SUMMARY
Proteolysis is a key regulatory process that promotes the (in)activation, translocation, and/or degradation of proteins. As such, there is considerable interest in methods to comprehensively characterize proteolytic pathways in biological systems. Here, we describe a robust and versatile proteomic platform that enables direct visualization of the topography and magnitude of proteolytic events on a global scale. We use this method to generate a proteome-wide map of proteolytic events induced by the intrinsic apoptotic pathway. This profile contained 91 characterized caspase substrates as well as 170 additional proteins not previously known to be cleaved during apoptosis. Surprisingly, the vast majority of proteolyzed proteins, regardless of the extent of cleavage, yielded persistent fragments that correspond to discrete protein domains, suggesting that the generation of active effector proteins may be a principal function of apoptotic proteolytic cascades.
INTRODUCTION
Proteases constitute 1–5% of eukaryotic genomes, with the human genome, in particular, encoding 566 predicted proteolytic enzymes (Puente et al., 2003). The physiological functions of proteases are essential in many physiological processes, including development (Matrisian and Hogan, 1990; Turgeon and Houenou, 1997), blood coagulation (Riewald and Ruf, 2001), and cell death (Alnemri, 1997), as well as many pathological events such as cancer (van Kempen et al., 2006) and infectious disease (Abdel-Rahman et al., 2004). Even the most well-studied proteolytic cascades remain only partially understood, and a large portion of human proteases are wholly uncharacterized with respect to endogenous substrates and biological functions. These gaps in our knowledge of protease biology have inspired the development of proteomic methods to profile protease-substrate relationships on a global scale (auf dem Keller et al., 2007). These efforts can be divided into three general categories. First are in vitro specificity profiling experiments such as peptide, phage, and bacterial display, in which a purified protease of interest is exposed to a large library of peptides/proteins to identify substrates (Harris et al., 2000; Ju et al., 2007; Kridel et al., 2001; Matthews and Wells, 1993). While these studies often yield valuable insight into the sequence specificity of proteases, interpretation of the biological significance of results is difficult given that the protease-substrate interactions occur in an artificial environment that differs substantially from natural biological systems. A second approach utilizes two-dimensional gel electrophoresis (2-DGE), where differences in the migration and intensity of cleaved substrates are detected by protein staining following activation or addition of a protease to a biological sample (Bredemeyer et al., 2004; Brockstedt et al., 1998; Gerner et al., 2000; Lee et al., 2004). This approach has the advantage of identifying substrates for proteases in endogenous settings. Although 2-DGE experiments and second-generation technologies built on this method have proven extremely valuable and are still in common practice, they suffer from issues of reproducibility, throughput, and sensitivity (Corthals et al., 2000; Gygi et al., 2000).
Neither peptide/protein display nor 2-DGE methods yields direct information on the sites of endogenous proteolytic cleavage. To address this limitation, a third set of proteomic technologies have emerged that use chemical labeling strategies to capture emergent N-termini from protease cleavage events (Dean and Overall, 2007; McDonald et al., 2005; Timmer and Salvesen, 2006; Van Damme et al., 2005). A number of variations on this technique have been introduced, including those that permit selective separation and/or enrichment of the cleaved N-terminal peptides (Dean and Overall, 2007; McDonald et al., 2005; Timmer et al., 2007). However, all such N-terminal labeling approaches possess drawbacks. Most notably, these methods, which profile only a single peptide from the C-terminal portion of cleaved proteins, do not provide any topographical information about proteolytic cleavage events. As such, no data are acquired on whether the cleaved portions of proteins remain intact or are further degraded. This is particularly problematic for N-terminal fragments of protease substrates, since robust and selective C-terminal labeling strategies have not yet been developed. Furthermore, the intact parent protein often goes undetected (due to N-terminal modifications that are prevalent on native proteins), and, therefore, the magnitude of proteolytic cleavage remains unknown. Finally, from a technical perspective, detection of cleavage is contingent on the identification of a single peptide, which, considering the small number of proteotypic peptides observed in most proteins [i.e., those peptides that can reliably be identified by LC-MS/MS; (Craig et al., 2005; Kuster et al., 2005)], likely results in substantial numbers of protease substrates remaining undetected.
Given the aforementioned limitations of current proteomic approaches, the challenge of comprehensively determining the magnitude and topography of protein cleavage events, which is essential to predict the functional consequences of proteolysis, still typically requires the time-consuming and costly process of generating multiple antibodies that recognize epitopes throughout the sequences of individual proteins. To address this problem on a global scale, we describe herein a robust, high-content proteomic platform to profile proteolytic events occurring in natural biological systems termed PROTOMAP (for PROtein TOpography and Migration Analysis Platform). We have applied this technology to the well-studied intrinsic apoptosis pathway in Jurkat T-cells, resulting in the identification of many established caspase-mediated proteolytic events, as well as over 150 additional proteins not previously known to be cleaved during apoptosis. PROTOMAP further yields a number of provocative conclusions about the general impact of proteolysis on the structural architecture of proteins in apoptotic cells.
RESULTS
PROTOMAP Methodology
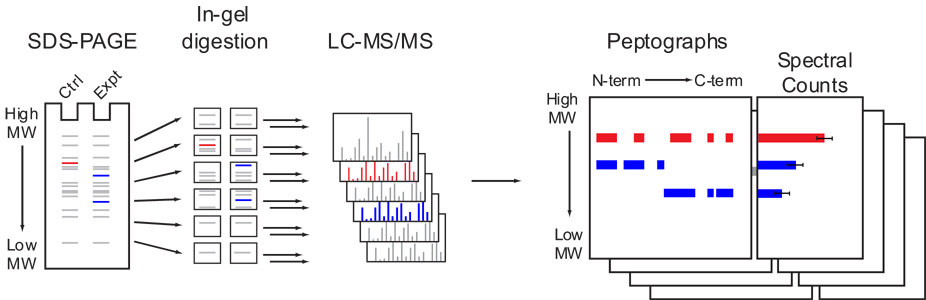
SDS-PAGE is a routine method for protein fractionation and serves the purpose of reducing sample complexity prior to MS analysis in proteomics investigations. SDS-PAGE also reveals information about the molecular mass of proteins; however, this information has rarely been systematically taken into account in large-scale proteomic experiments, even in cases where upfront SDS-PAGE fractionation steps were performed (Li et al., 2007; Lohaus et al., 2007; Shi et al., 2007). We therefore developed PROTOMAP with the goal of integrating SDS-PAGE migratory rates with sequence coverage and spectral count values acquired by LC-MS/MS to provide a rich set of data that could reveal global changes in the size, topography, and abundance of proteins in complex biological samples (Figure 1). To accomplish this integration and facilitate interpretation of results, we developed an algorithm that translates SDS-PAGE and LC-MS/MS data into a unique visual format for data representation, referred to as a peptograph. A typical PROTOMAP experiment begins with comparison of control and experimental samples by separation by one-dimensional (1D) SDS-PAGE. Each sample lane is then sliced into gel bands at fixed intervals (typically 0.5 cm), and these bands are digested with trypsin to release peptides that are subsequently analyzed by 1D reverse-phase LC-MS/MS. The resulting proteomic data from all of the gel bands are translated into peptographs, which plot sequence-coverage for a given protein in the horizontal dimension (N- to C-terminus, left to right) and SDS-PAGE migration in the vertical dimension (high to low molecular weight, top to bottom) (Figure 1). Control and experimental samples are distinguished in the peptograph by color (red and blue, respectively, with purple representing overlapping sequence coverage). Semi-quantitative analysis of each band is provided by “spectral-counting” (a common method for quantitation where spectra that match each peptide are counted to provide an estimate of their abundance) in a chart on the right side of each peptograph.
Figure 1. General methodological features of PROTOMAP.
Proteomes from control and experimental systems are separated by 1D SDS-PAGE and the gel lanes cut into bands at fixed intervals. Bands are digested with trypsin to release peptides that are analyzed by 1D reverse-phase LC-MS/MS. The resulting proteomic data are integrated into peptographs, which plot, in the left panel, sequence-coverage for a given protein in the horizontal dimension (N- to C-terminus, left to right) versus SDS-PAGE migration in the vertical dimension [high to low molecular weight (MW), top to bottom]. In the right panel, the peptograph also displays average spectral counts for each protein in each gel band. Proteins that undergo proteolytic cleavage are identified by shifts in migration from higher (parental, red) to lower (fragments, blue) MW species in control versus experimental systems. The sequence coverage shown in the peptograph provides a topographical map of the protein fragments that persist following proteolysis. The magnitude of proteolysis is estimated by comparing spectral counts of the parental protein in control versus experimental proteomes.
The comparative analysis of normal and apoptotic cells was expected to offer an excellent model system with which to test the sensitivity, precision, and utility of PROTOMAP for multiple reasons. First, caspase-mediated proteolytic cascades that mediate this process have been intensively studied and are known to generate numerous protein cleavage events (Fischer et al., 2003; Luthi and Martin, 2007; Timmer and Salvesen, 2006). Furthermore, the molecular pathways that contribute to apoptosis, an event of high relevance to many physiological and pathological processes are only partially understood (Abud, 2004; Cowan et al., 1984; Kerr et al., 1994).
Characterization of Established Proteolytic Markers of Apoptosis
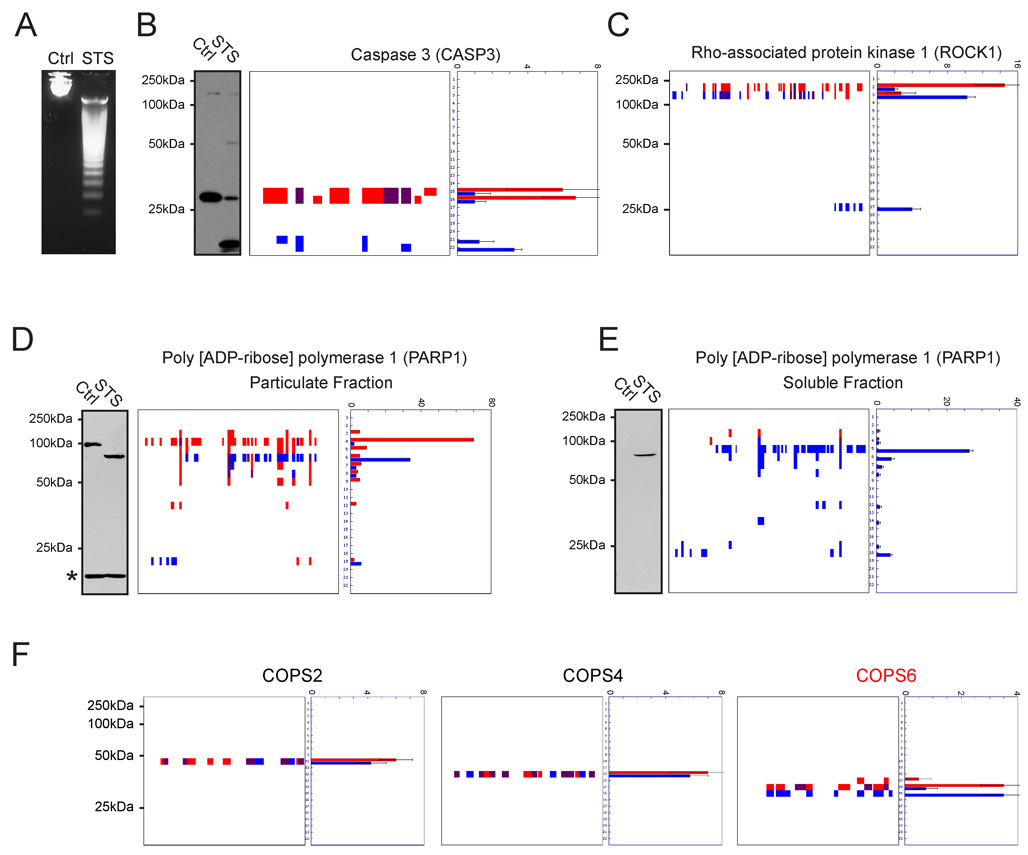
The intrinsic apoptosis pathway was induced in Jurkat T-cells by incubation with the pan-kinase inhibitor staurosporine (STS) for four hours. This time point was chosen because it represents an established midpoint in the Jurkat apoptosis time-course (Feng and Kaplowitz, 2002; Na et al., 1996), which we confirmed by monitoring DNA-fragmentation and caspase 3 activation (Figure 2A and B, respectively). Cells were lysed and the soluble proteomes (100 µg protein/sample) from control and apoptotic cells resolved by 1D-SDS-PAGE (10% polyacrylamide). Gel lanes were then cut into 22 bands (0.5 cm vertical length), and each band was subjected to in-gel trypsin digestion and analyzed by LC-MS/MS and the SEQUEST algorithm, which identifies peptides from their MS2 spectra (Eng et al., 1994). Data from the entire collection of gel-bands (176 bands in total: two samples with four replicates each) were converted to individual peptographs for each protein identified in the experiment (all peptographs can be viewed online at http://www.scripps.edu/chemphys/cravatt/protomap). Manual inspection of the resulting peptographs showed that known markers of apoptosis displayed expected patterns of cleavage, including caspase-3, a principal effector caspase (Cohen, 1997), which shifted from the 32 kDa pro-form in control samples (bands 15 and 16) to the active dimer comprised of 12 and 17 kDa species in apoptotic samples (bands 21 and 22, Figure 2B). Notably, even though the latter two species were not resolved on a 10% polyacrylamide gel, clear evidence of their mutual existence was provided by examination of the caspase-3 peptograph, which displayed sequence coverage spanning both the N- and C-terminal fragments of the enzyme in bands 21 and 22 of STS-treated cells (Figure 2B). Several other caspases were detected by PROTOMAP, including caspases 2, 6, 7, and 8, all of which underwent proteolytic cleavage in STS-treated cells (Supplemental Figure 1).
Figure 2. Established markers of apoptotic proteolytic pathways visualized by PROTOMAP.
Apoptosis was induced in Jurkat T-cells by treatment with staurosporine (STS) for 4 hrs, which produced DNA fragmentation (A) and caspase-3 cleavage (B). Cleavage of caspase-3 is evident from the peptograph (B, right panels), which identified peptides for the 32 kDa pro-form of this protease in control cells (red signals, bands 15 and 16), and peptides for the 12 and 17 kDa activated forms in STS-treated cells (blue signals, bands 21 and 22). Peptides detected in both samples are shown in purple. (C) Cleavage of ROCK1 (parent species, 158 kDa) to 130 and 28 kDa fragments. (D) and (E) Cleavage of PARP1 in STS-treated cells. Parental PARP1 was found in the particulate fraction of control cells (D) and, upon induction of apoptosis, was detected in the soluble fraction as a series of cleaved fragments (E). The asterisk in the PARP1 western blot of the particular fraction likely corresponds to antibody cross-reactivity with a background protein, since no spectral counts for PARP1 were detected in the corresponding gel band. Notably, a number of fragments of PARP1 that are not observed by immunoblotting were evident from the peptographs. This likely reflects the restricted epitopes recognized by anti-PARP1 antibodies. (F) Peptographs for representative members of the COPS family, showing selective cleavage of COPS6 in apoptotic cells, as previously demonstrated (Correia et al., 2007). Peptographs for all nine members of the COPS family can be found in Supplemental Figure 2. Spectral count data are represented as the mean +/− SEM for four independent experiments.
Caspase 3 is known to cleave the Rho-associated protein kinase, ROCK1, near the C-terminus at aspartate 1113 of a DETD consensus sequence (Coleman et al., 2001), releasing a 28 kDa autoinhibitory domain and generating a constitutively active 130 kDa form of the kinase (Jin and El-Deiry, 2005). The ROCK1 peptograph showed the parent protein migrating primarily in band 2 in control cells, corresponding to a molecular mass range of 150–200 kDa (full length ROCK1 is 158 kDa) (Figure 2C). Upon induction of apoptosis, two major fragments of ROCK1 were observed that corresponded to the larger N-terminal fragment in band 3 (mass range of 125–150 kDa), and smaller C-terminal fragment in band 17 (mass range of 25–30 kDa). Further examination of the peptographs pointed to D1113 as the implicit site of cleavage (Figure 2C). Notably, the parental and active kinase forms of ROCK1 were clearly resolved into separate gel bands (2 and 3 respectively), despite showing less than a 20% change in mass.
Another prototypical marker of apoptosis is cleavage of poly(ADP)ribose polymerase 1 (PARP1) (Zong et al., 2004). PARP1 is a 113 kDa enzyme involved in DNA repair, which is inactivated by caspase-3 during apoptosis. Under normal conditions, PARP1 is bound to DNA in the nucleus and therefore did not appear in the soluble fraction of control cells [although strong signals were observed for PARP1 in band 4 of the particulate fraction of control cells, corresponding to a mass range of 100–125 kDa (Figure 2D)]. Upon STS treatment, multiple cleaved fragments of PARP1 were released into the soluble fraction of Jurkat cells, including a strong 89 kDa fragment (band 5), which was confirmed by western blotting (Figure 2E).
To provide evidence that PROTOMAP can accurately discriminate cleaved from non-cleaved proteins, we examined peptographs for the eight subunits of the COP9 signalosome. The 39 kDa COPS6 subunit of this protein complex has recently been found to undergo caspase-mediated cleavage to generate a 36 kDa C-terminal fragment (Correia et al., 2007). Other signalosome subunits did not show evidence of proteolytic cleavage in this previous study. We observed essentially identical results in the PROTOMAP comparison of control and STS-treated Jurkat cells: all eight signalosome components were identified in bands consistent with their predicted molecular masses, but only COPS6 showed a shift in migration to a lower band in apoptotic cells (from band 14 to 15; Figure 2F and Supplemental Figure 2). This finding, in combination with the aforementioned data for ROCK1, underscores the remarkable resolution achievable by PROTOMAP, which can detect 10–20% changes in protein size across a large mass range (i.e., ~20–200 kDa).
Global Analysis of Proteolytic Events in Apoptosis
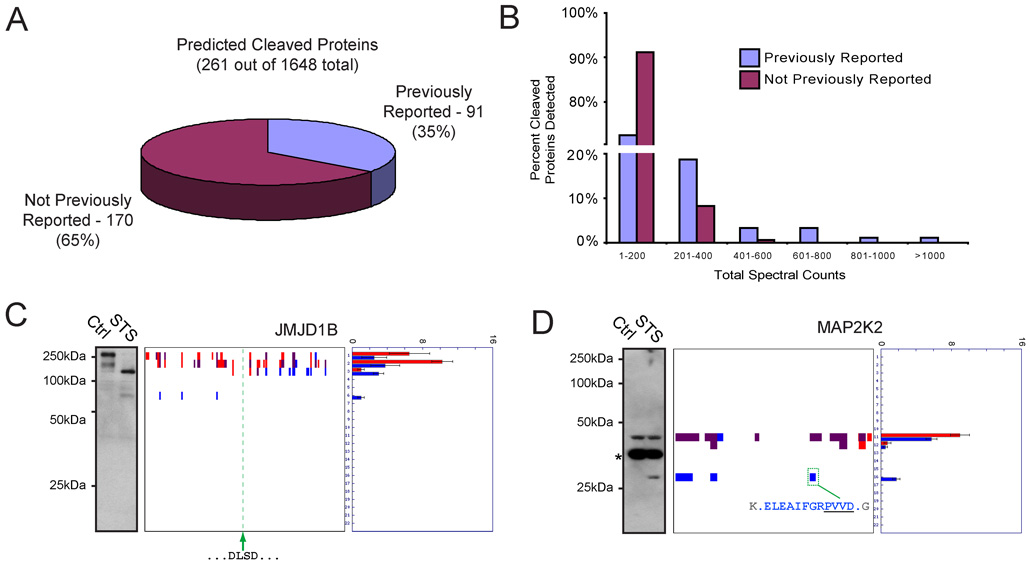
To comprehensively annotate proteins cleaved during apoptosis, we developed an algorithm termed PROTOSort to identify peptographs with altered signal intensities and/or gel migration patterns in control versus STS-treated cells (Supplemental Experimental Procedures). Using PROTOSort, we identified 261 proteins predicted to undergo cleavage or substantial down-regulation in apoptotic cells out of a total of 1648 proteins detected with sufficient spectral counts to permit quantitative analysis (a complete list of these predicted cleaved proteins is provided in Supplemental Table 1). No reduction in sensitivity was observed for detection of proteolytically cleaved proteins compared to non-cleaved proteins, as evidenced by the equivalent distribution of spectral counts for proteins from each class (Supplemental Figure 3).
Searches of the public literature and the CAspase Substrate dataBAse Homepage (“CASBAH”) (Luthi and Martin, 2007) revealed that 91 of the predicted cleaved proteins identified by PROTOMAP corresponded to known caspase substrates (Figure 3A and Supplemental Table 1). The large number of additional proteins that displayed altered migration patterns in STS-treated cells suggested that PROTOMAP uncovered many heretofore unknown proteolytic events in apoptosis. Proteins known to be cleaved in apoptosis were detected with high spectral-count values compared to previously unknown cleavage events (Figure 3B), suggesting that sensitivity has played a limiting role in the historical characterization of apoptotic proteolytic pathways. Western blotting analysis was used to confirm representative examples of predicted proteolytic events. JMJDB1, a putative histone demethylase, shifted from a parental 191 kDa protein (bands 1/2) in control cells to persistent 75 kDa N- and 100–125 kDa C-terminal fragments (bands 3 and 6, respectively) in STS-treated cells (Figure 3C). An equivalent migration pattern was observed for JMJDB1 by western blotting (Figure 3C). Similarly, the peptograph for MAP2K2, a 45 kDa member of the mitogen-activated protein kinase family, identified an N-terminal ~30 kDa persistent fragment in apoptotic cells (band 16), which was also observed by western blotting (Figure 3D).
Figure 3. Global analysis of cleaved proteins in apoptotic cells.
(A) A total of 261 predicted cleaved proteins were identified by PROTOMAP in apoptotic cells, 91 (35%) of which corresponded to established caspase substrates and/or proteins known to be proteolyzed during apoptosis. The remaining 170 proteins (65%) were not previously known to be cleaved during apoptosis. (B) Comparison of the spectral count values for previously known versus unknown cleaved proteins in apoptotic cells. Note that proteins with high spectral count values were predominantly from the former group. (C) and (D) Examples of predicted cleaved proteins that were confirmed by western blotting. In the case of JMJD1B (C), an implicit caspase cleavage sequence at amino acids 817–823 (tandem DLSD) could be identified that resided in between the persistent 75 kDa N- and 100–125 kDa C-terminal fragments. In the case of MAP2K2 (D), the explicit caspase cleavage site was identified by PROTOMAP at residue D284. The asterisk in the MAP2K2 western blot likely corresponds to antibody cross-reactivity with a background protein, as no spectral counts were detected for MAP2K2 in the corresponding gel band. Spectral count data are represented as the mean +/− SEM for four independent experiments.
Estimation of the Magnitude of Protein Cleavage Events
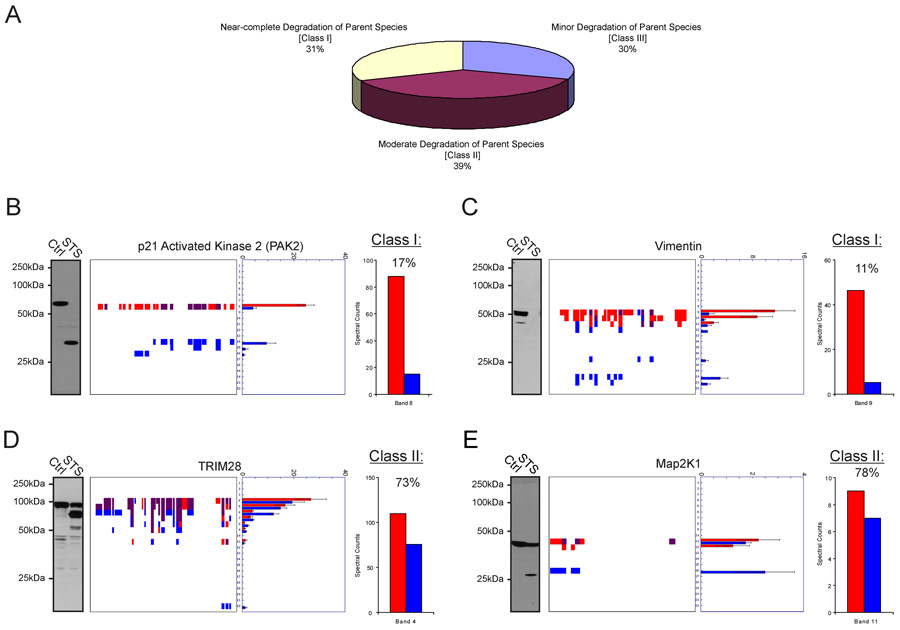
Numerous studies have confirmed the accuracy of spectral counting as an MS-based method for the relative quantitation of protein abundances in biological samples (Dong et al., 2007; Liu et al., 2004; Old et al., 2005). We therefore used this parameter to estimate the magnitude of protein cleavage events in apoptotic cells. The right panel of each peptograph reports average spectral count values for each protein in each band of the gel. Assuming that the slowest migrating species in each peptograph corresponds to the parental form of the protein [a premise that is supported by a global comparison of the predicted and measured masses for proteins identified by PROTOMAP (Supplemental Figure 4)], we then asked whether these values provide an accurate estimate of the extent of degradation of the parental protein in apoptotic cells. Cleaved proteins were divided into three general classes based on the predicted magnitude of cleavage of the parental species: Class I – near-complete degradation (< 20% spectral counts remaining in STS-treated cells); Class II – moderate degradation (20–80% spectral counts remaining in STS-treated cells); and Class III – minor degradation (> 80% spectral counts remaining in STS-treated cells). These classes were represented with similar frequencies, comprising 31, 39, and 30 percent of the predicted cleaved proteins, respectively (Figure 4A). Representative examples of class I and II proteins were examined by western blotting, which corroborated the predictions made by spectral counting. For two class I proteins, PAK2 and Vimentin, negligible signals were observed by western blotting for the parental protein in STS-treated cells (Figure 4B and C, respectively). In contrast, strong western signals were observed for the parental forms of the class II proteins TRIM28 and MAP2K1 in apoptotic cells (Figure 4D and E, respectively). It is also interesting to note that in three of the four cases (PAK2, Vimentin, and TRIM28), persistent protein fragments were detected by PROTOMAP that were not observed by western analysis. This finding likely reflects the restricted number of epitopes recognized by most antibodies and underscores the ability of PROTOMAP to provide a more complete topographical description of protein fragments in proteomes.
Figure 4. Estimation of the magnitude of protein cleavage events in apoptotic cells.
(A) Predicted cleaved proteins were divided into three general classes: I) near-complete (class I, < 20% spectral counts remaining in STS-treated cells), moderate (class II, 20–80% spectral counts remaining in STS-treated cells), and minor (class III, > 80% spectral counts remaining in STS-treated cells) degradation of the parent species. (B) and (C) Western blot confirmation that class I proteins PAK2 (B) and Vimentin (C) underwent near-complete degradation in apoptotic cells. (D) and (E) Western blot confirmation that the class II proteins TRIM28 (D) and MAP2K1 (E) were only partially degraded in apoptotic cells. The bar graph to the right of each peptograph represents total spectral counts detected for the parental species in control (red) and apoptotic (blue) samples. Spectral count data are represented as the mean +/− SEM for four independent experiments.
Generation of Detailed Topographical Maps of Cleaved Proteins
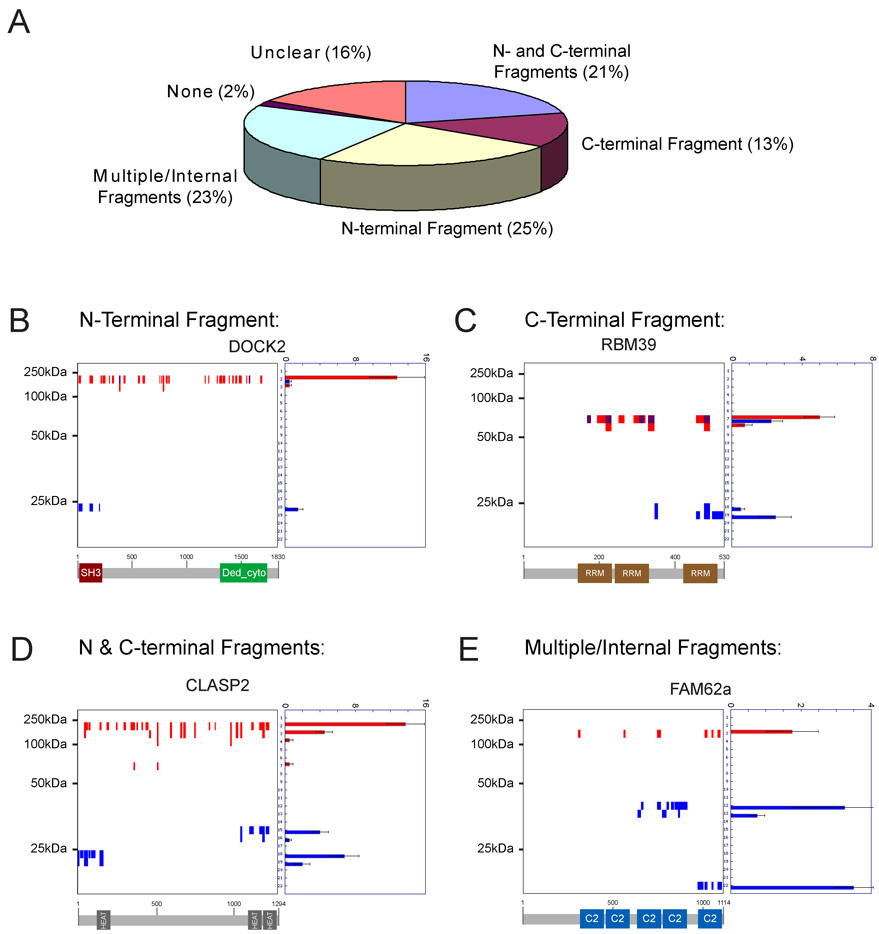
A striking feature of the PROTOMAP dataset was the prevalence of cleaved proteins that generated stable or persistent fragments in apoptotic cells. Indeed, a global analysis revealed that greater than 95% of the predicted cleaved proteins displayed at least one persistent fragment (Figure 5A). This high percentage was observed for all three classes of parental degradation (Supplemental Figure 5), indicating that even proteins whose parental forms underwent complete degradation tended to show persistent fragments. The persistent fragments observed in apoptotic cells derived from essentially all possible topographical classes: 1) N-terminal (25%) 2) C-terminal (13%), 3) N- & C-terminal (21%), and 4) internal (23%) (Figure 5A). Representative examples of each class are shown in panels B–E of Figure 5. Many of the persistent fragments mapped to domains that are known to form stably folded and functional modules, such as the SH3, HEAT, and C2 domains shown for the proteins DOCK2, CLASP2, and FAM62a, respectively (Figure 5B, D, and E, respectively, and Supplemental Table 1). Not all domains, however, were preserved in apoptotic cells, even in cases where the same domain was repeated multiple times in an individual protein (e.g., RBM39, which contains three RRM domains, only one of which persisted in apoptotic cells; Figure 5C). Clear instances were also found of persistent fragments that did not contain known protein domains, such as the C-terminal portion of the splicing factor SF3B2 (Supplemental Figure 8). It is interesting to speculate that these fragments may correspond to protein domains not yet recognized by current search algorithms. These data collectively suggest that apoptotic proteolytic cascades do not, in general, effect the complete degradation of proteins, but rather generate new forms of these proteins.
Figure 5. Visualization of the topography of protein cleavage events in apoptotic cells.
(A) The vast majority (> 95%) of cleaved proteins generated at least one persistent fragment in apoptotic cells. Essentially all possible topographical classes of fragments were observed: 1) N-terminal (25%) 2) C-terminal (13%), 3) N- & C-terminal (21%), and 4) internal (23%). “Unclear” refers to cleaved proteins with fragments that could not be obviously assigned to one of the other four fragment classes. (B)–(E) Peptographs for representative examples of N-terminal (B), C-terminal (C), N- and C- terminal (D), and multiple/internal (E) fragments. Predicted domain structures for each protein are shown below each peptograph to highlight the fact that most persistent fragments map to functional protein domains. Spectral count data are represented as the mean +/− SEM for four independent experiments.
Temporal Stability of Persistent Fragments
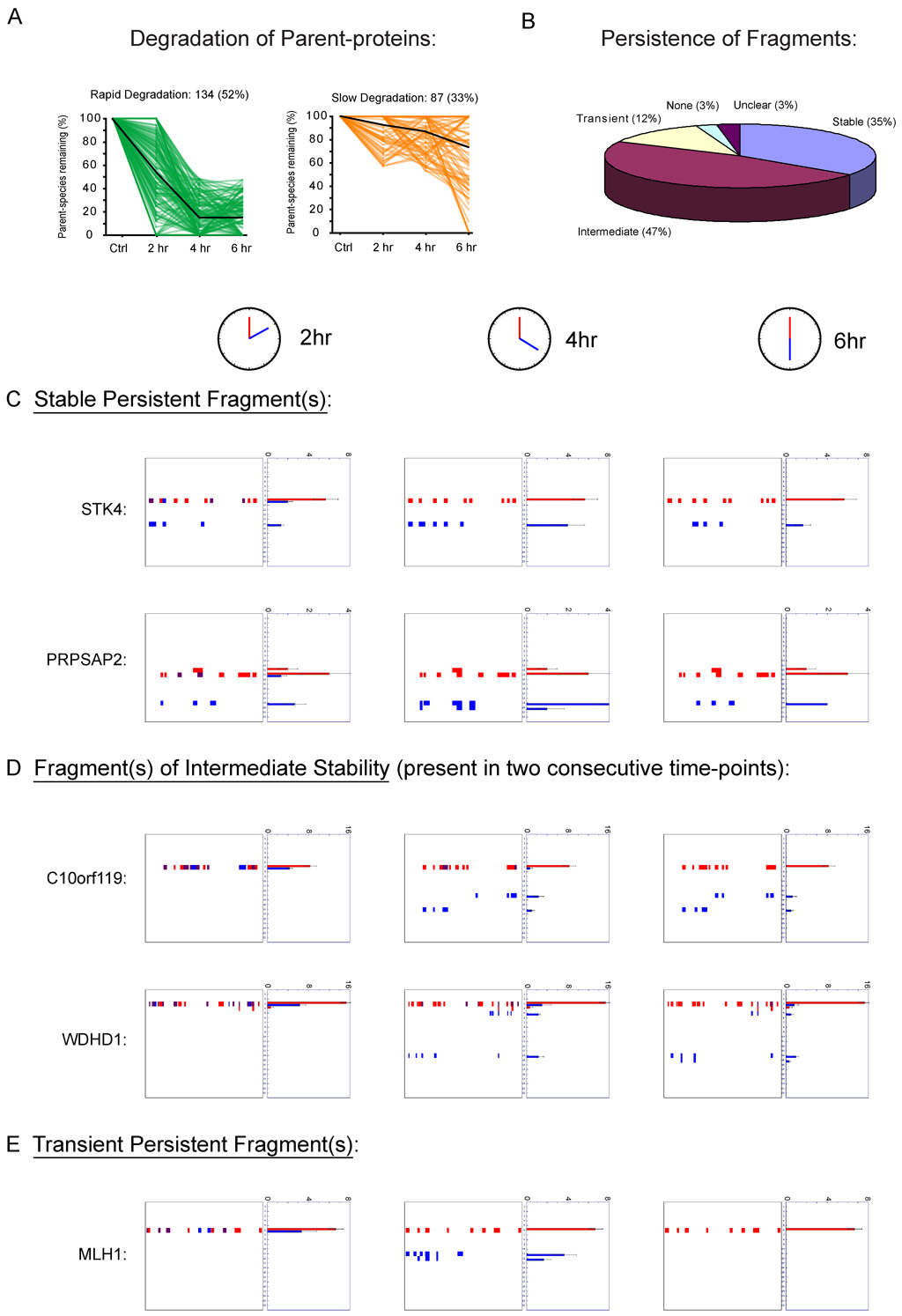
That persistent fragments were observed for many proteins whose parental species were completely degraded suggested that the life-time of these fragments is substantial, and that they may represent functional effectors rather than transient intermediates en route to total degradation. To more thoroughly investigate the temporal stability of persistent fragments, we analyzed cells at multiple time-points during the apoptotic cascade. PROTOMAP analyses were conducted at 2, 4, and 6 hours following staurosporine treatment, as these time points were found to bracket the early and late stages of apoptosis as judged by DNA fragmentation, caspase-3 activation, and PARP-1 cleavage (Supplemental Figure 7). The rate of degradation of proteins displaying persistent fragments varied widely, with roughly 52% and 33% of these proteins undergoing rapid and slow degradation, respectively (Figure 6A). More striking, however, was the stability of the persistent fragments of cleaved proteins. Indeed, more than one-third of these proteins displayed stable fragments that persisted at all three time-points (Figure 6B, C). The vast majority of the remaining proteins displayed fragments of intermediate stability that were detected at two consecutive time-points but absent from a third, thereby demonstrating lifetimes of at least 2 hours (Figure 6B, D). These intermediate fragments could be further subdivided into those that were observed early (2 and 4, but not 6 hr) versus late (4 and 6, but not 2 hr) in the time-course. The majority of intermediate fragments fell into the latter category, likely representing slowly accumulating fragments rather than fragments proceeding toward complete degradation. Only 12% of cleaved proteins displayed transient fragments that were detected exclusively at a single time-point (Figure 6B, E). Strikingly, many proteins that were rapidly degraded displayed stable fragments that persisted throughout all time-points (Figure 6C, Supplemental Table 3, Supplemental Figure 7C). In fact, stable persistent fragments were detected with overall higher frequency among the rapid- versus slow-degradation proteins (40% vs 24%, respectively; Supplemental Figure 7C). Collectively, these time-course data further support the premise that apoptotic proteolytic pathways commonly generate new, stable forms of proteins rather than effecting their total degradation and clearance.
Figure 6. Time-course analysis of proteolytic events in apoptotic cells.
(A) Rates of degradation of proteins could be divided into two general categories, rapid and slow, based on the quantity of parental species remaining at the 4 hr time-point following STS treatment (< 50% versus > 50% parental protein remaining, respectively). The time-course of degradation is shown for all proteins in these two categories, with the global averages indicated by dark lines. (B) Global analysis of the temporal stability of persistent fragments. Most proteins displayed fragments that were highly persistent, being detected at either all three time-points (C) or two consecutive time-points (D). A small fraction of proteins displayed transient persistent fragments (E) that were detected exclusively at a single time-point. See Supplementary Table 3 for a complete list of time-course profiles for cleaved proteins. Spectral count data are represented as the mean +/− SEM for three independent experiments.
Identification of Explicit Sites of Proteolytic Cleavage
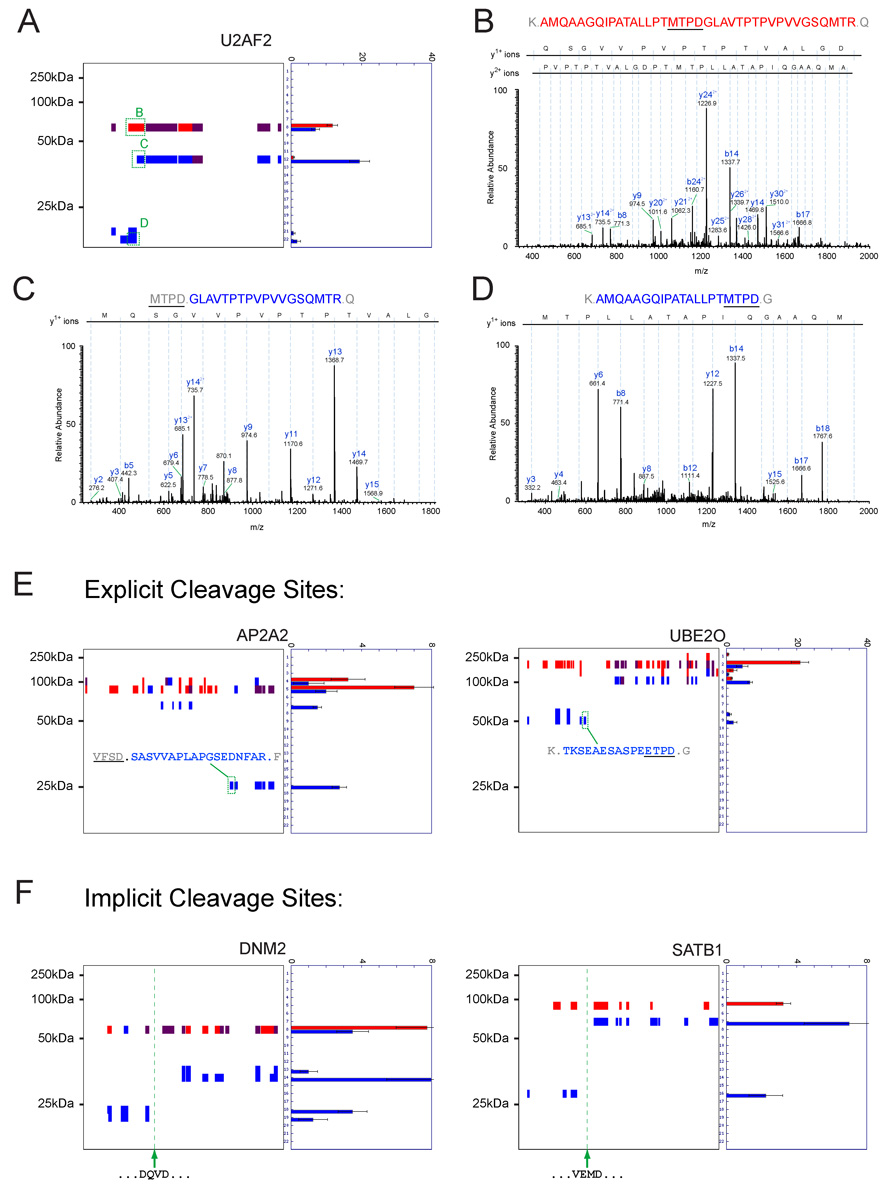
A prominent goal for technologies aiming to map proteolytic pathways is to determine the precise sites of protein cleavage (Schilling and Overall, 2007). Although PROTOMAP was not originally designed for the goal of detecting explicit sites of cleavage, thorough examination of our data revealed a surprising number of half-tryptic peptides where one terminus was defined by an aspartic acid residue that fell on the boundary of a persistent fragment. We detected 74 such peptides corresponding to 68 unique cleavage events in 61 proteins (Supplemental Table 4). A number of peptides spanning and bordering such scissile residues were hand-sequenced to ensure valid assignment. A representative example is shown in Figure 7 for the protein U2AF2, where a peptide spanning the cleavage site MTPD325 was detected in the parental protein species (Figure 7A and B), and peptides corresponding to both C- and N-terminal products of proteolysis were detected in bands 12 and 22, respectively (Figure 7A, C, and D). In most cases, it was possible to place the half-tryptic peptide precisely at the boundary of a persistent fragment, which bolstered confidence in the accuracy and validity of cleavage site assignment (Figure 7E; also see Figure 3D). Interestingly, in many instances where explicit cleavage sites were not detected, the topographical coverage provided by PROTOMAP was sufficient to identify implicit cleavage sites matching caspase consensus sequences. Indeed, literature searches revealed that several of these implicit cleavage events corresponded to validated sites of caspase-mediated proteolysis. Two representative examples, proteins DNM2 and SATB1, are shown in Figure 7F. Extrapolating from these results, candidate caspase proteolysis sites could also be predicted for other cleaved proteins (e.g., JMJD1B, Figure 3C).
Figure 7. Mapping precise sites of caspase-mediated proteolysis.
(A) Peptograph for the cleaved protein U2AF2 showing the locations of a peptide from the parent species that spans the exact site of cleavage (B), as well as two half-tryptic peptides from persistent C-terminal and N-terminal fragments (C and D, respectively). For each spectrum dominant diagnostic y- and b-ions are identified. All ions are in the +1 charge state unless otherwise indicated. (E) Additional representative examples of explicit caspase cleavage sites identified by PROTOMAP that map to the ends of N- or C-terminal persistent fragments (AP2A2 and UBE2O, respectively). (F) Representative examples of established caspase cleavage sites that can be implicitly assigned by PROTOMAP (DNM2 and SATB1, respectively). Spectral count data are represented as the mean +/− SEM for four independent experiments.
DISCUSSION
The large number of proteases encoded by the human genome underscores the pervasive role of proteolytic pathways in biological processes. A complete picture of endogenous substrates is, however, lacking for most, if not all human proteases. To address this challenge, we developed PROTOMAP, a conceptually simple and robust method that combines the exceptional resolution of intact proteins afforded by 1D-SDS-PAGE with the orthogonal separation and high sensitivity of shotgun LC-MS/MS. Information on the size of parent proteins and their proteolytic fragments is preserved by recording gel migration rates, which, when integrated with peptide sequence coverage and spectral count data acquired by LC-MS/MS, provides a holistic view of proteolytic cleavage events in native proteomes.
To evaluate the performance of PROTOMAP, we compared the proteomes of normal and apoptotic Jurkat T-cells. Apoptosis has been intensively investigated by previous proteomic methods (Brockstedt et al., 1998; Gerner et al., 2000; Schmidt et al., 2007; Thiede et al., 2006; Van Damme et al., 2005) and, thus, offered a good model system to evaluate the scope and sensitivity of PROTOMAP. PROTOMAP identified more than 250 cleaved proteins in apoptotic cells, nearly two-thirds of which corresponded to proteins not previously known to be proteolyzed during apoptosis. Confidence in the accuracy of cleaved protein assignments was bolstered by multiple lines of experimental evidence, including western blotting and explicit detection of many cleavage sites that match consensus sequences for caspases (Supplemental Table 4).
Perhaps the most striking feature of the PROTOMAP datasets is the remarkable number of cleavage events that were found to generate persistent protein fragments in apoptotic cells (Figure 5). We observed little correlation between the magnitude of cleavage of the parental protein and the likelihood of generating a persistent fragment (Supplemental Figure 5), which indicates that these fragments are not transient intermediates, but rather exist in apoptotic cells for time periods that extend well beyond the half-life of the parent protein. In further support of this notion, a time-course analysis revealed that the vast majority of fragments are stable for at least two hours, with many persisting for hours after complete degradation of their parent protein (Figure 6 and Supplemental Table 3) and well beyond the time it takes STS to kill the majority of Jurkat T-cells (~90% cell death by 4–6 hr post-treatment (Na et al., 1996)]. Interestingly, persistent fragments often corresponded to functional domains involved in a range of cellular processes, including enzymatic catalysis, protein-protein interactions, and protein-RNA/DNA interactions (Supplemental Table 1). These findings, in light of previous studies showing pro-apoptotic roles for fragments of proteins such as PAK2 and ROCK1 (Coleman et al., 2001; Rudel and Bokoch, 1997), suggest that the production of active (or dominant-negative) protein effectors may be a principal function of apoptotic proteolytic cascades. This could be mechanistically accomplished, at least in part, by the evolutionary introduction of caspase cleavage sites into interdomain linker regions, which are frequently unstructured and therefore readily accessible to proteases. One potential example of this phenomenon is PRPSAP2, a protein that shares 44% overall sequence identity with the purine-metabolizing enzyme phosphoribosylpyrophosphate synthetase 2 (PRPS2) (Supplemental Figure 6). High homology is observed throughout the two protein sequences, except for one insertion segment in PRPSAP2 (aa 218–248) that corresponds to an unstructured loop-region separating two domains (Supplemental Figure 6). PRPSAP2 was identified by PROTOMAP as a rapidly degraded protein in apoptotic cells that generates a highly stable N-terminal fragment (Figure 6C and Supplemental Figure 6). Interestingly, the precise site-of-cleavage was mapped to the caspase-consensus sequence DLVD223, which lies in the middle of the unstructured loop-region insertion. In contrast, PRPS2, which lacks this insertion, was not cleaved in apoptotic cells (Supplemental Figure 6).
An overview of the data provided by PROTOMAP reveals several advantages compared to previous methods for profiling proteolytic pathways in proteomes. First, unlike peptide labeling techniques, PROTOMAP provides a complete topographical description of the impact of proteolysis on protein structure, illuminating whether specific cleavage events generate persistent N- or C-terminal fragments (or both) and assigning predicted sizes to these products. The value of this high-content information is evident from a comparison of the data sets generated by PROTOMAP and N-terminal labeling. The most extensive N-terminal labeling study performed to date reported 50 proteins that underwent caspase-mediated cleavage in apoptotic cells (Van Damme et al., 2005). We identified 41 of these proteins by PROTOMAP, and, for 24, the peptographs showed clear evidence of protein cleavage (Supplemental Table 5). In many of these cases, PROTOMAP detected at least one persistent fragment with a mass consistent with the previously defined site of caspase-mediated proteolysis. However, in most instances, only a subset of the total possible persistent fragments was observed for the cleaved protein (i.e., either an N- or C-terminal fragment was detected, but not both) (Supplemental Table 5). PROTOMAP further revealed clear examples, such as the splicing factor SF3B2, where internal fragments were generated from multiple caspase-mediated cleavage events (Supplemental Figure 8), only one of which was detected by N-terminal labeling. These findings demonstrate that PROTOMAP discerns both the stability and structure of cleavage products of proteins, parameters that cannot be inferred from an analysis of sites of proteolysis alone. PROTOMAP also circumvents several limitations of other gel-based techniques for profiling proteolytic events. Specifically, PROTOMAP obviates the need for gel-based visualization of candidate cleavage events by protein staining, which contributed to substantial increases in sensitivity and proteome coverage. Indeed, several previous efforts using 1- and 2-DGE have collectively identified only 49 cleaved proteins in apoptotic cells (Brockstedt et al., 1998; Gerner et al., 2000; Thiede et al., 2005; Thiede et al., 2006).
In considering potential limitations of PROTOMAP and areas for future methodological improvement, one important challenge relates to the identification of explicit sites of proteolytic cleavage. Although we did not originally design PROTOMAP with the intention of detecting exact cleavage sites, we were pleasantly surprised that the extensive sequence coverage provided by this platform resulted in the identification of 61 distinct proteins for which one or more precise caspase cleavage sites were mapped in apoptotic cells (Supplemental Table 4). A substantial fraction of the caspase cleavage sites identified by PROTOMAP have not been previously reported (Supplemental Table 4). These proteolytic events thus serve as a rich source of information on the endogenous substrate profiles of caspases. In this context, comparison of our PROTOMAP data to a recent mRNA display study that mapped caspase substrates in vitro using recombinant proteins is illuminating (Ju et al., 2007). In at least two cases (GAPVD1 and TFG), the detailed topography provided by PROTOMAP permitted assignment of explicit cleavage sites to proteins that bore multiple consensus caspase cleavage sequences within the protein fragment profiled by mRNA display (Supplemental Figure 9). Thus, PROTOMAP was able to both confirm the physiological relevance of these proteolytic events in apoptotic cells and determine which of several candidate sites is endogenously cleaved. It is notable that the data for TFG fell far below our original spectral count threshold for quantitative analysis (11 total spectral counts for TFG; 30 total spectral counts for quantitative analysis), suggesting that PROTOMAP likely identified many additional low-abundance and legitimate cleavage events that, for the purpose of limiting false-positives, we have refrained from analyzing in this study. Indeed, over 100 additional putatively cleaved proteins were found in the sub-threshold data, including several established markers of apoptosis (e.g., caspase-8, STK3, and Cbl, Supplemental Table 2) (Graves et al., 1998; Widmann et al., 1998), as well as 20 additional explicit cleavage sites (Supplemental Table 4). N-terminal labeling studies have also uncovered several proteolytic events in apoptotic cells that are non-caspase-mediated (Enoksson et al., 2007), and it is possible that some of the cleaved proteins identified by PROTOMAP may result from the activity of other proteases activated during apoptosis (e.g., HtrA1, calpains, cathepsins). Collectively, these results indicate that further efforts to improve the sensitivity and refine the analysis of PROTOMAP data should yield an even larger number of proteins that can be quantitatively profiled, as well as provide enhanced sequence coverage to facilitate the mapping of explicit sites of proteolysis.
A second area of potential concern for PROTOMAP is the limit of resolution afforded by 1-DGE. Although very small changes in mass (< 5%) will likely prove difficult to detect, it is important to point out that PROTOMAP was able to identify proteins displaying 10–20% shifts in size across a large mass range (~20–200 kDa), indicating that excellent overall resolution can be achieved even when analysis is performed on a single 10% acrylamide SDS-PAGE gel. Further improvements in resolution of very large or small proteins could likely be achieved by varying the percentage of acrylamide in the SDS-PAGE gel. There are, of course, events besides proteolysis that can alter the migration of proteins by 1-DGE including phosphorylation, glycosylation, and alternative splicing. Considering that the vast majority of predicted cleaved proteins identified by PROTOMAP showed multiple lines of evidence indicative of proteolysis (e.g., conversion to one or more persistent fragments, reductions in the quantity of parental protein, and/or presence of an explicit caspase cleavage event), we do not believe that non-proteolytic events contributed significantly to the data acquired in this study.
In summary, PROTOMAP constitutes a robust and versatile strategy for the global analysis of proteolytic pathways in proteomes that complements and, in many ways surpasses, previously reported methods. A remarkable quantity of high-content information pertaining to proteolytic pathways in apoptotic cells was acquired by PROTOMAP, which in turn engenders hypotheses about the biochemical pathways that support this complex and important cellular process. Projecting beyond apoptosis, we believe that PROTOMAP will serve as a generally useful platform to characterize proteolytic pathways in a wide range of (patho)physiological processes, including tissue development, cancer, inflammation, and infectious disease.
EXPERIMENTAL PROCEDURES
Cell Culture and Induction of Apoptosis
Jurkat cells were grown under standard conditions and seeded to a density of 1 × 106 cells/ml prior to induction of apoptosis. Staurosporine (1 µM) was added and the cells were incubated for 2, 4, or 6 hrs at 37°C prior to lysis. See Supplemental Experimental Procedures for more detail.
Sample Preparation, SDS-PAGE, and LC-MS
100 µg of cytosolic protein was separated via a 10% SDS-PAGE gel and cut into 22 0.5 cm bands. Bands were subjected to in-gel trypsin digestion using standard procedures and resulting peptides were pressure-loaded onto a 100 µm (inner diameter) fused silica capillary column containing 10 cm of C18 resin. Peptides were eluted from the column using a 2-hour gradient with a flow-rate of 0.25µL/min directly into an LTQ ion trap mass spectrometer (ThermoFisher). The LTQ was operated in data-dependant scanning mode, with one full MS scan followed by seven MS/MS scans of the most abundant ions with dynamic exclusion enabled. See Supplemental Experimental Procedures for more detail.
Data Analysis
Raw MS/MS data was searched using the SEQUEST algorithm using a concatenated target/decoy variant of the human IPI database allowing for differential methionine oxidation and requiring static cysteine alkylation. SEQUEST data from each band were filtered and sorted with DTASelect with the following parameters: peptides were required to be tryptic on at least one terminus and the other terminal residue was allowed to be lysine, arginine, or aspartate. The minimum required deltaCN was 0.8 and peptides in the +1, +2 and +3 charge-states were required to have minimum XCorr values of 1.8, 2.5, and 3.5, respectively. Filtered proteomic data was organized and assembled into peptographs using three custom perl scripts. Using these methods our false-positive rate for peptides was found to be less than 0.01%. All of the custom scripts are available from our website (http://www.scripps.edu/chemphys/cravatt/protomap) and this analysis is described in full detail in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Andrew Su for programming assistance. This work was supported by National Institutes of Health (CA087660), the ARCS Foundation (G.M.S.), a Koshland Graduate Fellowship in Enzyme Biochemistry (G.M.S.), and the Skaggs Institute for Chemical Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Rahman HM, Kimura T, Hidaka K, Kiso A, Nezami A, Freire E, Hayashi Y, Kiso Y. Design of inhibitors against HIV, HTLV-I, and Plasmodium falciparum aspartic proteases. Biological Chemistry. 2004;385:1035–1039. doi: 10.1515/BC.2004.134. [DOI] [PubMed] [Google Scholar]
- Abud HE. Shaping developing tissues by apoptosis. Cell Death Differ. 2004;11:797–799. doi: 10.1038/sj.cdd.4401455. [DOI] [PubMed] [Google Scholar]
- Alnemri ES. Mammalian cell death proteases: a family of highly conserved aspartate specific cysteine proteases. J Cell Biochem. 1997;64:33–42. doi: 10.1002/(sici)1097-4644(199701)64:1<33::aid-jcb6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- auf dem Keller U, Doucet A, Overall CM. Protease research in the era of systems biology. Biological Chemistry. 2007;388:1159–1162. doi: 10.1515/BC.2007.146. [DOI] [PubMed] [Google Scholar]
- Bredemeyer AJ, Lewis RM, Malone JP, Davis AE, Gross J, Townsend RR, Ley TJ. A proteomic approach for the discovery of protease substrates. Proceedings of the National Academy of Sciences. 2004;101:11785–11790. doi: 10.1073/pnas.0402353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockstedt E, Rickers A, Kostka S, Laubersheimer A, Dorken B, Wittmann-Liebold B, Bommert K, Otto A. Identification of Apoptosis-associated Proteins in a Human Burkitt Lymphoma Cell Line. Cleavage of Heterogeneous Nuclear Ribonucleoprotein A1 by Caspase 3. J Biol Chem. 1998;273:28057–28064. doi: 10.1074/jbc.273.43.28057. [DOI] [PubMed] [Google Scholar]
- Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol. 2001;3:339–345. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- Correia JdS, Miranda Y, Leonard N, Ulevitch RJ. The Subunit CSN6 of the COP9 Signalosome Is Cleaved during Apoptosis. J Biol Chem. 2007;282:12557–12565. doi: 10.1074/jbc.M609587200. [DOI] [PubMed] [Google Scholar]
- Corthals GL, Wasinger VC, Hochstrasser DF, Sanchez J-C. The dynamic range of protein expression: A challenge for proteomic research. Electrophoresis. 2000;21:1104–1115. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1104::AID-ELPS1104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Cowan WM, Fawcett JW, O'Leary DD, Stanfield BB. Regressive events in neurogenesis. Science. 1984;225:1258–1265. doi: 10.1126/science.6474175. [DOI] [PubMed] [Google Scholar]
- Craig R, Cortens JP, Beavis RC. The use of proteotypic peptide libraries for protein identification. Rapid Communications in Mass Spectrometry. 2005;19:1844–1850. doi: 10.1002/rcm.1992. [DOI] [PubMed] [Google Scholar]
- Dean RA, Overall CM. Proteomics Discovery of Metalloproteinase Substrates in the Cellular Context by iTRAQTM Labeling Reveals a Diverse MMP-2 Substrate Degradome. Mol Cell Proteomics. 2007;6:611–623. doi: 10.1074/mcp.M600341-MCP200. [DOI] [PubMed] [Google Scholar]
- Dong M-Q, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR., III Quantitative Mass Spectrometry Identifies Insulin Signaling Targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Enoksson M, Li J, Ivancic MM, Timmer JC, Wildfang E, Eroshkin A, Salvesen GS, Tao WA. Identification of Proteolytic Cleavage Sites by Quantitative Proteomics. J Proteome Res. 2007;6:2850–2858. doi: 10.1021/pr0701052. [DOI] [PubMed] [Google Scholar]
- Feng G, Kaplowitz N. Mechanism of staurosporine-induced apoptosis in murine hepatocytes. Am J Physiol Gastrointest Liver Physiol. 2002;282:G825–G834. doi: 10.1152/ajpgi.00467.2001. [DOI] [PubMed] [Google Scholar]
- Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner C, Frohwein U, Gotzmann J, Bayer E, Gelbmann D, Bursch W, Schulte-Hermann R. The Fas-induced Apoptosis Analyzed by High Throughput Proteome Analysis. J Biol Chem. 2000;275:39018–39026. doi: 10.1074/jbc.M006495200. [DOI] [PubMed] [Google Scholar]
- Graves JD, Gotoh Y, Draves KE, Ambrose D, Han DKM, Wright M, Chernoff J, Clark EA, Krebs EG. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. The EMBO Journal. 1998;17:2224–2235. doi: 10.1093/emboj/17.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proceedings of the National Academy of Sciences. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proceedings of the National Academy of Sciences. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, El-Deiry WS. Overview of Cell Death Signaling Pathways. Cancer Biology & Therapy. 2005;4:139–163. doi: 10.4161/cbt.4.2.1508. [DOI] [PubMed] [Google Scholar]
- Ju W, Valencia CA, Pang H, Ke Y, Gao W, Dong B, Liu R. Proteome-wide identification of family member-specific natural substrate repertoire of caspases. Proceedings of the National Academy of Sciences. 2007;104:14294–14299. doi: 10.1073/pnas.0702251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JF, Winterford CM, Harmon BV. Apoptosis. Its significance in cancer and cancer therapy. Cancer. 1994;73:2013–2026. doi: 10.1002/1097-0142(19940415)73:8<2013::aid-cncr2820730802>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Kridel SJ, Chen E, Kotra LP, Howard EW, Mobashery S, Smith JW. Substrate Hydrolysis by Matrix Metalloproteinase-9*. J Biol Chem. 2001;276:20572–20578. doi: 10.1074/jbc.M100900200. [DOI] [PubMed] [Google Scholar]
- Kuster B, Schirle M, Mallick P, Aebersold R. Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- Lee AY, Park BC, Jang M, Cho S, Lee DH, Lee SC, Myung PK, Park SG. Identification of caspase-3 degradome by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 2004;4:3429–3436. doi: 10.1002/pmic.200400979. [DOI] [PubMed] [Google Scholar]
- Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-Scale Phosphorylation Analysis of alpha-Factor-Arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR. A Model for Random Sampling and Estimation of Relative Protein Abundance in Shotgun Proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lohaus C, Nolte A, Bluggel M, Scheer C, Klose J, Gobom J, Schuler A, Wiebringhaus T, Meyer HE, Marcus K. Multidimensional Chromatography: a Powerful Tool for the Analysis of Membrane Proteins in Mouse Brain. J Proteome Res. 2007;6:105–113. doi: 10.1021/pr060247g. [DOI] [PubMed] [Google Scholar]
- Luthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- Matrisian LM, Hogan BL. Growth factor-regulated proteases and extracellular matrix remodeling during mammalian development. Curr Top Dev Biol. 1990;24:219–259. doi: 10.1016/s0070-2153(08)60089-7. [DOI] [PubMed] [Google Scholar]
- Matthews DJ, Wells JA. Substrate Phage: Selection of Protease Substrates by Monovalent Phage Display. Science. 1993;260:1113–1117. doi: 10.1126/science.8493554. [DOI] [PubMed] [Google Scholar]
- McDonald L, Robertson DHL, Hurst JL, Beynon RJ. Positional proteomics: selective recovery and analysis of N-terminal proteolytic peptides. Nat Meth. 2005;2:955–957. doi: 10.1038/nmeth811. [DOI] [PubMed] [Google Scholar]
- Na S, Chuang T-H, Cunningham A, Turi TG, Hanke JH, Bokoch GM, Danley DE. D4-GDI, a Substrate of CPP32, Is Proteolyzed during Fas-induced Apoptosis. J Biol Chem. 1996;271:11209–11213. doi: 10.1074/jbc.271.19.11209. [DOI] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of Label-free Methods for Quantifying Human Proteins by Shotgun Proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- Riewald M, Ruf W. Mechanistic coupling of protease signaling and initiation of coagulation by tissue factor. Proceedings of the National Academy of Sciences. 2001;98:7742–7747. doi: 10.1073/pnas.141126698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel T, Bokoch GM. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- Schilling O, Overall CM. Proteomic discovery of protease substrates. Current Opinion in Chemical Biology. 2007;11:36–45. doi: 10.1016/j.cbpa.2006.11.037. [DOI] [PubMed] [Google Scholar]
- Schmidt F, Hustoft HK, Strozynski M, Dimmler C, Rudel T, Thiede B. Quantitative proteome analysis of cisplatin-induced apoptotic Jurkat T cells by stable isotope labeling with amino acids in cell culture, SDS-PAGE, and LC-MALDI-TOF/TOF MS. Electrophoresis. 2007;28:4359–4368. doi: 10.1002/elps.200700119. [DOI] [PubMed] [Google Scholar]
- Shi R, Kumar C, Zougman A, Zhang Y, Podtelejnikov A, Cox J, Wisniewski JR, Mann M. Analysis of the Mouse Liver Proteome Using Advanced Mass Spectrometry. J Proteome Res. 2007;6:2963–2972. doi: 10.1021/pr0605668. [DOI] [PubMed] [Google Scholar]
- Thiede B, Kretschmer A, Rudel T. Quantitative proteome analysis of CD95 (Fas/Apo-1)-induced apoptosis by stable isotope labeling with amino acids in cell culture, 2-DE and MALDI-MS. Proteomics. 2006;6:614–622. doi: 10.1002/pmic.200500120. [DOI] [PubMed] [Google Scholar]
- Thiede B, Treumann A, Kretschmer A, Söhlke J, Rudel T. Shotgun proteome analysis of protein cleavage in apoptotic cells. Proteomics. 2005;5:2123–2130. doi: 10.1002/pmic.200401110. [DOI] [PubMed] [Google Scholar]
- Timmer JC, Enoksson M, Wildfang E, Zhu W, Igarashi Y, Denault J-B, Ma Y, Dummitt B, Chang Y-H, Mast AE, et al. Profiling constitutive proteolytic events in vivo. Biochem J. 2007;407:41–48. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer JC, Salvesen GS. Caspase substrates. Cell Death Differ. 2006;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- Turgeon VL, Houenou LJ. The role of thrombin-like (serine) proteases in the development, plasticity and pathology of the nervous system. Brain Research Reviews. 1997;25:85–95. doi: 10.1016/s0165-0173(97)00015-5. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J, Gevaert K. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Meth. 2005;2:771–777. doi: 10.1038/nmeth792. [DOI] [PubMed] [Google Scholar]
- van Kempen LCL, de Visser KE, Coussens LM. Inflammation, proteases and cancer. European Journal of Cancer. 2006;42:728–734. doi: 10.1016/j.ejca.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Widmann C, Gibson S, Johnson GL. Caspase-dependent Cleavage of Signaling Proteins during Apoptosis. J Biol Chem. 1998;273:7141–7147. doi: 10.1074/jbc.273.12.7141. [DOI] [PubMed] [Google Scholar]
- Zong W-X, Ditsworth D, Bauer DE, Wang Z-Q, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.