Abstract
Nonalcoholic fatty liver disease (NAFLD) is a burgeoning health problem of unknown etiology that varies in prevalence among ethnic groups. To identify genetic variants contributing to differences in hepatic fat content, we performed a genome-wide association scan of nonsynonymous sequence variations (n=9,229) in a multiethnic population. An allele in PNPLA3 (rs738409; I148M) was strongly associated with increased hepatic fat levels (P=5.9×10−10) and with hepatic inflammation (P=3.7×10−4). The allele was most common in Hispanics, the group most susceptible to NAFLD; hepatic fat content was > 2-fold higher in PNPLA3-148M homozygotes than in noncarriers. Resequencing revealed another allele associated with lower hepatic fat content in African-Americans, the group at lowest risk of NAFLD. Thus, variation in PNPLA3 contributes to ethnic and inter-individual differences in hepatic fat content and susceptibility to NAFLD.
In humans, adipose tissue serves as a reservoir to limit the deposition of triglyceride (TG) in the liver and other metabolically active tissues1. The effectiveness of this buffer in protecting against the accumulation of fat in the liver varies widely among individuals: hepatic fat content ranges from less than 1% to more than 50% of liver weight in the general population2. The accumulation of excess TG in the liver, a condition known as hepatic steatosis (or fatty liver), is associated with adverse metabolic consequences, including insulin resistance and dyslipidemia3,4. In a subset of individuals hepatic steatosis promotes an inflammatory response in the liver, referred to as steatohepatitis, which can progress to cirrhosis and liver cancer3,5. Nonalcoholic fatty liver disease (NAFLD) is the most common form of liver disease in Western countries6. Approximately 10% of liver transplants performed in the United States are for cirrhosis related to NAFLD4.
Factors promoting deposition of fat in the liver include obesity, diabetes, insulin resistance, and alcohol ingestion3,6. The propensity to develop hepatic steatosis differs among ethnic groups, with African-Americans having the lower (24%) and Hispanics a higher (45%) frequency of the disorder than European-Americans (33%) in a large US urban population3. Hispanics also have a higher prevalence of steatohepatitis and cirrhosis, whereas African-Americans are less prone to develop liver failure2,7–9. The factors responsible for these ethnic differences in prevalence of hepatic steatosis and liver injury are not known.
To identify DNA sequence variations that contribute to inter-individual differences in NAFLD, we performed a genome-wide survey of nonsynonymous (NS) sequence variations in a multiethnic population-based study, the Dallas Heart Study10. We limited our analysis to NS sequence variations to screen directly the variants with a higher likelihood of affecting gene function. Hepatic fat content was measured in the Dallas Heart Study using proton magnetic resonance spectroscopy (1H-MRS), the most accurate, quantitative noninvasive method available2,11,12. Of the 12,138 NS variants assayed using chip-based oligonucleotide hybridization13, 9,229 exceeded the quality control threshold for the study (see METHODS) and were included in the analysis.
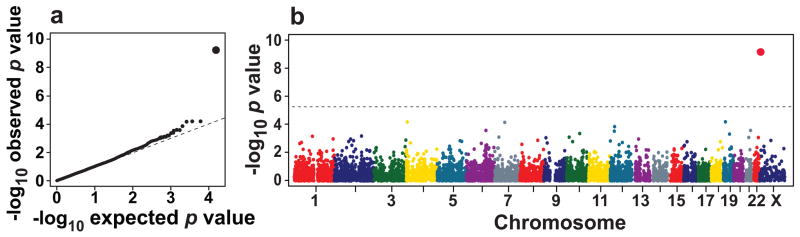
Each variant was tested for association with hepatic fat content in the 1,032 African-American, 696 European-American and 383 Hispanic study participants in the Dallas Heart Study who obtained 1H-MRS of the liver2. To maximize statistical power, the three ethnic groups were pooled and a global ancestry score (calculated using a panel of 2,270 ancestry informative SNPs) was included in the model to control for population stratification (see METHODS). The quantile-quantile plot of P-values showed no systematic deviation from the null distribution (Fig. 1a).
Figure 1.
Whole-genome scan of liver triglyceride content measured by proton magnetic resonance imaging in the Dallas Heart Study (n=2,111). (a) Quantile-quantile plot of P-values. (b) Scatter plot of P-values. The dashed line denotes the Bonferroni corrected significance threshold (P=5.4×10−6).
A single variant in PNPLA3 (rs738409) was strongly associated with hepatic fat content (P=5.9×10−10) (Fig. 1b). The variant is a cytosine to guanine substitution that changes codon 148 from isoleucine to methionine; this residue is highly conserved in vertebrates (Fig. 2a). PNPLA3 encodes a 481 amino acid protein of unknown function that belongs to the patatin-like phospholipase family14. The progenitor of this family, patatin, is a major protein of potato tubers and has nonspecific lipid acyl hydrolase activity15,16. None of the other NS sequence variants tested in the genome-wide scan exceeded the Bonferroni-corrected threshold for significance (P=5.4×10−6) (Fig. 1b).
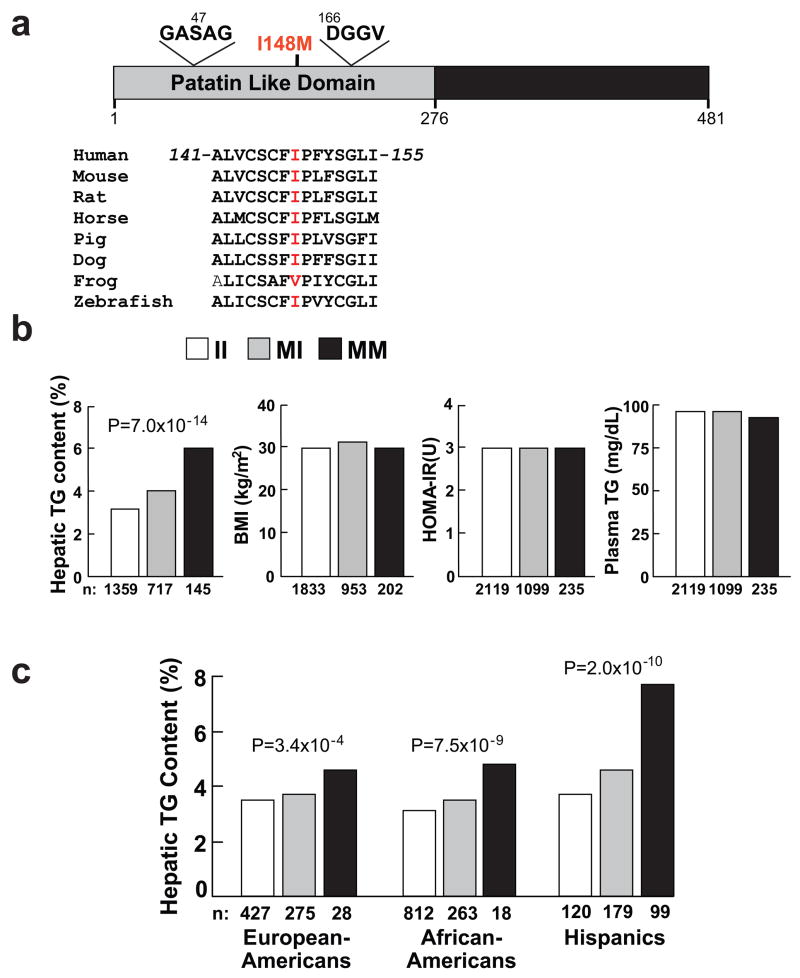
Figure 2.
Association between a sequence variant in PNPLA3 (I148M) and hepatic triglyceride (TG) content. (a) PNPLA3 is a 481 residue protein that contains a patatin-like domain at the N terminus (residues 10-179, UniProt http://pir.uniprot.org/uniprot/) with consensus sequences for a Ser-Asp catalytic dyad (Gly-X-Ser-X-Gly and Asp-X-Gly/Ala)14. The I148M substitution (rs738409) is located between the consensus sequences for the catalytic dyad and is highly conserved. (b) Median hepatic TG content, body mass index (BMI), homeostatic model assessment of insulin resistance (HOMA-IR) and plasma TG levels in the Dallas Heart Study. Values for hepatic fat content were compared using ANOVA. Age, gender, BMI, diabetes status, ethanol use and global ancestry were included as covariates in the model. (c) Median hepatic fat contents and PNPLA3-I148M genotypes in European-Americans, African-Americans and Hispanics in the Dallas Heart Study. Associations between hepatic fat content and PNPLA3-I148M genotypes were tested using ANOVA with age, local ancestry, gender, diabetes status, ethanol intake and BMI as covariates.
The association between PNPLA3-I148M and hepatic fat content remained highly significant (P=7.0 × 10−14) after adjusting for BMI, diabetes status, ethanol use, as well as global and local ancestry (Fig. 2b), and was apparent in all three ethnic groups (Fig. 2c and Supplementary Table 1 online). Thus, the association between rs738409 and hepatic fat content was not attributable either to the effect of known risk factors for liver fat accumulation or to population stratification.
The frequencies of the PNPLA3-148M allele were concordant with the relative prevalence of NAFLD in the three ethnic groups2; the highest frequency of the allele was in Hispanics (0.49), with lower frequencies observed in European Americans (0.23) and African-Americans (0.17). Accordingly, we examined the relationship between PNPLA3-I148M and evidence of hepatic inflammation, as indicated by release of liver enzymes into the circulation. A significant elevation in serum levels of alanine aminotransferase (ALT) was found in association with the PNPLA3-148M allele (P=3.7 × 10−4). Analysis of the three ethnic groups revealed that the association with ALT was limited to the Hispanics (P=1.3 × 10−5), the group with the greatest prevalence of hepatic steatosis and susceptibility to cirrhosis (Supplementary Table 1 online)2,7. The PNPLA3-148M allele was also associated with serum aspartate aminotransferase levels in Hispanics (P=0.002). These findings are consistent with our prior observation that a higher proportion of Hispanics with hepatic steatosis have evidence of hepatic inflammation7, and suggests that PNPLA3-148M allele adversely affects liver function.
Increased hepatic fat content is associated with insulin resistance and dyslipidemia [increased plasma levels of TG and lower levels of high density lipoprotein-cholesterol (HDL-C)], but the causal nature of these relationships remains poorly defined3. No association was found between the PNPLA3-148M allele and body mass index (BMI) or indices of insulin sensitivity, including fasting glucose and insulin (Supplementary Table 1 online) or homeostatic model assessment of insulin resistance (HOMA-IR) in the Dallas Heart Study (Fig. 2b). No associations were observed between PNPLA3 genotype and plasma levels of TG (Fig. 2b), total cholesterol, HDL-C or low density lipoprotein (LDL)-C (Supplementary Table 1 online). A corresponding analysis in a larger, biracial sample (n=14,821), the Atherosclerosis Risk in Communities Study (Supplementary Table 2 online)17, also revealed no association of PNPLA3-I148M with BMI, indices of insulin sensitivity, or plasma levels of TG or HDL-C (Supplementary Table 2 online). Based on the observed associations between both the SNP and hepatic fat and between hepatic fat and HOMA-IR in the Dallas Heart Study, the power to detect an association with HOMA-IR was >96% in the African-Americans and 91% in the European-Americans in ARIC.
The data from these studies indicate that the PNPLA3-148M allele is associated with a systematic increase in hepatic fat content but not with major alterations in glucose homeostasis or lipoprotein metabolism. Thus, increased hepatic fat content does not inevitably lead to insulin resistance, which is consistent with recent observations in some animal models18,19.
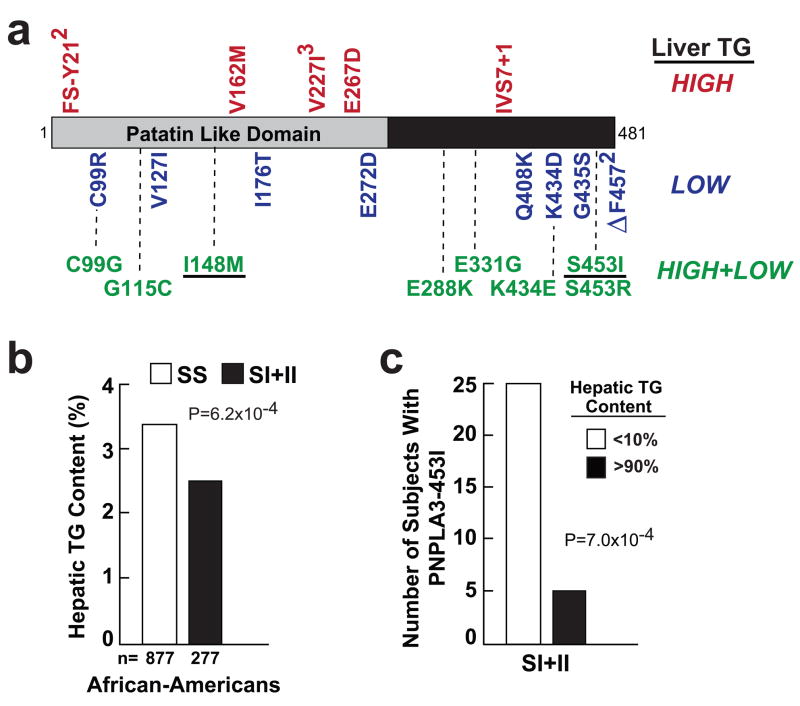
To determine if other sequence variations in PNPLA3 contribute to differences in hepatic fat content, we resequenced the coding region of PNPLA3 in the 80 men (32 African-Americans, 32 European-Americans, and 16 Hispanics) and 80 women who had the highest levels of hepatic fat in the Dallas Heart Study, and in a sex- and ethnicity-matched group with the lowest levels2. The number of individuals with NS variants found only in the high group (n=8) was similar to the number found only in the low group (n=9), but the three subjects with likely null mutations (Fs-Y21 and IVS7+1) were all in the high group (Fig. 3a), which is consistent with loss-of-function of PNPLA3 causing an increase in hepatic TG content.
Figure 3.
Identification of a PNPLA3 allele (S453I) associated with lower hepatic fat content in African-Americans in the Dallas Heart Study. (a) Exons and flanking introns of PNPLA3 were sequenced in the 32 European-American and African-American men and women and in the 16 Hispanic men and women with the lowest and highest hepatic triglyceride content determined using proton magnetic resonance imaging2. The NS variations identified in individuals in only the high, only the low and in both the high and low groups are shown. All the variants not found in both groups were present in only a single subject unless otherwise indicated. The rs numbers for polymorphisms and the oligonucleotides used for PCR-sequencing of the coding regions and are provided in Supplementary Tables 3 online and Supplementary Table 4 online. (b) Median hepatic TG content in African-Americans carrying the wild-type or PNPLA3-453I allele. (c) Number of individuals with PNPLA3-453I in the upper and lower deciles of hepatic fat content.
Eight variants were present in both the low and the high hepatic fat groups (Fig. 3a and Supplementary Table 3 online) and the six most common of these sequence variations were genotyped in the entire sample. One variant, PNPLA3-S453I, that was common in African-Americans (MAF=0.104) but rare in European-Americans (0.003) and Hispanics (0.008) (Supplementary Table 3 online) was associated with a significantly lower liver fat content. Median hepatic TG content was 18% lower in African-Americans with the PNPLA3-453I allele when compared to African-Americans homozygous for the wild-type allele (3.3% versus 2.7%, P=6.0 × 10−4) (Fig. 3b). Further evidence that the variant was associated with lower hepatic fat content was the finding that a significantly greater number of individuals with the PNPLA3-453I allele had a hepatic fat content in the lowest decile when compared to the highest decile of the population (Fig. 3b). No significant difference in the number of individuals identified in the extremes was found for any of the other SNPs (data not shown).
The effect of PNPLA3-S453I on hepatic fat content was independent of the PNPLA3-I148M polymorphism. Both variants were statistically significant when included in a multiple regression model and the S453I was significantly associated with hepatic fat in African-Americans homozygous for the PNPLA3-148I allele (data not shown). The identification of a second allele of PNPLA3 (i.e. S453I) that was independently associated with hepatic fat content further supports a role for PNPLA3 in determining hepatic TG levels, and indicates the presence of both loss-of-function and gain-of-function alleles at this locus. The mechanisms by which these alleles affect hepatic fat content are not known.
The frequencies of both PNPLA3-148M and of PNPLA3-453I in the three ethnic groups represented in the Dallas Heart Study are concordant with ethnic differences in hepatic fat content2. Exclusion of the individuals carrying either of these two alleles (148M and 453I) substantially attenuated the differences in hepatic fat content between the ethnic groups; regression analysis indicated that these two sequence variations accounted for 72% of the observed ethnic differences in hepatic fat content in the Dallas Heart Study. Thus, genetic variation in PNPLA3 accounts for a large fraction of the ethnic differences in the propensity to accumulate excess fat in the liver.
The physiological substrate(s) of PNPLA3 has not been defined. Expression of PNPLA3 is under metabolic control in adipose tissue and the liver; mRNA levels are low in the fasted state and increase dramatically with carbohydrate feeding20,21. PNPLA3 structurally resembles calcium-independent phospholipase A2 but the recombinant protein has low phospholipase activity when expressed in insect (Sf9) cells22. PNPLA3 has more robust activity against TG in vitro and can also transfer fatty acids to and from mono- and diacylglycerol22. It is not known if the major effect of PNPLA3 in the liver is to hydrolyze TG or to transfer fatty acids between lipids (transacetylation). Studies are in progress to determine the specific effects on lipid metabolism of the NS variants in PNPLA3 identified in this study.
Currently, we cannot accurately predict which individuals with fatty liver will develop steatohepatitis and progress to cirrhosis and liver failure. The finding that markers of liver inflammation (serum levels of liver-derived enzymes) were elevated in PNPLA3-148M carriers, which was also observed in an independent genome-wide association study23, suggests that this genetic variant may confer increased susceptibility to hepatic injury. Patatin-like phospholipase family members in other organisms are up-regulated in response to environmental insults24. The sequence variations we have identified in PNPLA3 may provide predictive information regarding the risk of developing hepatic steatosis and liver injury in response to environmental stresses such as caloric excess, infections, or drugs.
METHODS
Study populations
The Dallas Heart Study is a population-based probability sample of Dallas County. The sampling frame and the study design are described in detail in Victor et al.10. African-Americans were over-sampled (52% African American, self-identified as ‘black’, 29% European American, self-identified as ‘white’, 17% Hispanic self identified as ‘hispanic’ and 2% other ethnicities). The institutional review board of University of Texas Southwestern Medical Center approved the study and all study subjects provided written informed consent. Alcohol consumption was determined according to answers to previously validated questions2. Blood pressure, height, weight and BMI and calculated variables were measured as described10. Fasting blood samples were obtained from 3,551 subjects (ages 30–65) and 2,971 of these individuals completed a clinic visit; hepatic TG content was measured using 1H-MRS in 2,240 African-Americans, European-Americans and Hispanics7,12.
The association between PNPLA3-I148M and metabolic phenotypes were also examined in the Atherosclerosis Risk in Communities Study (ARIC), a large prospective study that focuses on cardiovascular disease in European-Americans and African-Americans. Details of the ARIC study design and the methods used to measure plasma lipid levels have been published previously17,25. The data used in this analysis was collected from the baseline examination.
Whole-genome association and other statistical methods
A genome-wide association analysis was performed using 12,138 NS sequence variations from dbSNP and the Perlegen SNP database (available on request). SNPs were assayed in 3,383 Africans-American, Caucasian and Hispanic participants of the Dallas Heart Study using high-density oligonucleotide arrays (Perlegen Sciences, Mountain View, CA). SNPs that met any of the following criteria were excluded (n=2,623): error probability > 20%, genotype call rate < 80%, or a significant deviation from Hardy-Weinberg Equilibrium (p-value <0.0001). Of the 9,515 SNPs that were successfully assayed, 286 were monomorphic in the Dallas Heart Study sample. The remaining 9,229 variants were tested for association with hepatic fat content in the 2,111 African-Americans, Caucasians and Hispanic subjects in the Dallas Heart Study who underwent 1H-MRS of the liver2 and in whom ancestry-informative SNPs had been assayed previously; global and local ancestries were inferred for each individual using STRUCTURE26 under a linkage model with 2,270 ancestry-informative SNPs27. We pooled all participants together and inferred global ancestry (the probability of an individual belonging to a given cluster) setting the number of clusters K equal to 3. Although the ancestry-informative SNP panel was primarily designed for African-Americans [the mean multipoint information content28 (IC̄) = 0.82], it was adequately informative in European-Americans (IC̄ = 0.63) and Hispanics (IC̄=0.66). We also inferred local ancestry as the probability that a particular genomic region belonged to a given cluster. The results were almost identical when ancestry adjustment was performed with the same SNPs using principal components analysis (data not shown).
The statistical significance of 9,229 SNPs in the whole genome association study was assessed using analysis of variance (ANOVA). To accommodate confounding factors, we included age, sex, and global ancestry as covariates in the model. The additive effect of each variant was tested by encoding the genotype variable as 0, 1, and 2. Since the distribution of hepatic fat levels is highly skewed, a power transformation (λ=1/4) was applied to the trait before the analysis. To account for multiple testing, we adjusted the significance threshold for the number of tests performed using the Bonferroni method. SNPs with a nominal P-value < 5.4×10−6 were considered significant on a genome-wide scale.
The association between PNPLA3 variants and hepatic fat content within each ethnic group was tested using ANOVA, including age, gender, BMI, diabetes status, ethanol use and local ancestry as covariates. Individuals whose genetic ancestry was not consistent with their self-reported ancestry (n = 11, 5, and 16 for African-Americans, European-Americans and Hispanics, respectively) and had a fractional ancestry that was more than 3 times the inter-quartile range below the 25th percentile for their reference group were excluded from the analysis. Because the distribution of hepatic TG content is skewed, we reported medians and inter-quartile ranges.
The association of PNPLA3-I148M with BMI, HOMA-IR and plasma TG levels was analyzed in the African-Americans, Caucasians and Hispanics together using the ANOVA including age, gender, and local ancestry as covariates. HOMA-IR was adjusted for BMI and plasma TG levels were adjusted for BMI and diabetes.
To determine the contribution of the sequence variation we identified in PNPLA3 to the ethnic differences in hepatic fat content, we examined the proportion of variance explained by ancestry (R1) using a linear model. We then determined the proportion of variance explained by ancestry after adjusting for the PNPLA3 genotypes (148M and 453I) (R2). The proportion of variance due to ancestry and explained by 148M and 453I was determined from (R1 - R2)/R1.
Resequencing PNPLA3
The exons and flanking introns of PNPLA3 were sequenced as described previously29 in the African-American, European-American and Hispanic men and women in the Dallas Heart Study with the highest and lowest hepatic TG content. Oligonucleotide primers used for sequencing are shown in Supplementary Table 4 online. All sequence variants identified were verified by manual inspection of the chromatograms and missense changes were confirmed by an independent resequencing reaction.
Genotyping assays
Fluorogenic 5′-nucleotidase assays were developed for PNPLA3-I148M and for the sequence variants identified in both the high and low hepatic TG groups in the resequencing experiments. Sequence variations in PNPLA3 were assayed using the TaqMan assay system (Applied Biosystems) on a 7900HT Fast Real-Time PCR instrument. Probes and reagents were purchased from Applied Biosystems.
Supplementary Material
Acknowledgments
We thank Tommy Hyatt, Joel Martin, Wendy Schackwitz, Anna Ustaszewska, Crystal Wright and the team at Perlegen Sciences for excellent technical assistance. We thank Kim Lawson for the statistical analysis of the data from the ARIC study. We thank Jay Horton and David Hinds for helpful discussions. We are grateful to the staff and participants of the Dallas Heart Study and the Atherosclerosis Risk in Communities Study for their contributions.
This work was supported by grants from the Donald W. Reynolds Foundation, the National Institutes of Health (RL1HL-092550, 1PL1DK081182 and HL-20948), NHLBI Program for Genomic Applications (HL-066681) and the U.S. Department of Energy (Contract DE-AC02-05CH11231).
Footnotes
AUTHOR CONTRIBUTIONS
H.H.H., J.C.C., E.B., L.A.P. and D.C. conceived, designed and directed the study; J.K., S.R., C.X. and A.P. performed and interpreted the genetic analysis. All the authors approved the final manuscript and contributed critical revisions to its intellectual content.
References
- 1.Wang MY, et al. Adipogenic capacity and the susceptibility to type 2 diabetes and metabolic syndrome. Proc Natl Acad Sci U S A. 2008;105:6139–44. doi: 10.1073/pnas.0801981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browning JD, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–95. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 3.Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–52. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCullough AJ. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin Liver Dis. 2004;8:521–33. viii. doi: 10.1016/j.cld.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Adams LA, et al. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–21. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 6.de Alwis NM, Day CP. Non-alcoholic fatty liver disease: the mist gradually clears. J Hepatol. 2008;48(Suppl 1):S104–12. doi: 10.1016/j.jhep.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Browning JD, Kumar KS, Saboorian MH, Thiele DL. Ethnic differences in the prevalence of cryptogenic cirrhosis. Am J Gastroenterol. 2004;99:292–8. doi: 10.1111/j.1572-0241.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 8.Clark JM, Brancati FL, Diehl AM. The prevalence and etiology of elevated aminotransferase levels in the United States. Am J Gastroenterol. 2003;98:960–7. doi: 10.1111/j.1572-0241.2003.07486.x. [DOI] [PubMed] [Google Scholar]
- 9.Caldwell SH, Harris DM, Patrie JT, Hespenheide EE. Is NASH underdiagnosed among African Americans? Am J Gastroenterol. 2002;97:1496–500. doi: 10.1111/j.1572-0241.2002.05795.x. [DOI] [PubMed] [Google Scholar]
- 10.Victor RG, et al. The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol. 2004;93:1473–80. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 11.Longo R, et al. Proton MR spectroscopy in quantitative in vivo determination of fat content in human liver steatosis. JMRI. 1995;5:281–285. doi: 10.1002/jmri.1880050311. [DOI] [PubMed] [Google Scholar]
- 12.Szczepaniak LS, et al. Magnetic resonance spectroscopy to measure hepatic triglyceride content: prevalence of hepatic steatosis in the general population. Am J Physiol Endocrinol Metab. 2004;288:462–468. doi: 10.1152/ajpendo.00064.2004. [DOI] [PubMed] [Google Scholar]
- 13.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–9. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 14.Wilson PA, Gardner SD, Lambie NM, Commans SA, Crowther DJ. Characterization of the human patatin-like phospholipase family. J Lipid Res. 2006;47:1940–1948. doi: 10.1194/jlr.M600185-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Rydel TJ, et al. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a ser-asp catalytic dyad. Biochem. 2003;42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- 16.Strickland JA, Orr GL, Walsh TA. Inhibition of Diabrotica larval growth by patatin, the lipid acyl hydrolase from potato tubers. Plant Physiol. 1995;109:667–674. doi: 10.1104/pp.109.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ARIC manual of operations: No 2, cohort component procedures. ARIC Coordinating Center, School of Public Health, University of North Carolina; Chapel Hill: 1987. [Google Scholar]
- 18.Minehira K, et al. Blocking VLDL secretion causes hepatic steatosis but does not affect peripheral lipid stores or insulin sensitivity in mice. J Lipid Res. 2008;49:2038–2044. doi: 10.1194/jlr.M800248-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monetti M, et al. Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab. 2007;6:69–78. doi: 10.1016/j.cmet.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Baulande S, Lassnier F, Lucas M, Pairault J. Adiponutrin, a transmembrane protein corresponding to a novel dietary - and obesity-linked mRNA specifically expressed in the adipose lineage. J Biol Chem. 2001;276:33336–33344. doi: 10.1074/jbc.M105193200. [DOI] [PubMed] [Google Scholar]
- 21.Lake AC, et al. Expression, regulation, and triglyceride hydrolase activity of adiponutrin family members. J Lipid Res. 2005;46:2477–2487. doi: 10.1194/jlr.M500290-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Jenkins CM, et al. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 23.Yuan X, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. doi: 10.1016/j.ajhg.2008.09.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rietz S, Holk A, Scherer GF. Expression of the patatin-related phospholipase A gene AtPLA IIA in Arabidopsis thaliana is up-regulated by salicylic acid, wounding, ethylene, and iron and phosphate deficiency. Planta. 2004;219:743–53. doi: 10.1007/s00425-004-1275-9. [DOI] [PubMed] [Google Scholar]
- 25.Brown SA, et al. Plasma lipid, lipoprotein cholesterol, and apoprotein distributions in selected U.S. communities. Arteriscler Thromb. 1993;13:1139–1158. doi: 10.1161/01.atv.13.8.1139. [DOI] [PubMed] [Google Scholar]
- 26.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–87. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith MW, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–13. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu X, et al. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005;37:177–81. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 29.Romeo S, et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.