Abstract
White adipose (fat) tissues regulate metabolism, reproduction and lifespan. Adipocytes form throughout life, with the most marked expansion of the lineage occurring during the postnatal period. Adipocytes develop in coordination with the vasculature, but the identity and location of white adipocyte progenitor cells are unknown. We used genetically marked mice to isolate proliferating and renewing adipogenic progenitors. We find that most adipocytes descend from a pool of these proliferating progenitors that are already committed either prenatally or early in postnatal life. These progenitors reside in the mural cell compartment of the adipose vasculature but not in the vasculature of other tissues. Thus, the adipose vasculature appears to function as a progenitor niche and may provide signals for adipocyte development.
How adipocytes (fat cells) develop is a fundamental biological question with important ramifications for human health and disease (1, 2). little is known about the identity, localization, or biological characteristics of endogenous adipocyte progenitors (2). These progenitors likely reside in the adipose stromal-vascular fraction (SVF), a heterogeneous mixture of cells operationally-defined by enzymatic dissociation of adipose depots followed by density separation from adipocytes (1, 3). Peroxisome proliferator-activated receptor gamma (PPARγ), a central regulator of fat formation, is necessary and sufficient for adipogenesis (4, 5). Thus, marking PPARγ-expressing cells in vivo might provide new insights into adipose lineage specification.
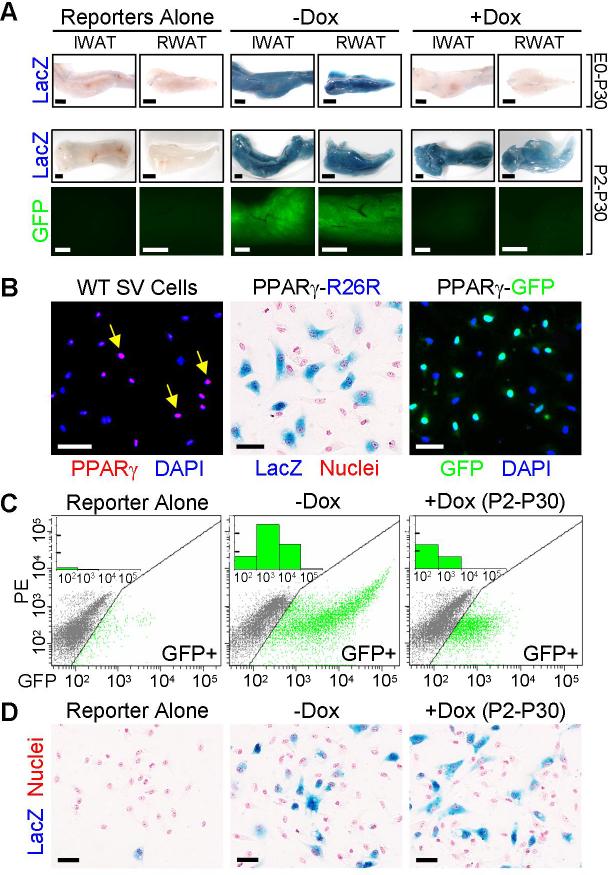
To mark and perform lineage analyses on PPARγ-expressing cells, we generated PPARγ-tet transactivator (tTA) (6) “knock-in” mice placing tTA under the control of the PPARγ locus (fig. S1). We introduced into these “PPARγ-tTA” mice two additional alleles: a tTA-responsive Cre allele (tetracycline response element-Cre; “TRE-Cre”) and an element that indelibly expresses lacZ in response to the Cre recombinase (ROSA26-flox-stop-flox-lacZ) (7, 8). With these genetic manipulations, we thereby created a “PPARγ-reporter” strain (“PPARγ-R26R” for PPARγ-Rosa26 reporter) in which the endogenous PPARγ promoter/enhancer induces expression of tTA, leading to Cre expression and an indelible lacZ marking of PPARγ-expressing cells and all descendants (fig. S1). The PPARγ-tTA strain functioned as expected; that is, it was active in adipose depots and repressed by doxycycline (Dox), establishing a tool to examine the adipose lineage (Fig. 1A, figs. S1 and S2).
Figure 1. PPARγ-expressing progenitors proliferate and maintain the precursor pool.
(A) PPARγ-tTA;TRE-Cre;R26R (PPARγ-R26R) or PPARγ-tTA;TRE-H2B-GFP (PPARγ-GFP) (bottom panels) mice were treated with or without Dox either from embryonic day 0 (E0) to postnatal day 30 (P30) (top row) or from P2 to P30 (middle and bottom rows) and then inguinal and retroperitoneal white adipose tissues (IWAT and RWAT respectively) were excised and examined for lacZ (blue) or GFP (green) expression. Left panels show equivalent depots of control mice containing either TRE-Cre;R26R or TRE-H2B-GFP.
(B) P30 stromal-vascular (SV) cells from wild-type (left panel), PPARγ-R26R (middle panel) and PPARγ-GFP (right panel) WAT were examined for expression of PPARγ (red) with immunocytochemistry (left panel) or for reporter expression. Left panel: Nuclei stained with DAPI (blue). Yellow arrows indicate cells that express PPARγ (purple). Middle panel: lacZ (blue), nuclei counterstained with nuclear fast red (red). Right panel: GFP (green), nuclei stained with DAPI (blue).
(C) Flow cytometry profiles of SV cells of untreated TRE-H2B-GFP (left) or PPARγ-GFP mice treated without (middle) or with Dox (right) from P2 to P30. X-axis is GFP fluorescent intensity. Y-axis is PE channel to help illustrate the distribution of GFP+ cells. Inset: X-axis is GFP fluorescent intensity; Y-axis is the cell count of the GFP+ cells per interval of fluorescent intensity (one unit = 1000). SV cells from TRE-H2B-GFP mice served as a gating control.
(D) SV cells removed from TRE-Cre;R26R (left panel) and PPARγ-R26R mice treated as indicated were isolated and stained with X-gal (blue) and nuclear fast red (red). Dox treatment did not alter the number or percentage of lacZ+ cells based upon statistical analysis of >2,000 cells counted in each group.
Scale bars: 2mm (A), 50μm (B, D)
To capture the rapid and dramatic expansion of the adipose lineage that occurs during the first postnatal month (1, 9), we Dox-treated the PPARγ-R26R mice starting at different days during this crucial window (fig. S3A). Interestingly, we found homogenous lacZ expression in P30 adipose depots that was not appreciably altered even when Dox administration began in the first postnatal days (Fig. 1A and fig. S3). This surprising result indicated that the vast majority of P30 adipocytes derived from a pre-existing pool of PPARγ-expressing cells, either adipocytes already present prenatally/early postnatally or proliferating precursors. Both interpretations conflict with previous data, however. The possibility that these cells are pre-formed adipocytes is incompatible with the proliferative increase that occurs over this time frame, while the notion that PPARγ-expressing cells are progenitors is inconsistent with cell culture studies (10, 11). To distinguish between the two possible interpretations, we examined the Dox-induced response of another reporter, TRE-H2B-GFP, that is stable in post-mitotic cells but, in contrast to the indelible lacZ marker, becomes diluted in proliferating cells following inhibition of the tet system (12, 13). Strikingly, Dox treatment (P2-P30) markedly reduced adipose depot and adipocyte GFP expression (Fig. 1A), indicating that PPARγ-expressing cells proliferate. Consistent with these data, ∼50% of adipocytes were labeled by bromodeoxyuridine (BrdU) when administered between P10 and P30 (fig. S4). The stability of lacZ marking together with the diminishing GFP expression indicate that adipose lineage cells, already instructed to express PPARγ prenatally, proliferate and are the major source of the spurt of adipocyte development observed in the first month of life.
The adipose stromal-vascular fraction (SVF) (fig. S5) is postulated to contain adipocyte progenitors (1, 14). We therefore investigated this location as a possible source from which the proliferating PPARγ-expressing cells characterized above may originate. Indeed we found that a subset of SV cells expressed immunocytochemically-detectable levels of PPARγ as well as the lacZ and GFP reporters (Fig. 1B and fig. S6). These SV resident PPARγ-expressing cells proliferate, as they incorporated BrdU after a brief 2-hour chase, even when the BrdU pulse-chase was initiated after 10 days of Dox pre-treatment to ensure that cells containing both GFP and BrdU expressed GFP prior to initiation of the brief BrdU pulse (fig. S7). In addition, GFP+ SV cells isolated by fluorescence-activated cell sorting (FACS) had considerable proliferative capacity (fig. S8). Further support for the in vivo proliferation of the GFP+ SV cells derives from flow cytometry profiles showing a Dox-induced (P2-P30) decrease in the number and fluorescent intensity of GFP+ SV cells (Fig. 1C and fig. S9). Notably, Dox did not reduce the number or percentage of lacZ+ SV cells, indicating that a pool of PPARγ-expressing cells remains in the SV compartment (Fig. 1D). Together these data indicate that the SV compartment of adipose depots contains PPARγ-expressing cells that divide, are mobilized from and also repopulate the SVF, and behave as an amplifying population that contributes to the adipocyte lineage.
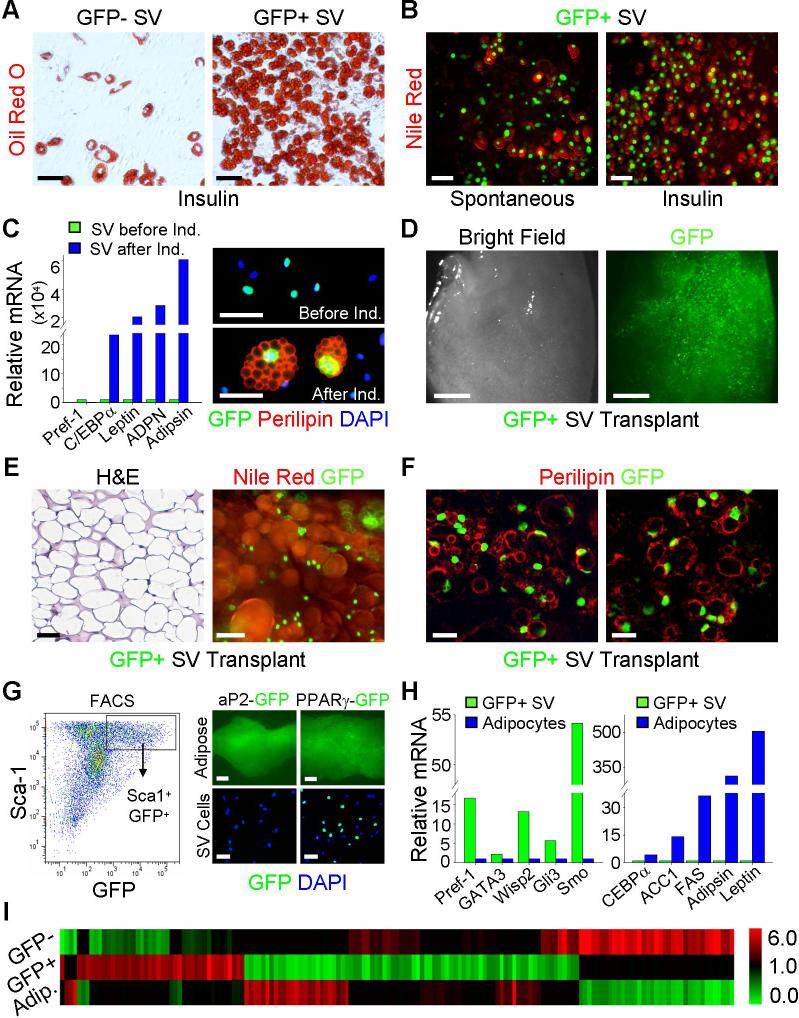
We assessed the adipogenic potential, in vitro and after transplantation, of FACS-isolated GFP+ SV cells (fig. S10). In culture, the sorted GFP+ SV cells underwent spontaneous and insulin-stimulated adipogenesis that was enhanced compared to GFP- SV cells (Figs. 2A, 2B and fig. S11). GFP+ SV adipogenesis mirrored the gene expression patterns described for preadipocyte cell line adipogenesis and the induced adipocytes expressed the perilipin protein with the appropriate subcellular distribution (Fig. 2C) (15). Moreover, freshly isolated GFP+ P30 SV cells transplanted into nude mice led to formation of an ectopic GFP+ depot, containing lipid-laden adipocytes that co-expressed GFP and perilipin (Figs. 2D, 2E and 2F). Thus these GFP+ SV cells have the proliferative and adipogenic properties expected of the endogenous progenitor population.
Figure 2. PPARγ-expressing SV cells are adipogenic and have a unique molecular signature.
(A) GFP- and GFP+ SV cells from PPARγ-GFP mice were sorted, plated, cultured to confluence and insulin-stimulated adipogenesis was examined with the lipid-specific stain Oil Red O (red).
(B) Sorted GFP+ SV cells were cultured in media or media supplemented with insulin and then stained with Nile Red, a lipid-specific fluorescent dye, to simultaneously visualize fat accumulation and GFP expression.
(C) Left panel: qPCR analysis of the indicated markers in sorted GFP+ cells before (green bars) and after (blue bars) insulin-stimulated adipogenesis. C/EBPα is an adipogenic transcription factor; leptin, adiponectin (ADPN), and adipsin are adipokines; Pref-1 is a preadipocyte marker whose expression inversely correlates with adipogenesis. Right panels: SV cells from P30 PPARγ-GFP adipose depots were examined for GFP (green) and perilipin (red, an adipocyte marker) expression both before (top) and after (bottom) adipogenic induction. Nuclei stained with DAPI (blue).
(D, E, F) FACS-isolated GFP+ SV cells were implanted into nude mice and the tissue that formed after 1 month was photographed with bright field (D, left panel) and fluorescent microscopy (D, right panel) and examined with H&E staining (E, left panel), GFP fluorescence and Nile Red staining (E, right panel), and GFP fluorescence and perilipin immunochemistry (F).
(G) Left panel: P30 PPARγ-GFP adipose depot SV cells were examined for Sca-1 and GFP expression with flow cytometry. The box indicates the Sca1+GFP+ double positive population. Right panels: IWAT (adipose, top) and SV cells (bottom) of aP2-GFP transgenics were analyzed for GFP expression, which was present in adipocytes but not SV cells. PPARγ-GFP serves as a control. Nuclei stained with DAPI (blue).
(H) qPCR analyses of the indicated markers of FACS-isolated GFP+ SV cells (green bars) and floated adipocytes (blue bars).
(I) GFP- SV cells, GFP+ SV cells and adipocytes were subjected to gene expression profiling. The heatmap illustrates 152 genes that differentiate GFP+ SV cells from the other populations. The red depicts a ≥2-fold increase in gene expression while the green depicts a ≤2-fold decrease in gene expression.
Scale bars: 50μm (A-C, E, G right panel bottom row), 2mm (D, G right panel top row), 20μm in confocal images (F).
To characterize the GFP+ SV progenitors and their relationship to other cells present in the adipose depot, we assessed cell surface marker expression using flow cytometry and FACS (fig. S10). The majority of GFP+ SV cells expressed Sca1 and CD34, but not CD105, CD45, TER-119, or Mac-1 (Fig. 2G and fig. S12). When these markers were used to positively or negatively select cells (and independently of the GFP reporter), we again isolated a subset of SV cells that generated a GFP+ ectopic adipose depot after transplantation (fig. S13). Some GFP+ SV cells could potentially be differentiated adipocytes that had yet to accumulate enough lipid to float during the density-based SV fractionation procedure. However, reporters driven by the promoter/enhancer of aP2 (16), a marker of adipocytes and a PPARγ target gene, displayed strong expression in adipose depots and adipocytes but not in SV cells, unlike the PPARγ reporters (Fig. 2G and fig. S14). Immunocytochemical analyses also showed that the GFP+ SV cells did not express perilipin, an adipocyte marker (Fig. 2C). In addition, the FACS-isolated GFP+ SV cells were molecularly distinct from adipocytes, expressing higher levels of the preadipocyte marker Pref-1, the adipogenic inhibitor GATA3, and targets of the anti-adipogenic Wnt (Wisp2) and Hedgehog (Smo, Gli3) pathways and much lower levels of numerous adipocyte markers (e.g., C/EBPα, FAS, leptin, etc.) (Fig. 2H and fig. S15A). Gene expression profiles further defined the GFP+ SV cells as a unique population within adipose tissues (Fig. 2I). Differentially expressed genes include: developmental transcription factors (e.g., goosecoid and Twist2); extracellular matrix genes (e.g., MMP3); anti-angiogenic factors (e.g., Stab1); and signaling cascade components (e.g., EGFR and FGF10) (fig. S15B). Thus, GFP+ SV cells are phenotypically distinct from adipocytes and other SV cells and have a unique molecular signature that allows prospective isolation for transplantation and further lineage analyses.
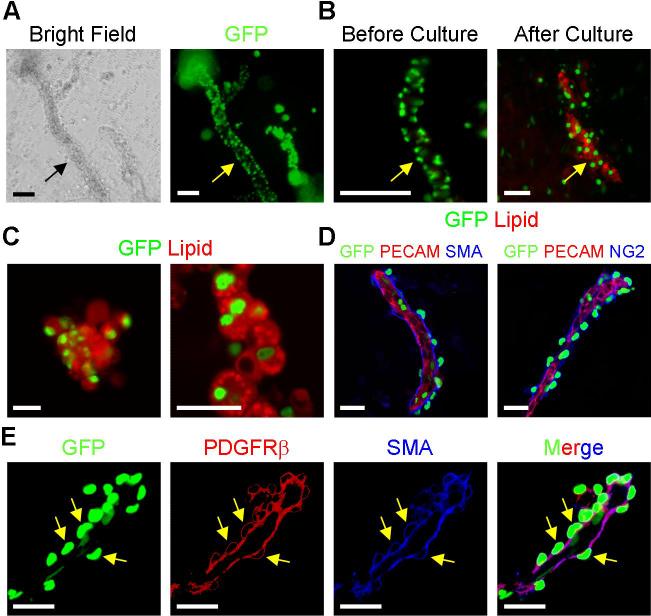
The local microenvironment (niche) is a crucial determinant of progenitor fate, function, and maintenance (17). In part due to the nature of the SV dissociation and isolation method, the anatomical location and neighboring cells of the SV adipocyte precursors are not known. To investigate the architecture of the SV compartment, we developed an SV particulate (SVP) isolation procedure designed to partially maintain the native SV structure while removing adipocytes that obscure visualization of the precursor location (fig. S16). In the SVPs, the majority of GFP+ cells were arrayed in tube-like structures (Fig. 3A). The GFP+ cells present in freshly isolated tubes did not contain lipid-droplets based upon inspection and lack of lipid staining (Figs. 3A and 3B). Organotypic cultures of SVPs led to formation of lipid-laden GFP+ adipocytes along the tubes, indicating that the tube-associated SVP GFP+ cells were adipogenic (Figs. 3B and 3C). Since the SVP tubes resembled blood vessels, we stained them with antibodies that recognize constituent cells of the vasculature including PECAM (endothelium) and three mural cell markers: SMA, PDGFRβ and NG2 (18). The SVP tubes expressed PECAM and were surrounded by cells that expressed SMA, PDGFRβ and NG2, indicating that they were vessels (Figs. 3D and 3E). Notably, GFP+ SVP cells expressed these mural cell markers (Figs. 3D and 3E). The notion that PPARγ might be expressed in a subset of mural cells is noteworthy as cultured mural cells, similar to mesenchymal stem cells, are multipotent and can be induced to undergo adipogenesis, chondrogenesis, osteogenesis and myogenesis and may provide a progenitor reservoir (18, 19).
Figure 3. SV particulate vessels contain GFP+ precursors that form adipocytes.
(A) SVP structures from P30 PPARγ-GFP mice were photographed with light (left panel) and fluorescent (right panel) microscopy. Arrows indicate SV tube.
(B) PPARγ-GFP SVP tubes were isolated and stained with the lipid-specific dye BODIPY either prior to culture (left panel) or after 3 days cultured on petri dishes in insulin (right panel). Arrows indicate SV tube.
(C) PPARγ-GFP SVP tubes were cultured in suspension. Formation of adipocytes that derive from the GFP+ tubes was assessed with BODIPY staining (red). GFP (green). Lipid droplets visualized with confocal microscopy (right panel).
(D) SVP isolates of P30 PPARγ-GFP mice were examined for expression of GFP (green) and the indicated endothelial (PECAM, red) and mural cell (SMA and NG2, blue) markers.
(E) PPARγ-GFP SVP vessel examined for expression of GFP (green), and the mural cell markers PDGFRβ (red) and SMA (blue). Yellow arrows indicate position of GFP+ nuclei within mural cells.
Scale bars: 50μm (A, B, left panels of C), 20μm in confocal images (right panels of C; D, E).
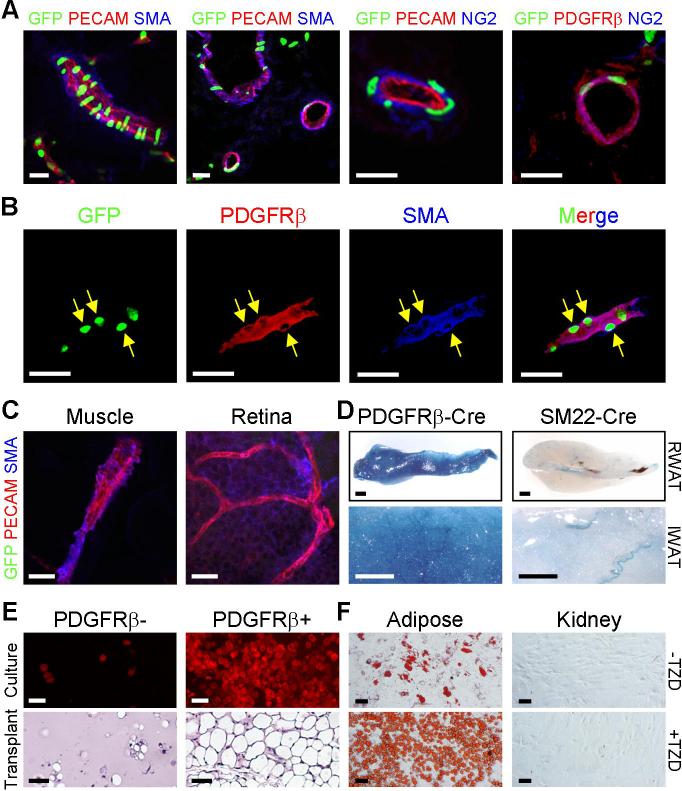
To investigate the distribution of the GFP+ progenitors within the mural cell compartment, we immunohistochemically-examined sections of freshly frozen PPARγ-GFP P30 adipose depots and other organs. In the adipose vasculature, we again observed co-localization of GFP and mural cell markers (Figs. 4A and 4B). The GFP+ vessels were of various sizes and disseminated throughout the depot (Fig. 4A). However, only a subset of mural cells within a vessel expressed GFP and some adipose vessels did not appear to harbor GFP+ progenitors (fig. S17). Strikingly, mural cells in other examined P30 tissues (including skeletal and cardiac muscles, kidney, retina, pancreas, spleen, lung, etc.) did not express the GFP reporter (Fig. 4C and fig. S18). In older animals (∼6 months), we did detect GFP in some small caliber PECAM-positive, SMA-negative adult skeletal muscle vessels (figs. S19 and S20). The majority of these adult skeletal muscle GFP+ cells expressed PECAM, and the cells were not adipogenic (fig. S20). Thus adipose depots appear to contain a unique population of progenitors present in the adipose depot mural cell compartment.
Figure 4. GFP+ cells are present in adipose depot mural cells.
(A) P30 PPARγ-GFP WAT was freshly frozen, cryo-sectioned, and examined with direct fluorescence for GFP and indirect immunofluorescence for the indicated endothelial (PECAM, red) and mural cell (SMA, blue; NG2, blue; PDGFRβ, red) markers.
(B) Cryo-section of a PPARγ-GFP adipose depot showing expression of GFP, PDGFRβ (red) and SMA (blue). Arrows indicate some mural cell nuclei that express GFP. (C) Muscle cryo-sections and retinal whole mount of PPARγ-GFP mice were examined for GFP, PECAM, and SMA as in (A, B). GFP was not expressed in mural cells of these tissues.
(D) RWAT (top panels, 5x) and IWAT (bottom panels, 20x) of P30 PDGFRβ-Cre;R26R and SM22-Cre;R26R mice were stained for β-galactosidase expression (blue).
(E) SV cells were isolated from P30 wild-type mice and sorted with a PDGFRβ antibody. Top panels: confluent PDGFRβ negative and positive cells were cultured in insulin and fat formation assessed with BODIPY (red). Bottom panels: PDGFRβ negative and positive cells were transplanted into nude mice, and the resultant tissues were sectioned and H&E stained.
(F) Adipose SVF and cells dissociated from kidney were sorted with a PDGFRβ antibody, and PDGFRβ positive cells were cultured in the absence (top) or presence of TZD (bottom).
Scale bars: 20μm in confocal images (A-C), 1mm (D), 50μm (E, F).
PDGFRβ marks mural cells and is required for their development (18). To explore the possibility that PDGFRβ-expressing cells were part of the adipocyte lineage, we X-gal stained adipose depots of P30 mice that contained both a PDGFRβ-Cre transgene (20), which expresses Cre in mural cells and other developing cells, and R26R. As a specificity control, we used SM22-Cre (21), a driver construct expressed in a subset of vascular smooth muscle cells. In these Cre-mediated lineage studies, we found that PDGFRβ-Cre generated strong and relatively homogenous lacZ expression throughout adipose depots in adipocytes and mural cells (Fig. 4D). In contrast, SM22-Cre did not, although lacZ was expressed in a distinct subset of adipose depot vessels (Fig. 4D).
To assess the adipogenic potential of PDGFRβ-expressing mural cells, we isolated PDGFRβ positive and negative cells from white adipose tissues and other organs by FACS, cultured them in insulin or transplanted them into nude mice (fig. S21). In both assays, the adipose depot PDGFRβ+ SV cells had high and substantially more adipogenic potential than PDGFRβ- SV cells (Fig. 4E); this adipogenesis was stimulated by thiazolidinediones (TZDs), diabetes drugs that activate PPARγ (22) (Fig. 4F). In contrast, PDGFRβ+ cells isolated from other organs did not display such potential and were unresponsive to TZDs (fig. S22 and Fig. 4F). Although we could identify sections that contained adipocytes in the non-adipose transplants, these adipocytes were GFP-negative (in contrast to adipocytes present in adipose depot SV PDGFRβ+ transplants) (fig. S22), apparently derived or recruited from host tissues. These data are consistent with the possibility that adipocyte progenitors reside as adipose depot mural cells with distinct properties such as adipogenic potential.
The intertwined epidemics of obesity and diabetes have led to a public health crisis that demands an improved understanding of adipocyte biology (2, 23). Yet the identity of the adipocyte progenitors and their precise location has remained elusive. Exploiting genetic reporters, we show that the pool of murine white adipocyte precursors has largely been committed prenatally or just after birth. These precursors divide, maintain the progenitor pool, and produce adipocytes. Some of these progenitors appear to be mural cells that reside in the vasculature of adipose tissues, results supported by early electron micrographic studies (24, 25). Thus the adipose vasculature appears to function as a progenitor niche and may provide signals for adipocyte development.
Several previous studies have documented an interplay between adipose tissue and the vasculature and shown that this interaction provides possible targets for obesity-diabetes therapies (26-29). The results described here add a new perspective to this interplay. In addition, they provide a foundation for further characterization of the adipose vascular niche and for prospective isolation of the adipocyte progenitors. Such experiments should help establish whether intervention in adipose lineage formation can be an effective therapeutic approach for obesity and diabetes.
Supplementary Material
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. Their manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Ailhaud G, Grimaldi P, Negrel R. Annu Rev Nutr. 1992;12:207. doi: 10.1146/annurev.nu.12.070192.001231. [DOI] [PubMed] [Google Scholar]
- 2.Gesta S, Tseng YH, Kahn CR. Cell. 2007;131:242. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Klaus S, Cassard-Doulcier AM, Ricquier D. J Cell Biol. 1991;115:1783. doi: 10.1083/jcb.115.6.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazar MA. Biochimie. 2005;87:9. doi: 10.1016/j.biochi.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 5.Farmer SR. Cell Metab. 2006;4:263. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kistner A, et al. Proc Natl Acad Sci U S A. 1996;93:10933. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu TS, Dandekar M, Monteggia LM, Parada LF, Kernie SG. Genesis. 2005;41:147. doi: 10.1002/gene.20110. [DOI] [PubMed] [Google Scholar]
- 8.Soriano P. Nat Genet. 1999;21:70. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 9.Cook JR, Kozak LP. Dev Biol. 1982;92:440. doi: 10.1016/0012-1606(82)90189-0. [DOI] [PubMed] [Google Scholar]
- 10.Altiok S, Xu M, Spiegelman BM. Genes Dev. 1997;11:1987. doi: 10.1101/gad.11.15.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosen ED, MacDougald OA. Nat Rev Mol Cell Biol. 2006;7:885. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 12.Kanda T, Sullivan KF, Wahl GM. Curr Biol. 1998;8:377. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 13.Tumbar T, et al. Science. 2004;303:359. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto TC, Lane MD. Crit Rev Biochem Mol Biol. 2005;40:229. doi: 10.1080/10409230591008189. [DOI] [PubMed] [Google Scholar]
- 15.Ntambi JM, Young-Cheul K. J Nutr. 2000;130:3122S. doi: 10.1093/jn/130.12.3122S. [DOI] [PubMed] [Google Scholar]
- 16.Graves RA, Tontonoz P, Platt KA, Ross SR, Spiegelman BM. J Cell Biochem. 1992;49:219. doi: 10.1002/jcb.240490303. [DOI] [PubMed] [Google Scholar]
- 17.Jones DL, Wagers AJ. Nat Rev Mol Cell Biol. 2008;9:11. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- 18.Armulik A, Abramsson A, Betsholtz C. Circ Res. 2005;97:512. doi: 10.1161/01.RES.0000182903.16652.d7. [DOI] [PubMed] [Google Scholar]
- 19.Dellavalle A, et al. Nat Cell Biol. 2007;9:255. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- 20.Foo SS, et al. Cell. 2006;124:161. doi: 10.1016/j.cell.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 21.Boucher P, Gotthardt M, Li WP, Anderson RG, Herz J. Science. 2003;300:329. doi: 10.1126/science.1082095. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann JM, et al. J Biol Chem. 1995;270:12953. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 23.Kopelman PG. Nature. 2000;404:635. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- 24.Iyama K, Ohzono K, Usuku G. Virchows Arch B Cell Pathol Incl Mol Pathol. 1979;31:143. doi: 10.1007/BF02889932. [DOI] [PubMed] [Google Scholar]
- 25.Cinti S, Cigolini M, Bosello O, Bjorntorp P. J Submicrosc Cytol. 1984;16:243. [PubMed] [Google Scholar]
- 26.Rupnick MA, et al. Proc Natl Acad Sci U S A. 2002;99:10730. doi: 10.1073/pnas.162349799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Nat Med. 2004;10:625. doi: 10.1038/nm1048. [DOI] [PubMed] [Google Scholar]
- 28.Kuo LE, et al. Nat Med. 2007;13:803. doi: 10.1038/nm1611. [DOI] [PubMed] [Google Scholar]
- 29.Nishimura S, et al. Diabetes. 2007;56:1517. doi: 10.2337/db06-1749. [DOI] [PubMed] [Google Scholar]
- 30.We thank S. Kennedy, T. Wang, R. Adams, R. Evans, S. Kernie, L. Monteggia, M. Osawa, R. Perlingeiro, Q. LaPlant, M. Iacovino as well as members of the Graff lab. JMG is a founder of Reata Pharmaceuticals, a privately-held company designed to address unmet needs in cancer, neurodegenerative, and inflammatory conditions. This work is supported by the NIH and NIDDK (1R01DK064261, 1R01DK066556) and also by the UTSW Excellence in Education Fund. WT, JMG, and UTSW may file a patent application on these studies.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.