SUMMARY
A key step in angiogenesis is the upregulation of growth factor receptors on endothelial cells. Here we demonstrate that a small regulatory microRNA, miR-296 has a major role in this process. Glioma cells and angiogenic growth factors elevate the level of miR-296 in primary human brain microvascular endothelial cells in culture. The miR-296 level is also elevated in primary tumor endothelial cells isolated from human brain tumors compared to normal brain endothelial cells. Growth factor-induced miR-296 contributes significantly to angiogenesis by directly targeting the hepatocyte growth factor-regulated tyrosine kinase substrate (HGS) mRNA, leading to decreased levels of HGS and thereby reducing HGS-mediated degradation of the growth factor receptors VEGFR2 and PDGFR-β. Furthermore, inhibition of miR-296 with antagomirs reduces angiogenesis in tumor xenografts in vivo.
Keywords: miRNA, angiogenesis, cancer, PDGFR, VEGFR, HGS
SIGNIFICANCE
The formation of new blood vessels by angiogenesis is essential for normal functions and involved in many disease states, including cancer. Control of the angiogenic switch in endothelial cells involves changes in levels of pro- and anti-angiogenic molecules acting in concert. Here we show that glioma- or growth factor-mediated induction of miR-296 in endothelial cells results in increased levels of pro-angiogenic growth factor receptors. These results indicate that miR-296 - belonging to the family of “angiomirs” - is functionally linked to the angiogenic phenotype and therefore provides new insights into the role of miRNA regulation in neovascularization. Further, manipulation of miR-296 levels may prove therapeutic in the large number of diseases where angiogenesis is a critical component.
INTRODUCTION
Angiogenesis is the formation of new blood vessels during growth and development as well as in disease-related processes like tumor growth, wound healing and restoring blood flow to tissues after injury (Folkman, 2007). De novo angiogenesis is a critical factor in cancer. For instance, malignant brain tumors are characterized by a marked increase in blood vessel formation, with tumor vessels having abnormal morphology which serves as a key feature in tumor grading (Brem et al., 1972; Folkerth, 2000). Increasing awareness of the importance of the vasculature in tumors has led to a focus on this as a therapeutic target (Kerbel and Folkman, 2002). The state of angiogenesis is a balance between pro- and anti-angiogenic molecules with a bias towards the proangiogenic mode (Jain, 2005). A common feature of angiogenic blood vessels is the high expression of pro-angiogenic growth factor receptors, such as platelet-derived growth factor receptor (PDGFR) and vascular endothelial growth factor receptor (VEGFR), which are targets of anti-angiogenic therapies (Batchelor et al., 2007; Shih and Holland, 2006). Further understanding of the orchestration of this angiogenic switch should help in the development of strategies to harness the dynamics of blood vessel formation in human health and disease. Recently, the discovery of microRNAs (miRNAs) has increased our knowledge regarding the complex control of gene expression. miRNAs comprise a large group of endogenous non-coding RNAs that can block mRNA translation and/or negatively regulate its stability (Ambros, 2004). At this time over 500 different miRNAs have been identified in human cells (Griffiths-Jones et al., 2006). Accumulating evidence indicates that regulation of miRNA levels is very important for proper growth and differentiation of many cell types and tissues (Bartel, 2004; Kloosterman and Plasterk, 2006; Krichevsky et al., 2003). It is also becoming clear that deregulated miRNA expression is a common feature of many human diseases, especially specific forms of cancer (Calin and Croce, 2006; Esquela-Kerscher and Slack, 2006; Ruvkun, 2006). Here, we aimed at identifying miRNAs that are critical to tumor angiogenesis.
RESULTS
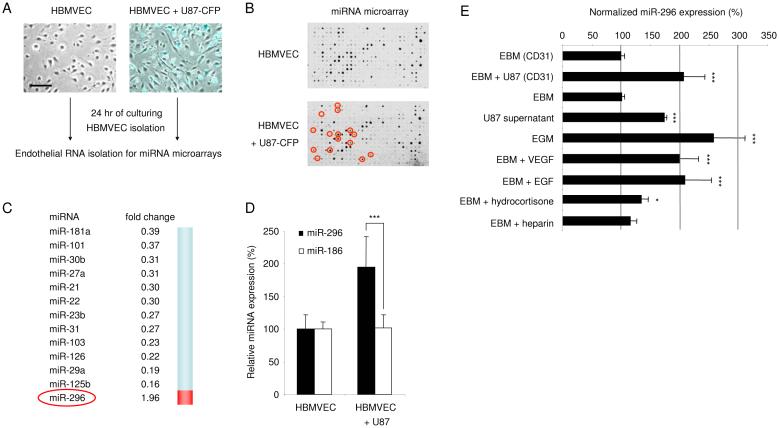
Since glioma cells have a high capacity to induce angiogenesis (Brem et al., 1972; Folkerth, 2000), we used them as a means to stimulate this process in normal endothelial cells in a co-culture system. Primary human microvascular endothelial cells isolated from normal human brain (HBMVECs; Cell Systems, ACBRI-376) were cultured in the presence or absence of human U87 glioma cells expressing the fluorescent protein Cerulean (CFP) in endothelial basal medium lacking additional angiogenic factors (EBM; Cambrex). Elongation of the endothelial cells was induced by the cancer cells as a first step in the activation of angiogenesis, as previously described (Khodarev et al., 2003) (Fig. 1A). After 24 hr of either culturing the endothelial cells alone or co-culturing them with human U87 glioma cells, the endothelial cells were isolated using CD31 magnetic beads (Dynal Biotech). The purity (>99%) of the endothelial cell preparation was confirmed by the absence of glioma cells expressing the CFP marker (data not shown). Total RNA was isolated from endothelial cells, and the small RNA fraction was hybridized to miRNA arrays containing probes for 407 mature miRNAs (as in (Krichevsky et al., 2003)) in order to identify differentially expressed miRNAs. Analysis of array hybridizations revealed eighty miRNAs expressed in HBMVECs at detectable levels (Fig. 1B and Supplementary Fig. S1), and confirmed the expression of a number of previously described miRNAs in endothelial cells (Kuehbacher et al., 2007; Poliseno et al., 2006; Suarez et al., 2007; Tuccoli et al., 2006). After exposure of HBMVECs to U87 glioma cells the expression levels of a number of miRNAs changed significantly. This suggests that glioma cells can influence miRNA expression in endothelial blood vessel cells (Fig. 1B and C). Most of the differentially expressed miRNAs were found to be down-regulated. One miRNA, miR-296, was identified and further confirmed by quantitative RT-PCR (qRT-PCR) analysis as up-regulated. We used miR-186 as a control miRNA and GAPDH as a normalization control, both of which were uniformly expressed in endothelial cells in the presence or absence of tumor cells (Fig. 1D). In this study we further investigated miR-296, since it was the only significantly up-regulated miRNA in the glioma-induced endothelial cells.
Fig. 1.
Glioma-induced disregulation of miRNAs in human brain endothelial cells (A) Primary human brain microvascular endothelial cells (HBMVEC) were cultured in the absence (left) or presence (right) of human U87-CFP glioma cells. Images were produced by using a combination of light and fluorescence microscopy, size bar 100 μm. (B) Array hybridization analysis of miRNAs extracted from CD-31+ cells, sorted from HBMVECs cultured without (upper array) or with (lower array) U87-CFP glioma cells for 24 hr. The density of the hybridization signals (black spots) reflects the relative expression level of particular miRNAs. Red circles indicate miRNAs which are significantly altered by co-culture with glioma cells. (C) A list of significantly decreased (fold change <0.5) or increased (fold change >1.9) miRNAs in HBMVECs exposed to U87-CFP glioma cells. (D) Overexpression of miR-296 was confirmed by qRT-PCR analysis. RNA extracted from CD31+ HBMVECs cultured in the absence or presence of U87 glioma cells was analyzed by qRT-PCR. The data were normalized to the level of GAPDH mRNA in each sample. (E) HBMVECs were cultured in the presence or absence of U87 glioma cells, isolated using CD31 beads, and subjected to miR-296 qRT-PCR. Alternatively, HBMVECs were cultured in various culture media and subjected to miR-296 qRT-PCR, the miR-296 levels were normalized to EBM (CD31). Error bars indicate S.D., *p < 0.05, ***p < 0.001, t test.
In order to determine what factors are responsible for the induction of miR-296, HBMVECs were cultured using various conditions after which RNA was isolated and the miR-296 expression levels were determined. Of note, miR-296 was not only induced in HBMVECs upon exposure of these cells to U87 glioma cells, but also by culture medium derived from U87 glioma cells and by EBM supplemented with angiogenic cocktail (EGM, containing hydrocortisone, EGF, FGF, VEGF, IGF, ascorbic acid, FBS, and heparin; Singlequots from Cambrex). Similar ∼2-fold induction of miR-296 in HBMVECs was achieved by EGM and glioma cell stimulation. Since U87-conditioned medium and EGM both contain many growth factors, HBMVECs were also stimulated with basal medium supplemented with VEGF or EGF, each resulting in significant miR-296 upregulation, in contrast to exposure to hydrocortisone and heparin which did not change levels (Fig. 1E).
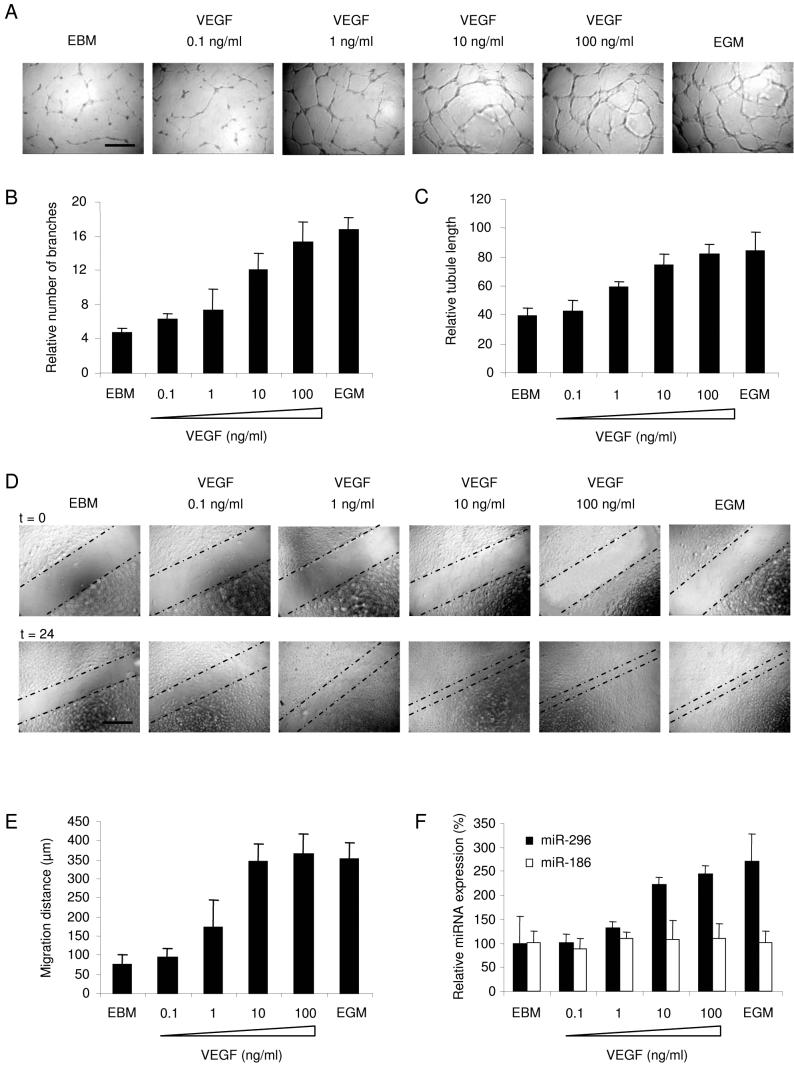
Since VEGF enhanced miR-296 expression in endothelial cells, we addressed VEGF function in angiogenesis in endothelial cell culture. First, HBMVECs were cultured In the presence of different amounts of VEGF in EBM on a Matrigel substrate to promote tubule formation for 24 hr. A clear dose-response effect of VEGF on endothelial tubule formation and branching was observed (Fig. 2A-C). In order to determine whether endothelial cell migration was also affected by VEGF, scratch assays were performed. HBMVECs were first cultured in EGM, then a spatula was used to scratch the monolayer, after which the cells were washed and cultured in EBM containing different amounts of VEGF, or in EGM, for 24 hr. VEGF significantly induced the migration of HBMVECs in a dose-response manner (Fig. 2D-E). To determine a correlation between VEGF-induced phenotype and miR-296 expression, HBMVECs were cultured for 24 hr in EBM containing different amounts of VEGF, or in EGM, after which RNA was isolated and the miR-296 expression levels were determined by qRT-PCR. Notably, 10 ng/ml of VEGF were sufficient to coordinately up-regulate miR-296 levels and induce endothelial cell migration and tubule formation in HBMVEC cultures (Fig. 2B, C, E, F).
Fig. 2.
VEGF-mediated induction of tubule formation, migration, and miR-296 expression (A) HBMVECs cells were cultured on Matrigel-coated plates in basal medium (EBM; Cambrex) only, basal medium supplemented with a cocktail of angiogenic factors (EGM), or different amounts of VEGF [size bar 300 μm], and analyzed for tubule branching (B) and tubule length (C) after 24 hr. (D) HBMVEC monolayer cultures were scratched and incubated as in (A). Directly after scratching (t = 0) and 24 hr later (t = 24) images were taken, size bar 300 μm. The dashed lines indicate the front of migration. (E) Quantitation of migration distance using MetaVue software. (F) HBMVECs were cultured as in (A) for 24 hr, after which RNA was isolated to determine the levels of miR-296 and miR-186 expression by qRT-PCR, The data were normalized to the level of GAPDH mRNA in each sample. Error bars indicate S.D.
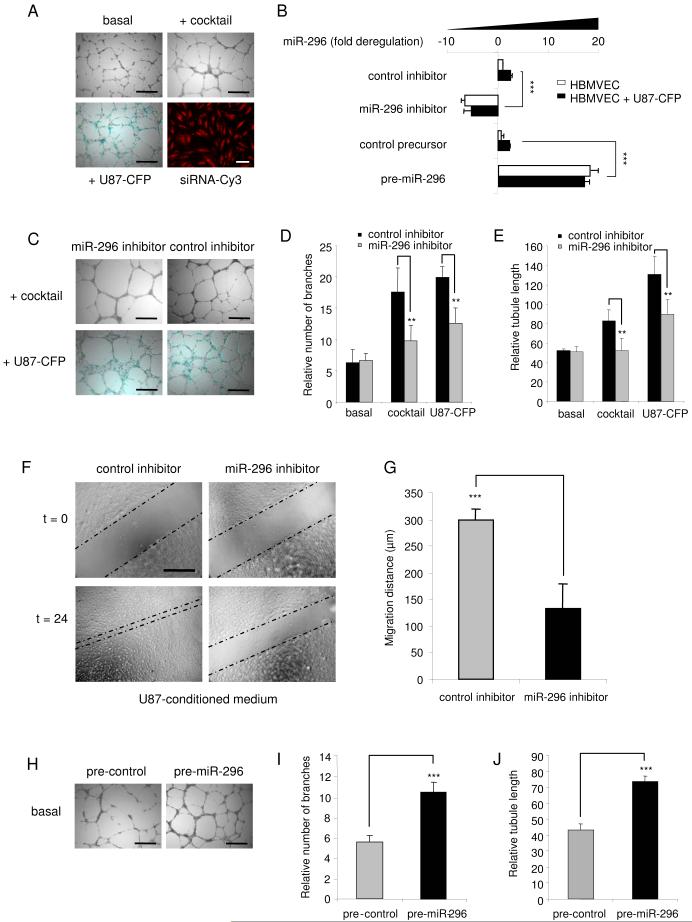
To address miR-296 function in angiogenesis, we examined effects of its inhibition on glioma-induced angiogenesis in cell culture. In these experiments, HBMVECs were cultured on Matrigel either in EBM alone, or with human glioma cells, or in EGM. Tubules were visualized by a combination of light and fluorescence microscopy. Endothelial cells were discriminated from CFP-expressing U87 glioma cells by fluorescence microscopy (Fig. 3A). miR-296 in endothelial cells was blocked by transfecting endothelial cells with a modified anti-sense miR-296 inhibitor (Ambion) prior to mixing the endothelial cells with U87 cells, with transfection efficiency being >99% as determined by transfection of siRNA-Cy3 (Alnylam) (Fig. 3A). Levels of miR-296 decreased 4-fold in HBMVECs in the presence of this inhibitor, as determined by qRT-PCR (Fig. 3B). At 48 hr after transfection of the miR-296 inhibitor or a non-related oligonucleotide of similar chemistry and 24 hr of culturing on Matrigel with or without U87-CFP cells, endothelial cells were analyzed for effects on tubule length and tubule branching (Fig. 3C). Downregulation of miR-296 resulted in a significant decrease in tubule branching (Fig. 3D) and total tubule length (Fig. 3E) in HBMVECs exposed to an angiogenic cocktail or U87 glioma cells as compared to unexposed cells, but had no significant effect on the viability of endothelial cells (Supplementary Fig. S2). U87 glioma cells induced tubule elongation to a greater extent than EGM, which may be due to other glioma cell induced effects, in addition to elevation of miR-296 (Fig. 3E). Similar inhibitory effects on tubule formation were observed when miR-296 was blocked in human umbilical vein endothelial cells exposed to an angiogenic cocktail (Supplementary Fig. S3), suggesting a general miR-296-mediated angiogenic mechanism operating in endothelial cells of different origins. In order to determine the effect of miR-296 inhibition on glioma-induced endothelial cell migration, scratch assays were performed. HBMVECs were transfected with the miR-296 inhibitor or control oligonucleotide. Twenty-four hr after transfection a scratch was made in the monolayer of HBMVECs cultured in U87-conditioned medium. Twenty-four hr after that the migration distance was analyzed by light microscopy and MetaVue software. Downregulation of miR-296 resulted in a significant decrease in U87-induced HBMVEC migration (Fig. 3F-G).
Fig. 3.
Angiogenesis co-culture assay: miR-296-mediated inhibition and induction of angiogenesis (A) HBMVECs cells were cultured on Matrigel-coated plates in basal medium (EBM; Cambrex) only, or basal medium supplemented with a cocktail of angiogenic factors (EGM), or with U87-CFP cells [size bar 300 μm]. Transfection efficiency of endothelial cells was determined (>99%) by using siRNA-Cy3 molecules [monolayer culture, size bar 50 μm]. (B) HBMVECs were transfected with anti-miR-296 inhibitor, pre-miR-296, or non-related control molecules and cultured in the presence or absence of U87-CFP cells. Inhibition and overexpression of miR-296 in CD31+-isolated endothelial cells was quantified by qRT-PCR. (C) HBMVECs were transfected with anti-miR-296 inhibitor or non-related control molecules and analyzed for tubule formation, size bar 300 μm. (D and E) Tubule formation was evaluated at 48 hr after transfection including 24 hr of culturing on Matrigel using the imaging program Image J. A significant decrease in tubule branching (D) and tubule length (E) was observed after transfection with the miR-296 inhibitor. (F) HBMVECs were transfected with anti-miR-296 inhibitor or non-related control molecules and analyzed for migration capacity. Inhibition of miR-296 resulted in a significant decrease in migration, as quantified in (G). (H) Overexpression of miR-296 at 24 hr after transfection of pre-miR-296 molecules resulted in increased angiogenesis in vitro [size bar 300 μm], as quantified by measuring tubule branching (I) and tubule length (J). Error bars indicate S.D., **p < 0.01 and ***p < 0.001, t test.
To investigate whether miR-296 acts as a dominant determinant of angiogenesis miR-296 precursor molecules or control precursor-like molecules (Ambion) were transfected into HBMVECs cultured in EBM medium. Transfection efficiency was >99% as analyzed by fluorescence microscopy using a precursor control molecule conjugated to a Cy3 fluorophor (Ambion) (data not shown). Overexpression of miR-296 in cells transfected with miR-296 precursors was confirmed by qRT-PCR (Fig. 3B). Twenty-four hr after transfection HBMVECs were plated on Matrigel in the absence of angiogenic stimuli. Pre-miR-296 stimulated tubule elongation and branching, in contrast to the control miRNA precursor (Fig. 3H-J). These effects were observed for up to at least 4 days after transfection of pre-miR-296 molecules into HBMVECs (Supplementary Fig. S4). Together these data support a positive regulatory role for miR-296 in the induction of angiogenesis, i.e. miR-296 inhibition reduces the angiogenic phenotype, while miR-296 overexpression increases it.
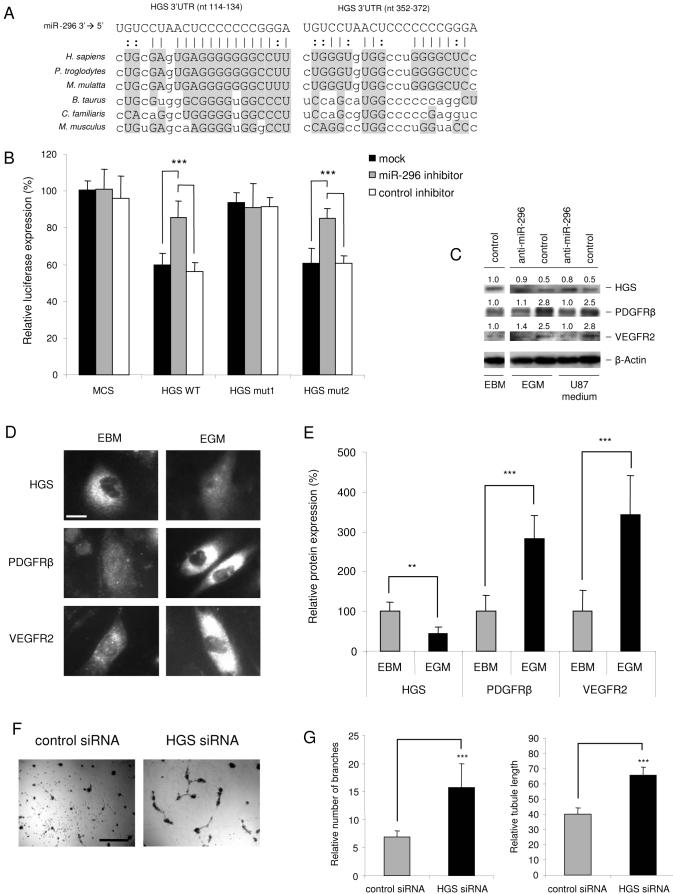
To identify target genes regulated by miR-296 and involved in control of angiogenesis, we next evaluated targets computationally predicted by publicly available algorithms (Griffiths-Jones et al., 2006; John et al., 2004; Krek et al., 2005; Lewis et al., 2005; Miranda et al., 2006). Interestingly, hepatocyte growth factor-regulated tyrosine kinase substrate (HGS) was one of the highest-scored (according to miRBase, http://microrna.sanger.ac.uk (Griffiths-Jones et al., 2006)) among the predicted miR-296 targets, with two potential conserved binding sites within its 3' UTR (Fig. 4A). The HGS protein is involved in the regulation of levels of growth factors receptors, such as PDGFR-β (Takata et al., 2000) and VEGFR2 (Ewan et al., 2006). Upon ligand-stimulation, phosphorylation, ubiquitination, and internalization of growth factor receptors, HGS mediates the sorting of these ligand/receptor complexes to lysosomes, where they are degraded (Bache et al., 2003; Ewan et al., 2006; Haglund et al., 2003; Raiborg et al., 2002; Stern et al., 2007; Takata et al., 2000). Elevated levels of growth factor receptors on the surface of endothelial cells, usually observed during tumor-induced angiogenesis, suggest a diminished role for HGS. In fact, silencing of HGS by siRNAs results in decreased degradation of growth factor receptors and hence their increased cellular levels (Bache et al., 2003; Stern et al., 2007). We hypothesized that miR-296 may down-regulate synthesis of HGS by direct binding to sites within the 3' UTR of its message, thereby facilitating accumulation of angiogenic growth factor receptors. To test this hypothesis, we constructed luciferase reporter vectors encoding the complete wild type 3' UTR of the HGS mRNA (HGS WT), as well as parallel control vectors containing mismatches in the predicted miR-296 binding sites (HGS mut1 and HGS mut2) or a luciferase vector containing no HGS UTR (MCS), and transfected them into miR-296-expressing HEK 293T cells (Miranda et al., 2006). Transfection of the HGS WT plasmid resulted in a decrease in luciferase expression (mock and control inhibitor) as compared to transfection of the MCS control (same conditions), supporting a negative effect of the endogenous miR-296 molecules on the HGS 3' UTR (Fig. 4B). Blockage of endogenous miR-296 molecules using anti-sense inhibitor (Ambion) resulted in a significant increase in HGS WT luciferase expression (Fig. 4B). Mutations in the predicted miR-296 binding site 1 (nt 114-134; HGS mut 1, Fig. 4A and 4B) caused a loss of luciferase sensitivity to miR-296 inhibition as compared to HGS WT, while HGS mut 2 with a mutation in miR-296 binding site 2 (nt 352-372) did not. These experiments validated the regulatory potential of miR-296 via the binding site 1, which has a better predicted base-pairing and is more conserved evolutionary relative to binding site 2, within the HGS 3'UTR.
Fig. 4.
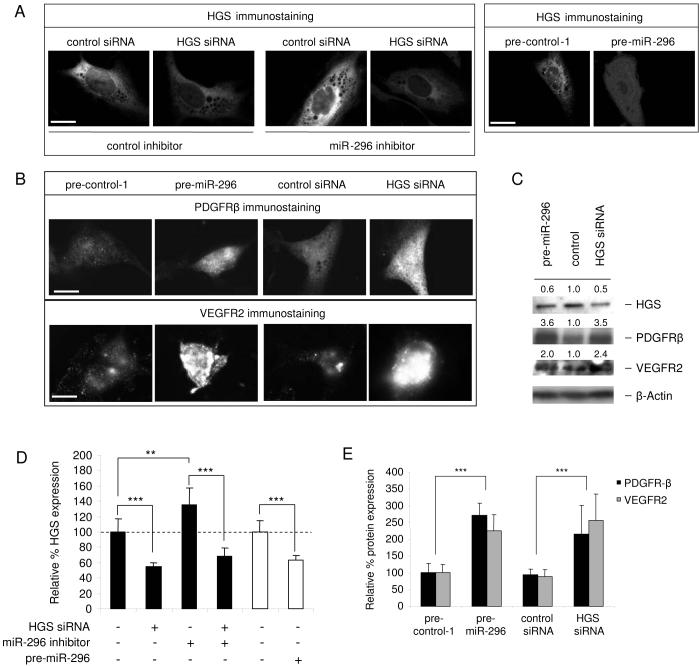
HGS is a direct target of miR-296 (A) Alignment of potential miR-296-binding sites in the 3' UTR of the HGS mRNA of different species. (B) pMir-Report vectors containing no 3'UTR (MCS), or containing the 3' UTR of the HGS mRNA (HGS WT) or mutated miR-296 binding sites (HGS mut1 and HGS mut2) and miR-296 or control inhibitors were co-transfected into HEK 293T cells. The inhibition of miR-296 by the anti-sense inhibitors resulted in a significant increase in luciferase signals of HGS WT and HGS mut2 but not HGS mut1 transfected cells. (C) Western blot analysis of HGS, PDGFR-β and VEGFR2 expression in HBMVECs cultured in basal medium (EBM) or HBMVECs stimulated by EGM or U87-conditioned medium. HGS expression decreased upon stimulation of HBMVECs, and PDGFR-β and VEGFR2 increased. Upon inhibition of miR-296 with anti-miR-296 molecules by transfection of HBMVECs stimulated by EGM or U87-conditioned medium, HGS expression increased, and PDGFR-β and VEGFR2 decreased. The relative blot intensities were quantified by using ImageQuant, the densitometric values normalized to β-Actin are indicated. (D)HGS, PDGFR-β and VEGFR2 expression levels under normal and angiogenic conditions. Representative images of HBMVEC immunostainings of HGS, PDGFR-β and VEGFR2 under normal (EBM) and angiogenic (EGM) conditions, size bar 20 μm. (E) Quantification of the HGS, PDGFR-β and VEGFR2 immunostaining levels, as represented in (D). Fluorescence signals were quantified using MetaVue software by analyzing at least 20 random cells per sample. (F) Silencing of HGS by siRNAs leads to increased HBMVEC tubule formation on Matrigel [size bar 300 μm], as quantified in (G) and (H). Error bars indicate S.D., **p < 0.01, ***p < 0.001, t test.
In order to confirm miR-296-mediated modulation of endogenous HGS in endothelial cells, HBMVECs were transfected with miR-296 inhibitor or control molecules and cultured in EGM or U87-conditioned medium. After 48 hr the cell lysates were analyzed for HGS expression levels by Western blotting. Inhibition of miR-296 resulted in increased levels of HGS (Fig. 4C). To determine whether HGS protein levels were reduced in angiogenic endothelial cells, HBMVECs were cultured in EBM, EGM, or U87-conditioned medium. After 24 hr, the HGS, PDGFR-β and VEGFR2 protein expression levels were determined by Western blotting and normalized using β-Actin levels. HBMVECs cultured in the angiogenesis-inducing (and miR-296-inducing) EGM or U87-conditioned medium exhibited decreased HGS expression levels, and increased PDGFR-β and VEGFR2 expression levels (Fig. 4C). In parallel, HGS, as well as PDGFR-β and VEGFR2 protein expression levels were determined under normal (EBM) and angiogenic (EGM) conditions by immunostaining. Immunostaining (Fig. 4D) and quantitation of the fluorescence intensities (Fig. 4E) showed that the HGS protein level is decreased at least 2-fold by angiogenic factors, whereas the PDGFR-β and VEGFR2 protein levels are increased about 3-fold. These results show that HGS is down-regulated under angiogenic conditions (in which miR-296 was up-regulated) with this reduction being prevented by blocking miR-296, and with HGS expression levels being inversely correlated with PDGFR-β and VEGFR2 levels. To further confirm the functional effect of HGS on angiogenesis, HGS siRNAs (Qiagen) or control siRNAs were transfected in HBMVECs. After 24 hr the transfected HBMVECs were cultured in EBM on Matrigel. Silencing of HGS resulted in increased tubule formation and branching (Fig. 4F-G), supporting critical involvement of HGS in the angiogenic process.
Importantly, anti-sense inhibition of miR-296 resulted in a moderate but significant increase in levels of endogenous HGS in HBMVECs and the effect was abolished by co-transfection with an siRNA directed against HGS, as shown by immunofluorescence visualization (Fig. 5A) and quantification of average fluorescence intensities (Fig. 5D). In addition, overexpression of miR-296 resulted in moderate but also significant downregulation of HGS expression (Fig. 5A and 5D). The knock-down of HGS by siRNA or pre-miR-296 was confirmed by Western blots (Fig. 5C). Notable, when HGS levels were decreased by either siRNA or pre-miR-296 transfection, a significant increase in PDGFR-β and VEGFR2 protein levels was observed by immunofluorescence staining (Fig. 5B and 5E), and confirmed by Western blots (Fig. 5C), consistent with downregulation of HGS by miR-296 which, in turn, reduces degradation of the growth factor receptors. Both down- and upregulation of miR-296 is considered to be physiologically relevant, since they affected the levels of downstream proteins (HGS, VEGFR2 and PDGFR-β) within the physiologically relevant range observed in angiogenic versus non-angiogenic conditions.
Fig. 5.
miR-296 affects HGS-modulated PDGFR-β and VEGFR2 expression (A) HGS immunostaining of HBMVECs after treatment with miR-296 inhibitors, anti-HGS siRNAs, and pre-miR-296 molecules, size bar 20 μm. (B) Immunostaining of PDGFR-β and VEGFR2 48 hr after transfection shows that transfection of pre-miR-296 molecules, as well as anti-HGS siRNAs, resulted in increased PDGFR-β and VEGFR2 expression in HBMVECs, size bar 20 μm. (C) Western blot analysis of HGS, PDGFR-β and VEGFR2 expressio in HBMVECs upon silencing of HGS by HGS siRNA or pre-miR-296. The relative blot intensities were quantitated by using ImageQuant, the densitometric values are indicated. (D) Quantification of the HGS immunostaining levels, as represented in (A). (E) Increased cellular levels of immunoreactive PDGFR-β and VEGFR2 seen in (B) were quantified using MetaVue software by analyzing at least 20 random cells per sample. Error bars indicate S.D., **p < 0.01, ***p < 0.001, t test.
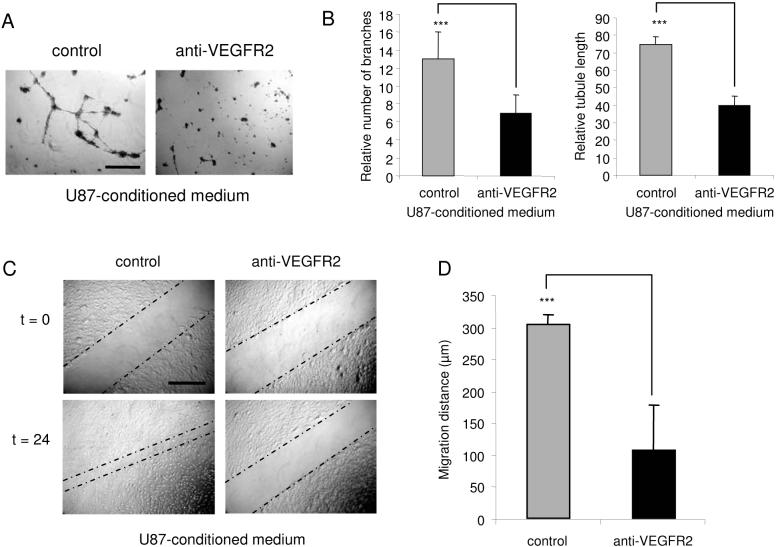
In order to confirm that HGS-regulated growth factor receptors are functional in glioma-induced angiogenesis, VEGFR2 receptor antibody blocking experiments were performed. HBMVECs were cultured in U87-conditioned medium. After 24 hr the HBMVECs were transferred onto Matrigel and cultured in U87-conditioned medium in the presence or absence of VEGFR2 blocking antibody (1 μg/ml). After 24 hr, a significant decrease in tubule length and branching was observed in the presence, as compared to the absence of these antibodies (Fig. 6A-B). In addition, scratch motility assays were performed using HBMVECs cultured in the presence or absence of VEGFR2 blocking antibody. After 24 hr a significant reduction in the migration distance was observed upon incubation of the HBMVECs in the presence of VEGFR2 antibody (Fig. 6C-D). These results indicate that functional VEGFR2 expression contributes to the angiogenic phenotype of HBMVECs stimulated by U87-conditioned medium.
Fig. 6.
Blocking of VEGFR2 reduces angiogenesis in vitro (A) HBMVECs were stimulated using U87-conditioned medium and cultured on Matrigel-coated plates in the presence or absence of VEGFR2 blocking antibody, size bar 300 μm. (B) After 24 hr VEGFR2 blocking significantly reduced tubule branching and tubule length. (C) HBMVECs cultured in a monolayer, scratched, and incubated in the presence or absence of VEGFR2 blocking antibody for 24 hr. VEGFR2 blocking resulted in a significant decrease in HBMVEC migration [size bar 300 μm], as quantified in (D). Error bars indicate S.D., ***p < 0.001, t test.
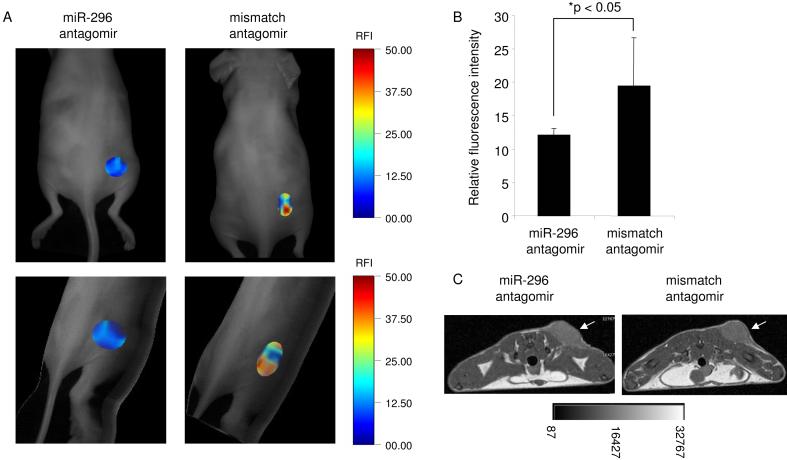
To analyze whether miR-296 affects glioma angiogenesis in vivo, a synthetic cholesterol-conjugated antagomir-296 antisense oligonucleotide was designed. U87 glioma cells were injected in the back flanks of nude mice. After 7 days of tumor growth, antagomir-296 or mismatch control antagomir were injected intravenously into these mice (100 μl of a 20 mg/ml stock diluted in PBS). At 4 days after injection of the antagomirs, AngioSense 750 (Visen Medical) was injected intravenously in order to quantify the neo-vascularization by fluorescence mediated tomography (Montet et al., 2007). The mice injected with the antagomir-296 showed a significant decrease in mean fluorescence intensity in the tumor as compared to the mice injected with the mismatch control (Fig. 7A-B). T1-weighted MR images were also acquired to show the tumor volume (Fig. 7C). These results demonstrate that inhibition of miR-296 affects glioma angiogenesis in vivo.
Fig. 7.
In vivo analysis of miR-296 inhibition on tumor neo-vascularization (A) Mice with subcutaneous U87 tumors (N = 6) were injected intravenously with miR-296 antagomirs or mismatch control antagomirs and 4 days later, the same mice were injected with Angiosense 750 and tumor vasculature was analyzed with FMT. Planar (top) and 3-D FMT images (bottom) are displaced in which the fluorescence signals are superimposed with the grayscale planar excitation light image of the mouse. (B) Quantitation of the mean fluorescence intensity measurements from tumors in (A) using OsiriX. Error bars indicate S.D., *p < 0.05, t test. (C) Transverse T1-weighted MR images (4.7T) acquired to show the tumor volume in (A), the white arrows indicate tumor mass
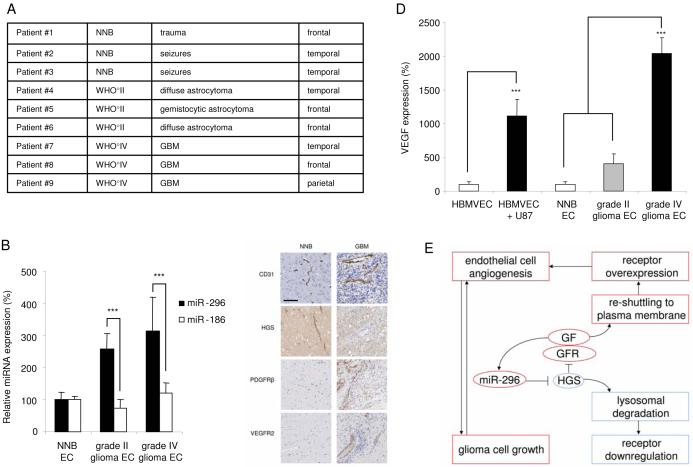
In order to establish the clinical relevance of miR-296 regulation in glioma angiogenesis, we performed experiments on human endothelial cells isolated from tumor tissue obtained from neurosurgical resections. Endothelial cells were isolated from blood vessels dissected from six glioma patients [three grade-IV gliomas (highly malignant) and three grade-II gliomas (less malignant)], all of which were highly angiogenic as confirmed by CD31 immunohistochemical analysis in pathological specimens (data not shown), as well as from three normal non-neoplastic human brain samples (Fig. 8A). miR-296 expression levels, determined by qRT-PCR, were elevated in tumor blood vessels relative to quiescent normal brain vessels (Fig. 8B), while control miR-186 showed no difference in expression levels. We further examined expression of HGS, PDGFR-β and VEGFR2 proteins in endothelial cells from human non-neoplastic brain and malignant gliomas. Immunohistochemical staining revealed that HGS expression was low and PDGFR- β and VEGFR2 were up-regulated in angiogenic glioma blood vessels relative to normal control vessels (Fig. 8C). Finally, using qRT-PCR, we determined mRNA levels of VEGF in endothelial cells isolated from non-neoplastic human brain and glioma tissues, as well as in HBMVECs cultured in the presence or absence of U87 glioma cells. A clear increase in VEGF mRNA was observed in tumor endothelial cells, and in U87-exposed HBMVEC cultures as compared to these controls (Fig. 8D), paralleling miR-296 expression levels. These data suggest that growth factor induced miR-296 expression observed in cultured brain endothelial cells parallels changes in miR-296 levels in angiogenic endothelial cells in brain tumors in vivo. Further, miR-296 downregulation of HGS signaling increases functional PDGFR-β and VEGFR2 in tumors, which is complemented by increased expression of VEGF.
Fig. 8.
Analysis of expression of miR-296 and its targets in tumor endothelial cells isolated from human gliomas (A) RNA was isolated from primary endothelial cell cultures (passage 0) prepared from normal human brain (NNB; N = 3), grade II astrocytomas (N = 3) and grade IV glioblastoma multiforme (N = 3). (B) RNA extracted from individual tumor endothelial samples was analyzed by qRT-PCR for expression levels of miR-296 and miR-186. All values were normalized to GAPDH mRNA levels in the same samples. (C) Immunohistochemical staining of brain blood vessels indicate increased and morphologically abnormal tumor blood vessels, as well as differential HGS, PDGFR-β, and VEGFR2 expression in non-neoplastic brain (NNB) and malignant glioma (GBM). Size bar 100 μm. (D) RNA extracted from individual tumor endothelial samples was analyzed by qRTPCR for expression levels of VEGF. All values were normalized to GAPDH mRNA levels in the same samples. (E) Schematic overview of the proposed angiogenic mechanism of miR-296. miR-296 is up-regulated in glioma endothelial cells and directly inhibits expression of HGS, thereby allowing the accumulation of growth factor receptors by attenuating their degradation. GF, growth factor; GFR, growth factor receptor. Error bars indicate S.D., ***p < 0.001, t test.
CONCLUSION
Altogether, these results support a role for miR-296 in promoting angiogenesis in tumors. We show that VEGF alone is capable of increasing miR-296 to levels equivalent to those reached with growth factor supplemented endothelial medium or glioma cells. This result points out an interesting feedback loop, whereby VEGF induces miR-296 expression, which targets HGS that, in turn, results in increased VEGFR2 and PDGFR-β protein levels and hence increased response to VEGF. In addition, EGF was also capable of inducing miR-296, suggesting a complex growth factor-growth factor receptor cross-talk mechanism, which combinatorially increases miR-296 levels.
MiR-296 was identified in a miRNA screen in normal human brain endothelial cells as being up-regulated in response to exposure to human brain glioma cells in culture. Down- and upregulation of miR-296 resulted in the inhibition and induction, respectively, of morphologic characteristics associated with angiogenesis of human endothelial cells. Sequence-specific inhibition of miR-296 by intravenous injection of cholesterol-conjugated antagomirs resulted in decreased neo-vascularization of tumors mice in mice. Upregulation of miR-296 expression was also demonstrated in tumor blood vessels isolated from human glioblastoma tumors. Altogether, these results support a role for increased miR-296 levels in promoting angiogenesis in tumors, according to the model proposed in Fig. 8E. It is likely that other endothelial miRNAs have related pro- or anti-angiogenic functions. A pro-angiogenic role for miR-296 was shown for HBMVECs as well as HUVECs; however, since miRNAs have multiple targets, miR-296 may also play important roles in other angiogenesis-unrelated physiological processes. We propose that in the endothelial context, miR-296 belongs to the family of “angiomirs”. Kuehbacher et al. showed that inhibition of let-7f and miR-27b also affected angiogenesis (Kuehbacher et al., 2007). Recently, miR-126 was found to play an important role in developmental angiogenesis and vascular integrity (Fish et al., 2008; Wang et al., 2008). We have detected several members of the let-7 family, as well as miR-27 and miR-126 in HBMVECs (Supplemental Fig. 1). Under conditions used in our study these miRNAs seemed down-regulated in HBMVECs exposed to glioma cells. Understanding functions of these miRNAs and their contribution to tumor angiogenesis will require further investigation. Thus, although several miRNAs have been implicated in endothelial processes in vitro (Chen et al., 2008; Kuehbacher et al., 2007; Lee et al., 2007; Poliseno et al., 2006; Suárez et al., 2007; Dews et al., 2006), and in vivo (Fish et al., 2008; Wang et al., 2008), miR- 296 plays a functional role in human tumor-induced angiogenesis in vitro and in vivo.
We have confirmed that the mRNA for HGS is a new target for miR-296. Our data suggest that miR-296 upregulation has a dominant down-tuning effect on HGS expression in angiogenic endothelial cells, albeit additional regulatory factors (including miRNAs) can not be excluded. Since HGS mediates the degradative sorting of PDGFR (Takata et al., 2000), as well as of VEGFR (Ewan et al., 2006) and EGFR (Bache et al., 2003; Stern et al., 2007), it seems likely that increased levels of these growth factor receptors on angiogenic blood vessels are due, at least in part, to the miR-296 downregulation of HGS expression.
In recent years a number of oncomirs (tumor-promoting miRNAs) and tumor suppressor miRNAs (tumor-inhibiting miRNAs) have been identified for various types of cancer cells [reviewed in (Esquela-Kerscher and Slack, 2006; Wurdinger and Costa, 2006). However, the delivery of inhibitors or precursor/mimics of such miRNAs to cancer cells in order to block tumor growth remains challenging. Tumor blood vessels offer an attractive additional target for the delivery of miRNA inhibitors or precursor/mimics in order to reduce tumor growth. Recent delivery configurations using siRNA-based and miRNA-inhibiting antagomirs proved effective for miRNA manipulation in vivo and blood vessels are a feasible target for such molecules in vivo (de Fougerolles et al., 2007; Krutzfeldt et al., 2005). Several siRNAs can target the vasculature resulting in alterations in blood vessel structure and function (Reich et al., 2003; Shen et al., 2006). These siRNAs are aimed at silencing single specific mRNA targets, but may have off-target side-effects (Jackson et al., 2003). miRNAs such as mir-296, however, can regulate a broad range of ‘natural’ mRNA targets (Miranda et al., 2006) and therefore might prove more efficient for fine-tuning cellular switch programs. Our results indicate that miR-296 is a critical component of the angiogenic process, possibly in a one-hit-multiple-target fashion (Wurdinger and Costa, 2006), and can give new insights into the role of miRNA regulation of neovascularization. Furthermore, manipulation of angiomir levels may prove therapeutic in the vast number of diseases where angiogenesis is a critical component.
EXPERIMENTAL PROCEDURES
Cells
Human brain microvascular endothelial cells (HBMVECs; Cell Systems ACBRI-376) and human umbilical vein endothelial cells (HUVECs; Cambrex) were cultured in EGM medium (Cambrex) for no more than 10 passages. U87 cells (U-87 MG; ATCC) and 293T cells were cultured in DMEM containing 10% FBS and antibiotics. U87-CFP cells were produced by stably transducing U87 cells with a CMV-controlled Cerulean expression cassette using a lentivirus vector (Rizzo et al., 2004).
Endothelial cell isolation
Endothelial cells were isolated from HBMVECs cultured alone or for 24 hr in the presence of glioma cells using CD31 Endothelial Cell Dynabeads (Dynal Biotech). Cells were trypsinized, and then trypsin was inactivated using trypsin neutralization solution (Cambrex). Single cell suspensions containing 1×106 cells were washed once in PBS and resuspended in 1 ml PBS to which 25 μl CD31 Dynabeads was added. The tubes were subjected to slow rotation for 20 min at 4°C. Subsequently, the tubes were placed in a magnet for 2 min and washed 3 × with 1 ml PBS containing 0.1% BSA. Isolated endothelial cells were plated in EGM medium and analyzed for the presence of U87-CFP contamination by using a combination of light and fluorescence microscopy. Only an occasional sporadic U87-CFP cell was observed, indicating a >99.9% purity of the endothelial cell population. Endothelial cells were also isolated from blood vessels dissected from three grade-IV gliomas and three grade-II gliomas, as well as from normal non-neoplastic human brain. All endothelial cell isolations from normal human brain and glioma tissue were performed using CD31 and VE-Cadherin MACS and >99% purity of cell isolations was confirmed by UEA-1 binding and Ac-DiI LDL uptake, as described previously (Miebach et al., 2006).
RNA isolation
Total RNA was isolated from endothelial cells passaged less than 7 times in culture and primary endothelial cells (passage 0) isolated from normal brain and glioma tissue. RNA isolation was carried out by adding 600 μl lysis buffer from the miRVANA miRNA isolation kit (Ambion) to the CD31-bead bound cells. The quality and quantity of the RNA was analyzed by photospectrometry and analytical 15% TBE-UREA gels (Biorad) after ethidium bromide staining.
miRNA inhibition and overexpression
For the inhibition of miR-296, 50 nM miR-296 inhibitor oligonucleotide (Ambion) or control oligonucleotides were transfected into HBMVECs or HUVECs using Lipofectamine 2000 (Invitrogen). For the overexpression of miR-296, 50 nM premiR-296, pre-control-Cy3, or pre-control-1 (Ambion) was transfected into HBMVECs. After 5 hr the cells were trypsinized, replated and cultured in EBM or EGM until further analysis.
In vitro glioma angiogenesis assay
HBMVECs or HUVECs (both passage <7) were cultured on Matrigel (Beckton Dickinson) in EBM (Cambrex) in the presence or absence of U87-CFP cells, or EGM (Cambrex). After 24 hr the cultures were analyzed by a combination of light and fluorescence microscopy. The experiments were performed in triplicate, repeated at least twice, and judged in a double-blind fashion by at least two observers. Three or more random pictures were taken of each culture using a digital camera system and the software program MetaVue was used, which were subsequently analyzed for total tubule length and number of tubule branches using the software program ImageJ.
In vitro migration scratch assay
Confluent HBMVECs (passage <7) were transfected using Lipofectamine2000 (Invitrogen) with miR-296 mimics, inhibitors or control oligonucleotides. At 5 hr after transfection, a cell scratch spatula was used to make a scratch in the cell monolayer after which the cell monolayers were rinsed and further incubated. Pictures of the scratches were taken by using a digital camera system coupled to a microscope. The cells were incubated for 24 hr and pictures of the scratches were taken by using a digital camera system coupled to a microscope. The software program MetaVue was used to determine the migration distance (μm) as the reduction of the width of the open area.
Luciferase miRNA target reporter assay
Total cDNA from HBMVECs was used to isolate the 3' UTR of HGS by PCR using the forward primer 5'HGS: TTGACTAGTCCCAGGCCATGCTCACGTCCGGAGTAACACTAC and the reverse primer 3'HGS: TTGAAGCTTGAAATACATTTTATTATCGCTGTACCATTCTGGGG. After digestion of the PCR product by SpeI and HindIII the HGS 3' UTR was cloned into the SpeI and HindIII sites of the multiple cloning site (MCS) of pMir-Report (Ambion) resulting in pMir-Report-3'HGS. Mutations were introduced in the potential miR-296 114-134 and 352-372 binding sites (Fig. 3A). The flanking primers that were used for the isolation of the HGS 3' UTR from the endothelial cDNA were combined with primers containing mutated target sequences. For the 5' (pMir-Report-3'HGS-mut114-134; denoted HGS mut1) miR-296 binding site primers mutP1AF: CAACCTGACTAAAACCCGGAACACCCCAAGCCCACCTCCCTTGTCCTCAG and mutP2AR: TTCCGGGTTTTAGTCAGGTTGAGAAGGGACACTACCGGCAGTAGAGGACA and for the downstream (pMir-Report-3'HGS-mut352-372; denoted HGS mut2) miR-296 binding site mutP1BF: CACAATGACACCTCCCCGAGCCTCTGCAGGGGCCTCTCTCGGCAGCCACA and mutP2BR: GCTCGGGGAGGTGTCATTGTGACACCACAGCCAGCTCACAGTGCGGCCAG. 293T cells were transfected with the pMir-Report vectors containing the HGS wild type and variant 3'UTRs and at 5 hr after transfection the cells were transfected again with 50 nM of miR-296 inhibitors (Ambion) or control oligonucleotides (Ambion). After 24 hr the cells were lysed and luciferase activity was measured using a luminometer. A plasmid containing an expression cassette for Renilla luciferase, pRenilla (Promega) was co-transfected and used to normalize the firefly luciferase values expressed from the pMir-Report constructs.
Immunofluorescence staining and Western blotting
Cell cultures were fixed using PBS containing 4% paraformaldehyde for 15 min at RT and - for HGS - permeabilized using PBS containing 1 % Triton-X100 for 5 min at RT. Non-specific antibody binding was blocked by incubating the cells in PBS containing 5% FCS for 30 min at RT. Mouse monoclonal anti-HGS antibodies (1:100, Alexis Biochemicals), anti-PDGFR-β (1:50, BD Biosciences), and anti-VEGFR2 (1:400, Abcam) antibodies were used to detect HGS, PDGFR-β, and VEGFR2, respectively. Donkey-anti-mouse-Cy3 (1:600, Jackson Immuno Labs, West Grove, PA) was used as secondary antibody. Immunofluorescence was analyzed using fluorescence microscopy. Immunohistochemical analysis of CD31 was performed as described (Miebach et al., 2006). For Western blotting cells were grown for 24 hr and lysed by standard procedure in RIPA buffer containing Protease Inhibitor Cocktail (Roche Diagnostics). Protein concentrations of total cell lysates were measured using Micro BCA Protein Assay Kit (Pierce Biotechnology) and 40 μg per lane of tissue culture lysate were resolved on SDS-PAGE gels (Invitrogen) followed by immunoblot detection and visualization with ECL Western blotting detection reagents (Pierce Biotechnology). The bands of the Western blots were quantitated using ImageQuant software (Molecular Dynamics). The intensities were normalized to anti-β-Actin levels and depicted as relative intensities. Immunobloting was performed with the following primary antibodies: mouse anti-HGS (1:1,000 Alexis Biochemicals), mouse anti-PDGFR-β (1:1000, Abcam), and mouse anti-VEGFR2 (1:1,000, Abcam), mouse anti-β-Actin (1:5,000; Abcam). In addition, www.proteinatlas.org was used as a source of immunohistochemical analysis of CD31, HGS, PDGFR-β, and VEGFR2 protein expression.
Quantitation of HGS, PDGFR-β and VEGFR2 expression in cells treated with siRNAs
The expression levels of HGS, PDGFR and VEGFR2 were measured by immunofluorescence intensity/cell. The experiments were performed in triplicate, repeated at least twice, and judged in a double-blind fashion by at least two observers. Twenty or more random pictures (1 cell per field) were taken using a Nikon fluorescent microscope system coupled to a digital camera analyzed using the software program MetaVue (Molecular Devices). The values of the HGS, PDGFR-β and VEGFR2 average intensities were averaged; the error bars denote S.D. and the t test was used to determine significant differences.
In vivo imaging of angiogenesis
All experiments on mice were approved by the Subcommittee on Research Animal Care at the Massachusetts General Hospital and were performed in accordance to their guidelines and regulations. One million U87 cells were mixed with 50 μl Matrigel (Beckton Dickinson) and directly injected subcutaneously in the back flanks of nude mice. After 7 days, each mouse was injected intra-ocular with 2 mg of miR-296 antagomirs or mismatch control antagomirs (100 μl of a 20 mg/ml stock diluted in PBS) (Regulus Therapeutics). Four days after injection of the antagomirs, 150 μl (2 nmol fluorochrome) of Angiosense 750 (Visen Medical, Woburn, MA) was injected intravenously. One hr later, coronal fluorescence mediated tomography (FMT) images were acquired using a continuous wavetype scanner capable of acquiring transillumination, reflectance and absorption data (Visen Medical). Prototypes of the device and reconstruction algorithms, as well as the imaging protocol, have been described previously (Montet et al., 2007; Ntziachristos et al., 2002; Montet et al., 2005; Grimm et al., 2005). Mice were also imaged with magnetic resonance (MR) using a 4.7T small animal MR scanner (Bruker BioSpin, Billerica, MA). The imaging protocol consisted of axial T1-weighted sequence with the following parameters: fast spin echo 635/15 msec (TR/TE), 4 averages, matrix slice 256×128, FOV 4.0 × 2.5 cm and slice thickness 1 mm. Sixteen slices with a slice thickness of 1 mm, field of view of 42 × 24 mm, and matrix of 128 × 64 were acquired.
ACKNOWLEDGEMENTS
We would like to acknowledge the Steve Kaplan Fellowship from the American Brain Tumor Association (TW), NIH-NCI P50 CA86355-04 (RW and XOB), NIHNCI P01 CA69246 (XOB), NIH-NINDS P30NS045776 (XOB), and the Brain Tumor Society (AMK). We thank Erdogan Erkan, Laurine Wedekind, and Peter Waterman for technical assistance and George S. Mack for critical reading of the manuscript. Jürgen Soutschek is an employee and shareholder of Regulus Therapeutics, the other authors declare to have no conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
miRNA microarray data The miRNA array data is deposited in the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/), accession number GSE13091.
Supplemental data The supplemental data include Supplemental Experimental Procedures and 4 Supplemental Figures, and can be found at www.cancercell.org.
Supplementary Material
REFERENCES
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bache KG, Raiborg C, Mehlum A, Stenmark H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J Biol Chem. 2003;278:12513–12521. doi: 10.1074/jbc.M210843200. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Batchelor TT, Sorensen AG, di Tomaso E, Zhang WT, Duda DG, Cohen KS, Kozak KR, Cahill DP, Chen PJ, Zhu M, et al. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11:83–95. doi: 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem S, Cotran R, Folkman J. Tumor angiogenesis: a quantitative method for histologic grading. J Natl Cancer Inst. 1972;48:347–356. [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Chen C, Chai H, Wang X, Jiang J, Jamaluddin MS, Liao D, Zhang Y, Wang H, Bharadwaj U, Zhang S, et al. Soluble CD40 ligand induces endothelial dysfunction in human and porcine coronary artery endothelial cells. Blood. 2008 doi: 10.1182/blood-2008-03-143479. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–5. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Ewan LC, Jopling HM, Jia H, Mittar S, Bagherzadeh A, Howell GJ, Walker JH, Zachary IC, Ponnambalam S. Intrinsic tyrosine kinase activity is required for vascular endothelial growth factor receptor 2 ubiquitination, sorting and degradation in endothelial cells. Traffic. 2006;7:1270–1282. doi: 10.1111/j.1600-0854.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–84. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkerth RD. Descriptive analysis and quantification of angiogenesis in human brain tumors. J Neurooncol. 2000;50:165–172. doi: 10.1023/a:1006499824379. [DOI] [PubMed] [Google Scholar]
- Folkman J. Angiogenesis: an organizing principle for drug discovery? Nat Rev Drug Discov. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm J, Kirsch DG, Windsor SD, Kim CF, Santiago PM, Ntziachristos V, Jacks T, Weissleder R. Use of gene expression profiling to direct in vivo molecular imaging of lung cancer. Proc Natl Acad Sci U S A. 2005;102:14404–14409. doi: 10.1073/pnas.0503920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerbel R, Folkman J. Clinical translation of angiogenesis inhibitors. Nat Rev Cancer. 2002;2:727–739. doi: 10.1038/nrc905. [DOI] [PubMed] [Google Scholar]
- Khodarev NN, Yu J, Labay E, Darga T, Brown CK, Mauceri HJ, Yassari R, Gupta N, Weichselbaum RR. Tumour-endothelium interactions in co-culture: coordinated changes of gene expression profiles and phenotypic properties of endothelial cells. J Cell Sci. 2003;116:1013–1022. doi: 10.1242/jcs.00281. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. Rna. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350–5. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Miebach S, Grau S, Hummel V, Rieckmann P, Tonn JC, Goldbrunner RH. Isolation and culture of microvascular endothelial cells from gliomas of different WHO grades. J Neurooncol. 2006;76:39–48. doi: 10.1007/s11060-005-3674-6. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Montet X, Figueiredo JL, Alencar H, Ntziachristos V, Mahmood U, Weissleder R. Tomographic Fluorescence Imaging of Tumor Vascular Volume in Mice. Radiology. 2007;242:751–758. doi: 10.1148/radiol.2423052065. [DOI] [PubMed] [Google Scholar]
- Montet X, Ntziachristos V, Grimm J, Weissleder R. Tomographic fluorescence mapping of tumor targets. Cancer Res. 2005;65:6330–6336. doi: 10.1158/0008-5472.CAN-05-0382. [DOI] [PubMed] [Google Scholar]
- Ntziachristos V, Tung CH, Bremer C, Weissleder R. Fluorescence molecular tomography resolves protease activity in vivo. Nat Med. 2002;8:757–60. doi: 10.1038/nm729. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- Reich SJ, Fosnot J, Kuroki A, Tang W, Yang X, Maguire AM, Bennett J, Tolentino MJ. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis. 2003;9:210–216. [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. Clarifications on miRNA and cancer. Science. 2006;311:36–37. doi: 10.1126/science.311.5757.36d. [DOI] [PubMed] [Google Scholar]
- Shen J, Samul R, Silva RL, Akiyama H, Liu H, Saishin Y, Hackett SF, Zinnen S, Kossen K, Fosnaugh K, et al. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 2006;13:225–234. doi: 10.1038/sj.gt.3302641. [DOI] [PubMed] [Google Scholar]
- Shih AH, Holland EC. Platelet-derived growth factor (PDGF) and glial tumorigenesis. Cancer Lett. 2006;232:139–147. doi: 10.1016/j.canlet.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Stern KA, Visser Smit GD, Place TL, Winistorfer S, Piper RC, Lill NL. Epidermal growth factor receptor fate is controlled by Hrs tyrosine phosphorylation sites that regulate Hrs degradation. Mol Cell Biol. 2007;27:888–898. doi: 10.1128/MCB.02356-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- Takata H, Kato M, Denda K, Kitamura N. A hrs binding protein having a Src homology 3 domain is involved in intracellular degradation of growth factors and their receptors. Genes Cells. 2000;5:57–69. doi: 10.1046/j.1365-2443.2000.00303.x. [DOI] [PubMed] [Google Scholar]
- Tuccoli A, Poliseno L, Rainaldi G. miRNAs regulate miRNAs: coordinated transcriptional and post-transcriptional regulation. Cell Cycle. 2006;5:2473–2476. doi: 10.4161/cc.5.21.3422. [DOI] [PubMed] [Google Scholar]
- Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–71. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdinger T, Costa FF. Molecular therapy in the microRNA era. Pharmacogenomics J. 2006 doi: 10.1038/sj.tpj.6500429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.