Abstract
Telomeres play an important role in protecting the ends of chromosomes and preventing chromosome fusion. We have previously demonstrated that double-strand breaks near telomeres in mammalian cells result in either the addition of a new telomere at the site of the break, termed chromosome healing, or sister chromatid fusion that initiates chromosome instability. In the present study, we have investigated the role of telomerase in chromosome healing and the importance of chromosome healing in preventing chromosome instability. In embryonic stem cell lines that are wild type for the catalytic subunit of telomerase (TERT), chromosome healing at I-SceI-induced double-strand breaks near telomeres accounted for 22 of 35 rearrangements, with the new telomeres added directly at the site of the break in all but one instance. In contrast, in two TERT-knockout embryonic stem cell lines, chromosome healing accounted for only 1 of 62 rearrangements, with a 23 bp insertion at the site of the sole chromosome-healing event. However, in a third TERT-knockout embryonic stem cell line, 10PTKO-A, chromosome healing was a common event that accounted for 20 of 34 rearrangements. Although this chromosome healing also occurred at the I-SceI site, differences in the microhomology at the site of telomere addition demonstrated that the mechanism was distinct from that in wild type embryonic stem cell lines. In addition, the newly added telomeres in 10PTKO-A shortened with time in culture, eventually resulting in either telomere elongation through a telomerase-independent mechanism or loss of the subtelomeric plasmid sequences entirely. The combined results demonstrate that chromosome healing can occur through both telomerase-dependent and independent mechanisms, and that although both mechanisms can prevent degradation and sister chromatid fusion, neither mechanism is efficient enough to prevent sister chromatid fusion from occurring in many cells experiencing double-strand breaks near telomeres.
Keywords: Chromosome healing, telomere, telomerase, chromosome fusion, chromosome instability
1. Introduction
Telomeres are DNA-protein complexes containing DNA repeat sequences that are added on to the ends of chromosomes by telomerase [1,2]. Telomeres serve multiple functions, including protecting the ends of chromosomes [3] and preventing chromosome fusion [4]. In humans, telomeres are maintained in germ line cells, but shorten with age in most somatic cells due to the lack of sufficient telomerase activity [4]. Telomere shortening in somatic cells eventually leads to replicative cell senescence due to the inability of the telomere to form a cap that protects the end of the chromosome [5–7]. Primary human fibroblasts that fail to senesce continue to show telomere shortening and eventually enter crisis, which involves increased chromosome fusion, aneuploidy, and cell death [8]. Thus, the ability to maintain telomeres is considered to be an important step in carcinogenesis, not only to avoid senescence, but also to avoid the extensive chromosome fusion during crisis [8,9]. Consistent with this hypothesis, human tumor cells commonly demonstrate the ability to maintain telomeres, either through expression of telomerase or through an alternative mechanism involving recombination [10,11].
Telomere loss results in chromosome instability both by allowing degradation of the ends of chromosomes and by promoting chromosome fusion. Chromosome instability due to telomere loss in yeast results mostly through degradation of the end of the chromosome [12]. Although degradation of the ends of chromosomes following telomere loss also occurs in mammalian cells [13], the resulting instability is less extensive than that resulting from chromosome fusion [14,15]. Fusion can occur between sister chromatids, or between different chromosomes if telomeres are lost on more than one chromosome [16]. Chromosome fusion results in chromosome instability through breakage/fusion/bridge (B/F/B) cycles, when chromosomes repeatedly fuse and break for many cell generations. B/F/B cycles involving sister chromatid fusions result in gene amplification and chromosome translocations [14,15], both of which are important chromosome aberrations associated with cancer [17,18].
Telomeres that have been lost can be restored, a process called chromosome healing. In yeast and Tetrahymena, chromosome healing can occur by de novo addition of telomeric repeat sequences on to the ends of chromosomes by telomerase [19,20]. De novo telomere addition by telomerase is inhibited by the Pif1 helicase, which has been proposed to be a mechanism for preventing telomerase from competing with double-strand break (DSB) repair mechanisms [21]. Chromosome healing has also been observed in humans, where it is responsible for terminal deletions resulting in genetic disease [22–24]. In addition, de novo addition of telomeres by telomerase has been demonstrated in vitro with human cancer cell extracts [25]. Despite this evidence that de novo addition of telomeres can occur in human cells, there is little evidence that chromosome healing occurs at most DSBs in mammalian cells, even when telomerase is expressed. In mammalian cells, telomeric repeat sequences are not found at most ionizing radiation or enzyme-induced DSBs [26–29], and the expression of telomerase has little effect on the cellular response to DSBs [30]. This lack of chromosome healing may be due to low levels of telomerase compared to DSB repair proteins, or because de novo telomere addition by telomerase in mammalian cells is actively suppressed, as it is in yeast [21].
Despite the lack of evidence for chromosome healing at most DSBs in mammalian cells, we have observed that DSBs near telomeres in mouse embryonic stem (ES) cells commonly result in either chromosome healing or sister chromatid fusion [13,31]. Because neither of these types of events have been observed at DSBs at interstitial sites, their occurrence at DSBs near telomeres is likely to be due to the proximity of the telomere. One possible explanation is that chromosome healing involves recombination between the DSB and existing telomeric repeat sequences. In fact, direct telomere addition to the ends of a transfected plasmid sequences have been observed [32] in a cell line that was subsequently demonstrated to be telomerase deficient and maintain telomeres through an alternative pathway [33]. Alternatively, de novo telomere addition by telomerase may be more efficient near telomeres. Studies in yeast have shown that de novo telomere addition by telomerase preferentially occurs near existing telomeric repeat sequences, which appear to serve as a localization site for telomere-associated proteins [19]. Sister chromatid fusion also may be more likely to occur near telomeres, since DSBs in subtelomeric regions in yeast are not efficiently repaired by nonhomologous end joining (NHEJ), and therefore result in gross chromosomal rearrangements [34]. In fact, telomeres may actively suppress the repair of DSBs to prevent chromosome fusion, since telomeric repeat sequences in yeast have been shown to suppress the activation of cell cycle checkpoints in response to DSBs [35]. In addition, the human TRF2 protein, which is required to prevent chromosome fusion, has been demonstrated to inhibit ATM, which is instrumental in activating the cellular response to DSBs [36]. This selective inhibition of DSB repair near telomeres may mean that chromosome healing is an important mechanism for preventing chromosome fusion and instability as a result of DSBs occurring within subtelomeric regions.
In view of the potential for chromosome healing to prevent chromosome instability due to telomere loss, we are investigating the mechanisms of chromosome healing and its regulation in mammalian cells. In the current study, we have conducted experiments to address whether chromosome healing in mammalian cells involves telomerase or recombination, and whether chromosome healing is important in preventing degradation and chromosome instability due to DSBs near telomeres. These experiments utilize mouse ES cell lines that contain selectable marker genes and a recognition site for I-SceI endonuclease that are located adjacent to a telomere. ES cell clones containing these telomeric selectable marker genes and I-SceI site were previously used to study chromosome healing and chromosome instability resulting from DSBs near telomeres [13,31]. Transgenic mice established from one of these ES cell clones have now been bred with mice containing a knockout of the TERT gene for the catalytic subunit of telomerase to generate ES cell lines that are wild type or homozygous for knockout of TERT. TERT knockout cell lines were chosen over knockdown of TERT using iRNA to insure complete elimination of telomerase activity. These wild type and TERT knockout ES cell lines were then used to determine the influence of telomerase on chromosome healing and sister chromatid fusion following I-SceI-induced DSBs near a telomere.
2. Materials and methods
2.1. Plasmids
The pPPT2-tel plasmid used for generation of the ES cell clone 10P and the 10P mice was created from the pNPT-tel plasmid [13] by replacement of the neo gene with the puromycinresistance (puro) gene [37]. pPPT2-tel therefore contains a puro gene with an HSV-tk promoter for positive selection in puromycin, a Herpes simplex virus thymidine kinase (HSV-tk) gene with a mouse pgk gene promoter for negative selection in ganciclovir, and 0.8 kb of telomeric repeats for seeding the formation of new telomeres following integration (Fig. 1).
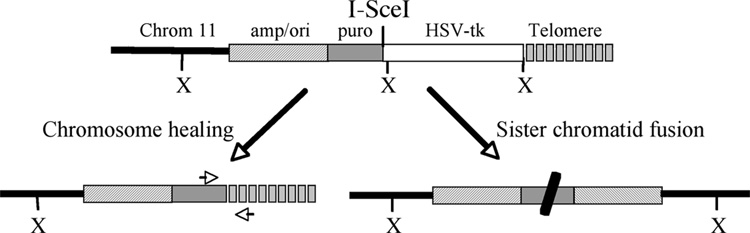
Fig. 1.
The structure of the telomeric plasmid sequences and the types of rearrangements resulting from I-SceI-induced DSBs in ES cell lines from 10P mice. The pPPT2-tel plasmid in ES cell clone 10P seeded the formation of a new telomere upon integration 1.4 Mb from the end of chromosome 11. The telomere contains an ampicillin-resistance gene and plasmid origin of replication (amp/ori), a puromycin-resistance gene (puro), an HSV-tk gene, an I-SceI endonuclease recognition site, and telomeric repeat sequences (telomere). The locations of the XbaI restriction sites used for Southern blot analysis are also shown. I-SceI endonuclease-induced DSBs near the telomere result in two types of events, chromosome healing involving the addition of telomeric repeat sequences at the site of the break (lower left), and inverted repeats resulting from sister chromatid fusion (lower right). PCR using primers specific for the puro gene and telomeric repeat sequences (arrows) are used to confirm the presence of telomere addition at the I-SceI site.
2.2. Establishment of ES cell clones and transgenic mice containing telomeric plasmid DNA
The methods for establishment and characterization of ES cell clones containing telomeric plasmids has been previously described [13,31]. The linearized pPPT2-tel plasmid containing the telomeric repeat sequences on one end was transfected into the ES cells, and clones containing stably-integrated plasmid were selected with puromycin. The puromycin-resistant ES cell clones were then screened by Southern blot analysis for clones in which a single copy of the plasmid had become the new telomere on a chromosome, as indicated by the large size of the band containing the telomeric repeat sequences. The telomeric integration site was then confirmed by fluorescent in situ hybridization, sequencing of the rescued plasmid sequences and adjacent cellular DNA, and Southern blot analysis following digestion with BAL31 nuclease [37]. The location of the newly seeded telomere was determined by sequence analysis of host DNA to be 1.4 Mb from the original end of chromosome 11 (Genbank Accession No. EF503725). The 10P transgenic mouse strain was established from the 10P ES cell clone in conjunction with the UCSF Transgenic Core Facility as previously described [38]. Briefly, ES cells were injected into blastocysts from C57BL/6J mice, and the resulting chimeric mice were bred with C57BL/6J mice to generate the 10P mouse strain containing the telomeric plasmid DNA. Polymerase chain reaction (PCR) with primers specific for the HSV-tk gene, and Southern blot analysis of tail DNA using a pNTPΔ probe, was employed to identify mice that contained the telomeric plasmid sequences, using protocols described for analysis of ES cell lines [31].
2.3. TERT knockout mice
TERT knockout mice were generated by homologous recombination in mouse embryonic stem cells. A genomic TERT fragment incorporating exons 3–8 was subcloned from an TERT BAC. The targeting plasmid was designed to delete exons 3–6, which includes much of the reverse transcriptase domain essential for mTert function. A puromycin positive selection cassette flanked by loxP sites was inserted in intron 2, and negative selection was achieved with a diphtheria toxin gene cassette. The targeting plasmid was transfected into J1 ES cells by electroporation. Puromycin-resistant ES colonies were analyzed by Southern blot analysis using both 5' and 3' probes to identify homologous recombinants. A correctly targeted ES clone was used to generate chimeric mice through blastocyst complementation and these chimeric mice were intercrossed with C57BL/6 mice to achieve germline transmission of the TERT null allele. Pups were screened for germline transmission by coat color and PCR for the targeted allele. To delete the puromycin cassette, TERT heterozygous mice were intercrossed with a CMV-Cre transgenic line and the offspring were screened by PCR (data not shown).
2.4. Isolation of ES cell lines from transgenic/knockout mice
The ES cell lines derived from the 10P × TERT knockout mice were established as previously described [39]. Briefly, blastocysts were collected by flushing out the uterine horns of 3.5-day postcoitum mice with M2 media (Sigma). The blastocysts were then transferred individually into 10-mm tissue culture wells containing a layer of PMEF feeder cells (Specialty Media) in ES media, and were cultured at 37 °C in a 5% CO2 humidified incubator for approximately 10 days. The inner-cell-mass-derived outgrowths were trypsinized and transferred into new tissue culture wells containing PMEF feeder cells. After 4 days in culture, the ES-cell-like clumps were selected, expanded, and frozen for future use. The ES cell lines were then screened for pPPT2-tel using primers specific for the HSV-tk gene [31], primers specific for wild type TERT (449 bp band using GCCAGCAATCAACTGACACTCG and ATGGGAACTTGGGAAGGAGAAGGG primers), and knockout TERT (545 bp band using GCCAGCAATCAAACTGACACTCG and AATGCTTGAGGGGAACCAGAGG primers). These ES cell lines are not isogenic, since the mice that they were established from have both 129 and C57BL/6 backgrounds.
2.5 Analysis of telomerase activity in ES cell clones
Telomerase activity was determined using the TRAPeze XL Telomerase Detection Kit (Millipore). The preparation of cell extracts from 1000 cells, and the assay procedure were performed according to manufacturer’s instructions. Individual sample values were determined using a Versa Fluor Fluorometer (BioRad).
2.6. Transfection with I-SceI and selection for loss of the HSV-tk gene
Expression of I-SceI endonuclease was achieved by transient transfection with the pCBASce plasmid. pCBASce contains the I-SceI gene with a chicken β-actin promoter, and has previously been shown to provide a high efficiency of cutting at I-SceI sites in mammalian cells [40]. Thirty micrograms of pCBASce was electroporated into 5 × 106 cells. The next day, the cells were plated at 2 × 105 cells per 100 mm tissue culture dish, and after culturing for 6 days to allow turnover of existing HSV-tk [41,42], selection was performed using medium containing 2 µM ganciclovir and 2 µg/ml puromycin. Individual colonies were then selected for analysis after approximately 2 weeks.
2.7. Southern blot analysis
Genomic DNA was purified by high-salt extraction. Cell pellets were washed and resuspended in STE (0.02 M Tris HCl, 0.01 M EDTA, 0.1 M NaCl, pH 8.5). Sodium dodecyl sulfate (Sigma) was then added to a final concentration of 0.5% and the lysed cells were heated at 60°C for 10 minutes. RNAse A (Sigma) was added to a final concentration of 50 µg/ml and the lysed cells incubated at 37°C for 4 hours, followed by the addition of Proteinase K (Sigma) to a final concentration of 32 µg/ml and incubation overnight at 37°C. 5M NaCl was then added to a final concentration of 1.4 M, and the samples chilled on ice for 30 minutes. The lysis solution was then centrifuged at 3,000 g, and the clear DNA solution removed using a wide-bore pipet. The DNA was precipitated with 0.7 volumes of isopropanol, and removed by capture with a sterile micropipet tip. The precipitated DNA was then added to a 1.5 ml Eppendorff tube containing 70% ethanol and pelleted at maximum speed with an Eppendorf 5415 centrifuge. After pouring off the ethanol solution, the DNA pellet is dried and resuspended in TE buffer (0.1 M NaCl, 0.01 M TrisHCl, pH 7.5).
Digestion of DNA with restriction enzymes and BAL31 nuclease was performed following the manufacturer’s recommendations (New England Biolabs). Standard agarose gel electrophoresis was performed as previously described [13]. PFGE was performed using 1% agarose in 0.5 × TBE (0.045M Tris-borate, 0.001M EDTA), at 150V, 10°C, and pulsed at 2 to 5 second intervals for PFGE. Southern blot analysis involved depurination of DNA by treatment with 0.25 M HCl for 20 minutes, denaturation in 0.5 M NaOH / 1.5 M NaCl for 30 minutes, neutralization in 1 M Tris/1.5 M NaCl pH 7.5 for 30 minutes, and transfer onto a charged nylon Hybond-N+ membrane (Amersham) in 10 X SSC (1.5 M NaCl / 0.15 M Na Citrate, pH 7.0) using a vacuum transfer apparatus (Pharmacia). Prehybridization for 30 minutes and hybridization overnight were performed at 65°C in roller bottles in hybridization buffer (0.25 M Na phospate buffer, pH 7.2, 1 mM EDTA, and 7% SDS. Probes were labeled with [α32P] dCTP (New England Nuclear) using a High Prime labeling kit (Roche). Filters were rinsed once briefly, and then washed twice for 30 minutes at 65°C in washing solution I (20 mM Na phosphate buffer, pH 7.2, 1mM EDTA, 5% SDS), and once at 65°C in washing solution II (20 mM Na phosphate buffer, pH 7.2, 1mM EDTA, 1% SDS). The probe consisted of the plasmid pNTPΔ, which contains the same sequences as the integrated pNPT-tel plasmid, except that it does not contain telomeric repeat sequences.
2.8. Cloning and sequence analysis of the sites of chromosome healing and fusion
Analysis of sites of chromosome healing was performed using PCR with one primer complementary to the puro gene (GTGGGCTTGTACTCGGTCAT) and one primer complementary to the telomeric repeat sequences (AACCCTAACCCTAACCCTAACC). PCR was performed as previously described [31] using an initial incubation for 2 min at 95°C, followed by 40 cycles of 95°C for 30 sec, 62°C for 30 sec, and 72°C for 30 sec, followed by one cycle of 95°C for 30 sec, 62°C for 30 sec, and 72°C for 2 min. Following their purification using a Qiaex II kit (Qiagen, Valencia, CA), the PCR products were sequenced directly by Molecular Cloning Laboratories (South San Francisco, CA), using the primer complementary to the puro gene.
The cloning of the inverted repeats resulting from sister chromatid fusion was achieved by plasmid rescue. Plasmid rescue was performed by first digesting the DNA with XbaI, followed by the circularization and concentration of the DNA as previously described [13]. The DNA was then used to transform STBL2 chemically competent bacteria (Gibco), and the circularized plasmid and adjacent cellular DNA was selected using ampicillin. The rescued sequences were then mapped using restriction enzymes and sequenced from various primers specific for plamsid and puro gene sequences.
2.9. Chromosome and telomere analysis
Chromosome analysis was performed following the addition of 0.1 µg/ml colcemid to the medium for 1.5 to 2 hours at 37 °C. The cells were then trypsinized, pelleted at 500xg for 5 minutes, the supernatant removed, 2 ml of 0.56% KCl added, and incubated for 30 minutes at 37 °C. The cells were then spread on a slide and dried at room temperature. Chromosome metaphase spreads and telomere analysis were performed as previously described [43], using the Telomere PNA FISH Kit/Cy3 (DAKO) using manufacturer’s instructions. Relative telomere fluorescence was determined on a minimum of 10 metaphase cells by quantitative fluorescence in situ hybridization (Q-FISH) [44] using the TFL-Telo Telomere Measurements and Analysis Software (http://www.flintbox.com). Image capture and analysis were performed following the protocols in the TFL-Telo User’s Manual. All images were captured during the same session to minimize variation in fluorescence intensity.
3. Results
3.1. Analysis of rearrangements in ES cell lines
The mouse ES cell lines used in this study were established from the 10P mouse strain that contains a puro gene for positive selection with puromycin, an HSV-tk gene for negative selection with ganciclovir, and an 18 bp I-SceI endonuclease recognition site for introducing DSBs, all of which are immediately adjacent to a telomere on chromosome 11 (Fig. 1). DSBs at the I-SceI site are introduced by transient transfection with an expression vector containing the I-SceI gene (see Materials and Methods), a method that is commonly used to study recombination in mammalian cells [27,29,45]. Selection for the loss of the telomeric HSV-tk gene on chromosome 11 is then performed using ganciclovir alone to isolate ganciclovir-resistant (ganr) subclones, or ganciclovir and puromycin together to isolate subclones resistant to both ganciclovir and puromycin (ganr/puror). The inclusion of puromycin is used to insure that the subclones that are selected have retained some portion of the telomeric plasmid sequences to allow for analysis of the type of rearrangements involved. Selection with puromycin also eliminates cells in the population that become resistant to ganciclovir due to silencing of the telomeric transgenes as a consequence of telomere position effect [38]. The ganr/puror subclones must retain at least one copy of the puro gene and its promoter, which begins approximately 30 bps proximal to the I-SceI site. As a result, the extent of degradation at the I-SceI site in ganr/puror subclones is limited, although with sister chromatid fusions, more extensive degradation is often observed on one of the two sister chromatids involved in the fusion.
Southern blot analysis following digestion with XbaI was used to screen for the types of rearrangements that occurred within the telomeric plasmid sequences on chromosome 11 in the 10P ES cell line. We have previously shown that regardless of whether telomere loss occurs spontaneously in human tumor cells, or through I-SceI-induced DSBs in mouse ES cells, the rearrangements observed primarily involve either chromosome healing or inverted repeats resulting from sister chromatid fusion [16]. Using this approach, I-SceI-induced chromosome healing in the ganr/puror 10P subclones can be detected by the loss of the original 3.0 and 4.3 kb fragments and the appearance of a large fragment containing a telomere (Fig. 1), which are very long in laboratory mice [46]. In contrast to chromosome healing, I-SceI-induced inverted repeats in the ganr/puror 10P subclones are detected by the replacement of the 3.0 and 4.3 kb fragments with a new fragment that varies in size depending on the extent of degradation prior to sister chromatid fusion (Fig. 1). We have rescued and analyzed 11 of these fragments from mouse ES cells and 8 from human tumor cells, and all 19 contained inverted repeats of the plasmid and cellular sequences at the end of the chromosome ([13], unpublished results, present study). Proof that these inverted repeats are formed through sister chromatid fusion has been demonstrated by the presence of anaphase bridges involving sister chromatids and the amplification of the subtelomeric DNA [13,15].
In addition to I-SceI-induced events, rearrangements resulting from spontaneous telomere loss, which are rare in wild type ES cells, can also be detected by this method. Unlike I-SceI-induced events, spontaneous telomere loss in ganr/puror subclones usually results in the loss or rearrangement of the 3.0 kb XbaI fragment containing the HSV-tk gene, but not the 4.3 kb XbaI fragment containing the I-SceI site and puro gene. This is because, based on size and random distribution of the breaks, spontaneous rearrangements of the HSV-tk gene are approximately 100-fold more likely to occur within the 3 kb XbaI fragment than within the 30 bp region between the puro gene promoter and the XbaI site in the 4.3 kb XbaI fragment. In contrast, I-SceI-induced rearrangements in ganr/puror subclones must always occur within this 30 bp region due to selection for the puro gene. Consistent with this conclusion, rescue and sequence analysis of a 4.3 kb XbaI fragment from a ganr/puror subclone that was missing the 3.0 kb XbaI fragment demonstrated that the 4.3 kb fragment still contained the I-SceI site (data not shown). Ganr/puror subclones that retain the original 4.3 kb XbaI band, but lose the 3.0 kb XbaI band, are therefore scored as rearrangements resulting from spontaneous telomere loss.
3.2. Analysis of ES cell lines with wild type TERT
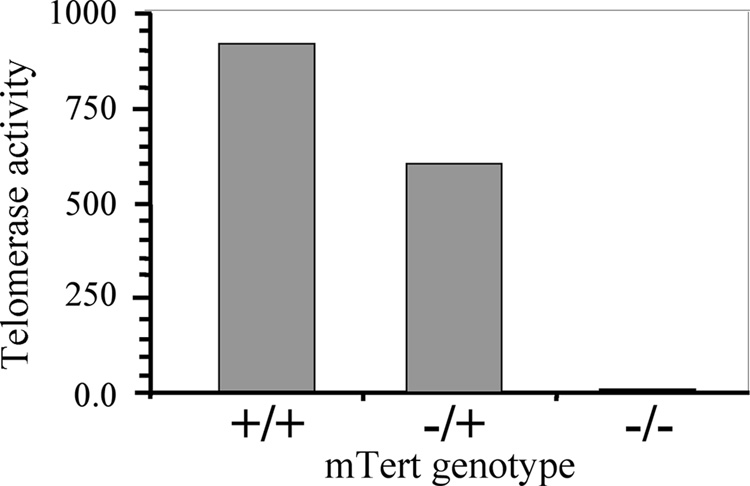
The ES cell lines used in this study were generated by breeding the 10P mouse strain with mice containing a knockout of exons 3 to 6 of TERT. The ES cell lines generated from blastocysts isolated from these mice were screened by PCR for the presence of the HSV-tk gene, the wild-type TERT gene, and the knocked-out TERT gene. Using this approach, we identified ES cell lines that contained the telomeric pPPT2-tel plasmid, and were wild type, heterozygous, or homozygous for the TERT knockout. Analysis of telomerase activity in the various ES cell lines was consistent with the PCR analysis for knockout of TERT, demonstrating the highest levels of telomerase activity in the wild-type cell lines, intermediate level of telomerase activity in the heterozygous cell lines, and no significant telomerase activity in the homozygous knockout cell lines (Fig. 2). Transient transfection of these ES cell lines with an expression vector containing the I-SceI gene typically resulted in a 10-fold increase in ganr/puror colonies, the background resulting primarily from silencing or point mutations, i.e., show no rearrangements. Southern blot analysis of the genomic DNA isolated from the I-SceI-induced ganr/puror sublclones generated from two of the ES cell lines with wild type TERT, 10PWT-C and 10PWT-F, gave results that were consistent with our earlier studies [13,31], i.e., the majority of events (22 of 35, 63%) involved chromosome healing that resulted in bands at the limit of resolution (Fig. 3, Table 1). There was no change in the lighter 2.3 kb and 4.4 kb control bands that result from hybridization of the mouse phosphoglycerate kinase (pgk) gene regulatory sequences in the plasmid probe to the endogenous pgk sequences. Digestion with EcoRI, which cuts on either side of the HSV-tk gene, showed that the band containing the HSV-tk gene was missing in all of the ganr/puror subclones (data not shown). The length of the newly added telomeres in the ganr/puror sublclones with the bands at the limit of resolution was analyzed by pulsed-field gel electrophoresis (PFGE), and found to be approximately 75 kb in length (data not shown), similar to that in the original 10P ES cell clone [37].
Fig. 2.
The level of telomerase activity in the ES cell lines isolated from embryos generated by crossing 10P mice with TERT knockout mice. The relative amount of telomerase activity in ES cell lines that are wild type, heterozygous, or homozygous for knockout of TERT was determined using the TRAPeze XL Telomere Detection Kit.
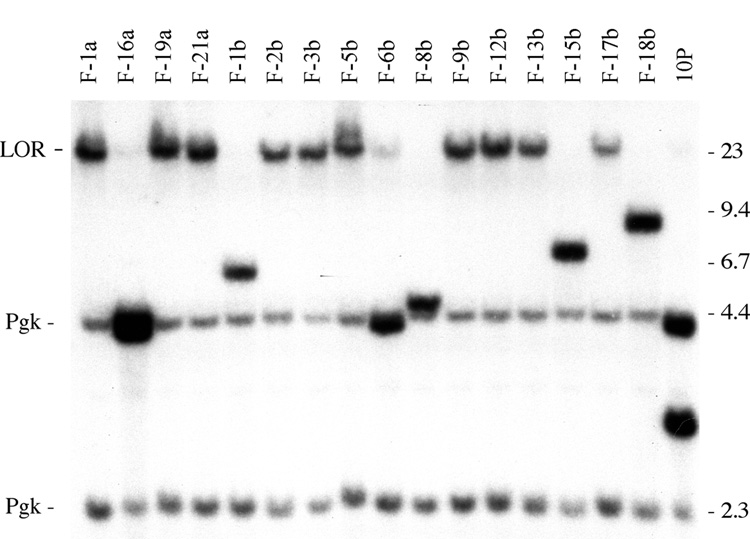
Fig. 3.
Analysis of rearrangements resulting from a DSB near a telomere in an ES cell line with wild type telomerase. Southern blot analysis of genomic DNA from I-SceI-induced ganr/puror subclones of the 10PWT-F ES cell line. DNA was digested with XbaI and hybridization was performed using the pNPTΔ plasmid as a probe (with no telomeric repeat sequences). Molecular weight markers consisting of Lambda bacteriophage HindIII fragments are indicated. The unrearranged telomeric plasmid sequences in the 10P cell line (10P) produce two plasmid-specific bands, a 3 kb band containing the HSV-tk gene, and a 4.3 kb band containing the internal portion of the plasmid and adjacent cellular DNA (see Fig. 1). In contrast, the I-SceI-induced ganr/puror subclones demonstrated either a large XbaI plasmid-specific band at the limit of resolution (LOR) as a result of chromosome healing, or variable-sized bands containing inverted repeats. The hybridization of the mouse pgk gene promoter and poly A addition sequences in the pNPTΔ plasmid probe to the endogenous mouse pgk gene results in two light bands of 2.3 and 4.4 kb that serve as loading controls.
Table 1.
Types of I-SceI-induced events in ES cell lines with wild type or knockdown TERT
| Cell Line | Chromosome Healing1 | Inverted Repeats2 |
|---|---|---|
| Wild type | ||
| 10PWT-F | 15 | 9 |
| 10PWT-C | 7 | 4 |
| Knockout | ||
| 10PTKO-H | 1 | 26 |
| 10PTKO-4B | 0 | 35 |
| 10PTKO-A | 20 | 14 |
Determined by PCR analysis of site of telomere addition
Bands of variable size and no telomere-specific PCR bands
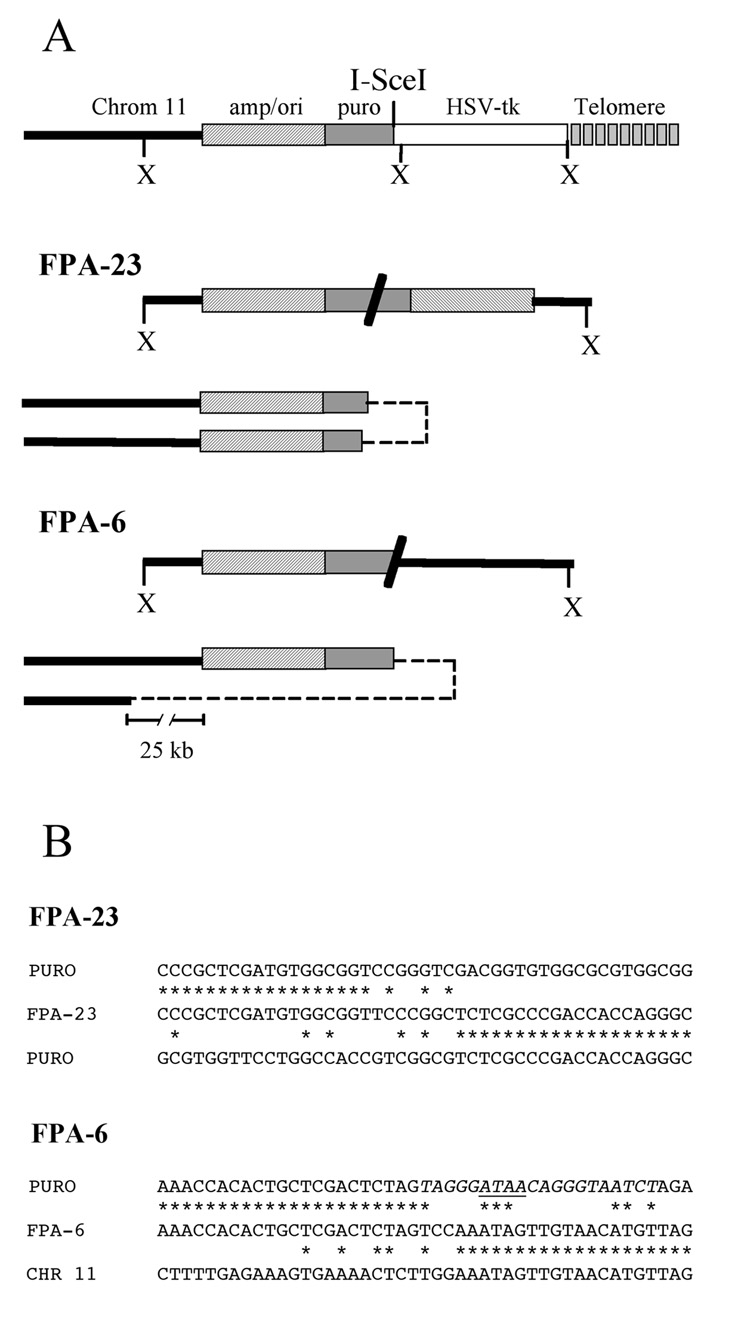
In addition to the large bands generated by XbaI digestion that are typical of chromosome healing, other I-SceI-induced ganr/puror subclones isolated from the ES cell lines with wild type TERT showed smaller bands (Fig. 3). Two of these subclones (F-16a and F-6b) had lost the 3.0 kb band, but retained the 4.3 kb band, as expected from spontaneous events. However, other subclones (F-1b, F-8b, F-15b, and F18b) contained bands that varied in size, which we have previously shown contain inverted repeats [13,16]. Consistent with our earlier results, the rescue of the fragments containing the plasmid and adjacent cellular DNA from two of the subclones with smaller bands demonstrated that they contained inverted repeats (Fig. 4a). In the ganr/puror subclone FPA-6, one end of the inverted repeat occurred in the puro gene promoter near the I-SceI site with the loss of 8 nucleotides, while the other end occurred within the cellular sequences 25 kb proximal to the telomeric plasmid sequences. Therefore, very little degradation occurred on one sister chromatid, while 29 kb of DNA was degraded from the end of the other sister chromatid. There were 2 bps inserted at the recombination site (Fig. 4b). In a second subclone, FPA-23, which was selected with ganciclovir alone, the formation of the inverted repeat occurred by recombination within the puro gene in both sister chromatids. The site of recombination occurred 710 bp from the I-SceI site on one sister chromatid and 450 bp on the other. Thus, degradation appeared to have occurred on both sister chromatids, although it cannot be completely ruled out that spontaneous telomere loss was not involved with subclones selected with ganciclovir alone. There were 7 bps inserted at the recombination site (Fig. 4b). As in our earlier studies, the results demonstrate that degradation commonly occurs prior to sister chromatid fusion, and that the extent of degradation can vary considerably, in some cases resulting in the complete loss of the plasmid sequences.
Fig. 4.
Analysis of the inverted repeats resulting from sister chromatid fusion in subclones from the 10PWT-F ES cell line with wild-type telomerase. The rescue of plasmid sequences and adjacent cellular DNA was performed using XbaI-digested genomic DNA. (A) The structure of the rescued XbaI fragments (above) and mechanism of formation by sister chromatid fusion (below) are shown for subclones FPA-6 and FPA-23. Restriction mapping and DNA sequence analysis demonstrated the presence of inverted repeats, either between sequences within the puro gene (FPA-23) or between the I-SceI site and cellular DNA located 25 kb from the integrated plasmid sequences (FPA-6). The locations of the various plasmid sequences and cellular DNA are indicated as shown in Fig. 1. The sites of end-to-end fusion of the sister chromatids is indicated by dashed lines. (B) The nucleotide sequence at the junction of the inverted repeats in subclone FPA-23 (FPA-23) is compared with the nucleotide sequences of the two puro genes (PURO). The nucleotide sequence at the junction of the inverted repeats in sublcone FPA-6 is compared with the nucleotide sequences of the puro gene (PURO) and genomic DNA from chromosome 11 (CHR 11). Homologous nucleotides (*), the I-SceI recognition site (italics), and the location of the 4 bp overhang generated by I-SceI (horizontal line), are shown.
3.3 Analysis of sites of chromosome healing in ES cells with wild type TERT
The presence of newly added telomeric repeat sequences on the end of the broken chromosome in the ganr/puror subclones was determined by PCR, using one primer specific for sequences within the puro gene, and the other primer specific for telomeric repeat sequences (Fig. 1). All but one of the subclones with bands at the limit of resolution produced PCR fragments that were of the size expected for direct telomere addition at the site of the I-SceI-induced DSB (data not shown). In contrast, no such PCR fragments were seen in subclones with smaller bands containing inverted repeats. DNA sequence analysis of the PCR fragments confirmed that in all but one of the subclones (21 of 22), telomeric repeat sequences were added directly at the site of the break, most often using microhomology as previously observed with ES clone A211 [31] (Fig. 5). The most common site for telomere addition involved a single bp of microhomology to telomeric repeat sequences with no loss of nucleotides from the 4 bp single-strand overhang generated by I-SceI (site “a”). The other two most common sites for telomere addition involve either 5 (site “b”) or 2 (site “c”) of microhomology to telomeric repeat sequences with the loss of either 4 or 1 of the nucleotides from the 4 bp I-SceI single-stranded overhang, respectively (Fig. 5). In addition, 5 other ganr/puror subclones with chromosome healing at the I-SceI site contained various point mutations at the site of telomere addition (i.e., bps that did not have homology to either the plasmid or telomeric repeat sequences), so that it was not possible to determine the exact sequence where chromsome healing occurred (data not shown). Only a single subclone showed significant degradation (25 bp) from the site of the I-SceI-induced DSB prior to telomere addition. This absence of degradation prior to telomere addition was not due to selection for the adjacent puro gene, because chromosome healing in all of the subclones selected with ganciclovir alone also occurred at the site of the I-SceI-induced DSB break (Fig. 5). Thus, unlike fusions, which are commonly accompanied by degradation of DNA at the site of the break, chromosome healing almost always occurs without degradation. Chromosome healing therefore occurs prior to, and prevents, degradation and sister chromatid fusion.
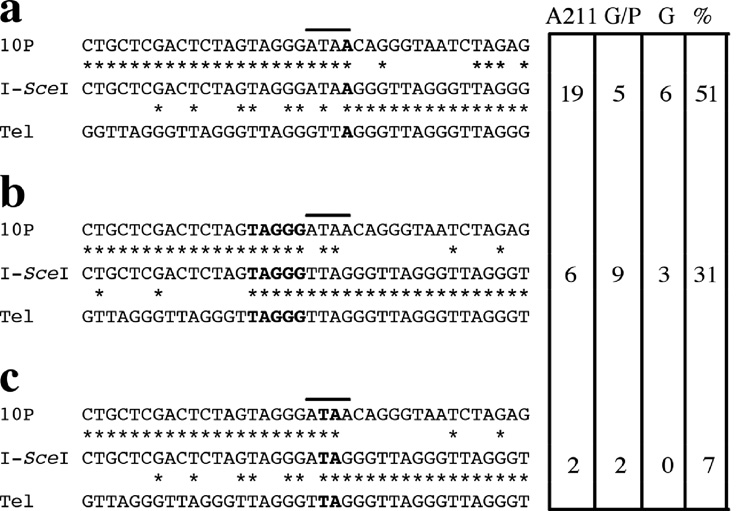
Fig. 5.
DNA sequence analysis of the sites of chromosome healing at the I-SceI-induced DSBs of ES cell lines with wild type telomerase. Comparisons are shown between the site of chromosome healing at the I-SceI site in the I-SceI-treated subclones (I-SceI), the sequence of the parental lines (10P), and telomeric repeat sequences (Tel). Homologous nucleotides (*), the location of the 4 bp overhang generated by I-SceI (horizontal line), and sequence microhomology at the site of telomere addition (bold) are shown. The number of chromosome healing events at the different sites and the percentage of the total events they represent are shown for ES cell clone A211, and the 10P wild type ES cell lines selected with both ganciclovir and puromycin (G/P) or ganciclovir alone (G). The total percentage does not equal 100% because five ganr/puror subclones and one ganr subclone that had point mutations at the I-SceI site during chromosome healing, and one ganr/puror subclone that had a 25 bp deletion prior to chromosome healing, are not included.
3.4. Analysis of ES cell lines with knockout of TERT
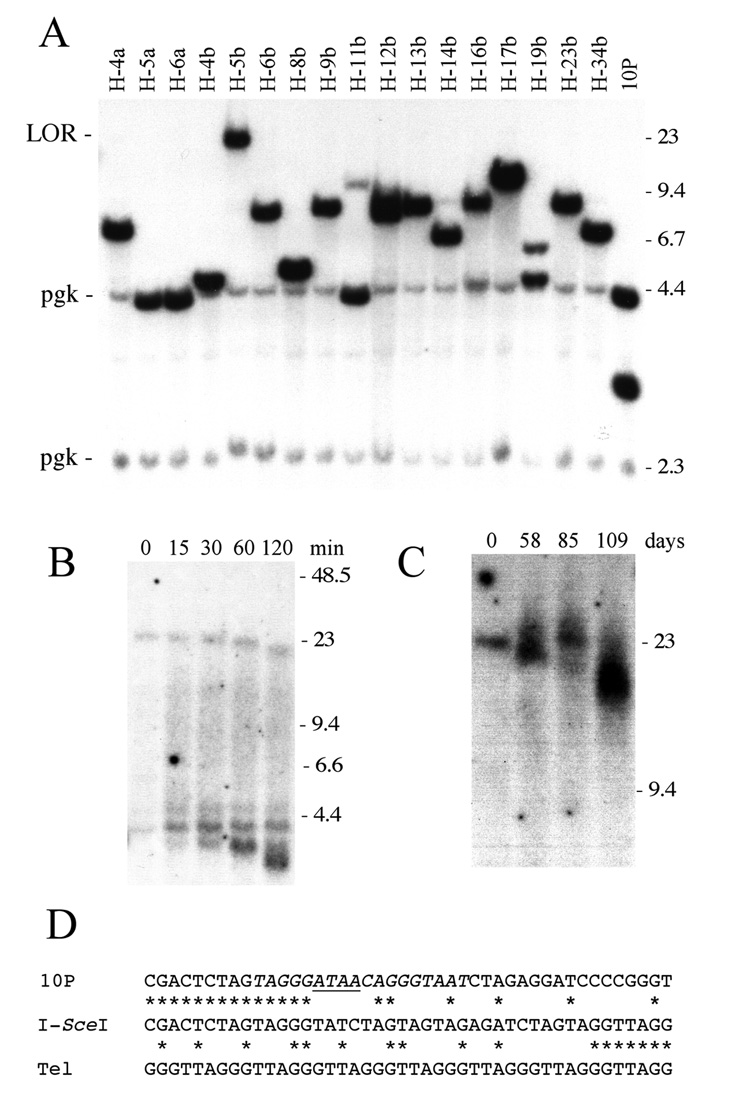
Southern blot analysis of ganr/puror sublclones isolated from the 10PTKO-H (Fig. 6A, Table 1) and 10PTKO-4B (Table 1) ES cell lines homozygous for knockout of the TERT gene showed that only one of 62 subclones (H-5b) contained a large XbaI band indicative of chromosome healing. Instead, most of the ganr/puror subclones contained the smaller bands typical of inverted repeats. In addition, three subclones (H-5a, H-6a, and H-11b) had rearrangements resulting from spontaneous telomere loss, as shown by the fact that they had lost the 3.0 kb XbaI band, but retained the 4.3 kb band. The analysis of chromosome healing in all of the subclones by PCR demonstrated that only subclone H-5b produced the band expected from the presence of a telomere at the site of the DSB (data not shown). Despite this fact, H-5b was confirmed to be homozygous for the knockout TERT by PCR and had no detectable telomerase activity (data not shown). PFGE following digestion with BAL31 nuclease also confirmed that the large XbaI band consisted of a terminal restriction fragment containing a telomere (Fig. 6B). Typical of terminal restriction fragments digested with BAL31 [31,47], upon digestion the band containing the telomere selectively decreased in size while other bands containing internal DNA fragments remained unchanged. The accumulation of small bands at the bottom of the gel during digestion with BAL31 result from the slower rate of degradation of subtelomeric DNA compared to telomeric DNA, as has been previously observed [31,47]. Although these results make it clear that chromosome healing occurred, unlike the newly formed telomeres in wild type cells that reach 75 kb in length, the newly added telomere in H-5b is only 25 kb in length (Fig. 6B) and becomes shorter and more heterogeneous in size upon further passage in culture (Fig. 6C). Moreover, unlike chromosome healing in ES cells with wild type TERT, DNA sequence analysis of the site of telomere addition demonstrated an insertion of 23 bp of DNA (Fig. 6D). Small insertions similar to this are commonly observed at DSBs repaired by NHEJ [27,28], suggesting a mechanism of telomere addition that is distinct from that in ES cell lines with wild type TERT.
Fig. 6.
Analysis of rearrangements resulting from a DSB near a telomere in the TERT knockout 10PTKO-H ES cell line. (A) Southern blot analysis of genomic DNA from I-SceI-induced ganr/puror subclones digested with XbaI and hybridized with the pNPTΔ plasmid probe. The limit of resolution (LOR), position of control bands for endogenous pgk sequences (pgk), and molecular weight markers are the same as in Fig. 3. (B) Demonstration of chromosome healing in the H-5b subclone by Southern blot analysis of DNA digested with BAL31 nuclease and separated by PFGE. DNA was digested with BAL31 nuclease for 0, 15, 30, 60 and 120 minutes, followed by digestion with XbaI. Hybridization was performed with the pNPTΔ plasmid probe. Terminal restriction fragments containing telomeres are selectively degraded by BAL31, while internal restriction fragments show no change in size. The bands that accumulate at the bottom of the figure are due to the slower rate of degradation of subtelomeric DNA compared to telomeric repeat sequences. (C) Analysis of changes in length of the telomere in the H-5b subclone during passage in culture by Southern blot analysis of DNA separated by PFGE. Genomic DNA from subclone H-5b that was isolated initially, and after 58, 85 and 109 days in culture, was digested with XbaI, and hybridization was performed using the pNPTΔ plasmid as a probe. (D) A comparison is shown between the site of chromosome healing at the I-SceI site in subclone H-5b (I-SceI), the sequence of the parental line (10P), and telomeric repeat sequences (Tel). Homologous nucleotides (*) and the location of the 4 bp overhang generated by I-SceI (horizontal line) are shown. A 23 bp DNA fragment was inserted at the site of telomere addition.
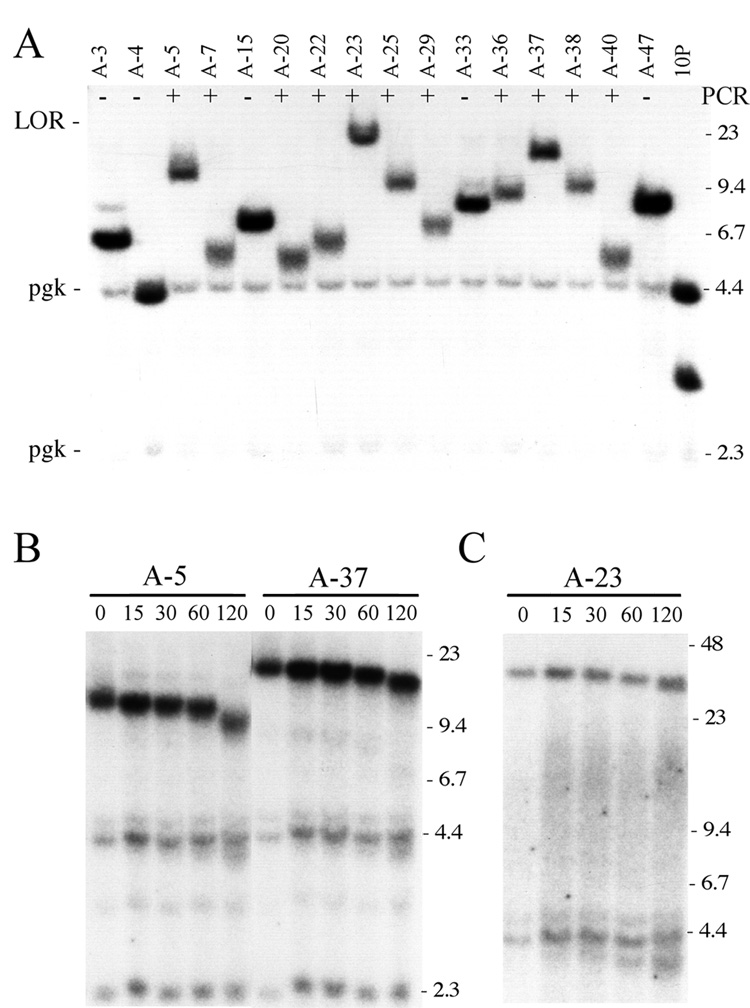
I-SceI-induced ganr/puror sublclones isolated from a third TERT knockout ES cell line, 10PTKO-A, also produced bands on Southern blots that were all smaller in size than those typical of chromosome healing in the wild type cell lines (Fig. 7A). However, unlike the other two TERT knockout ES cell lines, PCR analysis revealed that 20 of 34 (59%) of the ganr/puror subclones from 10PTKO-A produced bands indicating that chromosome healing had occurred at the I-SceI site (data not shown). Upon close inspection of the Southern blots, the subclones generating PCR bands were observed to be those with lighter bands (Fig. 7A). The presence of newly added telomeres on the end of the broken chromosome in 3 of these ganr/puror subclones was confirmed by BAL31 digestion using either conventional gel electrophoresis (Fig. 7B) or PFGE (Fig. 7C). The analysis of telomerase activity confirmed that these subclones lacked telomerase activity (data not shown). Thus, chromosome healing is common at the I-SceI-induced DSB in the 10PTKO-A ES cell line despite the absence of telomerase. However, unlike the newly added telomeres in ES cell lines with wild type TERT, these telomeres are variable in size and much shorter than 75 kb in length. In fact, the newly added telomeres in some subclones (e.g., A-7 and A-40) are only about 1 kb in length, as determined by subtracting length of the subtelomeric plasmid sequences (4.3 kb) from the size of the fragment containing the telomeric repeat sequences.
Fig. 7.
Rearrangements resulting from a DSB near a telomere in the TERT knockout 10PTKO-A ES cell line. (A) Southern blot analysis of genomic DNA from I-SceI-induced ganr/puror subclones digested with XbaI and hybridized with the pNPTΔ plasmid probe. The limit of resolution (LOR) and position of control bands for endogenous pgk sequences (pgk) are shown. Subclones that were positive (+) or negative (−) by PCR for chromosome healing are indicated. (B, C) Identification of terminal restriction fragments containing telomeres by Southern blot analysis of DNA digested with BAL31 nuclease. Genomic DNA from the A-5, A-23, and A-37 ganr/puror subclones was digested with BAL31 nuclease for 0, 15, 30, 60 and 120 minutes, followed by digestion with XbaI. The DNA was then separated by conventional agarose gel electrophoresis (B), or PFGE (C), and hybridization was performed with the pNPTΔ plasmid probe.
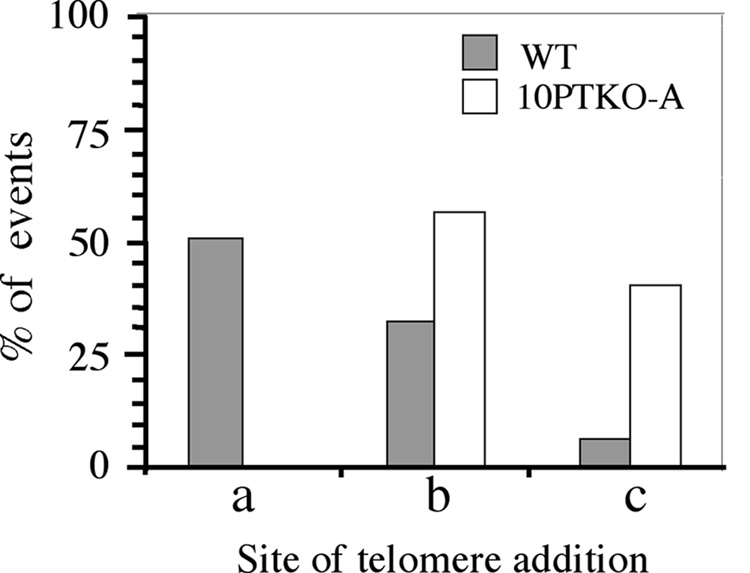
DNA sequence analysis of the PCR products generated from the 10P-TKO-A ganr/puror subclones confirmed that chromosome healing had occurred at the I-SceI site (Fig. 8). However, the preference for the site of addition of the newly added telomeric repeat sequences differed somewhat from that observed for the wild type ES cell clones or the original ES cell clone A211 [31]. A major site of addition of telomeric repeat sequences in wild-type ES cell clones, which involved 1 bp of homology without the loss of any nucleotides from the 4 bp overhang (site a), was not observed in the 10PTKO-A ES cell line. Instead, the addition of telomeric repeat sequences in 10PTKO-A occurred primarily at the two other sites at which chromosome healing occurred in the wild-type ES cell lines (sites b and c). As with the wild type ES cell lines, some sites of chromosome healing (2 of 20) contained point mutations that made the exact site of telomere addition difficult to determine. Regardless, it is clear from the difference in the preference in the site of telomere addition that the mechanism of chromosome healing in the TERT knockout 10PTKO-A ES cell line is distinct from that in ES cell lines with wild-type telomerase.
Fig. 8.
The preference of sites for chromosome healing in the TERT knockout 10PTKO-A ES cell line differs from that of ES cell lines with wild-type telomerase. The percent involvement of the three most common sites of chromosome healing in wild type ES cell lines (see Fig. 5) are compared with the percent of chromosome healing events at these same sites in the ganr/puror subclones of the 10PTKO-A ES cell line (n = 20).
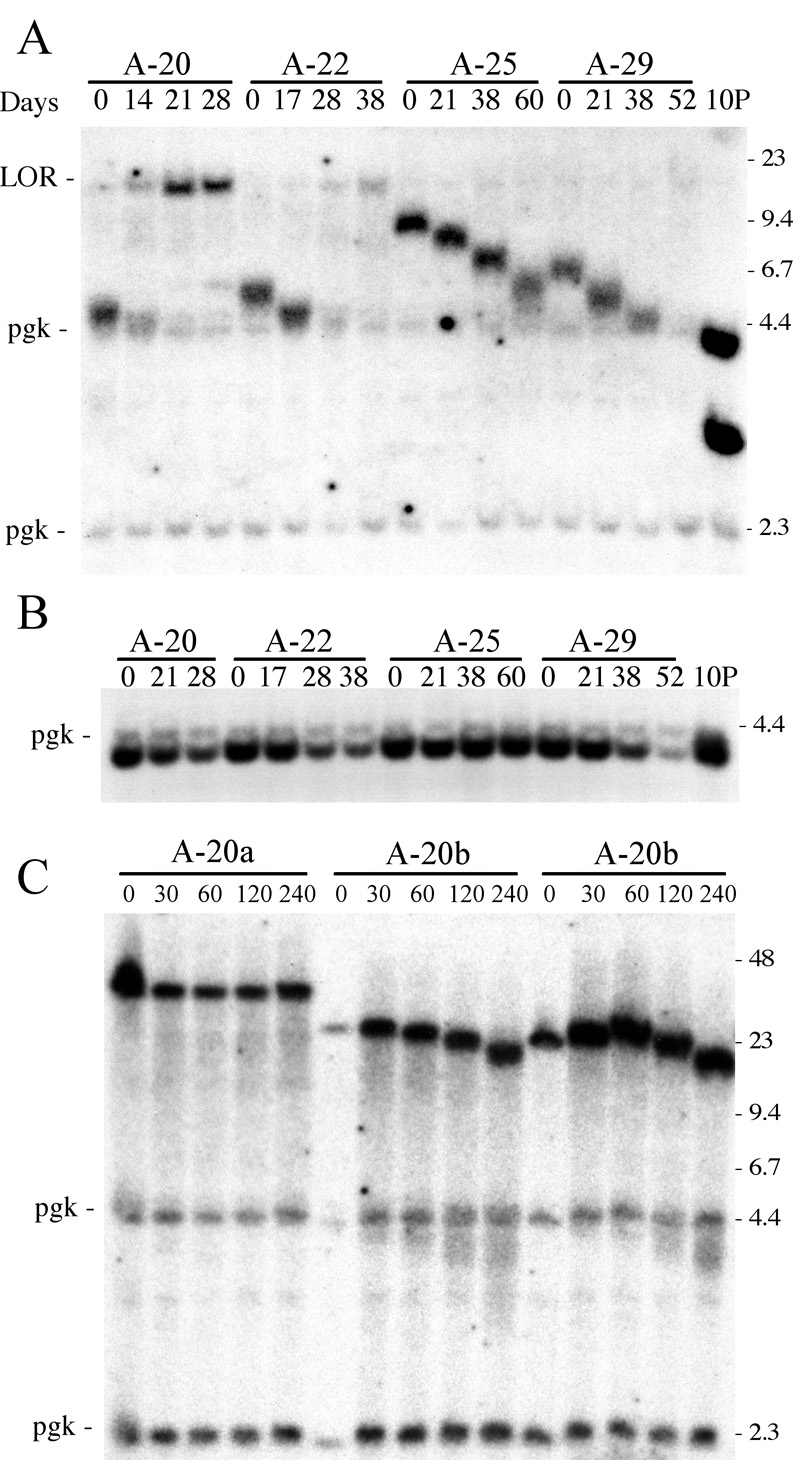
3.5. Analysis of telomere length in TERT knockout ES cell line 10PTKO-A
We next followed the changes in the length of the newly added telomeres in the ganr/puror subclones isolated from the 10PTKO-A ES cell line to learn more about the mechanism of telomere maintenance in this cell line. We previously showed that newly added telomeres in ES cells lines with wild-type telomerase increased in length with time in culture, eventually reaching a length of that in the parental cell line [31]. However, the newly added telomeres in the subclones from 10PTKO-A decreased in length with time in culture (Fig. 9). The rate of telomere shortening was estimated to be approximately 55 bp/cell division, similar to the 75 bp/cell division previously observed in Mus spretus embryo fibroblast cultures [48]. Moreover, upon reaching the size of the subtelomeric XbaI fragment without telomeric repeat sequences (4.3 kb) in subclones A-20, A-22, and A-29, the band disappeared and was replaced by a band at the limit of resolution (Fig. 9A), demonstrating that either telomere elongation or rearrangement of the fragment had occurred in many cells in the population. However, the lighter intensity of the new larger bands compared to the original bands indicated that many cells in the population had lost the subtelomeric plasmid sequences entirely. This was confirmed by Southern blot analysis following digestion with XbaI in combination with MluI, which cuts in the puro gene promoter 260 bp from the telomere and therefore removes the telomeric repeat sequences (Fig. 9B). The 4.1 kb XbaI/MluI band in subclones A-20, A-22, and A-29 decreased in intensity with time in culture, while no change in intensity was apparent in this band in subclone A-25, which had a longer telomere that continued to shorten during this period. These results demonstrate that severe telomere shortening results in the loss of the subtelomeric plasmid sequences in many cells in the population. The absence of gradual shortening of this band also demonstrates that subtelomeric DNA is rapidly degraded once the telomeric repeat sequences are no longer present to protect the end of the chromosome.
Fig. 9.
The dynamics of the changes in the length of the newly added telomeres in ganr/puror subclones of the TERT knockout 10PTKO-A ES cell line. (A) Genomic DNAs from the A-20, A22, A-25, and A-29 ganr/puror subclones grown for various days in culture were analyzed by Southern blot analysis. Digestion with XbaI demonstrates the gradual decrease in size of the terminal fragment containing the telomere with passage in culture, with the eventual disappearance of the band as it approaches the size of the 4.3 kb fragment without telomeric repeat sequences (lane 10P). The disappearance of the orignal band corresponds to the appearance of a new band at the limit of resolution, which varies in intensity in the different subclones. The limit of resolution (LOR), position of control bands for endogenous pgk sequences (pgk), and molecular weight markers are the same as in Fig. 3. (B) Digestion with XbaI in combination with MluI, which cuts off the telomeric repeat sequences, demonstrates the progressive loss of the plasmid sequences in many cells in the population as the telomere becomes critically short. Hybridization was performed using the pNPTΔ plasmid as a probe. The location of the 4.3 kb endogenous pgk band, which serves as a loading control, and the 4.4 kb lambda bacteriophage HindIII fragment, are shown. (C) Southern blot analysis was performed on genomic DNA from two second-generation subclones of 10PTKO-A that had elongated the elongated the newly added telomere. The DNA was digested with BAL31 nuclease for 0, 30, 60, 120 and 240 minutes, followed by digestion with XbaI and separation by PFGE. Hybridization was performed with the pNPTΔ plasmid probe.
While telomere shortening resulted in the loss of the plasmid sequences in many cells in the population, the new bands at the limit of resolution indicated that some cells in the 10PTKO-A subclones underwent rearrangements or telomerase-independent telomere elongation. The frequency at which the elongation of the terminal fragment occurred varied in the different subclones, being common in subclone A-20, but rare in subclone A-29. To explore the reason for the elongation of this fragment in more detail, we analyzed second-generation subclones of A-20 that were selected after 40 passages in culture, at which time the elongation had already occurred. As expected, these second-generation subclones showed either a large fragment at the limit of resolution or complete loss of the plasmid-specific bands (data not shown). Southern blot analysis using PFGE to determine the size of the fragments in three of these second-generation subclones, A-20a, A-20b, and A-20m, demonstrated that they were 34, 23, and 22 kb in length, respectively (Fig. 9C). The fragments in subclones A-20b and A-20m were sensitive to BAL31, proving they are still terminal fragments, and therefore contain elongated telomeres (Fig.9C). However, subclone, A-20a showed no sensitivity to BAL31, indicating that elongation of the fragment resulted from sister chromatid fusion or some other type of rearrangement. PCR analysis demonstrated that in all three subclones the DNA sequence at the site of chromosome healing remained the same as that found in the parental A-20 subclone (site c, Fig. 5), proving that the telomeric repeat sequences were still present. The results therefore demonstrate that rearrangement in subclone A-20a occurred prior to telomere loss, consistent with earlier studies showing that telomeric repeat sequences are commonly found at the sites of chromosome fusions following telomere shortening in human cells [49]. These results also demonstrate that telomere elongation in subclones A-20b and A-20m occurred prior to complete loss of the telomeric repeats, and therefore this telomere elongation did not involve chromosome healing. Whether the appearance of cells with an elongated telomere results from preferential elongation when the telomere becomes very short, as previously described for human cells that maintain telomeres through a telomerase-independent mechanism [33], or due to selection for cells in the population that had elongated the telomere prior to its becoming critically short, remains to be determined.
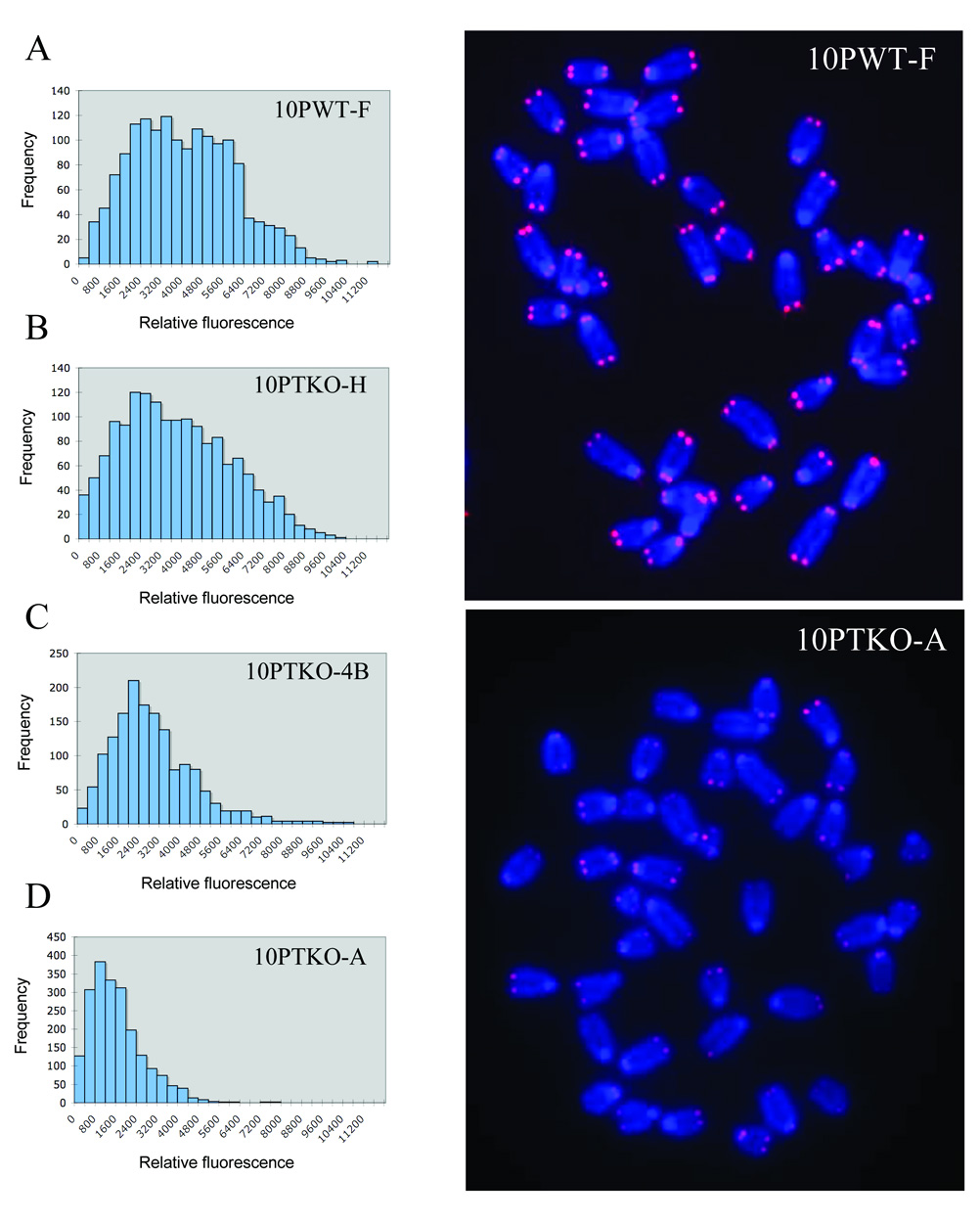
The above results demonstrate that clone 10PTKO-A can extend telomeres through a telomerase-independent mechanism. Previous studies have shown that TERT-deficient mouse embryo fibroblasts [50] and ES cells [47] that acquire the ability to maintain telomeres through alternative mechanisms after prolonged growth in culture typically show telomeres that are much shorter than telomeres in cells with wild type TERT. We therefore performed Q-FISH analysis to compare the relative telomere length in our TERT knockout ES cell lines with the relative telomere length in an ES cell line with wild type TERT (Fig. 10). Typical of telomeres in mouse cells with wild type TERT that vary in length from 10 to 80 kb [46], the telomeres in the 10PWT-F ES cell line with wild type TERT were highly heterogeneous in length (Fig. 10A). The telomeres in the TERT knockout ES cell line 10PTKO-H were also highly heterogeneous in length and only slightly shorter than the telomeres in 10PWT-F (Fig. 10B, Table 2). However, the telomeres in the knockout ES cell line 10PTKO-4B were considerably shorter than the telomeres in 10PWT-F (Fig. 10C, Table 2). Moreover, the telomeres in the knockout ES cell line 10PTKO-A were extremely short (Fig. 10D, Table 2), with many chromosomes showing no detectable telomeres. The 10PTKO-A ES cell line is therefore typical of other mouse embryonic fibroblasts and ES cell line that maintain telomeres through a telomerase-independent mechanism, which also contain very short telomeres [47,50].
Fig. 10.
Short telomeres in the 10PTKO-A TERT knockout ES cell line. Relative telomere intensity was analyzed by Q-FISH using a telomere-specific peptide nucleic acid (PNA) probe for (A) a wild type ES cell line, 10PWT-F, and the three TERT knockout ES cell lines (B) 10PTKO-H, (C) 10PTKO-4B, and (D) 10PTKO-A. Telomeres in the wild type ES cell line (upper right panel) are relatively homogenous and easily detectable on all chromosomes. In contrast, telomeres in 10PTKO-A (lower right panel) are relatively difficult to detect, with many chromosomes having no detectable signal.
Table 2.
Comparison of average telomere length in ES cell lines with wild type or Knockdown TERT
| Cell Line | Chrom Counted | Avg Tel Fluor | SD |
|---|---|---|---|
| Wild type | |||
| 10PWT-F | 392 | 3677 | 1976 |
| Knockout | |||
| 10PTKO-H | 393 | 3414 | 2094 |
| 10PTKO-4B | 395 | 2539 | 1637 |
| 10PTKO-A | 518 | 1263 | 1034 |
4. Discussion
The results presented here demonstrate for the first time that chromosome healing in mammalian cells can occur through more than one mechanism. One mechanism involves the de novo addition of telomeric repeat sequences by telomerase, as demonstrated by the fact that while chromosome healing is a common event in wild type ES cells (22 of 35 events), chromosome healing rarely occurs in the 10PTKO-H and 10PTKO-4B ES cell lines that are deficient in telomerase (1 of 62 events). The one chromosome-healing event that was observed in these two TERT knockout cell lines involved the insertion of a 23 bp sequence between the site of the DSB and the telomeric repeat sequences. The insertion of a small DNA fragment suggests a mechanism involving recombination, since small insertions are common at junctions repaired by NHEJ [26,28].
In addition to the mechanisms mentioned above, our results also show that some TERT knockout ES cell lines can efficiently perform chromosome healing through a telomerasein-dependent mechanism. This mechanism is active in the 10PTKO-A ES cell line, which unlike the TERT knockout ES cell lines 10PTKO-4B and 10PTKO-H, is very adept at chromosome healing despite the absence of telomerase activity (20 of 34 events). However, this mechanism does not appear to be active in ES cells with wild-type telomerase. In contrast to the wild-type ES cell lines, chromosome healing in 10PTKO-A was never observed to initiate from the terminal nucleotide of the 4 bp overhang generated by I-SceI (site a, Fig. 5), which was common in wild type ES cell lines. Instead, telomeres were often added at the site with 2 bp of homology (site c), which was rarely used in wild type ES cell lines. Therefore, although this alternative mechanism also utilizes microhomology for chromosome healing, its requirements for telomere addition are different from the telomerase-dependent mechanism. In view of the absence of telomerase activity, it is likely that chromosome healing in 10PTKO-A involves the addition of preexisting telomeric repeat sequences to the site of the DSB. This could occur by NHEJ, which commonly utilizes microhomology for end rejoining and the deletion of a small number of nucleotides [51,52], both of which were observed at sites of chromosome healing in 10PTKO-A. Consistent with our results with 10PTKO-A, we previously demonstrated the addition of telomeric repeat sequences to both ends of a plasmid sequence transfected into an immortal human cell line [32] that was later shown to maintain telomeres through a telomerasein-dependent mechanism [33]. Similar to the present study, this addition of telomeric repeat sequences also involved microhomology, with 2 bp of microhomology at one end of the plasmid, and 3 bp of microhomology at the other end.
An important difference between the chromosome healing in the wild type and telomerase-deficient ES cell lines is the length of newly added telomeres. The newly added telomeres in the wild type ES cell lines in this study are already 75 kb at the earliest time of analysis, similar to the length of the telomere in the parental ES cell lines. We estimate that less than 50 cell divisions had occurred during the period in culture required to grow sufficient cells to isolate genomic DNA for Southern blot analysis. Thus, an average of approximately 1,500 bp would have been added to the new telomere with each cell division, although initially the rate of telomere elongation may be even greater, since we previously observed that at later times the newly added telomeres can grow much more gradually [31]. Regardless, these results demonstrate that although mouse telomerase has poor processivity in cell lysates [53], it can be highly processive during the restoration of a lost telomere in living cells.
In contrast to ES cell lines with wild type telomerase, the newly added telomeres in the telomerase-deficient 10PTKO-A ES cell line are relatively short, with some being as short as 1 kb in length at the time of analysis. In addition, rather than being elongated, the telomeres shorten during passage in culture, which eventually results in the loss of the subtelomeric plasmid sequences in many cells in the population. However, the telomere is elongated in other cells, demonstrating that the 10PTKO-A ES cell line is capable of maintaining telomeres through a telomerase-independent pathway, although this pathway is relatively inefficient in protecting critically short telomeres compare to human cells that maintain telomeres through a telomerase-independent pathway [33]. This mechanism is not normally found in ES cell lines, because other studies have demonstrated that telomerase-deficient ES cell lines lack the ability to preferentially elongate shortened telomeres [54]. The acquisition of an alternative mechanism for telomere maintenance has previously been described for telomerase-deficient mouse ES cells [47]. The analysis of newly added telomeric DNA in one of the telomerase-deficient ES cell lines in this earlier study demonstrated that it was composed of both telomeric and subtelomeric DNA [31,47]. Although the newly added telomeres found at the site of chromosome healing in our study invariably consisted of telomeric repeat sequences, it cannot be ruled out that the telomeres added by chromosome healing or subsequent elongation also contain nontelomeric DNA sequences. This previous study also found that ES cell clones utilizing telomerase-independent telomere maintenance only appeared following a telomere-shortening-induced crisis after 450 cell divisions in culture. Why the 10PTKO-A ES cell line would have acquired an alternative mechanism for telomere maintenance after only a relatively few generations in culture is not known. One likely possibility is that it initially had short telomeres due to telomere shortening that occurred in vivo, which would have selected for cells that were capable of maintaining telomeres by alternative mechanisms. This telomere shortening in vivo could have occurred because the ES cell lines used in our study were derived from heterozygous TERT knockout mice. Mice that are heterozygous for TERT knockout experience gradual telomere shortening with each generation [55], although unlike homozgous TERT knockout mice, the heterozygous mice are able to maintain short telomeres [56]. Consistent with telomere shortening in vivo, two of the three TERT knockout cell lines used in our study had telomeres that were much shorter than telomeres in ES cells with wild type TERT.
Another important feature of chromosome healing is that in all but one subclone with a 25 bp deletion, it occurs at the location of the I-SceI-induced DSB. This contrasts with sister chromatid fusion, in which degradation of one or both sister chromatids commonly occurs. This degradation prior to sister chromatid fusion can be extensive, in one subclone reaching 30 kb in length. Why chromosome healing only occurs at the site of the initial I-SceI-induced DSB, and not at proximal sites following DNA degradation, is not known. However, it is not due to selection for the proximal puro gene, since the chromosome healing observed with selection with ganciclovir alone also occurred at the I-SceI site. Chromosome healing may therefore be in competition with other DNA repair mechanisms, which are associated with processing and degradation of DNA at the break site. If these other DNA repair enzymes encounter the DSB first, then telomerase or the telomerase-independent mechanism would be prevented from accessing the free end for telomere addition.
The fact that chromosome healing is a common event in our assay system would appear to contradict other studies that have concluded that chromosome healing is not a common event at DSBs generated by either I-SceI or ionizing radiation [26–29]. Even when chromosome healing is observed, the evidence suggests that it usually involves the capture of the ends of other chromosomes [57]. One possible explanation for the prevalence of chromosome healing at the I-SceI-induced DSB in our system is the proximity of the telomere. Studies in yeast have demonstrated that de novo telomere addition is much more likely to occur near pre-existing telomeric repeat sequences, which is independent of which side of the break the telomeric repeat sequences are located [19]. De novo telomere addition at DSBs at interstitial sites may also be inhibited as it is in yeast, where its inhibition by Pif1 is proposed to be a mechanism for preventing chromosome healing from interfering with DSB repair [21].
The different outcome of DSBs near telomeres compared to DSBs at most other locations may also reflect the fact that the proximity to a telomere may influence DSB repair. DSBs near telomeres in yeast have been show to be poorly repaired by NHEJ, and therefore result in complex chromosome rearrangements [34]. This deficiency in repair in subtelomeric regions may be due to the role of the telomere in preventing chromosome fusion, since telomeric repeat sequences in yeast have been shown to suppress the activation of cell cycle checkpoints in response to DSBs [35]. Similarly, the human TRF2 protein, which is required to prevent chromosome fusion, has been demonstrated to inhibit ATM, whose activation is instrumental in the cellular response to DSBs [36]. Alternatively, a deficiency in the repair of DSBs within subtelomeric DNA could also result from the heterochromatic structure of these regions ([58], unpublished results). Regardless of the cause, a deficiency in NHEJ within subtelomeric regions would make chromosome healing an important option for the cellular response to DSBs occurring near telomeres.
Our results also show that chromosome healing can prevent chromosome instability resulting from DSBs near telomeres. Because chromosome healing almost always occurs at the site of the break, it must precede and prevent degradation, sister chromatid fusion, and subsequent B/F/B cycles. The importance of sister chromatid fusion and B/F/B cycles resulting from telomere loss in DNA rearrangement in cancer is becoming increasingly evident. Telomerase deficient mice that also have a knockout in p53, and therefore are unable to eliminate cells with genomic instability, have a high frequency of carcinomas that contain chromosomes with rearrangements typical of B/F/B cycles [59–61]. In addition, telomere loss has been shown to play an important role in human cancer, and can continue to occur despite the expression of telomerase [62,63]. Chromosome healing could therefore limit the extent of the genomic instability commonly associated with human cancer, since chromosome healing has also been observed in human tumor cells in response to spontaneous [14,15] or I-SceI-induced (unpublished observation) telomere loss. However, like mouse ES cells, the efficiency of chromosome healing is not sufficient to prevent sister chromatid fusion and instability. Knowledge regarding the regulation of chromosome healing may therefore provide future approaches to limiting genomic instability in human cancer cell for the purpose of inhibiting tumor cell progression or adaptation to cancer therapy.
Acknowledgements
The work was supported by National Institute of Environmental Health Science Grant No. RO1 ES008427.
Abbreviations
- B/F/B
breakage/fusion/bridge
- DSB
double-strand break
- ES
embryonic stem
- ganr
ganciclovir-resistant
- ganr/puror
ganciclovir and puromycin-resistant
- HSV-tk
Herpes simplex virus-thymidine kinase
- NHEJ
nonhomologous end joining
- PCR
polymerase chain reaction
- PNA
peptide nucleic acid
- pgk
phosphoglycerate kinase
- PFGE
pulsed-field gel electrophoresis
- puro
puromycin-resistance
- Q-FISH
quantitative fluorescence in situ hybridization
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Ann. Rev. Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn EH, Greider CW, editors. Telomeres. Plainview, NY: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 4.de Lange T. Telomere dynamics and genome instability in human cancer. In: Blackburn EH, Greider CW, editors. Telomeres. Plainview, NY: Cold Spring Harbor Press; 1995. pp. 265–293. [Google Scholar]
- 5.Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 6.'Adda di Fagagna FdAdd, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 7.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol. Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 8.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harley CB. Telomeres and aging. In: Blackburn EH, Greider CW, editors. Telomeres. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1995. pp. 247–263. [Google Scholar]
- 10.Shay JW, Wrigh WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 11.Reddel RR. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. 2003;194:155–162. doi: 10.1016/s0304-3835(02)00702-4. [DOI] [PubMed] [Google Scholar]
- 12.Hackett JA, Greider CW. End resection initiates genomic instability in the absence of telomerase. Mol. Cell. Biol. 2003;23:8450–8461. doi: 10.1128/MCB.23.23.8450-8461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo AWI, Sprung CN, Fouladi B, Pedram M, Sabatier L, Ricoul M, Reynolds GE, Murnane JP. Chromosome instability as a result of double-strand breaks near telomeres in mouse embryonic stem cells. Mol. Cell. Biol. 2002;22:4836–4850. doi: 10.1128/MCB.22.13.4836-4850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouladi B, Miller D, Sabatier L, Murnane JP. The relationship between spontaneous telomere loss and chromosome instability in a human tumor cell line. Neoplasia. 2000;2:540–554. doi: 10.1038/sj.neo.7900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo AWI, Sabatier L, Fouladi B, Pottier G, Ricoul M, Murnane JP. DNA amplification by breakage/fusion/bridge cycles initiated by spontaneous telomere loss in a human cancer cell line. Neoplasia. 2002;6:531–538. doi: 10.1038/sj.neo.7900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murnane JP. Telomeres and chromosome instability. DNA Repair (Amst) 2006;5:1082–1092. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 17.Ingvarsson S. Molecular genetics of breast cancer progression. Sem. Cancer Biol. 1999;9:277–288. doi: 10.1006/scbi.1999.0124. [DOI] [PubMed] [Google Scholar]
- 18.Schwab M. Oncogene amplification in solid tumors. Sem. Cancer Biol. 1999;9:319–325. doi: 10.1006/scbi.1999.0126. [DOI] [PubMed] [Google Scholar]
- 19.Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerase α and δ. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- 20.Yu G, Blackburn EH. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J-Q, Monson EK, Teng S-C, Schultz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 22.Varley H, Di S, Scherer SW, Royle NJ. Characterization of terminal deletions at 7q32 and 22q13.3 healed by de novo telomere addition. Am. J. Hum. Genet. 2000;67:610–622. doi: 10.1086/303050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong AC, Ning Y, Flint J, Clark K, Dumanski JP, Ledbetter DH, McDermid HE. Molecular characterization of a 130-kb terminal microdeletion at 22q in a child with mild mental retardation. Am. J. Hum. Genet. 1997;60:113–120. [PMC free article] [PubMed] [Google Scholar]
- 24.Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett NA, King A, Higgs DR. The relationship between chromosome structure and function at a human telomeric region. Nat. Genet. 1997;15:252–257. doi: 10.1038/ng0397-252. [DOI] [PubMed] [Google Scholar]
- 25.Morin GB. Recognition of a chromosome truncation site associated with α-thalassaimia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 26.Honma M, Sakuraba M, Koizumi T, Takashima Y, Sakamoto H, Hayashi M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair (Amst) 2007;6:781–788. doi: 10.1016/j.dnarep.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Lin Y, Waldman AS. Capture of DNA sequences at double-strand breaks in mammalian cells. Genetics. 2001;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebuzzini P, Khoriauli L, Azzalin CM, Magnani E, Mondello C, Giulotto E. New mammalian cellular systems to study mutations introduced at the break site by nonhomologous end-joining. DNA Repair (Amst) 2005;4:546–555. doi: 10.1016/j.dnarep.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Sargent RG, Brenneman MA, Wilson JH. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latre L, Genesca A, Martin M, Ribas M, Egozcue J, Blasco MA, Tusell L. Repair of DNA broken ends is similar in embryonic fibroblasts with and without telomerase. Radiat. Res. 2004;162:136–142. doi: 10.1667/rr3203. [DOI] [PubMed] [Google Scholar]
- 31.Sprung CN, Reynolds GE, Jasin M, Murnane JP. Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1999;96:6781–6786. doi: 10.1073/pnas.96.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murnane JP, Yu L-C. Acquisition of telomere repeat sequences by transfected DNA integrated at the site of a chromosome break. Mol. Cell. Biol. 1993;13:977–983. doi: 10.1128/mcb.13.2.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murnane JP, Sabatier L, Marder BA, Morgan WF. Telomere dynamics in an immortal human cell line. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricchetti M, Dujon B, Fairhead C. Distance from the chromosome end determines the efficiency of double-strand break repair in subtelomeres of haploid yeast. J. Mol. Biol. 2003;328:847–862. doi: 10.1016/s0022-2836(03)00315-2. [DOI] [PubMed] [Google Scholar]
- 35.Michelson RJ, Rosenstein S, Weinert T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Q, Reynolds GE, Innes L, Pedram M, Jones E, Junabi M, Gao D-W, Ricoule M, Sabatier L, Van Brocklin H, Franc BL, Murnane JP. Telomeric transgenes are silenced in adult mouse tissues and embryo fibroblasts, but are expressed in embryonic stem cells. Stem Cells. 2007;25:3085–3092. doi: 10.1634/stemcells.2007-0478. [DOI] [PubMed] [Google Scholar]
- 38.Pedram M, Sprung CN, Gao Q, Lo AWI, Reynolds G, Murnane JP. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol. Cell Biol. 2006;26:1865–1878. doi: 10.1128/MCB.26.5.1865-1878.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagy A, Gertsenstein M, Vinersten K, Behringer R. Cold Spring Harbor Laboratory Press; 2003. Manipulating the mouse embryo: a laboratory manual. [Google Scholar]
- 40.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brisebois JJ, DuBow MS. Selection for spontaneous null mutations in a chromosomally-integrated HSV-1 thymidine kinase gene yields deletions and a mutation caused by intragenic illegitimate recombination. Mutat. Res. 1993;287:191–205. doi: 10.1016/0027-5107(93)90012-5. [DOI] [PubMed] [Google Scholar]
- 42.Murata S, Matsuzaki T, Takai S, Yaoita H, Noda M. A new retroviral vector for detecting mutations and chromosomal instability in mammalian cells. Muatat. Res. 1995;334:375–383. doi: 10.1016/0165-1161(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 43.Bai Y, Murnane JP. Telomere instability in a human tumor cell line expressing the NBS1 gene with mutations at sites phosphorylated by the ATM protein. Mol. Can. Res. 2003;1:1058–1069. [PubMed] [Google Scholar]
- 44.Poon SS, Lansdorp PM. Measurements of telomere length on individual chromosomes by image cytometry. Methods Cell Biol. 2001;64:69–96. doi: 10.1016/s0091-679x(01)64007-x. [DOI] [PubMed] [Google Scholar]
- 45.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zijlmans JMJM, Martens UM, Poon SSS, Raap AK, Tanke HJ, Ward RK, Lansdorp PM. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niida H, Shinkai Y, Hande MP, Matsumoto T, Takehara S, Tachibana M, Oshimura M, Lansdorp PM, Furuichi Y. Telomere maintenance in telomerase-deficient mouse embryonic stem cells: characterization of an amplified telomeric DNA. Mol Cell Biol. 2000;20:4115–4127. doi: 10.1128/mcb.20.11.4115-4127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prowse KR, Greider CW. Development and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl. Acad. Sci. USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hande P, Samper E, Lansdorp P, Blasco MA. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J. Cell Biol. 1999;144:589–601. doi: 10.1083/jcb.144.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 52.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prowse KR, Avilion AA, Greider CW. Identification of a nonprocessive telomerase activity from mouse cells. Proc Natl Acad Sci U S A. 1993;90:1493–1497. doi: 10.1073/pnas.90.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Kha H, Ungrin M, Robinson MO, Harrington L. Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc Natl Acad Sci U S A. 2002;99:3597–3602. doi: 10.1073/pnas.062549199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erdmann N, Liu Y, Harrington L. Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice. Proc Natl Acad Sci U S A. 2004;101:6080–6085. doi: 10.1073/pnas.0401580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrington L. Making the most of a little: dosage effects in eukaryotic telomere length maintenance. Chromosome Res. 2005;13:493–504. doi: 10.1007/s10577-005-0994-5. [DOI] [PubMed] [Google Scholar]
- 57.Slijepcevic P, Bryant PE. Chromosome healing, telomere capture and mechanisms of radiation-induced chromosome breakage. Int J Radiat Biol. 1998;73:1–13. doi: 10.1080/095530098142653. [DOI] [PubMed] [Google Scholar]
- 58.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat. Rev. Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 59.Artandi SE, Chang S, Lee S-L, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 60.Chang S, Khoo C, DePinho RA. Modeling chromosomal instability and epithelial carcinogenesis in the telomerase-deficient mouse. Cancer Biol. 2001;11:227–238. doi: 10.1006/scbi.2000.0374. [DOI] [PubMed] [Google Scholar]
- 61.Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- 62.Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal Cin P, Hoglund M, Mitelman F, Mertens F, Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc. Natl. Acad. Sci. USA. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo W-L, Ljung B-M, Chew K, Myambo K, Miranda M, Krig S, Garbe J, Stampfer M, Yaswen P, Gray JW, Lockett S. In situ analysis of genome instability in breast cancer. Nat. Genet. 2004;16:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]