was performed (α < 0.05). Statistically significant comparisons by logrank or Fisher’s Exact Test are indicated by * or **, respectively; the p-values are provided in the figures.
Protection against disease development upon virus challenge
Mice infected with lethaldose of virulent influenza A/H5N1 were monitored daily for signs of morbidity and mortality. Table 1 summarizes the results gathered from two independent trials and shows that in both trials the majority of H5N1 synDNA™-immunized animals remained asymptomatic after challenge (60% and 90%, respectively). Symptoms of disease i.e., hypothermia and clinical encephalitis, appeared on day 4–5 post-infection (p.i.) in both vaccinated and control groups (data not shown), similarly to prior studies we have performed using this virus isolate.3 For animals that did not survive viral challenge in any of the vaccinated groups, deaths occurred between day 7–8, which was 2–3 days after the appearance of symptoms. However when immunized with the H5N1 synDNA™ vaccine, animals that began to show hypothermia (0 to 13%) and encephalitic symptoms (1 to 13%) by day 4–5 p.i. survived the 21 day monitoring period and appear to have recovered by the end of the study period (22 days p.i.), as judged by weight and body temperature (Fig. 4 and Fig. 5). In contrast, although the overall survival rate of H5 synDNA™ vaccine-immunized animals appears similar to that of the H5N1 synDNA™ vaccine-immunized group (Table 1), fewer animals remained asymptomatic (26.5% and 42% in each trial). Among this group, hypothermia was more frequent and lasted >1 day (13%), as did encephalitic symptoms (13 to 37%).
Table 1.
Effect of lethal influenza A/H5N1 viral challenge on surviving mice
| No. of animals/total (%) | ||||||
|---|---|---|---|---|---|---|
| synDNA™ vaccines | Asymptomatic | Symptomatica | Survivors | Dead | ||
| Trial 1 | HY(1day) | HY(>1day) | E | |||
| H5N1 (n = 15) | 9 (60%) | 2 (13%) | - | 2 (13%) | 13 (87%) | 2 (13%) |
| H5 (n = 15) | 4 (26.5%) | 1 (6.5%) | 2 (13%) | 2 (13%) | 9 (60%) | 6 (40%) |
| N1 (n = 15) | - | 100% HY and E | - | 15 (100%) | ||
| Control (n = 15) | - | 100% HY and E | - | 15 (100%) | ||
| Trial 2b | HY(1day) | HY(>1day) | E | |||
| H5N1 (n = 19) | 13 (90%) | - | - | 1 (5%) | 14 (95%) | 1 (5%) |
| H5 (n = 19) | 6 (42%) | 1 (5%) | - | 5 (37%) | 12 (84%) | 3 (16%) |
| Control (n = 5) | - | 100% HY and E | - | 1 | ||
HY(1day), Hypothermia symptoms lasting 1 day; HY(>1day): Hypothermia symptoms lasting more than 1 day; E: Encephalitic symptoms.
Four mice/grp were sacrificed 1 week post-challenge for viral load analysis (see text).
Figure 4.
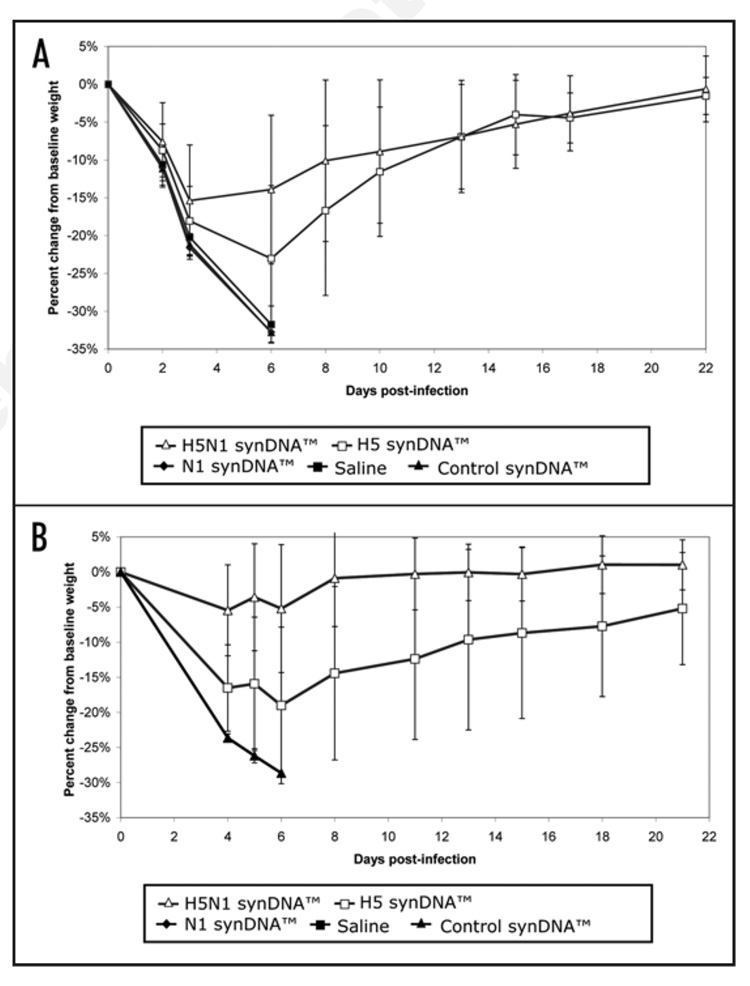
Percent change in body weight of synDNA™ vaccinated mice following challenge with influenza A/H5N1. In two independent trials of synDNA™ vaccine efficacy in mice (survival and details presented in Fig. 3), periodic recording of body weight was performed over the 21–22 day study period. Briefly, mice were vaccinated via intramuscular (i.m.) route with the indicated synDNA™ vaccine (50 µg per mouse) or, as controls, with saline or control synDNA™, on day -42, -28 and -14 relative to H5N1 challenge. Two weeks post-immunization (day 0), mice were challenged with a lethal dose of influenza A/H5N1(Vietnam/1203/04). Figure 4A and B represent the results of two independent vaccine efficacy trials, as described in Figure 3A and B, respectively. The percent change in body weight relative to baseline (day 0) was calculated for individual mice and the percent change averaged by group. Each data point represents the average of 15 mice as indicated for H5N1 synDNA™, H5 synDNA™, N1 synDNA™, control synDNA™ and saline groups.
Figure 5.
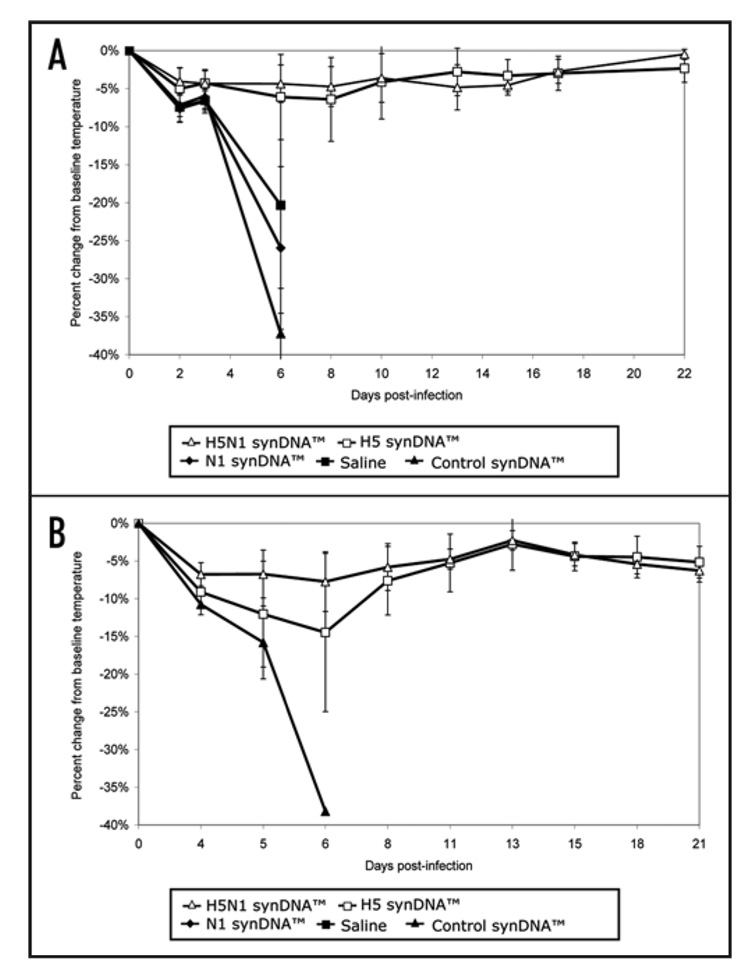
Percent change in body temperature of synDNA™ vaccinated mice following challenge with influenza A/H5N1. In two independent trials of synDNA™ vaccine efficacy in mice (survival and details presented in Fig. 3), periodic telemetric monitoring of body weight was performed over the 21–22 day study period. Briefly, mice were vaccinated via intramuscular (i.m.) route with the indicated synDNA™ (50 µg per mouse) or, as controls, with saline or control synDNA™, on day -42, -28 and -14 relative to H5N1 challenge. Two weeks post-immunization (day 0), mice were challenged with a lethal dose of influenza A/H5N1 (Vietnam/1203/04). Figure 5A and B represent the results of two independent vaccine efficacy trials, as described in Figure 3A and B, respectively. The percent change in body temperature relative to baseline (day 0) was calculated for individual mice and the percent change averaged by group. Each data point represents the average for H5N1 synDNA™ (n = 15), H5 synDNA™ (n = 15), N1 synDNA™ (n = 15), control synDNATM (n = 15, Trial 1 and n = 5, Trial 2) and saline groups (n = 15). Error bars represent standard deviations.
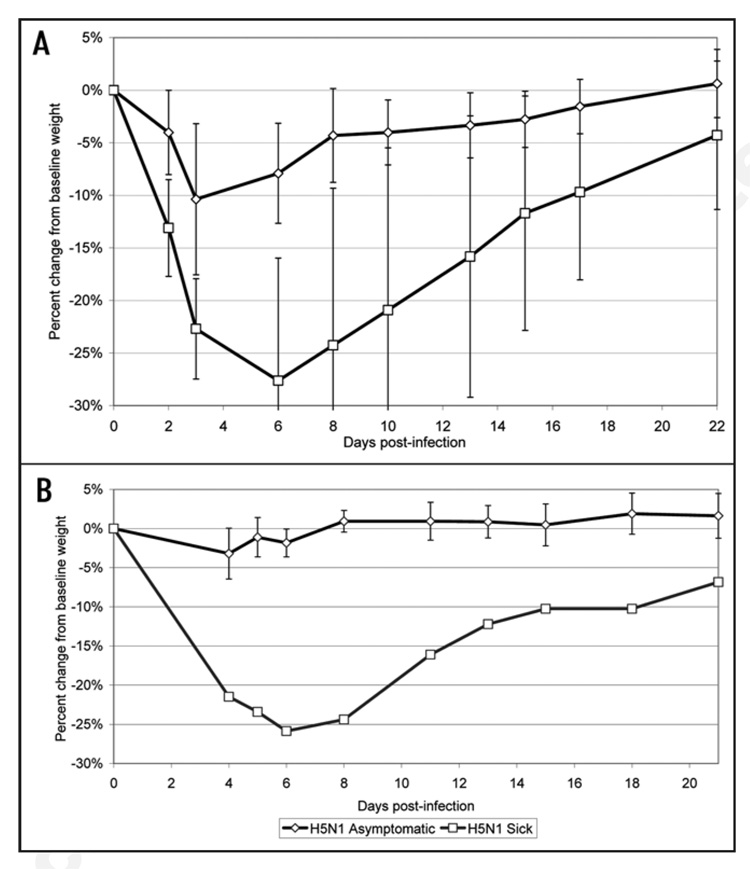
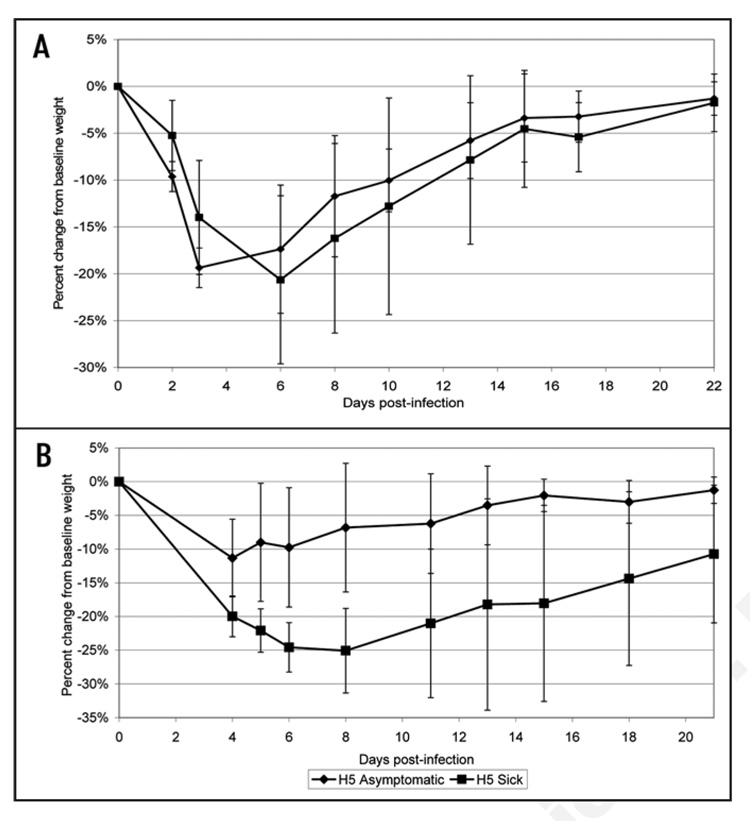
Analysis of the body weight and temperature data gathered throughout the challenge period shows that asymptomatic groups demonstrated no significant change in body temperature (data not shown). However in both trials, asymptomatic animals that had received the H5N1 synDNA™ vaccine exhibited less weight loss p.i. than sick animals (Fig. 6). This difference was not as marked in H5 synDNA™ vaccinated groups (Fig. 7); suggesting that the former vaccine was more efficient at modulating the development of influenza symptoms in mice than H5 synDNA™.
Figure 6.
Comparison of percent change in body weight between asymptomatic and sick mice following challenge with influenza A/H5N1. In two independent trials of H5N1 syn-DNA™ vaccine efficacy in mice (survival and details presented in Table 1), periodic recording of body weight was performed over the 21–22 day study period. Figure 6A and B represent the results of two independent vaccine efficacy trials. The percent change in body weight relative to baseline (day 0) was calculated for individual mice and the percent change averaged by group (asymptomatic versus sick). Each data point represents the average of (A) n = 9 mice for asymptomatic; n = 4 for sick, (B) n = 13 mice for asymptomatic; n = 1 for sick.
Figure 7.
Comparison of percent change in body weight between asymptomatic and sick mice following challenge with influenza A/H5N1. In two independent trials of H5 synDNA™ vaccine efficacy in mice (survival and details presented in Table 1), periodic recording of body weight was performed over the 21–22 day study period. Figure 7A and B represent the results of two independent vaccine efficacy trials. The percent change in body weight relative to baseline (day 0) was calculated for individual mice and the percent change averaged by group (asymptomatic versus sick). Each data point represents the average of (A) n = 4 mice for asymptomatic; n = 5 for sick, (B) n = 6 mice for asymptomatic; n = 6 for sick.
Induction of anti-HA immune responses
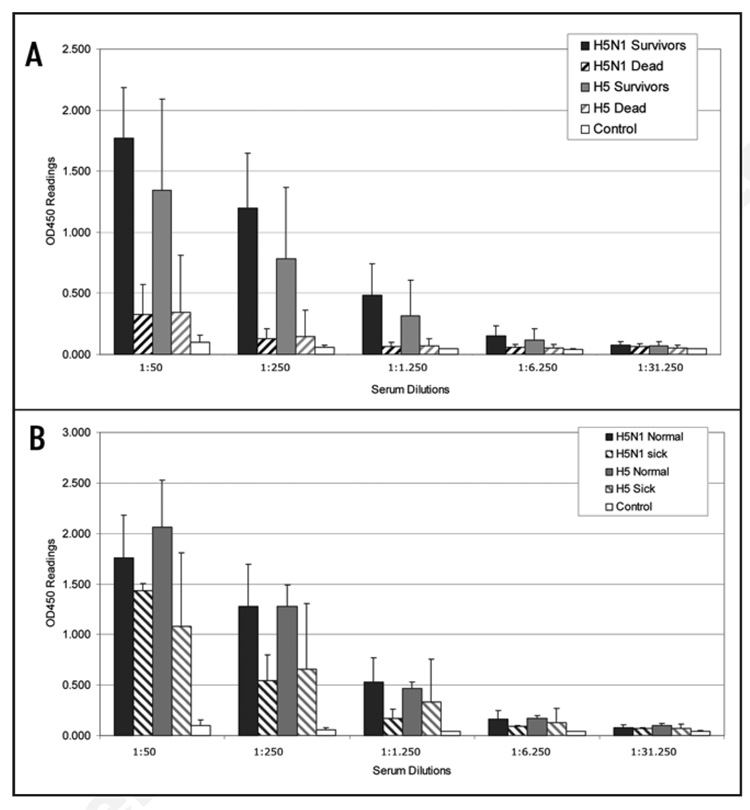
We then analyzed the levels of anti-H5 antibodies produced in synDNA™ vaccine-immunized mice in order to determine whether a correlation exists between survival after challenge and production of anti-H5 post-immunization. Sera were collected prior to influenza A/H5N1 virus challenge (at day 0, 2 weeks after the last vaccine booster) and analyzed for the presence of anti-H5 antibodies levels by ELISA. Figure 8A shows that, in both trials, all animals that survived the lethal influenza A/H5N1 virus challenge, whether immunized with the H5N1 or the H5 synDNA™ vaccine, had high anti-H5 antibody titers prior to challenge (solid bars) compared with controls. However, all the mice that later succumbed to the infection showed low anti-H5 antibody titers (hatched bars), albeit slightly higher titers than the control group. We then looked in more detail at the anti-H5 antibody titers in mice that survived the challenge without noticeable symptoms compared with mice that showed obvious signs of morbidity yet still recovered. Figure 8B shows the results of the ELISA study with respect to the disease data gathered after challenge, for both trials. It appears that the anti-H5 antibody titers in animals that showed signs of morbidity are slightly lower than in those that remained asymptomatic, however, this was not significant. This suggests that a specific anti-H5 antibody threshold must be attained to alleviate flu like symptoms in vaccinated mice.
Figure 8.
Serological analysis of synDNA™ vaccinated mice prior to H5N1 challenge (summary from trials 1 and 2). Mice were vaccinated via intramuscular (i.m.) route with the indicated synDNA™ (50 µg per mouse) or, as controls, with saline or control synDNA™, on day -42, -28 and -14 relative to H5N1 challenge. Serum from H5N1, H5 and control syn-DNA™ immunized mice was collected prior to challenge (on day 0, two weeks following third vaccination dose) and analyzed for the presence of anti-H5 antibodies by ELISA. Error bars represent standard deviations. (A) Anti-H5 antibody titers in mice that survived the lethal challenge with H5N1 virus are represented by solid bars for H5N1 synDNA™ (black; n = 27, see Table 1) and H5 synDNA™ (grey; n = 21, see Table 1). Hatched bars depict antibody titers in H5N1 and H5 synDNA™ immunized mice that did not survive the challenge (black, n = 3 and grey, n = 9, respectively. See Table 1). Anti-H5 antibody titers in mice immunized with control synDNA™ are represented by white solid bars (n = 7, for reagent sparing purposes anti-H5 antibody titers were assessed in only 6 animals from the control group in trial 1 and 1 in trial 2). (B) Comparative analysis of anti-H5 antibody titers in the pre-challenge sera of mice that survived the challenge without detectable infection symptoms (normal) and in morbid mice that recovered by the end of the trial (sick). N = 22, H5N1 normal; n = 5, H5N1 sick; n = 10, H5 normal; n = 11, H5 sick; n = 7, control (see Table 1).
Quantitative detection of H5N1 virus in organs from infected mice
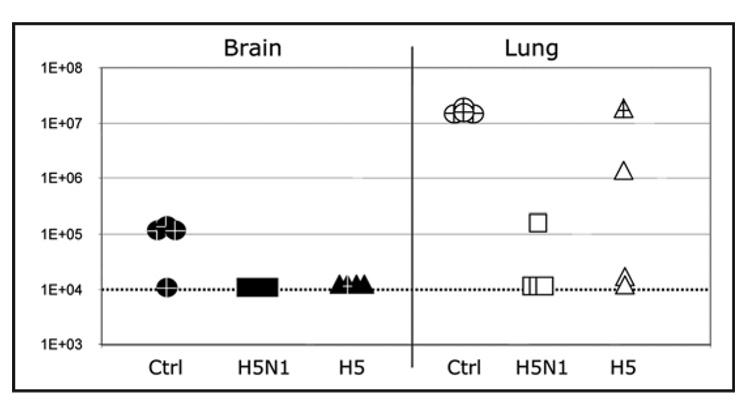
Next, viral titers in target organs from immunized mice post-challenge were studied (Trial 2, only). Four animals from each group were sacrificed on day 5 post-infection and both lung and brain tissues were collected. Viral titers in each specimen were determined by TCID50 assay (Fig. 9). In brain tissue of both H5N1 and H5 synDNA™ vaccine immunized mice, viral titers were below the detection limit of the assay (TCID50 <104/g tissue) compared with control mice where 3 out of 4 had an average 105 TCID50 per g of tissue. In 75% (3/4) of the lungs of mice immunized with H5N1 synDNA™ vaccine, virus was not detectable. However, for the single animal in this group that tested positive, the titer was ~100 times less than the control group and this animal showed no adverse symptoms of influenza infection. As for the lung samples from mice immunized with H5 synDNA™ vaccine all asymptomatic animals presented infectious viral titers significantly lower than in controls (up to 100 times less). The only mouse that showed morbidity symptoms post-challenge had a high viral titer similar to controls (open triangle). Interestingly, a single animal from the H5 synDNA™ vaccinated group showed a viral titer of 106 TCID50 per g of lung. However no virus was detected in the brain of this animal and at the time of euthanasia, no infection symptoms were detected. Together, these observations suggest that alone replication of the influenza virus in lungs to 106 TCID50 per g of tissue may not be sufficient to induce clinical disease symptoms.
Figure 9.
H5N1 virus level in the brain and lungs of synDNA™ vaccinated and virus challenged mice. In trial 2, mice were vaccinated via intramuscular (i.m.) route with the indicated synDNA™ (50 µg per mouse) or, as controls, with saline or control synDNA™, on day -42, -28 and -14 and challenged with a lethal dose of influenza A/H5N1 virus on day 0, as described in Figure 3B. At 5 days post-challenge, five mice per vaccination group were euthanized and organs collected for viral titration. Viral titer in brain (solid) and lung (open) tissues for individual mice in each H5N1 (square), H5 (triangle) and control (circle) synDNA™ immunized group are shown. Y axis represents H5N1 TCID50 per g of given tissue. Marked points (+) refer to mice that already presented signs of morbidity at day 5 post-challenge. The dotted line at TCID50 = 1E + 04 represents the limit of detection for our assay.
Discussion
The production of vaccines using inactivated or live attenuated virus are well established processes.29,30 However, the current conventional vaccine production capacity cannot fulfill the demand in the event of a potential pandemic.31 In addition, the ability of conventional production technologies, e.g., egg-based or cell-based technologies, to respond quickly should the virus strain change anti-genically during a pandemic, remains a major concern. Therefore, vaccines based on other platform technologies are being explored.7
The emergence of DNA vaccines as efficient alternatives to conventional vaccines in animals is well documented.8 The advantages of DNA vaccine include: (1) all DNA vaccines can be produced using the same process; (2) more rapid large scale production for potential pandemics is possible; and (3) DNA vaccine is highly stable and therefore, is less vulnerable to fluctuations in storage conditions—an important asset for vaccination campaigns in developing countries. In the case of influenza A/H5N1 virus, pDNA-based vaccines have been shown to induce both humoral and cellular responses, including cross-protection, in animal models.32–38 A major drawback, of pDNA vaccines, however, is the production source and method. The available technologies to produce backbone-free DNA products still rely on pDNA derived from bacteria.17–20 Thus, the removal of unwanted bacterial byproducts remains a critical quality control step, as with any therapeutics prepared from microorganisms. Recently, Vilalta et al., reported on the potency of a PCR-generated linear expression cassettes (LECs) against influenza A virus.39 However, scaling-up of PCR-based DNA production could face some challenges. We have addressed this concern by using a cell-free synthesis process (synDNA™) based on the isothermal DNA amplification process. While in principle similar to the PCR technology using a DNA polymerase to exponentially amplify a DNA template, the use of a mesophilic DNA polymerase without temperature cycling40,41 eliminates the need for specialized instrumentation and facilitates scaling-up production.
In order to illustrate the immunological activity of synDNA™ vaccine, we prepared constructs comprised solely of linear mammalian expression cassettes with no pDNA backbone sequences. Each construct was produced either as a single (H5 synDNA™) or two expression cassettes in tandem (H5N1 synDNA™). We then evaluated the efficacy of each synDNA™ construct in triggering a protective response to a lethal dose of virulent influenza A/H5N1 (Vietnam/1203/04) virus in mice. Our results indicate that the H5N1 synDNA™ and the H5 synDNA™ vaccines can protect the majority of treated mice (up to 93%). Interestingly, we also found that vaccination with the H5N1 synDNA™ vaccine and to a lesser extent, with the H5 synDNA™ vaccine, prevented the development of severe clinical disease. We had speculated that this difference may be attributed to variations in HA expression levels between the two constructs. However, in vitro expression experiments conducted in transiently transfected cells did not show significant differences in HA expression between H5N1 and H5 synDNA™ constructs. Alternatively, it is possible that the synDNA™-derived expression of HA protein alone is necessary for triggering a protective response, but not sufficient in some cases to reduce the viral load in certain tissues (such a lungs) below a critical titer necessary for preventing illness in mice. Since none of the mice immunized with the N1 synDNA™ vaccine survived the lethal challenge and because we did not detect any significant differences in the serological and organ virus titer profiles between mice immunized with the H5N1 synDNA™ and H5 synDNA™ vaccines, we speculate that the presence of the N1 expression cassette in our tandem synDNA™ vaccine H5N1 may modulate the severity of the infection symptoms. Our observations are in agreement with others suggesting that the production of antibodies against neuraminidase (NA) inhibit the enzyme activity of NA upon viral challenge thus reducing the severity of the disease.42–45
In summary, the present study shows that linear synDNA™ construct constitutes a viable alternative to pDNA in DNA vaccinations as well as conventional vaccines for preventing influenza A/H5N1-induced lethal disease in mice. Further studies are currently underway to better ascertain the mechanism of immunity triggered by synDNA™ vaccine as well as other genetic combinations in order to broaden the protection to encompass both seasonal and pre-pandemic influenza strains.
Materials and Methods
synDNA™ vaccine constructs
Construction of pCmv-H5 (CYGX-56) and pCmv-N1 (CYGX-57): Mammalian codon optimized H5 or N1 genes from influenza A/H5N1 (isolate A/Vietnam/1203/04, Accession# AAW807107 and AAT73329, respectively) were synthesized by GeneArt, AG (Regensburg, Germany). The sequences were additionally designed to have specific KpnI and XbaI sites at the 5' and 3' ends respectively, for directional cloning into the multiple cloning site of pCMV-MCS (Stratagene, La Jolla, CA, USA). Construction of pCmv-H5-Cmv-N1 (CYGX-10): CYGX-57 was digested with Not I and BamH I (New England Biolabs, Beverly, MA, USA). The 3.3 kbp fragment containing the expression cassette was subsequently end-filled with T4 DNA pol (New England Biolabs) as per manufacturers’ recommendations. This 3.3 kbp fragment was subsequently cloned downstream from the Cmv-H5 expression cassette in CYGX-56 previously digested with Pml I (New England Biolabs). Construction of pCmv-luciferase (CYGX-33): pGL3 (Promega Corp., Madison, WI, USA) was digested with Hind III and Xba I (New England Biolabs). The 1.69 kbp fragment encompassing the luciferase open reading frame was subsequently cloned into pCMV-MCS multiple cloning site using the same enzyme site as above.
synDNA™ vaccine production
The Cmv-driven expression cassettes46 H5N1 (~7 kbp), luciferase (~3.5 kbp), H5 (~4 kbp) and N1 (~3.3 kbp) (Fig. 1) were respectively excised from the plasmid vectors CYGX-10, CYGX-33, CYGX-56 and CYGX-57 using Not I restriction endonuclease (New England Biolabs), purified using Qiaquick™ Gel extraction kit (QIAGEN, Inc., Valencia, CA, USA), and circularized with T4 DNA ligase (New England Biolabs) as per manufacturer recommendations. Cell-free amplification of the re-circularized template was conducted using phi29 DNA polymerase (GE Healthcare, Piscataway, NJ, USA). Phi29 DNA polymerase is a single subunit, proofreading DNA polymerase isolated from the B. subtilis phage Φ29. Without the need for accessory proteins, this polymerase can perform ~104 polymerization cycles without dissociating from the template, incorporating on average ~70,000 nucleotides per enzyme/DNA binding event.47 Its high processivity and robust strand displacement activity, enables the process to easily reach amplification over 103-fold in 1 hr at constant temperature in vitro.40,41 The reported error rate of Phi29 DNA polymerase is 3–5 × 10−6 40,41,48 although up to 10−7 can be achieved (Kendirgi F, and Chen Y, unpublished results). According to the manufacturers’ recommendations, 10 ng of circular template (devoid of plasmid backbone) per reaction was mixed with the amplification cocktail mixes provided in a customized kit by GE Healthcare based on the Genomiphi™ HY DNA amplification kit. Following amplification, the polymerase was heat inactivated for 20 min at 65°C. The amplification concatamers were then digested into single linear expression units using the same restriction enzyme as above (~1 U/µg DNA) and purified to greater than 90% by a proprietary chromatography-based process.
The DNA concentration and purity was determined by OD260:280 ratio and integrity and homogeneity were assessed by densitometry (ImageQuant TL, GE Healthcare, Piscataway, NJ). All DNA material used in this study was >90% homogeneous and had an OD260:280 ratio between 1.80 and 1.98 (results not shown). Following purification, linear synDNA™ vaccines were recovered in isotonic citrate buffer (150 mM NaCl, 20 mM sodium citrate, pH 7) and used directly in immunization experiments described below.
Immunization and challenge
All animal studies were approved by the Institutional Animal Care and Use Committee at the University of Texas Medical Branch and were carried out according to NIH guidelines. Vaccination and implantation of transponders for telemetric temperature recording were carried out in the animal biosafety level (BSL)-2 facility at UTMB. The viral challenge was performed in the UTMB BSL-4 facility. Four-week-old female Balb/c mice (Harlan, Inc., Indianapolis, IN) weighting 12–20 g were vaccinated intramuscularly (i.m.) on day -42 with 50 µg (1 mg/ml) of the respective synDNA™ vaccines (25 µl/animal). As a control, isotonic citrate buffer (saline control) was injected by identical route as the vaccine. The animals received identical booster doses on days -28 and -14 and were then challenged intranasally (i.n.) with lethal doses of infectious virus (6.8 × 104 TCID50 in trial 1 and 5.3 × 103 TCID50 in trial 2; influenza A/H5N1 (Vietnam/1203/04; CDC Lot #2004706280) in 40 µl of PBS on day 0. As positive control for vaccine protection, survivors of previous H5N1-challenge (convalescent) were infected with H5N1 using half the virus dose of the other groups. Mouse serum samples were collected on day 0 for evaluation of anti-HA (H5) antibody levels.
Monitoring
Vaccination and H5N1 challenge studies were performed as detailed in the sections above. Animals were monitored for 21 days post-infection for death, and the development of encephalitis or paralysis. Standardized data recording was performed using the following definitions: encephalitis, development of discoordination, ataxia or transient seizures with retention of the ability to drink and feed; paralysis, hind limb (hemiplegic) or quadriplegic paralysis with the inability to reach the feeder or water bottle.3 The body temperatures and body weight were recorded via BMDS transponders using the DAS-6007 Probe (Bio Medic Data System, Inc., Seaford, DE) and using a scale tared to measure the animal’s body weight in grams, respectively. For the convalescent group, only daily survival was recorded.
Statistical analysis
Statistical analysis of survival for all groups over the 21 day monitoring period was performed using logrank test at a significant level of α < 0.05 in GraphPad® Prism (San Diego, CA). For pairwise comparison of the survival of treated and untreated (or mock-treated) groups Fisher’s Exact Test was performed at a significant level of α < 0.05 in GraphPad® Prism.
Enzyme linked immunosorbent assay (ELISA)
Microtiter plates (Costar 96 well assay plate) were coated with 100 ng of recombinant H5 protein (A/Vietnam/1203/04; Protein Sciences Corp., Meriden, CT) per well. Mouse serum samples collected on day 0 were serially diluted in PBST (phosphate-buffered saline (PBS); 0.05% Tween-20) added to the wells and incubated for 1.5 hrs at room temperature (RT). After extensive washes with PBST, adsorbed anti-H5 antibodies were detected using secondary goat anti-mouse IgG:horseradish peroxidase antibodies (The Jackson Lab, Bar Harbor, ME). Color development was then triggered with tetramethyl benzidin substrate solution and stopped by adding an equal volume of 2 molar H2SO4. Optical density was determined at 450 nm.
Transfection of cultured cells and western blotting
A549 human lung carcinoma cells (ATCC Cat. No. CCL-185, Manassas, VA) cultured in DMEM medium supplemented with 10% FCS (Invitrogen, Carlsbad, CA) were electroporated using GenePulser Xcell™ (BioRad). At 12–16 hrs following electroporation, cells were washed with ice cold PBS and directly lysed in SDS-PAGE loading buffer. Detection of HA and NA expression in the crude cells lysates was subsequently conducted, as follows. Lysates were resolved by SDS-PAGE and subsequently, proteins were transferred onto a nylon membrane (GE Healthcare), blocked with 5% BSA in PBST and subsequently incubated overnight at 4°C with anti-H5N1 goat polyclonal serum (1:500 in PBS containing 2.5% bovine serum albumin) or with affinity-purified rabbit anti-NA (1:500; Biodesign Int.). Following extensive washing with PBST, membranes were incubated a 1:7000 dilution of secondary alkaline phosphatase (AP)-conjugated donkey anti-goat or AP-donkey anti-rabbit antibodies (Promega Corp.,), followed by color development according to the manufacturer’s recommendations.
Animal tissue isolation and viral titer assessment
Organs collected at the indicated time points were sagittally sectioned in half. One half of each brain was homogenized in MEM containing 10% fetal bovine serum, to generate a 10% suspension (maintained at −80°C until further processing). The titer of infectious virus was determined, as follows.49 Median tissue culture infectious dose (TCID50) assay was performed using serial ten-fold dilutions of the tissue homogenates prepared in MEM without serum. Madin Darby canine kidney (MDCK) cells (ATCC) were grown to confluency in 96-well tissue culture plates, washed twice with 100 µl of Dulbecco’s Phosphate-Buffered Saline (DPBS), followed by inoculation of 100 µl of each tissue homogenate dilution into four replicate wells, or, as negative control, DPBS. Plates were incubated for 90 minutes at 37°C, 5% CO2, after which an additional 100 µl of MEM was added to each well. Plates were incubated for 4 days at 37°C, 5% CO2. HA assay was performed by removing 50 µl of supernatant from each well and transferring it to a 96-well plate, followed by addition of 50 µl per well of a 0.5% solution of horse erythrocytes suspended in DPBS w/Ca+ and Mg2+. Erythrocytes were allowed to settle and hemagglutination was documented for each replicate. For organ titrations, infectious virus titers were expressed in TCID50 dose per gram (g) of tissue.
Acknowledgments
The authors would like to thank Drs. Malcolm Skolnick and Cindee Ewell for their reviewing of the manuscript and Tera Guidry and John McHugh for their technical support. Dr. Slobodan Paessler was supported by faculty start-up funding provided by the Institute for Human Infections and Immunity at UTMB, a sponsored research agreement from CytoGenix, Inc and K08 Award #AI059491-01 from the National Institutes of Health. We also thank Jenna Linde for data entry and preparation of figures throughout the animal studies.
References
- 1.WHO. Cumulative number of confirmed human cases of avian influenza A/(H5N1) 2007 Reported to WHO: http://www.who.int/csr/don/en.
- 2.Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yun NE, Linde NS, Zacks MA, Barr IG, Hurt AC, Smith JN, Dziuba N, Holbrook MR, Zhang L, Kilpatrick JM, Arnold CS, Paessler S. Injectable peramivir mitigates disease and promotes survival in ferrets and mice infected with the highly virulent influenza virus, A/Vietnam/1203/04 (H5N1) Virol. 2008;373:198–209. doi: 10.1016/j.virol.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reece PA. Neuraminidase inhibitor resistance in influenza viruses. J Med Virol. 2007;79:1577–1586. doi: 10.1002/jmv.20951. [DOI] [PubMed] [Google Scholar]
- 5.Ilyushina NA, Hoffmann E, Salomon R, Webster RG, Govorkova EA. Amantadine-oseltamivir combination therapy for H5N1 influenza virus infection in mice. Antivir Ther. 2007;12:363–370. [PubMed] [Google Scholar]
- 6.Bresson JL, Perronne C, Launay O, Gerdil C, Saville M, Wood J, Hoschler K, Zambon MC. Safety and immunogenicity of an inactivated split-virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: phase I randomised trial. Lancet. 2006;367:1657–1664. doi: 10.1016/S0140-6736(06)68656-X. [DOI] [PubMed] [Google Scholar]
- 7.Subbarao K, Luke C. H5N1 viruses and vaccines. PLoS Pathog. 2007;3:40. doi: 10.1371/journal.ppat.0030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu MA, Wahren B, Karlsson Hedestam GB. DNA vaccines: recent developments and future possibilities. Hum Gene Ther. 2006;17:1051–1061. doi: 10.1089/hum.2006.17.1051. [DOI] [PubMed] [Google Scholar]
- 9.Weiner DB. Introduction to DNA vaccines issue. Vaccine. 2006;24:4459–4460. doi: 10.1016/j.vaccine.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Bergman PJ, Camps-Palau MA, McKnight JA, Leibman NF, Craft DM, Leung C, Liao J, Riviere I, Sadelain M, Hohenhaus AE, Gregor P, Houghton AN, Perales MA, Wolchok JD. Development of a xenogeneic DNA vaccine program for canine malignant melanoma at the Animal Medical Center. Vaccine. 2006;24:4582–4585. doi: 10.1016/j.vaccine.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 11.Listner K, Bentley LK, Chartrain M. A simple method for the production of plasmid DNA in bioreactors. Methods Mol Med. 2006;127:295–309. doi: 10.1385/1-59745-168-1:295. [DOI] [PubMed] [Google Scholar]
- 12.Boyle JS, Brady JL, Koniaras C, Lew AM. Inhibitory effect of lipopolysaccharide on immune response after DNA immunization is route dependent. DNA Cell Biol. 1998;17:343–348. doi: 10.1089/dna.1998.17.343. [DOI] [PubMed] [Google Scholar]
- 13.Liu L, Zhou X, Liu H, Xiang L, Yuan Z. CpG motif acts as a ‘danger signal’ and provides a T helper type 1-biased microenvironment for DNA vaccination. Immunology. 2005;115:223–230. doi: 10.1111/j.1365-2567.2005.02150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vabulas RM, Pircher H, Lipford GB, Hacker H, Wagner H. CpG-DNA activates in vivo T cell epitope presenting dendritic cells to trigger protective antiviral cytotoxic T cell responses. J Immunol. 2000;164:2372–2378. doi: 10.4049/jimmunol.164.5.2372. [DOI] [PubMed] [Google Scholar]
- 15.Paillard F. CpG: the double-edged sword. Hum Gene Ther. 1999;10:2089–2090. doi: 10.1089/10430349950017086. [DOI] [PubMed] [Google Scholar]
- 16.Chen ZY, He CY, Meuse L, Kay MA. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 2004;11:856–864. doi: 10.1038/sj.gt.3302231. [DOI] [PubMed] [Google Scholar]
- 17.Chen ZY, He CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 18.Moreno S, Lopez-Fuertes L, Vila-Coro AJ, Sack F, Smith CA, Konig SA, Wittig B, Schroff M, Juhls C, Junghans C, Timon M. DNA immunisation with minimalistic expression constructs. Vaccine. 2004;22:1709–1716. doi: 10.1016/j.vaccine.2003.09.051. [DOI] [PubMed] [Google Scholar]
- 19.Leutenegger CM, Boretti FS, Mislin CN, Flynn JN, Schroff M, Habel A, Junghans C, Koenig-Merediz SA, Sigrist B, Aubert A, Pedersen NC, Wittig B, Lutz H. Immunization of cats against feline immunodeficiency virus (FIV) infection by using minimalistic immunogenic defined gene expression vector vaccines expressing FIV gp140 alone or with feline interleukin-12 (IL-12), IL-16, or a CpG motif. J Virol. 2000;74:10447–10457. doi: 10.1128/jvi.74.22.10447-10457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schakowski F, Gorschluter M, Junghans C, Schroff M, Buttgereit P, Ziske C, Schottker B, Konig-Merediz SA, Sauerbruch T, Wittig B, Schmidt-Wolf IG. A novel minimal-size vector (MIDGE) improves transgene expression in colon carcinoma cells and avoids transfection of undesired DNA. Mol Ther. 2001;3:793–800. doi: 10.1006/mthe.2001.0322. [DOI] [PubMed] [Google Scholar]
- 21.Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, Chittaganpitch M, Waicharoen S, Nguyen DT, Nguyen T, Nguyen HH, Kim JH, Hoang LT, Kang C, Phuong LS, Lim W, Zaki S, Donis RO, Cox NJ, Katz JM, Tumpey TM. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol. 2005;79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dybing JK, Schultz-Cherry S, Swayne DE, Suarez DL, Perdue ML. Distinct pathogenesis of hong kong-origin H5N1 viruses in mice compared to that of other highly pathogenic H5 avian influenza viruses. J Virol. 2000;74:1443–1450. doi: 10.1128/jvi.74.3.1443-1450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zitzow LA, Rowe T, Morken T, Shieh WJ, Zaki S, Katz JM. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J Virol. 2002;76:4420–4429. doi: 10.1128/JVI.76.9.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leneva IA, Roberts N, Govorkova EA, Goloubeva OG, Webster RG. The neuraminidase inhibitor GS4104 (oseltamivir phosphate) is efficacious against A/Hong Kong/156/97 (H5N1) and A/Hong Kong/1074/99 (H9N2) influenza viruses. Antiviral Res. 2000;48:101–115. doi: 10.1016/s0166-3542(00)00123-6. [DOI] [PubMed] [Google Scholar]
- 25.Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley AJ, Kawaoka Y. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol. 1999;73:3184–3189. doi: 10.1128/jvi.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz JM, Lu X, Frace AM, Morken T, Zaki SR, Tumpey TM. Pathogenesis of and immunity to avian influenza A H5 viruses. Biomed Pharmacother. 2000;54:178–187. doi: 10.1016/S0753-3322(00)89024-1. [DOI] [PubMed] [Google Scholar]
- 27.Bright RA, Cho DS, Rowe T, Katz JM. Mechanisms of pathogenicity of influenza A (H5N1) viruses in mice. Avian Dis. 2003;47:1131–1134. doi: 10.1637/0005-2086-47.s3.1131. [DOI] [PubMed] [Google Scholar]
- 28.Rowe T, Cho DS, Bright RA, Zitzow LA, Katz JM. Neurological manifestations of avian influenza viruses in mammals. Avian Dis. 2003;47:1122–1126. doi: 10.1637/0005-2086-47.s3.1122. [DOI] [PubMed] [Google Scholar]
- 29.Cox MM. Cell-based protein vaccines for influenza. Curr Opin Mol Ther. 2005;7:24–29. [PubMed] [Google Scholar]
- 30.Gerdil C. The annual production cycle for influenza vaccine. Vaccine. 2003;21:1776–1779. doi: 10.1016/s0264-410x(03)00071-9. [DOI] [PubMed] [Google Scholar]
- 31.Poland GA, Jacobson RM, Targonski PV. Avian and pandemic influenza: an overview. Vaccine. 2007;25:3057–3061. doi: 10.1016/j.vaccine.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 32.Epstein SL, Tumpey TM, Misplon JA, Lo CY, Cooper LA, Subbarao K, Renshaw M, Sambhara S, Katz JM. DNA vaccine expressing conserved influenza virus proteins protective against H5N1 challenge infection in mice. Emerg Infect Dis. 2002;8:796–801. doi: 10.3201/eid0808.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jimenez GS, Planchon R, Wei Q, Rusalov D, Geall A, Enas J, Lalor P, Leamy V, Vahle R, Luke CJ, Rolland A, Kaslow DC, Smith LR. Vaxfectin(trade mark)-Formulated Influenza DNA Vaccines Encoding NP and M2 Viral Proteins Protect Mice against Lethal Viral Challenge. Hum Vaccin. 2007;3 doi: 10.4161/hv.3.5.4175. [DOI] [PubMed] [Google Scholar]
- 34.Kodihalli S, Goto H, Kobasa DL, Krauss S, Kawaoka Y, Webster RG. DNA vaccine encoding hemagglutinin provides protective immunity against H5N1 influenza virus infection in mice. J Virol. 1999;73:2094–2098. doi: 10.1128/jvi.73.3.2094-2098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kodihalli S, Kobasa DL, Webster RG. Strategies for inducing protection against avian influenza A virus subtypes with DNA vaccines. Vaccine. 2000;18:2592–2599. doi: 10.1016/s0264-410x(99)00485-5. [DOI] [PubMed] [Google Scholar]
- 36.Laddy DJ, Yan J, Corbitt N, Kobasa D, Kobinger GP, Weiner DB. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine. 2007;25:2984–2989. doi: 10.1016/j.vaccine.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharpe M, Lynch D, Topham S, Major D, Wood J, Loudon P. Protection of mice from H5N1 influenza challenge by prophylactic DNA vaccination using particle mediated epidermal delivery. Vaccine. 2007;25:6392–6398. doi: 10.1016/j.vaccine.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Tompkins SM, Zhao ZS, Lo CY, Misplon JA, Liu T, Ye Z, Hogan RJ, Wu Z, Benton KA, Tumpey TM, Epstein SL. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg Infect Dis. 2007;13:426–435. doi: 10.3201/eid1303.061125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vilalta A, Jimenez G, Rusalov D, Planchon R, Lalor P, Carner K, Chaplin JA, Komai M, Manthorpe M, Kaslow DC, Rolland A. Vaccination with polymerase chain reaction-generated linear expression cassettes protects mice against lethal influenza a challenge. Hum Gene Ther. 2007;18:763–771. doi: 10.1089/hum.2007.009. [DOI] [PubMed] [Google Scholar]
- 40.Esteban JA, Salas M, Blanco L. Fidelity of phi 29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J Biol Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
- 41.Nelson JR, Cai YC, Giesler TL, Farchaus JW, Sundaram ST, Ortiz-Rivera M, Hosta LP, Hewitt PL, Mamone JA, Palaniappan C, Fuller CW. TempliPhi, phi29 DNA polymerase based rolling circle amplification of templates for DNA sequencing. Biotechniques. 2002:44–47. [PubMed] [Google Scholar]
- 42.Johansson BE, Bucher DJ, Kilbourne ED. Purified influenza virus hemagglutinin and neuraminidase are equivalent in stimulation of antibody response but induce contrasting types of immunity to infection. J Virol. 1989;63:1239–1246. doi: 10.1128/jvi.63.3.1239-1246.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson BE, Kilbourne ED. Dissociation of influenza virus hemagglutinin and neuraminidase eliminates their intravirionic antigenic competition. J Virol. 1993;67:5721–5723. doi: 10.1128/jvi.67.10.5721-5723.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu M, Fang F, Chen Y, Wang H, Chen Q, Chang H, Wang F, Zhang R, Chen Z. Protection against avian influenza H9N2 virus challenge by immunization with hemagglutinin-or neuraminidase-expressing DNA in BALB/c mice. Biochem Biophys Res Commun. 2006;343:1124–1131. doi: 10.1016/j.bbrc.2006.03.088. [DOI] [PubMed] [Google Scholar]
- 45.Chen Z, Kadowaki S, Hagiwara Y, Yoshikawa T, Matsuo K, Kurata T, Tamura S. Cross-protection against a lethal influenza virus infection by DNA vaccine to neuraminidase. Vaccine. 2000;18:3214–3222. doi: 10.1016/s0264-410x(00)00149-3. [DOI] [PubMed] [Google Scholar]
- 46.Papadakis ED, Nicklin SA, Baker AH, White SJ. Promoters and control elements: designing expression cassettes for gene therapy. Curr Gene Ther. 2004;4:89–113. doi: 10.2174/1566523044578077. [DOI] [PubMed] [Google Scholar]
- 47.Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 48.Salas MdV M, Lazaro JM, Blanco L. In: Phi29 DNA Polymerase, a Potent Amplification Enzyme. Demidov VVaB NE, editor. DNA Amplification Norfolk, U.K.: Horizon Bioscience; 2004. [Google Scholar]
- 49.WHO. Geneva, Switzerland: WHO Manual on Animal Influenza Diagnosis and Surveillance. 2002:1–105.