Abstract
SRC-3/AIB1 is a master growth coactivator and oncogene, and phosphorylation activates it into a powerful coregulator. Dephosphorylation is a potential regulatory mechanism for SRC-3 function but the identity of such phosphatases remains unexplored. Herein, we report that using functional genomic screening of human Ser/Thr phosphatases targeting SRC-3’s known phosphorylation sites, the phosphatases PDXP, PP1 and PP2A were identified to be key negative regulators of SRC-3 transcriptional coregulatory activity in steroid receptor signalings. PDXP and PP2A dephosphorylate SRC-3 and inhibit its ligand-dependent association with estrogen receptor. PP1 stabilizes SRC-3 protein by blocking its proteasome-dependent turnover through dephosphorylation of two previously unidentified phosphorylation sites (Ser101 and S102) required for activity. These two sites are located within a degron of SRC-3, and are primary determinants of SRC-3 turnover. Moreover, PP1 regulates the oncogenic cell proliferation and invasion functions of SRC-3 in breast cancer cells.
Introduction
Steroid receptors are ligand-regulated transcription factors that control a diverse array of normal physiological processes and disease states including cancer (Mangelsdorf et al., 1995; Tsai and O’Malley, 1994). Estrogen receptor (ER) is a well-known steroid receptor that plays a critical role in breast cancer development, and its functions are primarily mediated by steroid receptor coactivator 3 (SRC-3/AIB1/ACTR/p/CIP/TRAM-1/RAC3)(Anzick et al., 1997; Chen et al., 1997; Glass and Rosenfeld, 2000; Halachmi et al., 1994; Li et al., 1997; McKenna et al., 1999; McKenna and O’Malley, 2002; Takeshita et al., 1997; Torchia et al., 1997).
SRC-3 is a member of the SRC family that also includes SRC-1 (Onate et al., 1995) and SRC-2/TIF2/GRIP1 (Hong, Kohli et al. 1996; Voegel, Heine et al. 1996). SRC-3 was discovered to be overexpressed or amplified in a majority of breast tumors (Anzick, Kononen et al. 1997). Later studies using mouse models further defined SRC-3 as an important oncogene (Kuang, Liao et al. 2004; Torres-Arzayus, Font de Mora et al. 2004) and a master regulator with many other functions (Wu, Qin et al. 2002; Louie, Zou et al. 2004; Zhou, Yan et al. 2005; Yan, Yu et al. 2006; Yu, York et al. 2007). SRC-3’s distinct functions are regulated by chemically diverse posttranslational modification (PTM) “codes” including phosphorylation, methylation, acetylation, SUMOylation, ubiquitination and many other modifications (Chen et al., 1999; Feng et al., 2006; Font de Mora and Brown, 2000; Naeem et al., 2007; Wu et al., 2006; Wu et al., 2007; Wu et al., 2005). Deciphering SRC-3’s PTM functional codes is crucial for understanding the phenotypic biology of SRC-3 (Lonard et al., 2007).
SRC-3 protein is phosphorylated in cancer cells. Six Ser/Thr phosphorylation sites have been reported previously, which are targeted by different kinases and respond to a variety of stimuli such as steroid hormones, cytokines, and growth factors (Wu, Smith et al. 2005; Zheng, Wu et al. 2005; Wu, Feng et al. 2007). Phosphorylation at distinct sites results in specific coactivator complex formation which in turn determines differential activation of target genes (Wu, Smith et al. 2005; Wu, Feng et al. 2007). Moreover, phosphorylation converts SRC-3 from an inactive state into a potent transcriptional activator; during this process, it also promotes SRC-3 ubiquitination and ultimately leads to its degradation (Wu, Smith et al. 2005; Lonard and O’Malley B 2007; Wu, Feng et al. 2007). Interestingly, the oncogenic potential of SRC-3 was reported to be closely linked to the phosphorylation state of SRC-3 (Wu et al., 2005).
Protein phosphorylation is a reversible PTM and the dynamic interplay between kinases and phosphatases determines the steady-state of phosphorylation and activation of a protein. Following completion of a phospho-dependent stimulation, phosphatases are likely to restore a cell to a pre-stimulated homeostatic state. Ser/Thr phosphatases are mainly responsible for removal of phosphate from phosphorylated Ser/Thr. They are classified into three structurally distinct families: PPM, PPP, and FCP/SCP (Cohen, 2003; Gallego and Virshup, 2005). The members of PPM and FCP/SCP families are generally Mg2+-dependent phosphatases while the PPP family is represented by PP1 and PP2A; the latter do not require Mg2+ for their activities and are sensitive to the phosphatase inhibitor okadaic acid (Cohen, 2003).
In contrast to the well-studied regulation of SRC-3 phosphorylation by kinases, virtually nothing is known about the roles that phosphatases play in the regulation of SRC-3 function. Using a functional genomic approach, we identified and characterized three critical phosphatases, PDXP, PP1 and PP2A, that were observed to target either known or two new phosphorylation sites of SRC-3. Importantly, these two phosphorylation sites in SRC-3 are located in a 26S proteasome degron that directs its degradation. These three phosphatases also appear important for ER/SRC-3 signaling, regulating SRC-3 activity by either inhibiting interactions between ER and SRC-3 or blocking 26S proteasome-dependent turnover of SRC-3. Our results provide essential evidence for the key role of phosphatases in ER/SRC-3 signaling.
Results
A Functional Genomic Screen Identifies Phosphatases Targeting the Known Phosphorylation Sites of SRC-3 and Important for Regulating Its Functions
To identify phosphatases which can dephosphorylate SRC-3, we took a functional genomic approach: screening a Ser/Thr phosphatase library (Lin, Duan et al. 2006) available in the human genome database. In 293T cells, we carried out transient transfections of expression vectors for each phosphatase and SRC-3 as well as ER. Using well-characterized phosphorylation specific antibodies against five Ser and one Thr phosphorylation site of SRC-3, we identified 15 candidate phosphatases: PPM1H, PSPH, PDXP, TAPP2C, SCP1, SCP2, SCP4, FCP1, PP1, PP2A, PP2B, PP5, PPM1G, PPM1A, and PPM1B. Each candidate was capable of dephosphorylating a certain subset of SRC-3’s six known phosphorylation sites (data not shown).
To assess the functional activities of these 15 candidate phosphatases in SRC-3-mediated ER signaling, we utilized 3 types of transcriptional luciferase reporter gene assays to eliminate the effect on ER and general transcription factors. The results from reporter gene assays uncovered inhibitory functions of multiple candidate phosphatases (Figure S1A, B).
PDXP, PP1 and PP2A appeared to inhibit ERE-luc activities in a dose-dependent manner in the presence both of ER and SRC-3 but showed no appreciable effect on ER activity in the absence of SRC-3 (data not shown) or on Gal4-VP16 activation of pG5-luc (Figure S1B). Therefore, these results suggested that PDXP, PP1 and PP2A could play specific roles in dephosphorylating SRC-3 and inhibiting its transcriptional activities.
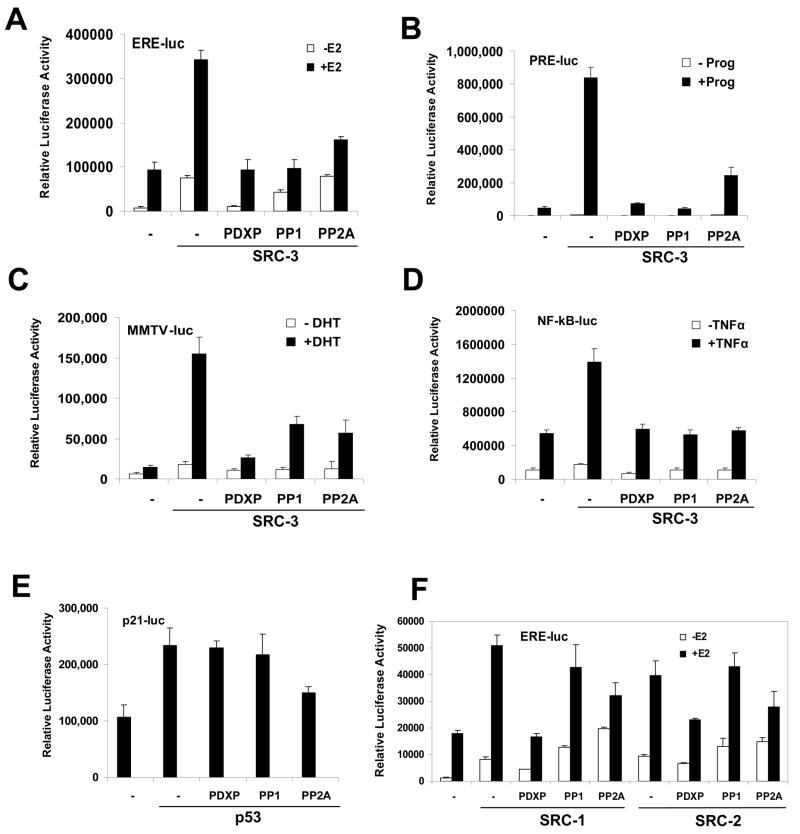
Since SRC-3 was identified as a coactivator for steroid receptors such as ER, PR and AR, we examined effects of the three phosphatases PDXP, PP1 and PP2A on PR and AR luciferase reporter activities in addition to ER (Fig 1A). As shown in Fig 1B and C, PDXP, PP1 and PP2A also inhibited SRC-3 coactivation of PR and AR, suggesting the important role of phosphorylation/dephosphorylation in SRC-3 functions in steroid signaling. SRC-3 also is known to be involved in other signaling pathways. One example is enhancement of NF-κB target gene activities (Wu et al., 2002). We carried out NF-κB luciferase reporter assays and found that PDXP, PP1 and PP2A all were able to inhibit the SRC-3-enhanced reporter activities (Fig 1D). Next we asked whether these phosphatases have some selectivity and fail to inhibit certain transcription factor activations. Using p53’s target gene p21 promoter-driving luciferase reporter (p21-luc), cotransfection of either PDXP or PP1 did not exhibit a significant effect on the reporter activity (Fig 1E), while PP2A appeared partially to repress this reporter. This result was consistent with a previous finding that human SRC-3 is not a coactivator for p53 (Lee et al., 1999).
Figure 1. Ser/Thr Phosphatases PDXP, PP1 and PP2A Inhibit the Transcriptional Activities of SRC-3.
(A) PDXP, PP1 and PP2A inhibit SRC-3 coactivation in the Estrogen Responsive Element (ERE) luciferase reporter. ERE-luc luciferase assays were carried out with ER only, or together with SRC-3, and an additional phosphatase as indicated, in the presence (+E2) or absence (−E2) of estradiol. Error bars indicate the standard error of the mean.
(B) PDXP, PP1 and PP2A inhibit SRC-3 coactivation in the Progesterone Responsive Element (PRE) luciferase reporter. Same as (A) except using PR, PRE-luc and progesterone (Prog). Error bars indicate the standard error of the mean.
(C) PDXP, PP1 and PP2A inhibit SRC-3 coactivation in the Androgen Responsive Element luciferase reporter (MMTV-luc). Same as (A) except using AR, MMTV-luc and dihydrotesterone (DHT). Error bars indicate the standard error of the mean.
(D) PDXP, PP1 and PP2A inhibit SRC-3 coactivation in the NF-κB responsive luciferase reporter (NF-κB-luc). Similar to (A) except using NF-κB-luc and TNFα (TNFα). Error bars indicate the standard error of the mean.
(E) PDXP and PP1 show no inhibition of p21 luciferase reporter (p21-luc) activated by p53. Same as (D) except using p21-luc and p53 (p53). Error bars indicate the standard error of the mean.
(F) Differential effects of PDXP, PP1 and PP2A on SRC-1 and SRC-2 coactivation activities. Same as (A) except using SRC-1 and SRC-2. Error bars indicate the standard error of the mean.
We next assessed whether there are similar effects of PDXP, PP1, and PP2A on other members of the SRC family including SRC-1 and SRC-2. PDXP and PP2A inhibited SRC-1 or SRC-2 coactivation to varying degrees (Figure 1F), but inhibition by PP1 phosphatase appeared to be selective for SRC-3 (Figure 1A), indicating an interesting differential regulation of the SRC family member activities by PP1.
PDXP and PP2A Dephosphorylate SRC-3 and Inhibit the Interaction between ER and SRC-3
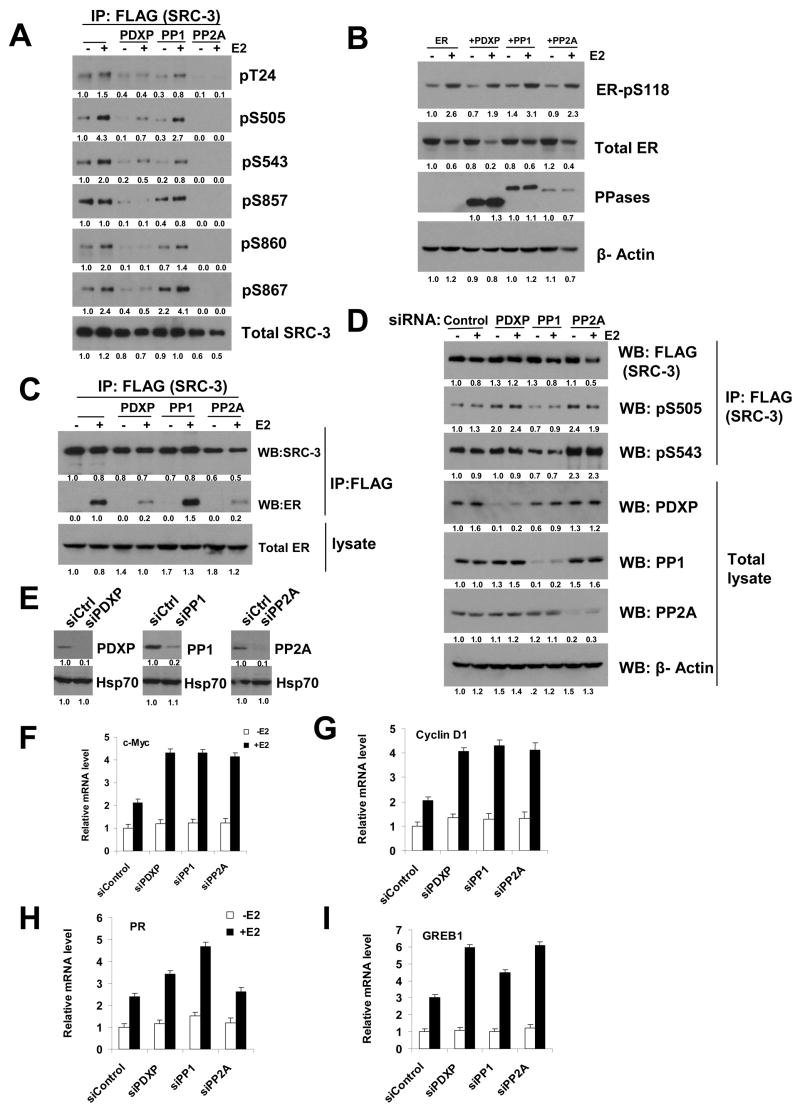
To investigate the molecular mechanisms of the negative effects of PDXP, PP1 and PP2A on SRC-3 activity, these 3 phosphatases along with ER and SRC-3 were expressed in 293T cells. As reported previously (Wu, Qin et al. 2004; Zheng, Wu et al. 2005), treatment of cells with E2 results in an increase in the phosphorylation levels of SRC-3 (Figure 2A). In the presence of PDXP and PP2A, the phosphorylation levels at all 6 sites dramatically decreased, while in the presence of PP1 the decreases in SRC-3 phosphorylation levels occurred moderately and more selectively for Thr24, Ser505 and Ser543 (Figure 2A).
Figure 2. Characterization of Phosphatases PDXP, PP1, and PP2A in ER/SRC-3 Signaling.
(A) PDXP, PP1, and PP2A dephosphorylate SRC-3 at the majority of known phosphorylation sites. Expression plasmids for ER and FLAG-SRC-3, together with PDXP, PP1A or PP2A were transfected into 293T cells. After treatment of cells with E2, cell lysates were immunoprecipitated (IP) with FLAG antibodies followed by loading the same amount of IP products for SDS-PAGE and Western blotting using SRC-3 phospho-specific antibodies as indicated. Numbers below the blots are quantifications for relative abundance of proteins.
(B) PDXP, PP1, and PP2A do not dephosphorylate ER-pSer118. Total cell lysates in (A) were analyzed by Western blotting using phospho-specific antibodies for ER Ser118 (ER-pS118), anti-ER antibodies for total ER levels (total ER), FLAG antibody for each phosphatase (PPases).
(C) PDXP and PP2A inhibit the ligand-dependent association between ER and SRC-3. As described in (A), immunoprecipitated SRC-3 was loaded in the same amount for SDS-PAGE followed by Western blotting using the indicated antibodies.
(D) siRNA knockdown of PDXP and PP2A increases SRC-3 phosphorylation levels. siRNAs to PDXP, PP1, PP2A or control siRNA were transfected into cells followed by either direct Western blotting of total cell lysates or IP-Western analysis using phospho-antibodies as indicated.
(E) siRNA knockdown of PDXP, PP1, or PP2A in ZR75-1 cells. Western blot analysis was performed using the indicated antibodies.
(F) siRNA knockdown of PDXP, PP1, or PP2A increases c-Myc mRNA levels. After siRNA knockdown of each phosphatase shown in (E), cells were treated with E2 for 2 hrs before examination of c-Myc mRNA levels by real time RT-PCR. Error bars indicate the standard error of the mean.
(G) (H) (I) Effects of siRNA knockdown of PDXP, PP1, or PP2A on cyclin D1 (G), PR (H) and GREB1 (I) mRNA levels. Same as (F) except examination of indicated mRNAs. Error bars indicate the standard error of the mean.
In the same experiment, we assessed the phosphorylation status of ER. As shown in Figure 2B, estrogen increased the phosphorylation level (~2.5-fold) at the ER Ser118 site although the total ER protein levels decreased due to ligand-dependent protein turnover, as reported previously (Lonard et al., 2000). In the presence of PDXP, PP1 or PP2A, both ER-pS118 levels and ER protein turnover appeared similar to those in the absence of these phosphatases (Figure 2B).
It was reported that mutation of SRC-3 phosphorylation sites can decrease interactions between ER and SRC-3 (Wu, Qin et al. 2004). We, thus, tested whether PDXP, PP1 and PP2A inhibited the ligand-dependent interaction between ER and SRC-3 using co-immunoprecipitation experiments. In the presence of PDXP or PP2A, this interaction was significantly impaired while PP1 had no effect on the interaction (Figure 2C). This result suggests that inhibition of luciferase reporter activities by PDXP and PP2A is, at least partially, a consequence of inhibition of the hormone-dependent interaction between ER and SRC-3.
siRNA Knockdown of PDXP, PP1 and PP2A Increases SRC-3 Phosphorylation Levels and Target Gene Expression
We next carried out experiments to reduce the phosphatases PDXP, PP1 and PP2A in cells. As shown in Figure 2D, siRNAs to PDXP, PP1 or PP2A were capable of efficiently knocking down the level of each phosphatase. Knockdown of PP2A resulted in an increase in phosphorylation of SRC-3 at S543 and S505. Knockdown of PDXP appeared to have a similar effect but to a lesser degree than PP2A. Knockdown of PP1 did not show an appreciable increase of SRC-3 phosphorylation levels at these sites.
Furthermore, we examined the effects of knockdown of PDXP, PP1 and PP2A on ER/SRC-3 target gene expression, including c-Myc, cyclin D1, PR and GREB1. As expected, in breast cancer cell line ZR75-1, c-Myc and cyclin D1 mRNA levels were increased ~2-fold after siRNA knockdown of PDXP, PP1 or PP2A (Figure 2F, G). Interestingly, estrogen target genes PR and GREB1 responded to siRNA knockdown of either PDXP, PP1 or PP2A in a differential manner: the PR mRNA level was elevated markedly upon E2 treatment after siRNA knockdown of PP1, while the GREB1 mRNA levels increased more under knockdown of PDXP and PP2A than with knockdown of PP1 (Fig 2H, I).
PP1 Stabilizes SRC-3 Protein by Blocking its Proteasome-Dependent Turnover
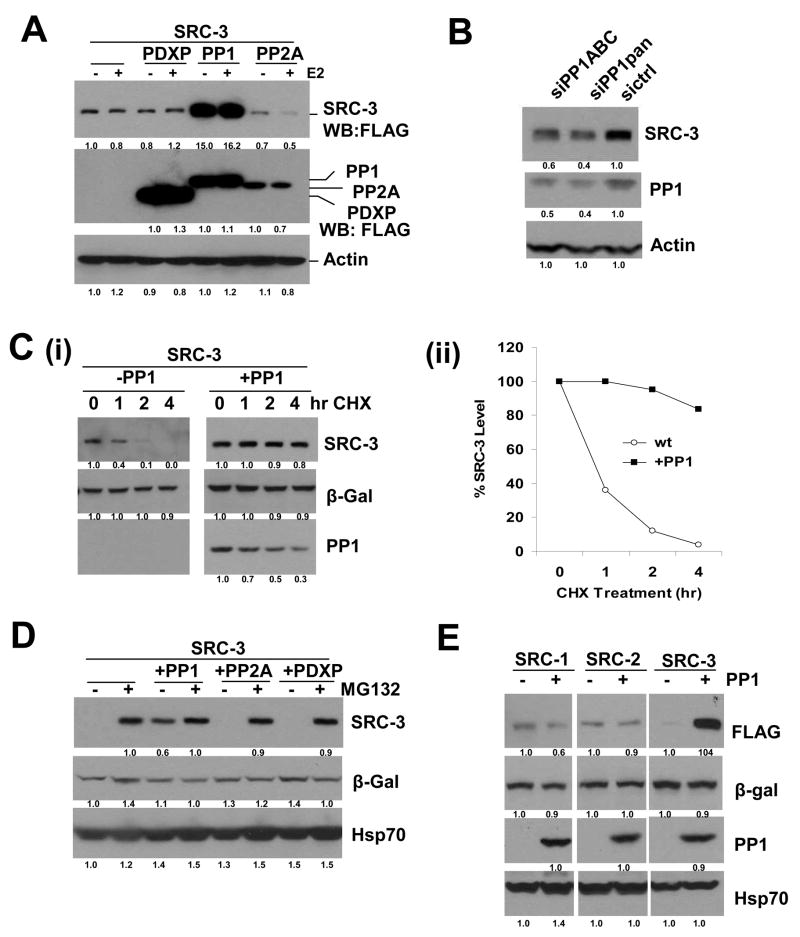
Since we did not observe a defective interaction of ER and SRC-3 in the presence of PP1, we were intrigued as to how PP1 inhibits SRC-3 signaling. When examining the total protein levels of SRC-3, surprisingly, we found a robust increase of SRC-3 in the presence of PP1; in contrast, there was no change of SRC-3 level in the presence of either PDXP or PP2A (Figure 3A). In the human genome there are 3 PP1 proteins with nearly 90% conserved amino sequence identities (Cohen, 2002). Partial functional redundancy of the three isoforms makes it extremely difficult to analyze knockdown of a single isoform. When knocking down all three PP1 isoforms simultaneously, SRC-3 protein levels were reduced (Figure 3B). In addition, we observed a dramatic increase in high molecular weight species of SRC-3, suggesting a likely accumulation of poly-ubiquitinated SRC-3 in the presence of PP1 (Figure S4). We next examined whether this increase in SRC-3 protein level is due to stabilization of SRC-3 protein by PP1. Cycloheximide (CHX) treatment experiments were performed to compare the SRC-3 protein stability in the presence or absence of PP1. Treatment of cells with CHX up to 4 hrs led to a decrease in SRC-3 protein levels, but this decrease was not seen in the presence of PP1 (Figure 3C), indicating indeed that PP1 stabilizes SRC-3 protein. We further asked whether this stabilization of SRC-3 protein is caused by inhibiting proteasome-dependent protein turnover. Treatment of cells with the proteasome inhibitor MG132 results in a robust increase of SRC-3 protein level, as previously reported (Li, Wu et al. 2007). In contrast to the presence of PDXP and PP2A, in the presence of PP1, proteasome-dependent stabilization of SRC-3 was virtually lost, indicating that stabilization of SRC-3 by PP1 is proteasome-sensitive (Figure 3D). In the same experiments, β-Gal and Hsp70 protein levels, employed as exogenous and endogenous loading control proteins, respectively, remained relatively constant. In addition, we examined the effects of PP1 on SRC-1 and SRC-2 proteins, and importantly, as shown in Figure 3E, PP1 did not affect SRC-1 and SRC-2 protein levels, suggesting that the stabilization by PP1 is specific to SRC-3.
Figure 3. PP1 Stabilizes SRC-3 Protein by Blocking its Proteasome-Dependent Turnover.
(A) PP1 increases SRC-3 protein levels. FLAG-tagged SRC-3 and each phosphatase were expressed in 293 cells in the presence or absence of E2. SRC-3 protein levels in total cell lysates were analyzed by Western blot using anti-FLAG antibodies.
(B) siRNAs targeting all three PP1 isoforms decrease SRC-3 levels. Two different siRNA pools against PP1 (siPP1ABC or siPP1pan) were transfected into 293T cells followed by Western blotting with the indicated antibodies.
(C) PP1 blocks SRC-3 protein turnover. Cycloheximide (CHX) treatment experiments were performed in the presence (+) or absence (−) of PP1 co-transfected with SRC-3 and β-Gal followed by Western blotting using the indicated antibodies (i), and quantification (ii).
(D) PP1 blocks SRC-3 proteasome-dependent turnover. Expression plasmids for SRC-3 and β-Gal, or combined with PP1, PP2A, or PDXP were transfected into 293 cells. After treatment of cells with MG132 (2.5 uM), cell lysates were immunoblotted using the indicated antibodies.
(E) PP1 does not stabilize SRC-1 and SRC-2 proteins. FLAG-tagged SRC-1, SRC-2, or SRC-3 were expressed in the presence or absence of PP1 followed by Western blotting.
SRC-3 is a shuttling protein between nucleus and cytoplasm (Amazit et al., 2007; Qutob et al., 2002). Previous studies demonstrate that all SRC-3 proteasome-dependent protein turnover occurs in the nucleus and cytoplasmic localization of SRC-3 mutants also prevents proteasome-dependent turnover (Li et al., 2007). Thus, we asked whether PP1 alters SRC-3 subcellular localization. Immunofluorescence staining revealed that both SRC-3 and ER remained in the nucleus under a complete medium culture condition when PP1, PDXP and PP2A were present (Figure S5).
Identification of a Phospho-26S Proteasome Degron in SRC-3
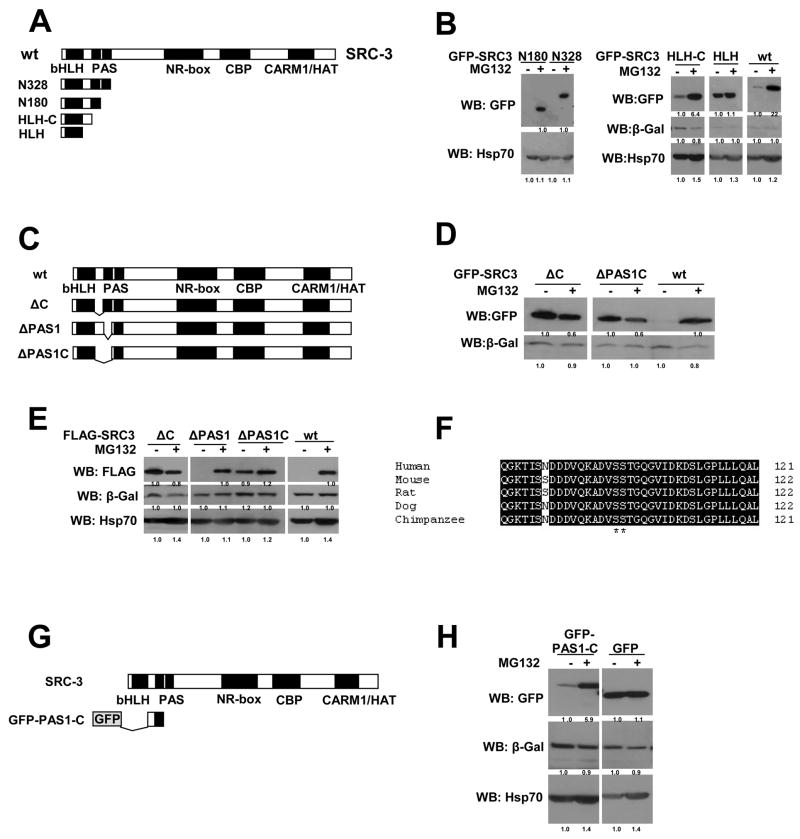
To further study the molecular mechanism of inhibition of SRC-3 proteasome-dependent turnover by PP1, we needed to define the region in SRC-3 that is a primary determinant for its response to MG132 in the nucleus. To answer this question, a series of truncation mutants of SRC-3 were constructed with a GFP-tag, including N328, N180, HLH-C and HLH (Figure 4A). GFP-SRC-3 wt responded to MG132 in the same manner as the untagged or FLAG-tagged (Figure 4B, E). Using these GFP-SRC-3 mutants, we were able to define a critical region of SRC-3 for response to MG132 that was located between the N-terminal bHLH and PAS domains (Figure 4B). This region is at the “C” terminus of the bHLH domain and therefore designated as the “C” region (Figure 4A).
Figure 4. Identification of a 26S Proteasome Degron in SRC-3 Protein.
(A) Schematic representation of SRC-3 truncation mutants.
(B) The “C” region between the bHLH and PAS domains determines SRC-3’s response to the proteasome inhibitor MG132. GFP-tagged SRC-3 mutants or wt as indicated were expressed in 293 cells with a β-gal control vector followed by treatment with MG132 and Western blot analysis using an anti-GFP antibody to detect SRC-3.
(C) Schematic representation of SRC-3 deletion mutants.
(D) Similar to (B) except examining the deletion mutants instead.
(E) Similar to (D) but using FLAG-tagged SRC-3 deletion mutants instead of GFP-tagged mutants.
(F) Sequence alignment of SRC-3’s degron among different species. Numbers at the right are the amino acid residue positions in each full-length protein.
(G) Schematic of a short SRC3 degron containing peptide fused to GFP (GFP-PAS1-C).
(H) The degron region is transferable and able to drive GFP degradation. GFP alone or GFP-PAS1-C was expressed in 293 cells followed by Western analysis using the indicated antibodies.
To validate that this “C” region is crucial for SRC-3’s response to MG132, we constructed another series of SRC-3 mutants which contained different deletions that surround the “C” region under the context of the full-length SRC-3 protein (Figure 4C). Consistent with results from the truncation mutants above (Figure 4A, B), deletion of the “C” region alone (ΔC), or “C” together with PAS1 resulted in an inhibition of SRC-3 proteasome-dependent turnover in either of the GFP-tagged mutants (Figure 4D) or FLAG-tagged mutants (Figure 4E). Thus, our results confirmed that the SRC-3 “C” region indeed is a critical determinant for its proteasome-dependent turnover. Sequence alignment of the SRC-3 “C” region among different species indicates that it is highly conserved (Figure 4F).
Importantly, we tested whether the SRC-3 “C” region is transferable. We generated a fusion protein which was comprised of GFP and a short SRC-3 peptide containing the “C” region (Figure 4G). GFP alone was not responsive to MG132, suggesting its degradation occurs in a proteasome-independent fashion, whereas the fusion protein became responsive to MG132 (Figure 4H). This result shows that the SRC-3 “C” region is able to bring another diverse protein into the proteasome for subsequent degradation. Immunofluorescence analysis revealed that GFP alone and GFP-PAS1-C were present in both the nucleus and cytoplasm (data not shown).
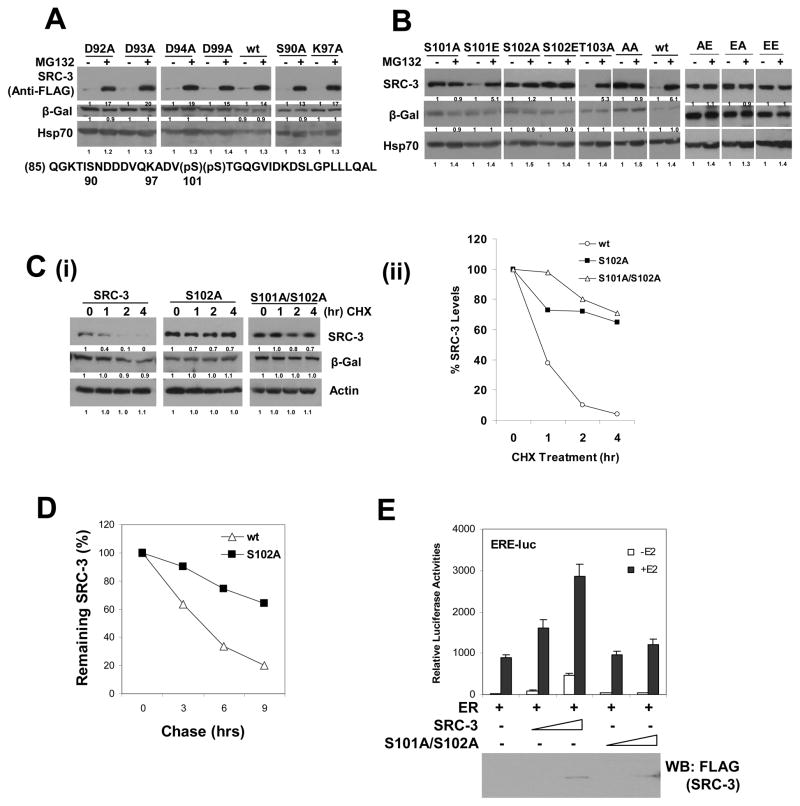
The “C” region contains 37 amino acid residues and some of the residues are potential targets for posttranslational modifications or other interactions; we therefore determined the key residues in this region by generating mutations of each candidate residue to Ala, including D92A, D93A, D94A, D99A, S90A, K97A, S101A, S102A and T103A. We tested these mutants for MG132-dependent stabilization. As shown in Figure 5A, B, only S101A and S102A mutants, which mimic loss of phosphorylation, lost responses to MG132; all other mutants were still sensitive to MG132, as was that of wild-type. Additional mutations between G104 and L121, or before S90 residues did not affect MG132 responses (data not shown). Because Ser is a typical target for phosphorylation, mutations for S101 and S102 were generated to include S101E and S102E, which mimic phosphorylation states; different combinations of double mutations were assayed (Figure 5B). S101E mutant, like wt, remained responsive to MG132, suggesting that this site is an authentic charged phosphorylation site. Intriguingly, S102E also blocked a response to MG132, likely due to the fact that phosphorylation of S102 provides a docking site for S101 phosphorylation, and/or glutamine acid (E) can not fully mimic the phosphorylation status at this site. As a consequence, all double mutants behaved similar to the S102 mutant, showing no response to MG132 (Figure 5B).
Figure 5. Functional Identification of Two Phosphorylation Sites (Ser 101, 102) in SRC-3’s Degron.
(A) Normal responses to MG132 of some single point mutations in SRC-3’s degron. Each FLAG-tagged SRC-3 wt and single residue mutant with a change to Ala as indicated was expressed in 293 cells and treated with MG132 prior to Western analysis. Sequence below is from SRC-3’s degron.
(B) Ser101 and Ser102 are two phosphorylation sites in SRC-3. Single amino acid residue Ser101 or Ser102 was mutated to Ala (A) or Glu (E) as indicated. When both residues S101 and S102 were mutated to Ala and/or Glu, they are indicated as AA, AE, EA, and EE, respectively. Each mutant was expressed in 293 cells and treated with MG132 prior to Western analysis.
(C) Ser101 and Ser102 mutations to Ala stabilize SRC-3. CHX treatment experiments were performed as described in the Experimental Procedures using SRC-3 wt, S102A, and S101A/S102A mutants (i). The quantification of this data is shown in panel (ii).
(D) Pulse-Chase analysis of SRC-3 wt and the S102A mutant. The experiment was performed in 293 tetracycline-inducible stable cell lines for FLAG-tagged SRC-3 wt or S02A mutant.
(E) SRC-3 S101A/S102A mutant is inactive in coactivation. Transient transfection of increasing amounts of either SRC-3 wt or its S101A/S102A mutant in ERE-luc reporter assays. Expressed protein levels were examined by Western blot shown below. Error bars indicate the standard error of the mean.
Next we examined whether the S101 and S102 mutant proteins become stable, as was seen in the case of the ΔC mutant. CHX experiments were performed, and as expected, indeed the double mutant S101A/S102A, single mutant S102A (Figure 5C), and S101A (data not shown) were more stable than wild type. To further substantiate our results, we performed additional pulse chase experiments for S102A that confirmed our conclusion (Figure 5D). Immunofluorescence analysis indicated that all of these point mutations were nuclear localized and substantiated further the critical role of S101 and S102 in SRC-3’s 26S proteasome degron (Figure S6). We additionally assessed the transcriptional coactivation capacity of the SRC-3 S101A/S102A mutant. When SRC-3 wt and its S101A/S102A mutant proteins were expressed at the same level in ERE-luc reporter assays, the S101A/S102A mutant lost its coactivation capacity; in contrast, wt SRC-3 stimulated transcription in a dose dependent manner (Figure 5E).
PP1 Stabilizes SRC-3 Protein through Dephosphorylation of SRC-3 at Ser 101 and Ser 102
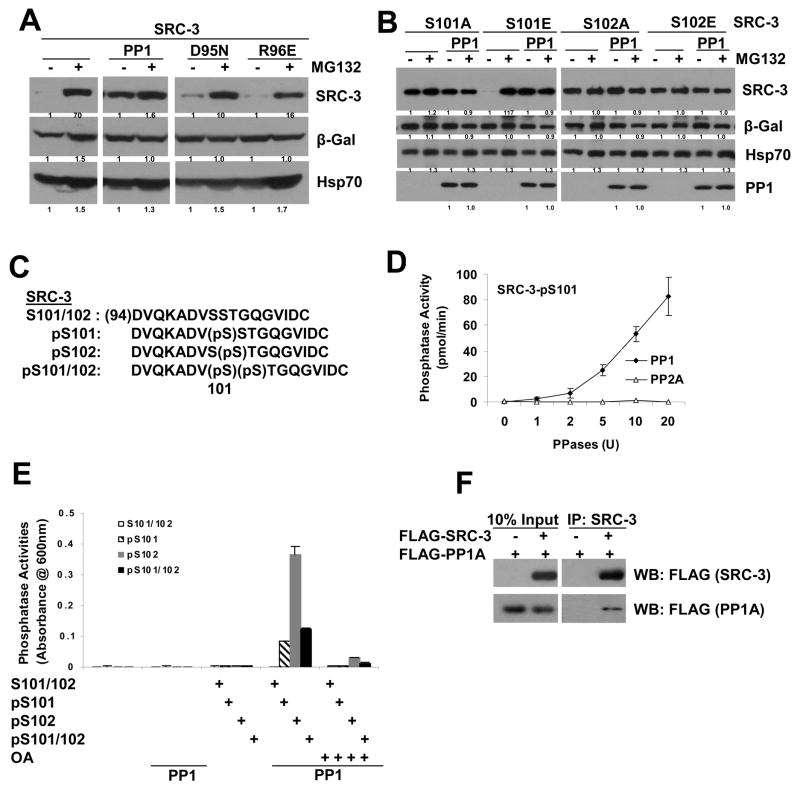
Since the effect of PP1 on SRC-3 protein stability resembles the SRC-3 S101A and S102A mutants, we examined whether there are additive effects between PP1 and S101A/S102A mutants. Similar to what was shown previously, PP1, but not its catalytic mutants (D95N and R96E)(Zhang et al., 1996), blocks SRC-3’s response to MG132 (Figure 6A). PP1 had no additional effect on SRC-3 S101A, S102A and S102E mutant protein levels in the presence or absence of MG132 (Figure 6B). PP1 enhanced the S101E protein level without MG132, which likely is due to dephosphorylation of S102 (Figure 6B). These results suggest that PP1 and SRC-3 S101 and S102 mutants have overlapping effects on SRC-3 protein stability.
Figure 6. PP1 Stabilizes SRC-3 Protein through Dephosphorylation of SRC-3 Ser101 and Ser102 Sites.
(A) PP1 but not its catalytic mutants (D95N, R96E) prevent SRC-3 from proteasomal degradation. The experiments were performed as described in the Experimental Procedures.
(B) PP1 stabilizes SRC-3 primarily through the S101 and S102 sites. Overlapping effects of PP1 on SRC-3 stability, and SRC-3 S101A or S102A mutants. Each SRC-3 S101 and S102 mutant was expressed with or without PP1 in the presence or absence of MG132, followed by Western analysis with the indicated antibodies.
(C) Sequences of SRC-3 phosphorylated peptides containing S101 and S102. Peptides with no phosphorylation (S101/102) as a control, with phospho-S101 (pS101), phospho-S102 (pS102), or phosphorylated at both S101 and S102 (pS101/102), are indicated.
(D) PP1, but not PP2A, dephosphorylates the SRC-3 phospho-S101 peptide in vitro. In vitro phosphatase assays were performed as described in the Experimental Procedures. Error bars indicate the standard error of the mean.
(E) PP1 dephosphorylates both SRC-3 pS101 and pS102 in vitro. SRC-3 control peptide (S101/102), or phospho-peptides (pS101, pS102 or pS101/102) were used in in vitro phosphatase assays with recombinant PP1 protein as described in (D) in the presence or absence of the PP1 inhibitor, okadaic acid (OA). Error bars indicate the standard error of the mean.
(F) Co-immunoprecipitation of PP1 and SRC-3. Expression vectors for FLAG-PP1A and FLAG–SRC-3 were transfected into 293 cells. Cell lystates were immunoprecipitated by SRC-3 antibodies followed by Western blot using FLAG antibodies.
We next asked whether PP1 stabilization of SRC-3 results from “direct” dephosphorylation of the S101 and S102 sites. We first took a biochemical approach, designing the four peptides covering SRC-3 S101 and S102: a control peptide with no phosphorylation residue at these sites (S101/S102), a peptide with only S101 phosphorylated (pS101), a peptide with only S102 phosphorylated (pS102), and a peptide with both S101 and S102 phosphorylated (pS101/102) (Figure 6C). Using these peptides, we performed in vitro phosphatase assays to examine whether PP1 is able to dephosphorylate the pS101 or pS102 sites. As shown in Figure 6D, PP1, but not PP2A, dephosphorylated pS101 in a phosphatase dose-dependent manner. Similar results were observed by using pS102 or pS101/102. As a control, PP2A was able to dephosphorylate its own substrate peptide (data not shown). These dephosphorylations were dramatically reduced upon addition of okadaic acid, an inhibitor of PP1. Intriguingly, dephosphorylation of pS102 appeared stronger than pS101 and pS101/102 by PP1 (Figure 6E). This result suggests that PP1 has the capacity to directly dephosphorylate SRC-3’s phosphorylated S101 and S102 sites.
In addition, we examined the interaction between PP1 and SRC-3 using co-immunoprecipitation. After expression of Flag-tagged SRC-3 and PP1A in 293 cells, co-IP experiments demonstrated that PP1 and SRC-3 were physically associated with each other (Fig 6F).
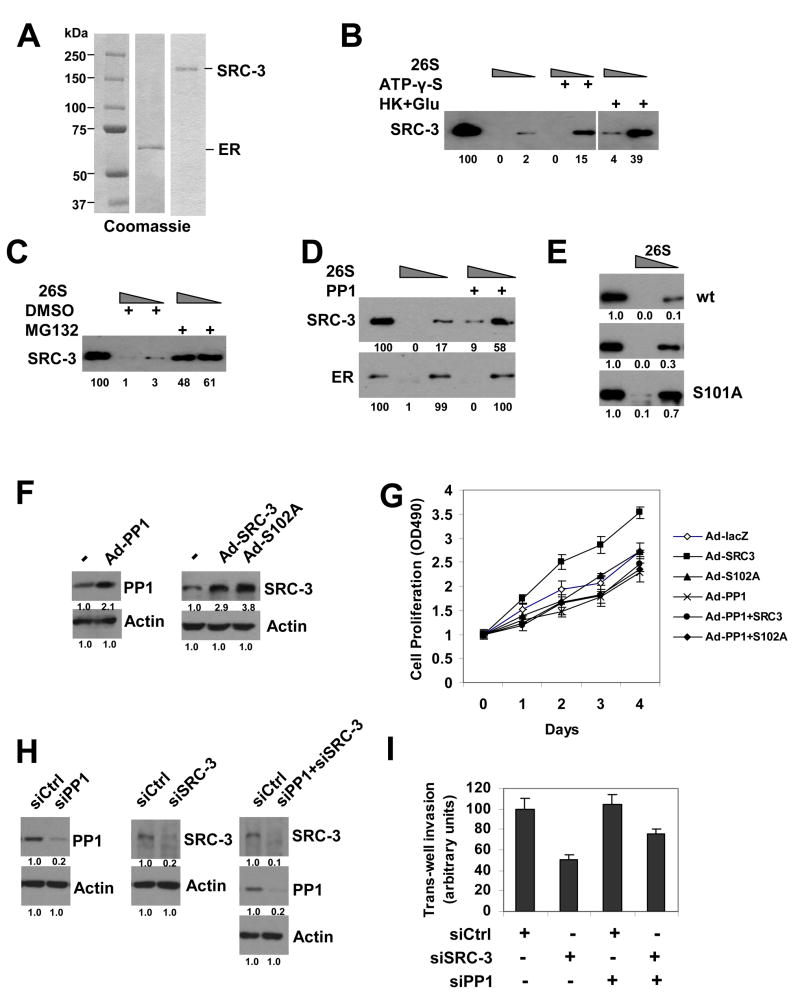
Further, we tested whether an in vitro cell-free system was able to recapitulate the SRC-3 stability phenomenon in cells. By using purified recombinant SRC-3 and ER proteins (Figure 7A), and 26S proteasome fractions, we indeed observed in vitro degradation of SRC-3 protein by the 26S proteasome and dramatic inhibition by the proteasome inhibitor MG132 (Figure 7C). We went on to show that the non-hydrolysable ATP analog ATP-γ-S or depletion of ATP by hexokinase and glucose inhibited the degradation of SRC-3 by the 26S proteasome (Figure 7B). Importantly, treatment of SRC-3 with PP1 inhibited degradation of SRC-3 while ER degradation by proteasomes was not effected (Figure 7D). We also tested whether phosphorylation-defective mutant S101A or phosphorylation-mimic mutant S101E can be degraded to the same extent by the proteasome pathway. As shown in Figure 7E, SRC-3 S101A mutant was more resistant to degradation by 26S than wt counterpart or S101E mutant. Our data clearly suggest that inhibition of 26S proteasome-dependent degradation of SRC-3 by PP1 occurs at least in part by dephosphorylation of SRC-3 at S101 and further suggests that it acts by a direct mechanism.
Figure 7. PP1 Regulates SRC-3 Proteasome-Dependent Degradation In Vitro and Oncogenic Functions in Cancer Cells.
(A) SDS-PAGE analysis of purified recombinant ER and SRC-3 proteins by Coomassie staining. The left lane indicates protein markers in kDa.
(B) ATP-dependent degradation of SRC-3 by 26S proteasomes in vitro. Increasing amounts of 26S proteasomes containing ATP were incubated with recombinant SRC-3 protein in the presence of ATP-γ-S, or hexokinase and glucose (HK+Glu), as indicated, for 10 min followed by immunoblotting. Numbers under the blot are quantifications for relative amounts of remaining SRC-3. The left lane in each panel shows the SRC-3 input before degradation.
(C) In vitro inhibition of 26S proteasomal degradation of SRC-3 by MG132.
(D) Inhibition of 26S proteasomal degradation of SRC-3, but not ER, protein by PP1. SRC-3 and ER proteins were pre-treated with PP1, followed by 26S degradation analysis.
(E) SRC-3 phosphorylation-defective mutant S101A is more resistant to 26S proteasomal degradation than its wt and phosphorylation-mimic mutant S101E. Immunopurified SRC-3 wt, S101E, and S101A proteins were subjected to 26S degradation assays.
(F) Ectopic expression of PP1, SRC-3 or SRC-3 S102A mutant in ZR75-1 cells. Western blot was performed after infection of adenovirus expression vectors for PP1A (Ad-PP1), SRC-3 (Ad-SRC-3), or SRC-3 S102A mutant (Ad-S102A) in ZR75-1 for 2 days.
(G) PP1 inhibits SRC-3 stimulated-cell proliferation. ZR75-1 cells were infected with adenovirus vectors expressing either control lacZ (Ad-lacZ), SRC-3 wt (Ad-SRC3), SRC-3 S102A mutant (Ad-S102A), PP1A (Ad-PP1), PP1A and SRC-3 (Ad-PP1+SRC3), or PP1A and S102A (Ad-PP1+S102A). Cell proliferations were determined each day. See Experimental Procedures for details. Error bars indicate the standard error of the mean.
(H) siRNA knockdown of PP1, SRC-3, or both PP1 and SRC-3 in the invasive breast cancer cell line MDA-MB-231. Protein levels were detected by Western blot after transfection siRNAs for two days using antibodies as indicated.
(I) Trans-well matrigel cell invasion assays after knockdown SRC-3 and/or PP1. MDA-MB-231 cells were transfected with control siRNA (siCtrl), siSRC-3, siPP1, or both siPP1 and siSRC-3. for two days, followed by trans-well matrigel invasion assays as described in Experimental Procedures. Error bars indicate the standard error of the mean.
PP1 Inhibits Oncogenic Functions of SRC-3 in Breast Cancer Cells
Importantly, we carried out experiments to assess the biological consequences of PP1 and SRC-3. Using the breast cancer cell line ZR75-1, adenovirus-mediated expression of PP1, SRC-3 wt or its PP1 target site mutant S102A enhanced each of the two protein levels in the cells (Fig 7F). Subsequently we examined cell proliferation under each condition. SRC-3 wt increased ZR75-1 cell proliferation but not that of the mutant S102A. Introducing PP1 and SRC-3 together resulted in an inhibition of SRC-3-enhanced cell proliferation; PP1 did not affect SRC-3 S102A mutant (Fig 7G). These results indicate the essential role of PP1-regulated dephosphorylation of the S102 site in SRC-3 physiological functions.
Finally, we examined the effect of siRNA knockdown of PP1 on SRC-3 mediated cancer cell invasion potential. siRNA knockdown of SRC-3 in the invasive breast cancer cell line MDA-MB-231 resulted in a marked reduction of trans-well matrigel invasion to about 50%, but under siRNA knockdown of PP1 conditions, this effect was less remarkable (Fig 7H, I). Taken together, these results suggest that dephosphorylation of SRC-3 by PP1 plays an important role in regulating oncogenic SRC-3 metastasis functions.
Discussion
Reversible phosphorylation is a key posttranslational modification that regulates many essential cellular processes. In steroid signaling, phosphorylation contributes to both ligand-dependent and ligand-independent activation of target genes (Denner et al., 1990; Wu et al., 2005). Treatment of cells with the tumor promoter okadaic acid, an inhibitor of PP1 and PP2A, stimulates multiple steroid receptor-mediated transcriptional reactions such as with the progesterone receptor, estrogen receptor, and retinoic acid receptor (Denner et al., 1990; Lefebvre et al., 1995; Lu et al., 2003). Furthermore, steroid receptor coregulators, as mediators of steroid receptor function, are both targets and propagators of posttranslational modification codes (Lonard and O’Malley B, 2007). Phosphorylation plays a key role in regulating a coregulator’s function, e.g., hypophosphorylated SRC-3 often exists in an “inactive” state while phosphorylated SRC-3 becomes an active protein (Lonard and O’Malley B, 2007; Wu et al., 2007; Wu et al., 2004).
Using a functional genomic approach, we systemically identified and studied protein phosphatases involved in the regulation of SRC-3 phosphorylation and its functional activity. In an initial library screening, fifteen phosphatases were discovered to dephosphorylate SRC-3; interestingly, the majority of the phosphatases appeared to inhibit SRC-3 activity. We were surprised that so many phosphatases participated in SRC-3 dephosphorylation and altered its functions. However, it is known that coactivators mediate the effects of many different signaling pathways for activation of a variety of gene expressions and coactivator phosphorylation is likely to serve as a primary molecular basis for coactivator specificity (Wu, Qin et al. 2004; Wu, Smith et al. 2005; Lonard and O’Malley B 2007). It is conceivable that during transcription, dephosphorylation of key coactivators occurs during coactivator complex disassembly, degradation and/or recycling in gene expression programs. Therefore, multiple different phosphatases could contribute to a wide variety of regulatory processes in living cells. It is likely that we have greatly underestimated the importance and specificity of phosphatases in the regulation of coactivator phosphorylation and activity.
In this study, three essential phosphatases PDXP, PP1 and PP2A were confirmed and substantiated to play multiple key roles in regulating SRC-3 functions. PDXP was reported as a phosphatase for dephosphorylation of pyridoxal 5-prime-phosphate (PLP), an active form of vitamin B6 (Jang et al., 2003), but its functions in other signaling pathways are largely unknown. PP2A is a well-characterized tumor suppressor, and its loss of function often appears to be cooperative with oncoproteins in tumorigenesis (Janssens et al., 2005; Mumby, 2007). It also regulates other signaling cascades such as Ca(2+)-dependent gene expression (Anderson et al., 2004). In this study, we demonstrated that PP2A is involved in dephosphorylation and inactivation of the oncoprotein, SRC-3.
We were initially puzzled that PP1 strongly inhibited SRC-3 activity but only weakly dephosphorylated SRC-3 (Figure 1; 2). Further studies revealed that PP1 blocked SRC-3 proteasome-dependent turnover by specific dephosphorylation of only two phosphorylation sites S101 and S102 (Figure 3A, C, D; Figure 6D, E). These results provide a mechanism for a phosphatase function in estrogen signaling and gene transcription, and are consistent with prior findings that coregulators must be capable of proteasome-dependent turnover to maintain their functional activities (Lonard et al., 2000; Perissi et al., 2004). Recent studies have shown that PP1 also regulates the stability of other signaling proteins such as TIM protein in the Drosophila circadian clock (Fang et al., 2007) and the β-catenin degradation complex in Wnt/β-catenin signaling (Luo et al., 2007). PP1 activity also has been reported to prevent oncogenic transformation on occasion (Liu et al., 2006). PP1 is comprised of a complex of enzymes, encoded by 3 highly conserved catalytic subunit genes, interacting with a large variety of targeting subunits that direct PP1 to each specific substrate (Ceulemans and Bollen, 2004). A unique feature is that PP1 can be regulated by many small phosphoprotein inhibitors such as DARPP-32. Interestingly, progesterone-facilitated sexual receptivity is mediated by phosphorylation of DARPP-32 at Thr-34, a key site in DARPP-32 activation that in turn inhibits PP1 activity (Mani et al., 1994; Mani et al., 2000). This serves as an example of coordinated activation of steroid hormone signaling by the simultaneous activation of kinases and/or inhibition of the phosphatase PP1, eventually leading to phosphorylation of steroid receptors and/or the coactivator SRC-3.
One of the most interesting aspects of PP1 regulation of SRC-3 function is the targeting of two phosphorylation sites in the 26S proteasome degron of SRC-3. SRC-3 protein degradation is subject to regulation by the ubiquitin/26S proteasome (Lonard et al., 2000; Wu et al., 2007). However, the molecular basis by which the ubiquitinated proteins are recognized and destroyed by the 26S proteasome is largely unknown. In this study, we identified a primary determinant for its proteasome-dependent degradation (Figure 4, 5). More importantly, this degron was regulated by phosphorylation, which provides an opportunity for small molecule targeting drugs. Given the fact that SRC-3/AIB1 is overexpressed in a majority of breast cancers and is one of the key contributors to tumorigenesis, the phospho-degron within this important oncoprotein offers a potential therapeutic target for intervention in SRC-3 oncogene activity.
It has been reported that overexpresion of SRC-3/AIB1 and HER-2/neu contribute to tamoxifen therapeutic resistance in breast cancer patients (Osborne et al., 2003). We hypothesize that PP1 may be involved in an initial stage of inactive SRC-3 protein stabilization and overexpression in breast cancer. Subsequent HER-2 overexpression then could lead to activation of multiple kinase signalings and enhancement of phosphorylation and activation of SRC-3, eventually turning SRC-3 into a potent oncogenic regulator. Finally, our study provides a further example of the elegant signaling equilibrium inherent to key regulatory proteins in mammalian cells.
Experimental Procedures
Human Ser/Thr Phosphatase Library Screening
293T cells seeded in 10 cm plates were transfected with 6 ug of an SRC-3 expression plasmid, 3 ug of an ER plasmid and 3 ug of the vector encoding each human Ser/Thr phosphatase from a previously described expression library (Lin et al., 2006). Two days later, cell lysates were immunoprecipitated with anti-FLAG-M2 agarose beads followed by SDS-PAGE and Western blotting analysis with anti-phospho-specific SRC-3 antibodies as previously described (Zheng et al., 2005).
Luciferase Reporter Gene Assays
One day prior to transfection, HeLa cells were plated in 24-well plates in phenol red-free DMEM medium containing 5% charcoal-stripped serum (Lonard, Nawaz et al. 2000). Expression vectors for ERα, SRC-3, each phosphatase, and an ERE-luciferase reporter gene were transfected with TransIT-LT1 (Mirus) into HeLa cells. One day later, cells were treated with 10 nM E2 overnight before measuring luciferase activities (Li, Wu et al. 2007). SRC-1, -2 along with ERα and each phosphatase, were also examined. Likewise, PR and PRE-luc, or AR and MMTV-luc were assayed with each phosphatase in the same manner with treatment of 1 nM progesterone or 10 nM DHT, respectively. Experiments with NF-κB-luc were treated with TNFα (10 ng/ml) for 4 hr prior to luciferase assays (Wu, Qin et al. 2004). Experiments with p21-luc and p53 have been described previously (Lonard, Nawaz et al. 2000). Gal4-VP16 and pG5-luciferase containing multiple Gal4 binding sites in its promoter were assayed directly without E2 treatment.
Immunoprecipitation and Western Blot
As previously described (Wu, Qin et al. 2004; Li, Wu et al. 2007), 293T cells were seeded in medium containing 5% charcoal-stripped fetal bovine serum. After transfection with expression plasmids for SRC-3 in combination with either PDXP, PP1 or PP2A for 2 days, cells were treated with 10−7 M E2 for 1 hr followed by disrupting cells in lysis buffer (20 mM Tris-HCl, 125 mM NaCl, 0.5% NP-40, 2 mM EDTA) supplemented with phosphatase and protease inhibitor cocktails (Roche, Indianapolis, IN). Cell lysates were immunoprecipitated with EZview red anti-FLAG M2 affinity gel for 2 hrs at 4 °C with constant rotation and then the beads were washed 4 times with the above buffer. Finally the beads were directly boiled in Laemmili sample buffer prior to separation by 4–20% SDS-PAGE Novex gels (Invitrogen). Western blotting was performed by first blocking nitrocellulose membranes with 5% nonfat milk in PBS-T buffer prior to adding phospho-specific SRC-3 antibodies or other antibodies as indicated. Image J software (NIH, US) was used for quantification of Western blots. Antibodies used in Western blotting were: anti-ER-phospho-S118 (Santa Cruz), anti-ER (Thermo Scientific, Fremont, CA), anti-FLAG-HRP (Sigma), monoclonal anti-β-Actin-HRP (Sigma), anti-PP1a and PP1 (Santa Cruz), monoclonal anti-PP2A (Upstate) which recognizes both of PP2Ac isoforms, anti-β-Gal (Roche), monoclonal anti-Hsp70 (BD Biosciences), anti-α-tubulin (Santa Cruz), anti-SRC-3/AIB1 and anti-GFP (BD Biosciences).
siRNA Knockdown
The siRNAs for PDXP, PP1, PP2A and control siRNAs were purchased from Dharmacon as ON-TARGET plus SMART pools. siRNAs were transfected with TransIT-TKO Transfection Reagent (Mirus) according to manufacturer’s instructions. Three days after transfection, cell lysates were analyzed for each protein. siRNAs for pan PP1, which target all three PP1 isoforms, were from Santa Cruz. For the experiments examining the effects of siRNA knockdown of PDXP, PP1, and PP2A on SRC-3 phosphorylation, one day after transfection of each siRNAs, SRC-3 and ER expression plasmids were transfected into cells for an additional two days. Subsequently, cell lysates were immunoprecipitated with anti-FLAG-M2 beads followed by SDS-PAGE and Western blot using the indicated antibodies.
Real-Time RT-PCR (RT-qPCR)
MCF-7 or ZR75-1 cells were cultured in 5% charcoal-stripped serum before transfection with siRNAs against each phosphatase. Three days after transfection, cells were treated with estradiol (10−7 M) for the indicated times. Subsequently, total RNA was isolated using TRIzol (Invitrogen) and TaqMan Gene Expression Assays (Applied Biosystems) were performed in a 7500 real time PCR machine (Applied Biosystems) to analyze c-Myc, cyclin D1, PR and GREB1 gene expression. mRNA levels were normalized to 18S as the endogenous control. Each TaqMan probe for its respective target gene mRNA and One-Step RT-PCR master mix were also from Applied Biosystems. Each target was measured in triplicate.
Protein Stability Analysis
Protein decay was studied by either cycloheximide (CHX) treatment experiments or pulse chase assays, both of which have been described previously (Li, Wu et al. 2007). CHX treatment experiments were performed using 0.5 mM CHX (Sigma) for up to 4 hrs after 2 days post-transfection of cells with the indicated expression plasmids. Cell lysates were analyzed by SDS-PAGE and Western blot. Quantification was conducted using Image J software (NIH, US). Proteasome-dependent protein degradation was analyzed by treatment of cells with either the proteasome inhibitor MG132 at 2.5 uM (Sigma) or DMSO vehicle control for overnight, followed by SDS-PAGE and Western blot analysis.
In Vitro Protein Degradation by 26S Proteasomes
This in vitro assay was performed using the purified recombinant SRC-3 protein and mammalian 26S proteasome fraction (BostonBiochem). Some experiments were conducted in the presence of 70 uM ATP-γ-S (EMD Biosciences) or hexokinase (20 units/ml) (Sigma) and glucose (70 mM). The reactions were carried out at 30 °C for 10 min. FLAG-tagged SRC-3 protein was baculovirus-expressed and purified from Sf9 cells (Baculovirus/Monoclonal Antibody Core Facility at Baylor College of Medicine), while ERα was from Invitrogen. SRC-3 S101A and S101E mutant proteins were expressed in 293 cells and partially purified by immunoprecipitation using anti-FLAG-M2 agarose beads.
Phosphorylated Peptide Synthesis and In Vitro Phosphatase Assays
Non-phosphorylated control peptide or phosphorylated peptides derived from SRC-3 with either phospho-S101 and/or S102 were chemically synthesized by Abgent (San Diego, CA). The peptide sequences are as follows: S101/102: DVQKADVSSTGQGVIDC; pS101: DVQKADV(pS)STGQGVIDC; pS102: DVQKADVS(pS)TGQGVIDC; pS101/102: DVQKADV(pS)(pS)TGQGVIDC. Purified PP1 and PP2A proteins were purchased from NEB and Upstate, respectively. In vitro Ser/Thr phosphatase assay was performed as previously described (Lu et al., 2004) using the Serine/Threonine Phosphatase Assay System (Promega). Each reaction was incubated at 30 °C for 20 min. Free phosphate released by dephosphorylation was bound by molybdate and measured by O.D. absorbance at 600 nm. Free phosphate standards also were measured in the experiment to quantify the phosphatase activity.
Trans-well Matrigel Invasion Assays
MDA-MB-231 cells in 6-well plates in complete growth medium were transfected with 40 nM of siRNAs against SRC-3 and/or PP1(Darhmacon). Two days after transfection, cells were detached and transferred to Trans-well Matrigel Invasion chambers (BD Biosciences) following manufacturer’s protocols. Cells were allowed to migrate and invade through matrigel membrane for one day before fixing with 4% formaldehyde and stained with crystal violet. The cells on the apical side of each insert were scraped off by Q-tips. The number of cells that had migrated through the matrigel membrane was counted under a microscope. Values in controls were arbitrarily set at 100.
Cell Proliferation Assays
Adenovirus vectors for expression of SRC-3 wt, its S102A mutant and lacZ were constructed using ViraPower Adenoviral Expression System (Invitrogen) according to the manufacturer’s instruction. Adenovirus expressing PP1A was purchased from Vector Biolabs. One day after adenovirus infection of ZR75-1 cells, the cells were seeded into 96-well plates in triplicate with 2000 cells/well. Cell proliferations were assayed every day using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega). Relative values of cell proliferation in day 0 were set at 1.
Immunofluorescence
As described previously (Li, Wu et al. 2007), GFP-SRC-3, its mutants, and GFP-ER were directly visualized 2 days after transfection of corresponding expression plasmids into HeLa cells, while FLAG-tagged SRC-3 and its mutants were immunostained with primary anti-FLAG antibody (Sigma) and secondary goat anti-mouse Alexa 488 or 555 (Molecular Probes). DAPI was used to stain the nucleus.
Repetitions of Experiments
Experiments in all figures were repeated at least three times each.
Data Quantification
Quantification of Western blots was performed using the Image J software (NIH, US) following the instructions provided. The numbers for quantification were relative abundance of proteins in all cases; experiments were presented for the untreated or starting materials as 1 or 100.
For additional materials and methods, see the Supplemental Data.
Supplementary Material
Acknowledgments
This work was supported by NIH grants (HD08188 and NURSA) and Welch Foundation. We thank Ray-Chang Wu and David Lonard for providing reagents, and Charles Foulds for critical reading of the manuscript.
References
- Amazit L, Pasini L, Szafran AT, Berno V, Wu RC, Mielke M, Jones ED, Mancini MG, Hinojos CA, O’Malley BW, Mancini MA. Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation. Mol Cell Biol. 2007;27:6913–6932. doi: 10.1128/MCB.01695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KA, Noeldner PK, Reece K, Wadzinski BE, Means AR. Regulation and function of the calcium/calmodulin-dependent protein kinase IV/protein serine/threonine phosphatase 2A signaling complex. J Biol Chem. 2004;279:31708–31716. doi: 10.1074/jbc.M404523200. [DOI] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Cohen PT. Protein phosphatase 1--targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- Cohen PTW. Overview of protein serine/threonine phosphatases. In: Arino J, Alexander DR, editors. Protein Phosphatases. Heidelberg, Germany: Springer-Verlag; 2003. pp. 1–20. [Google Scholar]
- Denner LA, Weigel NL, Maxwell BL, Schrader WT, O’Malley BW. Regulation of progesterone receptor-mediated transcription by phosphorylation. Science. 1990;250:1740–1743. doi: 10.1126/science.2176746. [DOI] [PubMed] [Google Scholar]
- Fang Y, Sathyanarayanan S, Sehgal A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–1518. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Yi P, Wong J, O’Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font de Mora J, Brown M. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol Cell Biol. 2000;20:5041–5047. doi: 10.1128/mcb.20.14.5041-5047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Protein serine/threonine phosphatases: life, death, and sleeping. Curr Opin Cell Biol. 2005;17:197–202. doi: 10.1016/j.ceb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- Jang YM, Kim DW, Kang TC, Won MH, Baek NI, Moon BJ, Choi SY, Kwon OS. Human pyridoxal phosphatase. Molecular cloning, functional expression, and tissue distribution. J Biol Chem. 2003;278:50040–50046. doi: 10.1074/jbc.M309619200. [DOI] [PubMed] [Google Scholar]
- Janssens V, Goris J, Van Hoof C. PP2A: the expected tumor suppressor. Curr Opin Genet Dev. 2005;15:34–41. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Lee SK, Kim HJ, Kim JW, Lee JW. Steroid receptor coactivator-1 and its family members differentially regulate transactivation by the tumor suppressor protein p53. Mol Endocrinol. 1999;13:1924–1933. doi: 10.1210/mend.13.11.0365. [DOI] [PubMed] [Google Scholar]
- Lefebvre P, Gaub MP, Tahayato A, Rochette-Egly C, Formstecher P. Protein phosphatases 1 and 2A regulate the transcriptional and DNA binding activities of retinoic acid receptors. J Biol Chem. 1995;270:10806–10816. doi: 10.1074/jbc.270.18.10806. [DOI] [PubMed] [Google Scholar]
- Li C, Wu RC, Amazit L, Tsai SY, Tsai MJ, O’Malley BW. Specific amino acid residues in the basic helix-loop-helix domain of SRC-3 are essential for its nuclear localization and proteasome-dependent turnover. Mol Cell Biol. 2007;27:1296–1308. doi: 10.1128/MCB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci U S A. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, et al. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Wang RH, Berndt N. Protein phosphatase 1alpha activity prevents oncogenic transformation. Mol Carcinog. 2006;45:648–656. doi: 10.1002/mc.20191. [DOI] [PubMed] [Google Scholar]
- Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- Lonard DM, Nawaz Z, Smith CL, O’Malley BW. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol Cell. 2000;5:939–948. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Lu Q, Surks HK, Ebling H, Baur WE, Brown D, Pallas DC, Karas RH. Regulation of estrogen receptor alpha-mediated transcription by a direct interaction with protein phosphatase 2A. J Biol Chem. 2003;278:4639–4645. doi: 10.1074/jbc.M210949200. [DOI] [PubMed] [Google Scholar]
- Lu X, Bocangel D, Nannenga B, Yamaguchi H, Appella E, Donehower LA. The p53-induced oncogenic phosphatase PPM1D interacts with uracil DNA glycosylase and suppresses base excision repair. Mol Cell. 2004;15:621–634. doi: 10.1016/j.molcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Luo W, Peterson A, Garcia BA, Coombs G, Kofahl B, Heinrich R, Shabanowitz J, Hunt DF, Yost HJ, Virshup DM. Protein phosphatase 1 regulates assembly and function of the beta-catenin degradation complex. EMBO J. 2007;26:1511–1521. doi: 10.1038/sj.emboj.7601607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SK, Allen JM, Clark JH, Blaustein JD, O’Malley BW. Convergent pathways for steroid hormone- and neurotransmitter-induced rat sexual behavior. Science. 1994;265:1246–1249. doi: 10.1126/science.7915049. [DOI] [PubMed] [Google Scholar]
- Mani SK, Fienberg AA, O’Callaghan JP, Snyder GL, Allen PB, Dash PK, Moore AN, Mitchell AJ, Bibb J, Greengard P, O’Malley BW. Requirement for DARPP-32 in progesterone-facilitated sexual receptivity in female rats and mice. Science. 2000;287:1053–1056. doi: 10.1126/science.287.5455.1053. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Mumby M. PP2A: unveiling a reluctant tumor suppressor. Cell. 2007;130:21–24. doi: 10.1016/j.cell.2007.06.034. [DOI] [PubMed] [Google Scholar]
- Naeem H, Cheng D, Zhao Q, Underhill C, Tini M, Bedford MT, Torchia J. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol. 2007;27:120–134. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SA, Wong J, Allred DC, Clark GM, Schiff R. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95:353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Qutob MS, Bhattacharjee RN, Pollari E, Yee SP, Torchia J. Microtubule-dependent subcellular redistribution of the transcriptional coactivator p/CIP. Mol Cell Biol. 2002;22:6611–6626. doi: 10.1128/MCB.22.18.6611-6626.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita A, Cardona GR, Koibuchi N, Suen CS, Chin WW. TRAM-1, A novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Wu H, Sun L, Zhang Y, Chen Y, Shi B, Li R, Wang Y, Liang J, Fan D, Wu G, et al. Coordinated regulation of AIB1 transcriptional activity by sumoylation and phosphorylation. J Biol Chem. 2006;281:21848–21856. doi: 10.1074/jbc.M603772200. [DOI] [PubMed] [Google Scholar]
- Wu RC, Feng Q, Lonard DM, O’Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Wu RC, Qin J, Hashimoto Y, Wong J, Xu J, Tsai SY, Tsai MJ, O’Malley BW. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) Coactivator activity by I kappa B kinase. Mol Cell Biol. 2002;22:3549–3561. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O’Malley BW. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Wu RC, Smith CL, O’Malley BW. Transcriptional regulation by steroid receptor coactivator phosphorylation. Endocr Rev. 2005;26:393–399. doi: 10.1210/er.2004-0018. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Z, Brew K, Lee EY. Mutational analysis of the catalytic subunit of muscle protein phosphatase-1. Biochemistry. 1996;35:6276–6282. doi: 10.1021/bi952954l. [DOI] [PubMed] [Google Scholar]
- Zheng FF, Wu RC, Smith CL, O’Malley BW. Rapid estrogen-induced phosphorylation of the SRC-3 coactivator occurs in an extranuclear complex containing estrogen receptor. Mol Cell Biol. 2005;25:8273–8284. doi: 10.1128/MCB.25.18.8273-8284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.