Abstract
Transcript elongation is a critical step in the production of mature messenger RNAs. Many factors have been identified that are required for transcript elongation, including Spt5. Studies in yeast determined that spt5 is required for cell viability, and analyses in Drosophila indicate Spt5 is localized to sites of active transcription suggesting it is required generally for transcription. However, the requirement for spt5 for cell viability in a metazoan organism has not been addressed. We determined that zebrafish foggy/spt5 is required cell-autonomously for the posterior migration of facial branchiomotor neurons from rhombomere 4 (r4) into r6 and r7 of the hindbrain. These genetic mosaics also give us the unique opportunity to determine if spt5 is required for mRNA transcription equivalently at all loci by addressing two processes within the same cell — neuronal migration and cell viability. In a wild-type host, spt5 null facial branchiomotor neurons survive to at least 5 days post fertilization (dpf) while failing to migrate posteriorly. This indicates that spt5-dependent transcript elongation is required cell autonomously for a complex cell migration, but not for the survival of these same cells. This work provides evidence that transcript elongation is not a global mechanism equivalently required by all loci, and may actually be under more strict developmental regulation.
Keywords: facial branchiomotor neurons, migration, transcript elongation, spt5
Introduction
Embryonic development depends on proper transcription and translation of a myriad of genes required for cell viability, morphogenesis, and induction and realization of cell fates. Transcription is a multi-step process that involves recruitment of RNA polymerase to a promoter by specific and general initiation factors, escape from the promoter, RNA chain elongation, and termination of the nascent transcript (Lee and Young, 2000). Transcript initiation at individual loci is regulated by factors that either promote or repress transcription of genes, and a number of genes identified in genetic screens for developmental defects regulate transcript initiation. While most work has focused on regulation of transcript initiation, it has recently become clear that transcript elongation is another step at which gene expression can be developmentally regulated. Recent work in the zebrafish has shown that the transcript elongation factors foggy/spt5 and pandora/spt6 are required for embryo survival, gene expression, and acquisition of particular neuronal cell fates (Guo et al., 2000; Keegan et al., 2002).
Genetic evidence in yeast first indicated that spt5 plays a role in transcript elongation, and biochemical analysis showed that Spt5 directly binds to the large subunit of RNA polymerase II (polII) in vitro via four KOW homology domains (Hartzog et al., 1998). The regulation of transcript elongation by Spt5 can be either positive or negative depending on the cellular context. For instance, Spt5 promotes transcript elongation in limiting nucleotide concentrations (Wada et al., 1998) or in response to heat shock stimulation (Andrulis et al., 2000; Keegan et al., 2002; Jennings et al., 2004). However, Spt5 is also a member of the DSIF (DRB sensitivity inducing factor) complex that is required for the elongation inhibitory activity of the ATP analogue 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) (Wada et al., 1998; Yamaguchi et al., 1999). While these experimental conditions reveal positive and negative elongation functions for Spt5, the in vivo context of positive versus negative control is still unknown.
Two in vivo studies in Drosophila melanogaster demonstrate that Spt5 colocalizes with actively transcribing phosphorylated RNA polymerase II (Pol II) to hundreds of sites on polytene chromosomes (Andrulis et al., 2000; Kaplan et al., 2000). This has been considered evidence that Spt5 has a general positive effect on virtually all active transcription; however the negative elongation effect appears to be less general. The first allele of foggy/spt5 to be described in the zebrafish, m806, a point mutant in the C terminus of the protein abolishes the negative effect on elongation in vitro while leaving its positive function intact (Guo et al., 2000). foggy/spt5m806 mutant embryos have a specific reduction in dopamine-secreting neurons in the hypothalamus and related neurons in the eye, and a corresponding increase in the number of serotonin-producing neurons in the hypothalamus. Recently, a point mutation in the Drosophila spt5 that primarily affects the negative transcript elongation function revealed a specific maternal requirement for spt5 to repress transcription of the pair-rule genes even-skipped and runt (Jennings et al., 2004).
The zebrafish foggy/spt5 null mutant exhibits a combination of specific phenotypes including heart defects and a number of neuronal defects (Keegan et al., 2002; this work). If spt5 is required equivalently at all loci, then one would expect the null phenotype, which reflects loss of positive as well as negative transcript elongation, to have severe and potentially early embryonic defects. The null phenotype has led to the important question of how loss of a general transcript elongation factor can lead to specific pheonotypes. One possibility is that spt5 is required for elongation at many or all loci; the specific phenotypes in particular tissues of zygotic spt5 mutants may be due to different rates of cell proliferation or to varying rates of spt5 mRNA and/or protein turnover in different tissues. spt5 mRNA is expressed maternally, and by this model the tissues most affected in spt5-/- embryos would be those that deplete this maternal expression the fastest. Cells lacking spt5 function would be predicted to die from lack of transcription at loci required to maintain cellular homeostasis. A second possibility is that the requirement for the positive effect of spt5 on elongation is not general, but is preferentially required by specific loci and specific developmental contexts. By this second hypothesis, one might expect to identify cells that require spt5 function for distinct aspects of their development, but not for their survival.
We identified an allele of foggy/spt5, fh20, in a genetic screen to identify genes that control the posterior migration of facial branchiomotor neurons in the zebrafish hindbrain. In wild-type zebrafish embryos, facial branchiomotor neurons are born in rhombomere 4 (r4) of the developing hindbrain and subsequently migrate caudally to reside in r6 and r7. A number of genes have been identified that are required for this migration to take place, some of which regulate transcription of unknown target genes (reviewed in Chandrasekhar, 2004). The identification of facial branchiomotor neuron migration defects in foggy/spt5 mutant embryos allowed us to test the hypothesis that spt5 is not required equivalently for production of all transcripts by addressing two processes within a single cell type. Using mosaic analysis we show that the positive effect of foggy/spt5 on elongation is required cell-autonomously for facial branchiomotor neurons to migrate posteriorly from r4, but is not required within these same cells for their survival. These data suggest that foggy/spt5 is not required equivalently for production of all mRNA transcripts within a cell and provides further evidence that transcript elongation via spt5 is a step at which control of gene expression may be developmentally regulated.
Results
fh20 is required for facial branchiomotor neuron migration and exhibits pleiotropic defects
We identified the fh20 mutant in a genetic screen for defects in facial branchiomotor neuron migration in the isl1-GFP transgenic line that expresses GFP in a subset of cranial neurons (Higashijima et al., 2000). fh20 mutant embryos display a pleiotropic morphological phenotype by 48 hpf which includes developmental delay first evident at about 26 hpf, severely reduced pigmentation, and a poorly differentiated heart tube accompanied by pericardial edema (Fig.1B). The somites of fh20 mutant embryos initially form correctly, but subsequently degenerate between 36 and 48 hpf. By 48 hpf, TUNEL staining revealed widespread neural cell death in fh20-/- embryos compared to wild-type (Fig. 1C, D). fh20-/- embryos die by 4 dpf, while heterozygous embryos have no detectable morphological phenotype (data not shown).
Fig. 1.
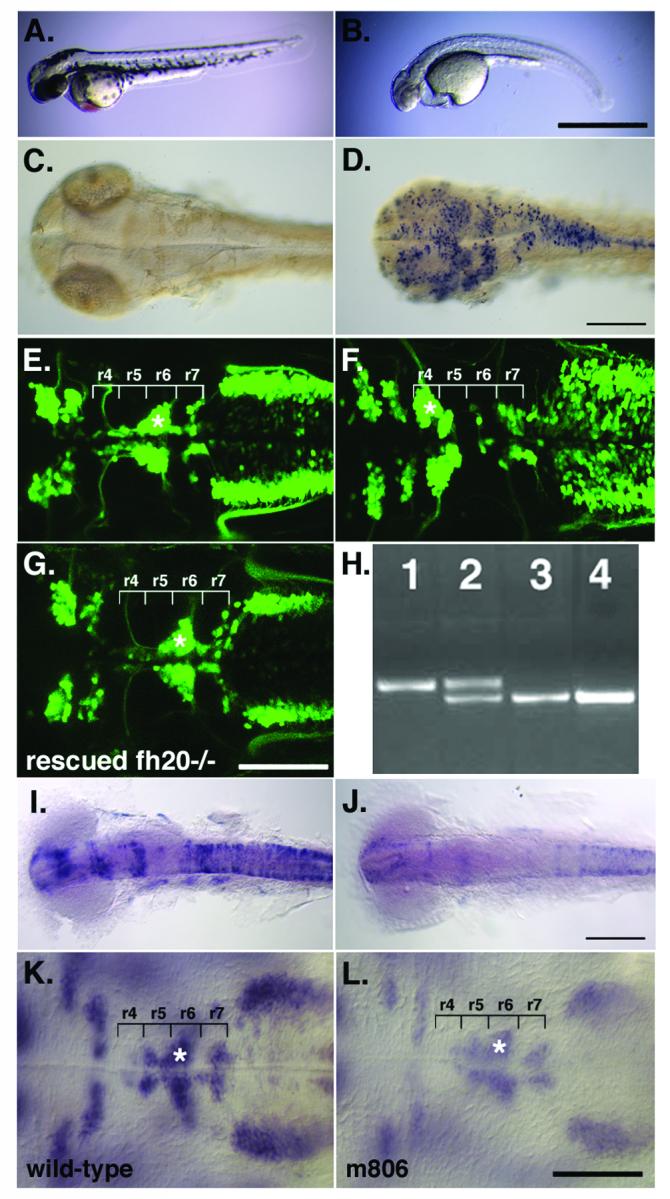
fh20-/- mutant embryos exhibit multiple defects including failure of facial branchiomotor neuron migration. Anterior is to the left in all panels. (B) The morphological phenotype of fh20-/- embryos includes developmental delay and failure of tail elongation, absence of pigment formation, failure of the ventral retina to close, heart tube hypotrophy accompanied by pericardial edema, and degeneration of somites. The mutation is lethal at about 4 dpf. TUNEL labeling indicates that apoptotic cell death is increased in (D) fh20-/- compared to (C) wild-type. (F) Facial branchiomotor neurons (asterisks) fail to migrate posteriorly from r4 in isl1-GFP transgenic fh20-/- embryos compared to (E) wild-type. (G) Full-length wild-type spt5 mRNA injected into fh20-/-embryos rescues facial branchiomotor neuron migration. (H) PCR genotypes for an absolutely linked microsatellite marker (30020R9) in (1) homozygous wild-type, (2) fh20+/-, (3) fh20-/-, and (4) the rescued fh20-/- embryo shown in (G). In situ hybridization for ngn1 expression indicates a decrease in intensity that correlates with the fh20 mutation; (I) wild-type, (J) fh20-/-. (K-L) Facial branchiomotor neurons, identified by islet1 in situ hybridization, migrate normally in the m806 point mutant of foggy/spt5 that abolishes the negative effect on transcript elongation. Scalebars = 900 μm (A, B) and 100 μm (C-L).
Facial branchiomotor neurons in wild-type isl1-GFP transgenic embryos migrate posteriorly from r4 to establish clusters in r6 and r7, leaving behind them axons that exit from lateral r4 to extend into the second branchial arch and innervate jaw support muscles (Higashijima et al., 2000; Fig. 1E, asterisk). In fh20 mutant embryos, facial branchiomotor neurons fail to migrate from r4, instead forming a characteristic “footprint” of facial branchiomotor neuron cell bodies in r4 and anterior r5 (Fig. 1F, asterisk). The facial branchiomotor axons collect at lateral r4 where they exit the hindbrain correctly, but fail to innervate their targets, presumably due to an absence of second arch-derived cartilages and muscles (data not shown).
In situ hybridization determined that expression of the genes dlx2, eng3, hoxb1a, hoxb4, EphA4, and myoD is indistinguishable between wild-type and fh20 mutant embryos suggesting that establishment of regional specification is unaffected. Additionally, expression of the neurogenic delta genes, deltaA, deltaB, and deltaD, is comparable in mutant and wild-type embryos. However, reduction of ngn1 mRNA expression in approximately 27% of embryos from an fh20+/- incross (24/86) was detected as early as the 8-somite stage, and this difference was maintained as late as the 18-somite stage (Fig. 1I-J). When these embryos were genotyped for the tightly linked marker 30020R9, we found that 19/24 faintly expressing embryos were genotypically fh20-/- while only 4/62 darkly expressing embryos were fh20-/-, indicating a strong correlation between ngn1 expression intensity and genotype (p<0.001). However, no neurogenic phenotype was apparent in fh20-/- embryos.
fh20 is a hypomorphic splice allele of zebrafish foggy/spt5
We placed fh20 on linkage group 15 between microsatellites z20993 and z7871, an interval that includes the transcript elongation factor, foggy/spt5. We tested whether fh20 is an allele of foggy/spt5 by complementation to the sk8 null allele of spt5 (Keegan et al., 2002) and rescue with wild-type spt5 mRNA. Crosses of fh20+/- to sk8+/- fish yielded 23 phenotypic mutants out of 97 embryos (23.7%) showing the two mutants fail to complement, and fh20 is likely an allele of foggy/spt5 (data not shown). We generated full-length spt5 mRNA for microinjection into one-cell stage embryos from an fh20+/-;isl1-GFP intercross to address whether wild-type spt5 mRNA can rescue fh20 mutant embryos. Seventeen percent (8/47) of phenotypically wild-type embryos (Fig. 1G) were genotypically fh20-/- as determined by PCR amplification of a tightly linked microsatellite marker (Fig. 1H), indicating wild-type spt5 mRNA rescues the fh20 phenotype.
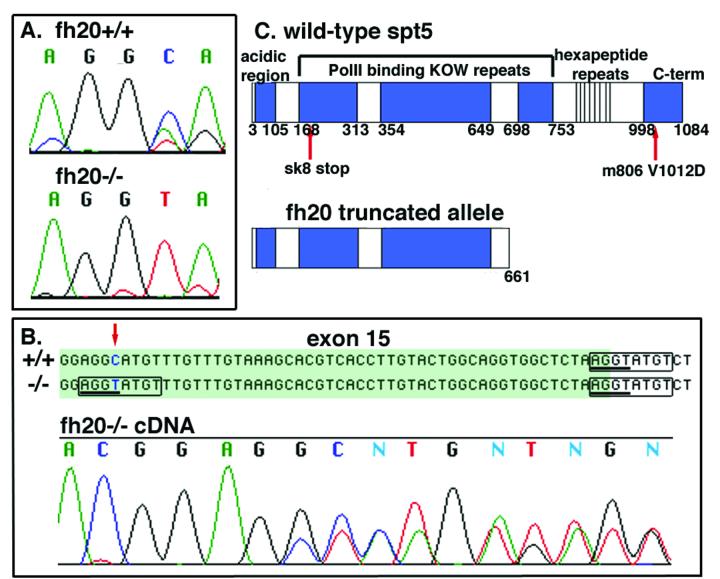
The protein structure of spt5 consists of an amino terminal acidic region, four conserved KOW Pol II binding repeats, and a carboxy-terminal hexapeptide repeat region. Genomic sequence from wild-type and fh20 mutant products identified a single base transition from C to T (Fig. 2A) that creates a conserved AGGT splice donor sequence and makes the eight nucleotide region around the change identical to the endogenous exon 15 splice donor which appears 50 bp downstream. Homozygotes express both correctly and incorrectly spliced products, as evidenced by a double trace from directly sequenced cDNA, indicating that the fh20 allele is a hypomorphic splice mutation that causes premature splicing from the middle of exon 15 to the splice acceptor of exon 16 (Fig. 2B). Thus the close proximity of the two splice donor sites leads to a hypomorphic mix of correctly and incorrectly spliced mRNA and a hypomorphic phenotype. The single base transition in a correctly spliced mRNA is a silent mutation, but the incorrectly spliced mRNA results in a frameshift that subsequently truncates the protein at an in-frame stop codon after amino acid 661, N-terminal to the fourth RNA Pol II binding repeat (Fig. 2C).
Fig. 2.
fh20 is a hypomorphic cryptic splice allele of foggy/spt5. (A) Genomic sequence analysis indicates the fh20 allele is a C-T transition corresponding to position 1904 of the spt5 mRNA. (B) This base change (red arrow) results in the creation of an AGGT consensus splice donor sequence (underlined), and the eight base region including the change (box in fh20) matches eight bases around the endogenous splice donor (boxes in wt and fh20) for exon 15 (shaded green). Sequence analysis of fh20-/- cDNA shows a double trace indicating that correctly and incorrectly spliced mRNA transcripts are present in the same fh20 mutant. Thus the fh20 allele is a hypomorph. (C) The misspliced mRNA results in a frameshift, and the protein terminates at amino acid (∼676), prior to the fourth KOW RNA pol II binding motif.
The fh20 allele of foggy/spt5 is expected to affect both the positive and negative aspects of transcript elongation since it is truncated in the region required for RNA Pol II binding, and the phenotype closely resembles that of the null allele. To determine if the facial branchiomotor neuron migration defects observed in fh20 mutants are primarily due to loss of positive or negative regulation, we examined facial branchiomotor neurons in the hypomorphic foggy/spt5m806 allele that is defective in the negative aspect of transcript elongation (Guo et al., 2000). Facial branchiomotor neurons in foggy/spt5m806 mutant embryos, detected by in situ hybridization for islet1 mRNA, migrate normally into r6 and r7 (Fig. 1K-L) indicating that facial branchiomotor neuron migration is primarily dependent on the positive transcript elongation function of spt5.
foggy/spt5 is required cell-autonomously for facial branchiomotor neuron migration but not for neuron survival
foggy/spt5 is expressed ubiquitously in the developing embryo and is presumably required for the elongation of a number of mRNA transcripts. To address whether spt5 is required cell-autonomously within facial branchiomotor neurons or in the environment of these cells as they migrate posteriorly through the hindbrain, we made genetic mosaics at gastrula stage (Fig. 3A) and analyzed the transplant hosts by confocal microscopy at 48 hpf. In these mosaics, donor-derived cells are red, and donor-derived motor neurons, which also express GFP, are yellow. Wild-type facial branchiomotor neurons placed in a wild-type host migrate normally through the hindbrain to take up residence in r6 and r7 (Fig. 3B; n=14/14). Facial branchiomotor neurons from a wild-type donor placed in a foggy/spt5fh20 mutant host migrate further posteriorly than do foggy/spt5fh20 cells (Fig. 3C; n=14/20), though less completely than in wt to wt control mosaics. This partial non-autonomous effect is likely due in part to the community effect we described previously where facial branchiomotor neurons migrate more completely in larger numbers than they do as individuals (Cooper et al., 2003). In contrast, foggy/spt5fh20 mutant facial branchiomotor neurons placed in a wild-type host fail to leave r4 but project axons into the periphery and look otherwise normal (Fig. 3D; n=11/11). These data suggest that spt5 is required primarily cell-autonomously for facial branchiomotor neuron migration.
Fig. 3.
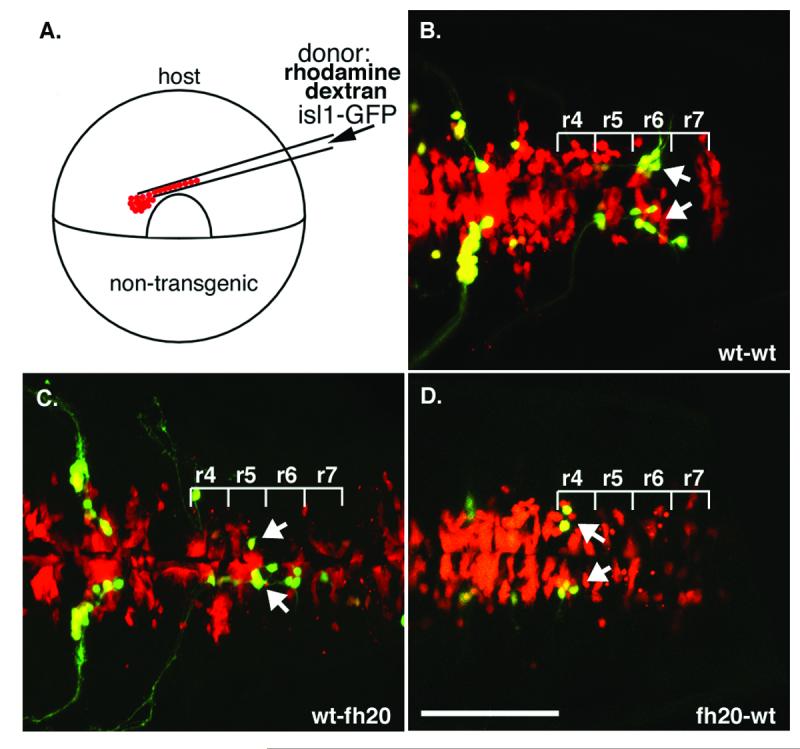
foggy/spt5 is required cell-autonomously for facial branchiomotor neuron migration. (A) Genetic mosaics were generated by transplanting cells from rhodamine dextran-labeled isl1-GFP transgenic donor cells to unlabeled hosts at gastrula stage (6 hpf), and confocal images were collected at 36 hpf. Donor-derived cells are red, and donor derived motor neurons, which express GFP, are yellow. (B) Wild-type facial motor neurons (arrow) migrate posteriorly in a wild-type host; n=14/14. (C) Wild-type facial motor neurons (arrows) also migrate posteriorly from r4 in a foggy/spt5fh20 mutant host, though to a lesser extent than in wild-type to wild-type control mosaics; n=14/20. (D) foggy/spt5fh20 facial motor neurons (arrow) fail to migrate from r4 in a wild-type host; n=11/11. Scalebar = 100μm.
Since foggy/spt5fh20 mutant embryos die by 4 dpf and exhibit widespread neuronal cell death by 48 hpf (Fig. 1D), it seemed possible that the cell-autonomous migration defect might reflect a role for spt5 in facial branchiomotor neuron survival, which would be expected to affect the neurons’ ability to migrate. We analyzed hindbrains of wild-type hosts containing foggy/spt5fh20 mutant donor-derived facial branchiomotor neurons at 5 dpf and observed that not only did foggy/spt5fh20 motor neurons survive in wild-type hosts, but they also remain in r4 four days after their wild-type counterparts have migrated posteriorly (Fig. 4B; n=12/13). Thus the cell-autonomous effect on facial branchiomotor neuron migration revealed by the hypomorphic foggy/spt5fh20 allele does not reflect a delay to migration or an effect on cell survival.
Fig. 4.
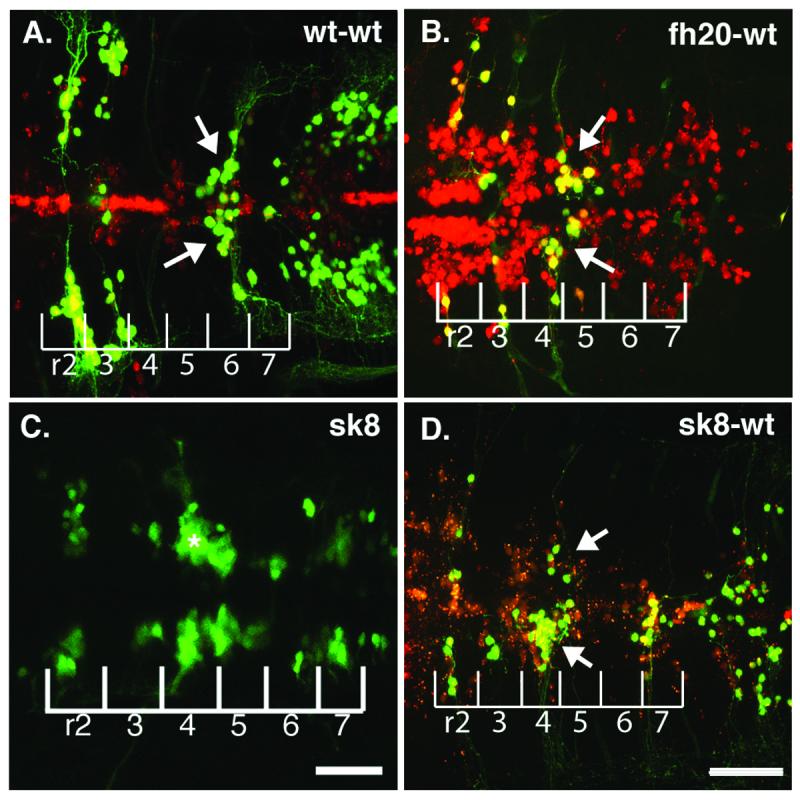
spt5 is not required for facial branchiomotor neuron survival to at least 5 dpf (A) wild-type transplant hosts containing wild-type donor cells (red) and donor-derived motor neurons (yellow) at 5 dpf. Wild-type facial motor neurons (arrow) in a wild-type host migrate posteriorly, differentiate and extend normal axonal processes from r4. (B) Hypomorphic foggy/spt5fh20 facial motor neurons (arrow) placed in a wild-type host fail to migrate but are still detectable by GFP expression and have normal morphology with axons extending from r4 at 5 dpf; n=12/13. (C) The isl1-GFP motor neuron phenotype of the foggy/spt5sk8 null allele at 48 hpf demonstrates that facial motor neurons fail to migrate posteriorly from r4 (asterisk). (D) foggy/spt5sk8 null facial motor neurons (arrow) placed in a wild-type host fail to migrate normally but have axons extending from r4 at 5 dpf; n=8/8. Scalebars = 40μm. Scalebar in D also corresponds with A and B.
Since the fh20 allele is hypomorphic, it remained possible that spt5 is in fact required for cell survival, but that genes required for survival are less sensitive to loss of spt5-dependent transcription than are genes required for facial branchiomotor neurons to migrate out of r4. To distinguish these hypotheses, we used the null sk8 allele of foggy/spt5 (Keegan et al., 2002) to perform genetic mosaic experiments similar to those performed using the fh20 allele. Facial branchiomotor neurons in 48 hpf foggy/spt5sk8 mutant embryos fail to migrate posteriorly from r4 (Fig. 4C) and appear indistinguishable from the hypomorphic fh20 facial branchiomotor neuron phenotype (Fig. 1F). foggy/spt5sk8 null cells were placed in wild-type hosts, and the host embryos were raised to 5 dpf. Confocal imaging of dissected fixed host brains revealed that foggy/spt5sk8 null facial branchiomotor neurons persist in the hindbrain and have axons that extend into the periphery (Fig. 4D; n=8/8). The presence of foggy/spt5 null facial branchiomotor neurons with normal morphology in r4 suggests that spt5 is required for neuronal migration but not for survival of these same cells at least until 5 dpf.
Discussion
Previous work has suggested that spt5 is required broadly for transcript elongation in yeast and Drosophila. Co-immunofluorescence in Drosophila showed that Spt5 colocalizes at sites of elongating phosphorylated Pol II on polytene chromosomes and that Spt5 is recruited to heat shock genes following heat shock (Andrulis et al., 2000; Kaplan et al. 2000). While this evidence has been used to suggest that Spt5 is required generally for transcript elongation, no loss of function study in Drosophila, or any other metazoan has provided evidence that spt5 is indeed required equivalently for all mRNA production. Zebrafish foggy/spt5 mutant embryos have multiple defects but survive until 4 dpf. One explanation for the pleiotropy of the foggy/spt5 mutant phenotype is maternal perdurance and the non-equivalent loss of maternal spt5 in different tissues. Maternal spt5 mRNA persists until 10 hpf (Keegan et al., 2002), and the stability of spt5 protein has not been determined. While this is a plausible explanation for certain aspects of the foggy/spt5 phenotype, it fails to explain the effects that we observe within individual cells — that spt5 is required for a complex migration behavior evident at 2 dpf but is not required for the survival of these same cells until at least 5 dpf. The cell autonomous failure of migration suggests that there is a deficit in the production of spt5-dependent transcripts, but the viability of these cells suggests that transcripts required autonomously for facial branchiomotor neuron cell viability are spt5-independent until at least 5 dpf.
It remains plausible that rather than affecting specific transcripts required for neuronal migration, spt5 increases the overall efficiency of transcript elongation, and that neuronal migration is more sensitive to this effect than neuronal survival. In addition to spt5, the genes spt4 and spt6 also code for transcript elongation factors (Hartzog et al., 1998). The zebrafish mutant or morpholino phenotype of spt4 is unknown, but the spt6-/- morphological phenotype closely resembles that of spt5 mutants (Keegan et al., 2002). The observation that loss of function of spt5 and spt6 together results in morphological and heat shock induction defects more severe than loss of either single gene suggests neither alone is a complete loss of transcript elongation. This is not likely due to a simple functional redundancy since spt5 mRNA does not rescue the spt6 mutant phenotype, nor does spt6 mRNA rescue loss of spt5 (Keegan et al., 2002). It is also important to note that spt6 has additional chromatin remodeling characteristics distinct from the function of spt5 that might account for its enhancement of the spt5-/- phenotype (Bortvin and Winston, 1996).
Recent work suggested that the negative elongation effect of spt5 is required at distinct loci (Guo et al., 2000; Jennings et al., 2004) possibly in a manner specific to cis-acting regulatory features rather than transcript characteristics such as length or GC content (Jennings et al., 2004). Although specific targets of Spt5 in zebrafish have not been identified, our genetic mosaic results looking at migration versus survival of facial branchiomotor neurons suggest that the positive effect on transcript elongation is also more specific than previously thought. Several genes have been identified that are required cell-autonomously for facial branchiomotor neuron migration in vertebrates and might be good candidates for Spt5 targets. These include the transcription factors hoxb1 (Studer et al., 1996; Goddard et al., 1996; McClintock et al., 2002; Cooper et al., 2003; Arenkiel et al., 2004), pbx4 (Cooper et al., 2003), nkx6.1 (Muller et al., 2003), and ebf1 (Garel et al., 2000). Of these only the functions of hoxb1a and pbx4 have been shown to be required for facial branchiomotor neuron migration in zebrafish (Cooper et al., 2003). In situ hybridization to investigate hoxb1a expression showed no difference between wild-type and foggy/spt5 mutants (data not shown), though the broad expression of hoxb1a throughout r4 precludes the ability to investigate expression specifically within facial branchiomotor neurons.
Holstege et al. (1998) showed that not all transcriptional machinery components are required equivalently for the production of mRNAs. A comparison of microarray expression data between yeast mutants for various transcription initiation components revealed that most are general in that they are required for the production of an equivalent set of transcripts that also require RNA polymerase II. Surprisingly, however, several transcriptional components were required for the expression of only a relatively small subset of these genes. For instance, a number of pheromone responsive genes depend on the mediator component Srb5, and the cyclin dependent kinase, Srb10, appears to repress genes regulating nutrient starvation response. Our results are consistent with a model in which transcript elongation components function with some degree of specificity on downstream genes. A similar microarray comparison in either Drosophila or zebrafish might uncover a surprisingly specific requirement for the positive and negative effects of spt5 on distinct subsets of developmentally regulated genes. Since Spt5 has been seen to localize globally to sites of active transcription (Andrulis et al., 2000; Kaplan et al., 2000), this specificity could be conferred by biochemical modifications to the hexapeptide repeat region or physical interactions with Spt5 that regulate its activity rather than subcellular localization. Further biochemical and cell biological analyses of spt5 will likely reveal the mechanisms by which transcriptional components can exert specific regulation over gene expression.
Experimental Procedures
Fish husbandry
Zebrafish and embryos were raised at 28.5°C and staged as previously described (Kimmel et al., 1995).
Genetic Screen, Linkage Analysis, and Cloning
The fh20 allele was identified in a forward genetic screen of the isl1-GFP transgenic zebrafish line (Higashijima et al., 2000) by utilizing the early pressure (EP) method (Johnson et al., 1995). To map the fh20 mutation, heterozygous females were crossed to wild-type males from the polymorphic Wik strain, and embryos were raised to adulthood. Linkage mapping was initiated in pools of 24 wt and fh20-/- gynogenetic diploid embryos generated by EP; this placed fh20 on chromosome 15. Further fine mapping using diploid mutant embryos localized fh20 to a 0.4 cM region between z20993 (4 recombinants/2334 meioses) and z7871 (2/682) containing the spt5 candidate gene.
Two overlapping fragments of spt5 cDNA (1.7 kb 5′ and 2.2 kb 3′) were amplified from wild-type and fh20 mutant embryos by RT-PCR. Direct sequencing of gel-purified PCR products was performed in both directions.
Embryo Rescue and Complementation Analysis
Full-length spt5 mRNA for injections was generated using the SP6 mMessage mMachine kit (Keegan et al., 2002) following manufacturers instructions (Ambion). Injections were performed into dechorionated 1-2 cell stage embryos of an fh20+/-; isl1-GFP intercross. Embryos were raised to 48 hpf and sorted based on the extent of facial branchiomotor neuron migration. These embryos were subsequently lysed, and DNA was used to genotype individual embryos by PCR amplification of a microsatellite marker that is tightly linked to the fh20 locus (0/3677). The microsatellite was identified from sequencing data produced by the Zebrafish Sequencing Group at the Sanger Institute and annotated at the Ensembl Zebrafish Genome Browser (v16.2.1) (30020R9For 5′- TTTTGTTAGTAGTTGTTTTTGGTC-3′; 30020R9Rev 5′-ATTCTGTCCACACAAATTCTTTAT-3′). We also performed complementation analysis by crossing an fh20+/- male to an sk8+/- female.
Genetic mosaics
Embryos from either an fh20+/-;isl1-GFPtg/+ or an sk8+/-;isl1-GFPtg/+ intercross were used for donors and hosts to generate genetic mosaics. Donor embryos were injected with 2.5% lysinated rhodamine dextran (10 kD MW Fluor-Ruby, Molecular Probes) in 0.2 M KCl. Transplants were performed at shield stage (6 hpf) as in Moens and Fritz (1999). All confocal analysis was performed as in Cooper et al (2003). The image of the sk8 to wild-type transplant host in figure 4D had faint GFP expression. Enhancement of the GFP channel caused bleed-though into the rhodamine channel resulting in yellow-red donor cells.
Acknowledgements
The authors would like to thank Hitoshi Okamoto and Shin-ichi Higashijima for the isl1-GFP transgenic line. Fish heterozygous for the null sk8 allele of foggy/spt5 and the construct used to generate full-length wild-type spt5 mRNA were a generous gift from Deborah Yelon. Wild-type and hypomorphic mutant embryos for the m806 allele were kindly provided by Su Guo. We are grateful for excellent fish care and technical assistance provided by Sean Rhodes and Jennifer Stout. We would also like to thank members of the Moens lab for insightful discussions and critical reading of the manuscript. This work was support by (CBM), PHS NRSA T32 GM07270 from NIGMS (KLC), and the FHCRC Summer Undergraduate Internship Program (JA). CBM is an investigator with the Howard Hughes Medical Institute.
Grant Sponsor: NIH NICHD; Grant number: HD37909
References
- Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel BR, Tvrdik P, Gaufo GO, Capecchi MR. Hoxb1 functions in both motoneurons and in tissues of the periphery to establish and maintain the proper neuronal circuitry. Genes Dev. 2004;18(13):1539–1552. doi: 10.1101/gad.1207204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272(5267):1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A. Turning heads: development of vertebrate branchiomotor neurons. Dev Dyn. 2004;229(1):143–161. doi: 10.1002/dvdy.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, Leisenring WM, Moens CB. Autonomous and nonautonomous functions for Hox/Pbx in branchiomotor neuron development. Dev Biol. 2003;253(2):200–213. doi: 10.1016/s0012-1606(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Garel S, Garcia-Dominguez M, Charnay P. Control of the migratory pathway of facial branchiomotor neurones. Development. 2000;127(24):5297–5307. doi: 10.1242/dev.127.24.5297. [DOI] [PubMed] [Google Scholar]
- Goddard JM, Rossel M, Manley NR, Capecchi MR. Mice with targeted disruption of Hoxb-1 fail to form the motor nucleus of the VIIth nerve. Development. 1996;122(10):3217–3228. doi: 10.1242/dev.122.10.3217. [DOI] [PubMed] [Google Scholar]
- Guo S, Yamaguchi Y, Schilbach S, Wada T, Lee J, Goddard A, French D, Handa H, Rosenthal A. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20(1):206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95(5):717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20(9):2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BH, Shah S, Yamaguchi Y, Seki M, Phillips RG, Handa H, Ish-Horowicz D. Locus-specific requirements for Spt5 in transcriptional activation and repression in Drosophila. Curr Biol. 2004;14(18):1680–1684. doi: 10.1016/j.cub.2004.08.066. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Africa D, Horne S, Postlethwait JH. Half-tetrad analysis in zebrafish: mapping the ros mutation and the centromere of linkage group I. Genetics. 1995;139(4):1727–1735. doi: 10.1093/genetics/139.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Morris JR, Wu CT, Winston F. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000;12:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan BR, Feldman JL, Lee DH, Koos DS, Ho RK, Stainier DYR, Yelon D. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development. 2002;129:1623–1632. doi: 10.1242/dev.129.7.1623. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genetics. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Kheirbek MA, Prince VE. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development. 2002;129(10):2339–54. doi: 10.1242/dev.129.10.2339. [DOI] [PubMed] [Google Scholar]
- Moens CB, Fritz A. Techniques in neural development. Methods Cell Biol. 1999;59:253–272. doi: 10.1016/s0091-679x(08)61829-4. [DOI] [PubMed] [Google Scholar]
- Muller M, Jabs N, Lorke DE, Fritzsch B, Sander M. Nkx6.1 controls migration and axon pathfinding of cranial branchio-motoneurons. Development. 2003;130(23):5815–5826. doi: 10.1242/dev.00815. [DOI] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identify and abnormal migration of motor neurons in mice lacking Hoxb-1. Development. 1996;125(6):1025–1036. [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Chaleff DT, Valent B, Fink GR. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984;107(2):179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]