Abstract
Chromosomal translocations involving the mixed lineage leukemia (MLL) gene are associated with aggressive acute lymphoid and myeloid leukemias. These translocations are restricted to an 8.3-kb breakpoint region resulting in fusion of amino terminal MLL sequences in frame to 1 of more than 60 different translocation partners. The translocations consistently delete the plant homeodomain (PHD) fingers and more carboxyl terminal MLL sequences. The function of the PHD fingers is obscure and their specific role in transformation has not been explored. Here we show that inclusion of the PHD fingers in the MLL fusion protein MLL-AF9 blocked immortalization of hematopoietic progenitors. Inclusion of 2 or more PHD fingers reduced association with the Hoxa9 locus and suppressed Hoxa9 up-regulation in hematopoietic progenitors. These data provide an explanation for why MLL translocation breakpoints exclude the PHD fingers and suggest a possible role for these domains in regulating the function of wild-type MLL.
Introduction
Balanced translocations involving mixed lineage leukemia (MLL) are common in human acute myeloid leukemia (AML) and acute lymphoid leukemia (ALL).1 MLL positively regulates Hox and other target gene expression through SET domain–dependent histone H3 lysine 4 methyltransferase activity.2,3 Like MLL, MLL fusion proteins bind directly to Hox loci and up-regulate their expression.2–5 Deregulated expression of Hox genes including Hoxa9 and the Hox cofactor Meis1 appears to be central for MLL fusion protein mediated transformation.6–8
Translocations in MLL occur within an 8.3-kb breakpoint cluster region (BCR) resulting in fusion of approximately 1400 amino acids of N-terminal MLL to 1 of more than 60 fusion partners.9 The BCR region has been extensively studied and contains several structural elements including Alu elements, topoisomerase II cleavage sites, and DNase I hypersensitive sites. These along with error-prone nonhomologous end joining have been proposed as mechanisms for MLL translocations.10 It is also possible that there is a functional reason for the location of the breaks. For example, sequences immediately 5′ to the BCR encode the CXXC domain, which must be retained intact for MLL-fusion protein transformation.11,12 Notably, sequences 3′ to the BCR, encoding the PHD fingers, are consistently deleted in leukemogenic fusion proteins. The function of PHD fingers in general is obscure. Some have been reported to mediate protein-protein interactions such as binding to methylated histones or other chromatin associated proteins.13,14 For example, HCF-1 binds to the HBM region between the bromodomain and PHD#4 of MLL and modulates transcription through E2F interactions.15–17 In addition, the nuclear cyclophilin Cyp33 has been reported to bind the PHD fingers leading to enhanced binding of HPC2 and BMI-1 to the CXXC domain.18,19
The consistent deletion of the PHD fingers in balanced translocations, the reports of interactions of the PHD fingers with corepressors,20 the disrupted MLL architecture in MLL partial tandem duplications in AML21 and the finding that MLL partial deletion of exons 7 and 8, which encode the first PHD finger, is associated with T-ALL22 all raise the possibility that the domains play a negative regulatory role in MLL function. Understanding the contribution of the PHD domain would provide insights into not only the location of breaks in the BCR but also the possible functions of the enigmatic PHD domains in normal and leukemogenic MLL function. Here we assess the role of the PHD fingers in transcriptional regulation and test the significance of inclusion of the PHD fingers, located downstream of the BCR, on MLL-fusion protein mediated transformation.
Methods
All animal studies were approved by the University of Michigan Committee on Use and Care of Animals and Unit for Laboratory Medicine. For complete materials and methods information, see Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results and discussion
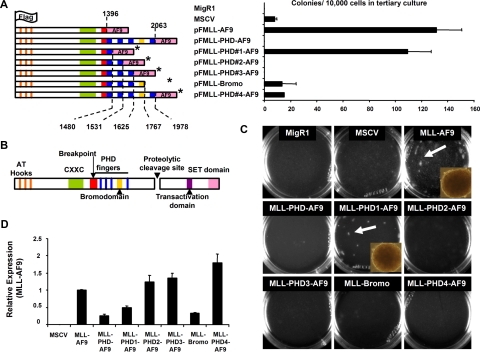
The effect of the PHD fingers on MLL fusion protein–mediated transcriptional regulation and transformation was assessed by engineering MSCV-based MLL-AF9 fusion constructs with various combinations of the 4 MLL PHD fingers and bromodomain (Figure 1A). Equal expression of retroviral constructs was confirmed in transiently transfected 293 cells by quantitative polymerase chain reaction (qPCR; Figure S1). Western blotting showed the fusion proteins were expressed at the predicted molecular weights (Figure S2). As evidence of MLL protein complex integrity the MLL fusions were found to coimmunoprecipitate Menin (Figure S3). Retrovirus was packaged and used to transduce bone marrow from 6- to 8-week-old C57/BL6 mice after 5-fluorouracil treatment as previously described.23,24 Transduced cells were cultured in methylcellulose in the presence of IL-3, IL-6, GM-CSF, SCF, and G418. After 8 days in culture, methylcellulose cultures were harvested and reseeded for secondary and tertiary cultures. While colonies were observed in all infection conditions, only MLL-AF9 and MLL-PHD#1-AF9–transduced cells formed a significant number of colonies in tertiary cultures (Figure 1A,C). These colonies displayed a compact morphology characteristic of MLL fusion protein mediated immortalization (Figure 1C insets). Notably, transduction with an MLL-AF9 construct that contained only the second PHD finger successfully immortalized mouse bone marrow (Figure S4). Compact colonies were not seen in cells transduced with other constructs, which showed colonies of diffuse differentiating cells in primary and secondary cultures that were largely absent in tertiary cultures (Figure 1A,C). The few colonies present in tertiary plating, with the exception of MLL-AF9 and MLL-PHD#1-AF9, consisted of differentiated cells (Figure 1A). Finally, expression of MLL-fusion proteins was confirmed in the transduced bone marrow by real-time PCR using primers selective for the exogenous transcripts (Figure 1D).
Figure 1.
The PHD fingers of MLL inhibit MLL-AF9 mediated transformation of mouse bone marrow. (A) MLL-AF9 and MLL-PHD-AF9 fusion constructs were engineered into the MSCV retroviral vector and transduced into mouse bone marrow. The final amino acid number of MLL in each construct is indicated with a dotted line. *indicates the absence of amino acids 1407 to 1432. Domains are colored-coded according to panel B. Bar graph indicates numbers of colonies per 10 000 cells plated in tertiary colony assays of transduced cells. Error bars indicate SD from duplicate experiments. (B) Schematic representation of wild-type MLL with color-coded domain structure. (C) Colony assays are shown after tertiary plating. Dense colonies were visible after tertiary plating only in MLL-AF9 and MLL-PHD#1-AF9–transduced cells. Representative dense transformed colonies are shown in insets. Micrographs were obtained using Olympus IX50 microscope, 10×/0.30 UPlanF1 Ph1 objective lens, Olympus DP70 camera, Olympus DP70 acquisition software, and Olympus DP contoller software (all Olympus, Center Valley, PA). (D) Real-time PCR detected expression of MLL-AF9 fusion constructs from cDNA generated from cells collected after the first round of colony assays. Expression is shown relative to MLL-AF9. Error bars indicate SD of experiments performed in triplicate.
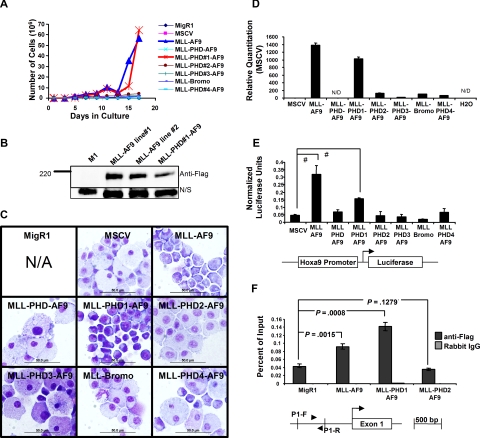
Liquid cultures of MSCV transduced bone marrow cells maintained in the presence of IL-3, SCF, and G418 provided further support for the growth inhibitory effects of the PHD fingers. Bone marrow cells transduced with MLL-AF9 and MLL-PHD#1-AF9 displayed a marked proliferative advantage compared with transductions with MSCV constructs containing 2 or more PHD fingers (Figure 2A). Western blotting for the flag tagged proteins in cell lines established from bone marrow transduced with MLL-AF9 and MLL-PHD#1-AF9 confirmed fusion protein expression (Figure 2B). Cells transduced with MLL-AF9 fusion constructs containing 2 or more PHD fingers failed to proliferate and became quiescent after 2 weeks in culture. Longer incubation resulted in cell death. Wright-Giemsa staining of cells after 15 days in culture (when significant differences in proliferation were observed; Figure 2A) revealed a more blastlike morphology of cells transduced with MLL-AF9 and MLL-PHD#1-AF9 compared with the other transductants, which showed full differentiation and were composed predominantly of macrophages (Figure 2C).
Figure 2.
Inclusion of the PHD fingers in MLL-AF9 leads to hematopoietic cell differentiation and reduced Hoxa9 expression. (A) Transduced mouse bone marrow was grown in liquid culture under G418 selection and in the presence of SCF and IL-3. A significant growth advantage was observed in MLL-AF9 and MLL-PHD#1-AF9–transduced cells. (B) MLL-AF9 and MLL-PHD#1-AF9 fusion proteins were detected by Western blot in cell lines generated from transduced bone marrow. N/S indicates Nonspecific band serving as loading control. (C) Morphology of transduced bone marrow was examined by Wright-Giemsa staining. Inclusion of 2 or more PHD fingers induced a more differentiated phenotype. Scale bars = 50 μm. N/A indicates not applicable. Micrographs were obtained using Olympus BX51 microscope, 100×/1.40 PLAN APO oil immersion objective lens, Olympus DP70 camera, Olympus DP70 acquisition software, and Olympus DP controller software. (D) Hoxa9 expression was examined using real-time PCR and cDNA from cells collected after the first round of colony assays. Significant suppression of Hoxa9 is detected after transduction with fusion constructs containing 2 of more PHD fingers. Error bars indicate SD from experiment performed in triplicate. N/D indicates not detected. (E) Luciferase assay showing significant transactivation by MLL-AF9 and MLL-PHD1-AF9. Assays were performed in transiently transfected 293 cells in serum starved growth conditions. A schematic of the Hoxa9-luciferase reporter used in the assay is shown. One of 3 representative experiments is shown. #P < .001 determined using Student t test. (F) Chromatin immunoprecipitation assay was performed in transiently transfected 293 cells. Immunoprecipitations were performed with Flag antibodies or normal rabbit IgG. Binding was detected by real-time PCR with the P1 primer set (▶, ◀) amplifying the promoter of HOXA9. Statistical significance was determined using Student t test. One of 3 representative experiments is shown. Similar results were obtained using a separate primer set amplifying the HOXA9 promoter.
Inclusion of the PHD fingers appeared to inhibit the ability of MLL-AF9 to activate transcriptional targets. Analysis by qPCR of cells after the first round of replating revealed a robust activation of the endogenous Hoxa9 locus by both MLL-AF9 and MLL-PHD#1-AF9 (Figure 2D) but not in cells transduced with MLL-AF9 constructs containing 2 or more PHD fingers. Furthermore, luciferase assays performed using a Hoxa9-LUC construct for a direct analysis of transactivation potential of MLL fusion proteins showed that only MLL-AF9 and MLL-PHD1-AF9 could significantly activate the Hoxa9 promoter (Figure 2E). Our experiments suggest that the differences between the activities of the fusion constructs are not simply the result of differences in protein stability (Figures S2,S3). One explanation for this would be that the PHD fingers have or recruit corepressor activity.18 However, experiments with tethering GAL4 fusions of the various PHD domains (not shown as well as Zeleznik-Le25) show neither an activating nor repressing effect of the PHD domains in isolation. However, chromatin immunoprecipitation experiments did reveal that, despite comparable levels of expression (Figures S2,S3), less MLL-PHD2-AF9 protein was bound to the HOXA9 locus than MLL-AF9 and MLL-PHD1-AF9, suggesting that inclusion of 2 or more PHD fingers alters the efficiency or stability of target loci binding (Figure 2F).
While we do not have a complete understanding of the role of the MLL PHD fingers, our data along with previous work suggest that translocations occur within the BCR region so that the DNA binding activity of the DNA methyltransferase homology region is preserved and the transcriptional suppression activity of the PHD domain is deleted. The PHD fingers may mediate transcriptional suppression by interaction with corepressors that directly repress transcription or destabilizing MLL-PHD-AF9 binding to target loci. Of note, we cannot completely exclude a mechanism of transformation inhibition where the PHD fingers function in trans. Identification of PHD interacting proteins is likely to shed light on the mechanisms of transcription suppression and inhibition of transformation. This will provide insights into how the normal function of MLL is regulated and how this process is perturbed in acute leukemia.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health grants RO1CA78815 and RO1CA92251 (J.L.H.) and T32 HL07622 (A.G.M.), and a Specialized Center of Research grant from the Leukemia & Lymphoma Society (White Plains, NY).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.G.M. designed research, analyzed data, and wrote the manuscript; D.G. and A.M.U. generated reagents; and J.L.H. designed research, analyzed data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jay L. Hess, MD, PhD, M5240 Medical Sciences I, 1301 Catherine Avenue, Ann Arbor, MI 48109-0602; e-mail: jayhess@umich.edu.
References
- 1.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura T, Mori T, Tada S, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 4.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 5.Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rozovskaia T, Feinstein E, Mor O, et al. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4: 11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- 8.Zeisig BB, Milne T, Garcia-Cuellar MP, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broeker PL, Super HG, Thirman MJ, et al. Distribution of 11q23 breakpoints within the MLL breakpoint cluster region in de novo acute leukemia and in treatment-related acute myeloid leukemia: correlation with scaffold attachment regions and topoisomerase II consensus binding sites. Blood. 1996;87:1912–1922. [PubMed] [Google Scholar]
- 10.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst) 2006;5:1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slany RK, Lavau C, Cleary ML. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi X, Hong T, Walter KL, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wysocka J, Swigut T, Xiao H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 15.Takeda S, Chen DY, Westergard TD, et al. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 2006;20:2397–2409. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 17.Yokoyama A, Wang Z, Wysocka J, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fair K, Anderson M, Bulanova E, Mi H, Tropschug M, Diaz MO. Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol Cell Biol. 2001;21:3589–3597. doi: 10.1128/MCB.21.10.3589-3597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci U S A. 2003;100:8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz DC, Friedman JR, Rauscher FJ., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD, bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caligiuri MA, Schichman SA, Strout MP, et al. Molecular rearrangement of the ALL-1 gene in acute myeloid leukemia without cytogenetic evidence of 11q23 chromosomal translocations. Cancer Res. 1994;54:370–373. [PubMed] [Google Scholar]
- 22.Lochner K, Siegler G, Fuhrer M, et al. A specific deletion in the breakpoint cluster region of the ALL-1 gene is associated with acute lymphoblastic T-cell leukemias. Cancer Res. 1996;56:2171–2177. [PubMed] [Google Scholar]
- 23.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 24.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeleznik-Le NJ, Harden AM, Rowley JD. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci U S A. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.