Abstract
We describe the synthesis of peptide nucleic acid (PNA)-titanium dioxide (TiO2) nanoconjugates and the several novel methods developed to investigate the DNA hybridization behaviors of these constructs. PNAs are synthetic DNA analogs resistant to degradation by cellular enzymes, which hybridize to single strand DNA (ssDNA) with higher affinity than DNA oligonucleotides, invade double strand DNA (dsDNA), and form different PNA-DNA complexes. Previously, we developed a DNA-TiO2 nanoconjugate capable of hybridizing to target DNA intracellularly in a sequence-specific manner, with the ability to cleave DNA when excited by electromagnetic radiation, but susceptible to degradation which may lower its intracellular targeting efficiency and retention time. PNA-TiO2 nanoconjugates described herein hybridize to target ssDNA, oligonucleotide dsDNA, and supercoiled plasmid DNA under physiological-like ionic and temperature conditions, enabling rapid and inexpensive, sequence-specific precipitation of nucleic acids in vitro. When modified by the addition of imaging agents or peptides, hybridization capabilities of PNA-TiO2 nanoconjugates are enhanced which provides essential benefits for numerous in vitro and in vivo applications. The series of experiments shown here could not be done with either TiO2-DNA nanoconjugates or PNAs alone, and the novel methods developed will benefit studies of numerous other nanoconjugate systems.
Keywords: titanium dioxide, peptide nucleic acid, nanoparticle, DNA, hybridization
Introduction
During their lifetime, organisms often acquire unwanted foreign or mutated DNA that may negatively affect their health. Traditional modes of diagnosis are often unable to detect the presence of deleterious DNA; additionally, common treatments for such diseases do not address the underlying cause of disease—changes at the level of the genome, and therefore do not discriminate well between target and healthy cells. This lowers the therapeutic efficacy of such conventional treatments. New types of early-detection imaging agents and sequence-specific gene therapies are needed to diagnose and remove unwanted DNA in diseased cells, without affecting healthy, neighboring cells. Imaging and elimination of unwanted genes and gene products have been major goals of molecular biology over the last few decades, and a sudden proliferation of different RNA interference techniques [reviewed in (1–3)] attest to that trend. As a result of recent advancements in nanotechnology, biologists now have access to materials with novel properties which emerge only at the nano scale, enabling innovative imaging and therapeutic approaches [4–6]. In our laboratory we have previously synthesized a DNA-TiO2 nanoconjugate which is suitable for imaging and inflicting inducible, sequence-specific cleavage of unwanted target DNA [7].
TiO2 nanoparticles smaller than 20 nm covalently attached to single stranded DNA (ssDNA) via the bidentate enediol ligand dopamine [6,7] are able to bind to complementary DNA sequences. Hybridization between the oligonucleotide of nanoconjugates and complementary DNA inside cells can occur, leading to sequence specific retention of DNA-TiO2 nanoconjugates in the nucleus or mitochondria of a cell [7,8]. The TiO2 nanoparticle has the ability to serve as a multi-modal imaging agent as it can be conjugated to imaging molecules such as gadolinium compounds—used as contrast agents for MRI [9,10] and optically fluorescent agents that are ortho-substituted enediol ligands [5,6,11].
Although DNA nanoconjugates may serve as possible vehicles to image and remove unwanted DNA, their targeting efficiency and intracellular retention may be lowered by cellular factors, such as degradation by intracellular nucleases. To address these potential problems and improve the stability of hybridization with target sequences, we developed a peptide nucleic acid (PNA)-TiO2 nanoconjugate which may possess advantages over its DNA-TiO2 counterpart. PNAs are a class of DNA analogue that contain the same bases as DNA, but instead of a phosphate diester backbone, possess an achiral polyamide backbone consisting of N-(2-aminoethyl) glycine units [12]. These characteristics provide PNAs with benefits over DNAs for many applications. For example, advanced non-ionic oligonucleotides such as PNAs possess far better intracellular stability and specificity [13], are more resistant to nuclease and protease attack [14], and have a lower affinity toward DNA binding proteins [15] than DNA. Due to their neutrally-charged backbones and standard bases, PNAs can bind different nucleic acids with high affinity and form various complexes with complementary DNA [16]. PNAs are able to form PNA/RNA [17], PNA/DNA [18], and PNA/PNA duplexes [19], as well as PNA/DNA/PNA triplexes [20]. A PNA/DNA duplex has a higher melting temperature than a DNA/DNA duplex; moreover, modified homopyrimidine bisPNAs and homopyrimidine bisPNA-peptide conjugates containing a mixed base extension of the Watson-Crick polypyrimidine strand have shown the remarkable ability to invade linear double stranded (dsDNA) [21,22]. Prior to this study, mixed base PNAs have not been reported to invade dsDNA, but mixed base PNAs are capable of end invasion of DNA duplexes [23]. Additionally, homothymine PNA-anthraquinone conjugates have demonstrated the ability to photoinduce cleavage on the displaced DNA strand [21]. Therefore, PNA-TiO2 nanoconjugates can be expected to bind target nucleic acid sequences with higher avidity than DNA-TiO2 nanoconjugates, while being resistant to nuclease attacks. Presence of the TiO2 nanoparticle, on the other hand, is expected to provide versatility to the nanoconjugate that far surpasses the benefits of free PNAs. Instantaneous combining of PNA carrying nanoparticles with a medley of peptides with cell type specific, cell penetrating and subcellular localization capabilities, allows for fine tuned targeting not possible for free PNAs. Ability of nanoparticles to anchor optically fluorescent molecules or agents with high magnetic resonance contrast is also unique to TiO2 nanoparticle conjugated PNAs. Finally, nanoparticle bound PNAs can also be used as a means for rapid and inexpensive, sequence specific nucleic acid precipitation in vitro.
This study was aimed at characterization of the hybridization abilities of PNA-TiO2 nanoconjugates, with or without addition of imaging agents and peptides. The presence of the TiO2 nanoparticle, to which the PNA is firmly tethered, prevents PNA-TiO2 nanoconjugates from migrating through gels; moreover absorption of TiO2 in the wavelength range of nucleic acids [6] prevents spectrophotometry approaches to study PNA-TiO2/DNA hybridization. Therefore, instead of gel electrophoresis and/or spectrophotometry we had to develop alternative approaches to evaluate hybridization properties of these nanoconjugates. It is likely that the approaches developed for this study could benefit investigation of other types of nucleic acid-nanoparticle nanoconjugates. The results of this study demonstrate that PNA-TiO2 nanoconjugates hybridize to target DNA in a sequence-specific manner, engage in strand exchange and end invasion, and invade supercoiled plasmid DNA containing a mixed base target under physiological-like ionic and temperature conditions.
Materials and Methods
PNAs, DNA Oligonucleotides, Nanoconjugates, and Plasmids
All nucleic acid sequences used within this study are depicted in Figure 1a. PNA containing a sequence of a segment of ribosomal 18S rDNA gene was synthesized with a dopamine conjugated via succinic acid to the N terminal end (Biosynthesis). PNA-TiO2 nanoconjugates were synthesized by covalently linking 3nm TiO2 nanoparticles to the PNA in a 1:1 molar ratio as described previously for DNA oligonucleotides [7]. Under these conditions some nanoparticles without PNA and some with more than one PNA molecule per nanoparticle can be expected to occur, in addition to one nanoparticle:one PNA nanoconjugate species. The DNA oligonucleotides used to prepare target oligonucleotide dsDNA “dsr18” and PNA complementary molecular beacon “r18ASMB1” (with 6-Carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein as a fluorophore and Dabcyl as a quencher) were obtained from Sigma Genosys. A non-complementary dsDNA oligonucleotide “dsFM20” was prepared from “bFM20” and “aFM20AS” oligonucleotides and a non-complementary molecular beacon “MS5MB1” were obtained from the same source. All DNA oligonucleotides were kept as 100 mM stock solutions in TE buffer (10mM TrisCl 1mM EDTA pH=8) at −80°C.
Figure 1.
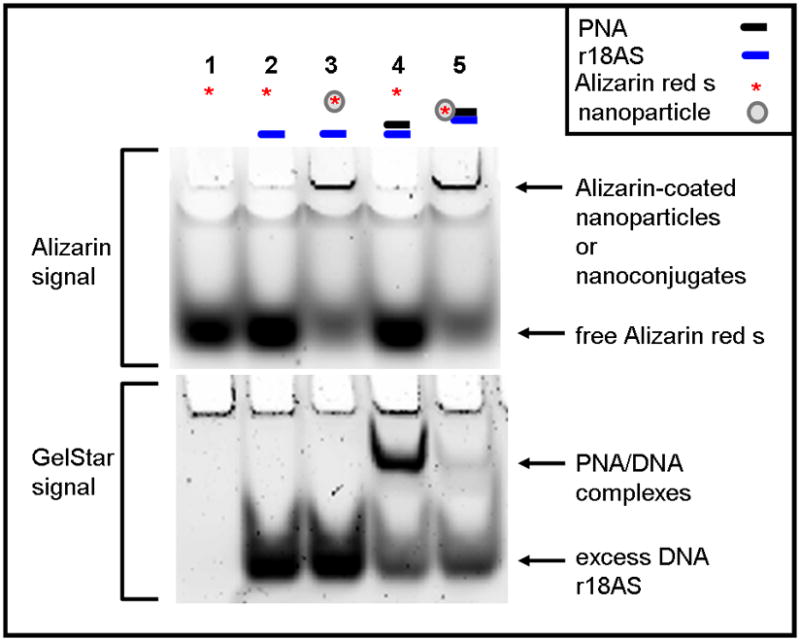
PNA-TiO2 nanoconjugates can be successfully conjugated and withstand incubation at 95°C. Lanes 1–4 contained control reactions: lane 1 is free Alizarin red s; lane 2 is a mixture of Alizarin red s and complementary oligonucleotide DNA; lane 3 is Alizarin red s conjugated nanoparticle (without PNA) and complementary oligonucleotide DNA; lane 4 contains a mixture of PNA (PNA-r18S), complementary oligonucleotide (r18AS) and Alizarin red s. The test sample (lane 5) contains a mix of Alizarin red s-coated TiO2-PNA and threefold molar excess of complementary r18AS oligonucleotide DNA. All samples were heated to 95°C for 2 minutes and slowly cooled to room temperature over 3 hours to allow annealing. After cooling, samples were run on a 16% polyacrylamide gel for approximately two hours and imaged for Alizarin red s, without separation of gel from the glass plates that contained it. The same gel was taken out of the glass plates, stained with GelStar to visualize DNA and DNA-PNA hybrids and then re-imaged.
A short peptide segment of epidermal growth factor (pEGF) (N term - RRRHIVRKRTLRR - C term) (Sigma-Genosys) containing a nuclear localization signal (NLS) or Alizarin red s (Fluka) was conjugated to PNA-TiO2 nanoconjugates to cover approximately 5% (9 molecules) and 10% (17 molecules) of the nanoparticle surface respectively. Three nanometer TiO2 nanoparticles have approximately 173 Ti atoms on the surface, each providing a potential binding site for one peptide or Alizarin red s. Conjugation of these nanoparticle modifiers was confirmed by a shift in peak absorbance wavelength of the nanoconjugates compared to nanoparticles alone. In addition, Alizarin red s is a fluorescent molecule (excitation 530–560 nm, emission 580 nm) and can be monitored using a fluoroimager (Typhoon Trio Variable Mode Imager).
Nanoparticles of colloidal TiO2 were prepared as has been described in detail elsewhere [7]. The size of nanoparticles used in these experiments was 3 nm, which renders them avid in binding with dopamine-modified PNAs. At neutral pH (6–8), TiO2 nanoparticles can be precipitated by centrifugal forces greater than 0.2 g.
The plasmid pKaede-MN1 (Marine Biological Laboratory) was used to investigate the ability of nanoconjugates to invade plasmid DNA. The oligonucleotides r18Sclone and r18ASclone (Figure 1a) formed a dsDNA insert that was cloned into EcoR1/Xho1 sites of this plasmid, producing the recombinant pKaede-MN1-R18.
Gel Electrophoresis
Hybridization reactions containing Alizarin red s, ssDNA (67.0 μM), and either PNAs or PNA-TiO2 nanoconjugates (33.5 μM) were heated to 95°C for 2 min and cooled to room temperature over 3 hours. Reactions were separated on 8–16% polyacrylamide gels (acrylamide/bisacrylamide 19:1, EMD Chemicals Inc.) in 1x TBE buffer (Sigma) for 2–3 hours. Nanoparticles were visualized in wells of the gel, still contained within glass plates, by monitoring Alizarin red s fluorescence by a fluoroimager Typhoon Trio Variable Mode Imager. DNA bands were visualized on the same gel after its removal from the glass plates and staining with GelStar (Cambrex). PNAs do not absorb this dye, as well as several other DNA intercalating dyes [24], therefore they were visualized only in the context of hybrids with DNA.
Nucleic Acid Hybridization Monitoring Using a Real Time PCR Apparatus
Hybridization reactions were monitored using a 7300 Real Time PCR System (Applied Biosystems) and the following hybridization-dissociation protocol: 95°C for 15 seconds, cooling to 4°C, incubation at 4°C for 30 seconds, and gradual reheating to 95°C over approximately 1 ¼ hours. HEX signal (for molecular beacons) or Power Sybr Green (Applied Biosystems) signal (for dsDNA) was recorded incrementally (approximately every 20 seconds) during the reheating phase. Hybridization and invasion reactions were also performed at 37–38°C for 20 minutes, with HEX or Power Sybr Green signal being recorded incrementally throughout (approximately every 60 seconds). The average fluorescence measurement during this time was reported. All hybridization reactions involving molecular beacons (0.33 μM) contained nanoconjugates or free PNAs (0.66 μM) in the presence of 10 mM sodium phosphate buffer (NaH2PO4). Unless otherwise noted, experiments utilizing Power Sybr Green (Applied Biosystems) contained nanoconjugates or PNAs (1.0 μM) and complementary dsDNA oligonucleotides (0.5 μM). Fluorescence intensity values were calculated by automated Applied Biosystems software, as well as dissociation curves and derivative dissociation curves.
Plasmid Invasion
Nanoparticles and nanoconjugates used in plasmid invasion studies were partially coated with Alizarin red s (surface coverage was 10% or 17 molecules of Alizarin red s per nanoparticle). Plasmid precipitation was done by taking advantage of the fact that in neutral pH buffers 3 nm TiO2 nanoparticles can be precipitated by centrifugation at 0.2 g. Nanoparticles or nanoconjugates (115 nM) were mixed with supercoiled plasmid DNA (57.5 nM) in sodium phosphate buffer with 137 mM sodium and incubated 2 hours at 37 °C to allow for dsDNA invasion. Centrifugation for 10 min at 0.2 g followed; the pellet was washed by the same buffer and re-precipitated. Following a second wash, samples were loaded onto a gel and subjected to electrophoresis. Due to large size of the plasmid DNA, force of electrophoresis separated plasmid molecules from nanoconjugates, allowing the plasmids to enter into the gel.
Statistical Analyses
Statistical significance of differences were determined through ANOVA analyses followed by Tukey Honest Significant Difference multiple comparisons tests using Systat 10.2 statistical analyses software (Systat Software Inc.). The bar graphs show means of three independent experiments with standard error (SE) and statistical significance; the latter indicated by * as described in each figure legend.
Results
Conjugation of PNA-TiO2 nanoconjugates and hybridization to complementary DNA
To demonstrate the conjugation efficiency of the PNA-TiO2 nanoconjugates, we used to our advantage the fact that TiO2 nanoparticles do not enter into polyacrylamide gels during typical electrophoresis [7]. To image the nanoparticles we conjugated to them the fluorescent agent Alizarin red s [5], whereas nucleic acids (ssDNA and ssDNA/PNA hybrids) were stained by GelStar dye after the electrophoresis. All nucleic acids used throughout this study are listed in Table 1. Hybridization reactions and controls were run on a polyacrylamide gel and imaged first for Alizarin red s, then stained with GelStar to visualize DNA (Figure 1). In lane 4: with 1:2 molar ratio of PNA and complementary oligonucleotide, a PNA/DNA band can be visualized, while no such band is notable in lane 5: with 1:2 ratio of PNA-TiO2 and complementary oligonucleotide. The vast majority of PNA in that lane is conjugated to the nanoparticle and therefore is not free to enter the gel. Hence, conjugated TiO2-PNAs withstand incubation at 95°C.
Table 1.
Nucleic acid sequences used in this study.
| Sequence Name | Sequence (beginning with N-term or 5′ end) |
|---|---|
| - PNA free and/or on nanoparticles | TTTCCTTGGATGTGGT |
| - r18S Oligonucleotide (homologous to PNA sequence) | TTTCCTTGGATGTGGT |
| - r18AS Oligonucleotide (complementary to PNA sequence) | ACCACATCCAAGGAAA |
| - FM20 Oligonucleotide | TTGCTTGGTAGACCAGGCTG |
| - FM2CAS Oligonucleotide | CAGCCTGGTCTACCAAGCAA |
| - r18AS Molecular Beacon (homologous to r13AS, complementary to r18S and PNA) | [MHEX]CCCCACCACATCCAAGGAAAGGGG[MDAB] |
| - MS5 Molecular Beacon (non-complementary beacon) | [MHEX]CCCCGAGAGAGAGAGAGAGAGAGAGGGG[MDAB] |
| - sense strand of r18S clone | TCGAGTTTCCTTGGATGTGGTG |
| - antisense strand of r18AS clone | AATTCACCACATCCAAGGAAAC |
To investigate the ability of PNA-TiO2 nanoconjugates to hybridize to complementary target DNA, we used molecular beacons containing either a complementary or non-complementary DNA sequence in its loop (Figure 2a). Changes in molecular beacon fluorescence, due to hybridization to the nanoconjugate, were monitored using a standard dsDNA dissociation protocol (Applied Biosystems); anticipated results are shown schematically in Figure 2a. When the molecular beacon exists alone (Figure 2a, top panel) or does not contain a complementary target to the nanoconjugate (Figure 2a, bottom panel), the molecular beacon exists primarily in the closed position at temperatures below the melting temperature of the stem and low fluorescence values can be expected. In either of these cases, a sigmoid increase in fluorescence will occur when temperatures are raised above the melting temperature of the molecular beacon stem and the beacon transforms to the open position. Conversely, when the molecular beacon contains a complementary target, hybridization of the nanoconjugate will increase the presence of the open species of the molecular beacon and raise fluorescence values at lower temperatures (Figure 2a, middle panel). The addition of free TiO2 nanoparticles (Figure 2b, blue curve) had little effect on the shape of the curve produced by molecular beacon alone (Figure 2b, black curve). The addition of PNA-TiO2 nanoconjugates complementary to the molecular beacon resulted in a more than six-fold increase at initial fluorescence measurement at 14°C (Figure 2b, purple curve); as expected, this ratio is eventually lost at temperatures melting the molecular beacon stem. None of these alterations in fluorescence curves were observed when the same dissociation analyses were performed using a molecular beacon with a non-complementary target DNA sequence in the molecular beacon loop (Figure 2c). On the other hand, the presence of increasing concentrations of competitor—non-labeled oligonucleotide complementary to PNA (with a sequence identical to the loop of the molecular beacon) sequesters PNA-TiO2 nanoconjugates and reduces the observed changes in fluorescence due to hybridization between the molecular beacon and PNA-TiO2 nanoconjugate (Figure 2d).
Figure 2.
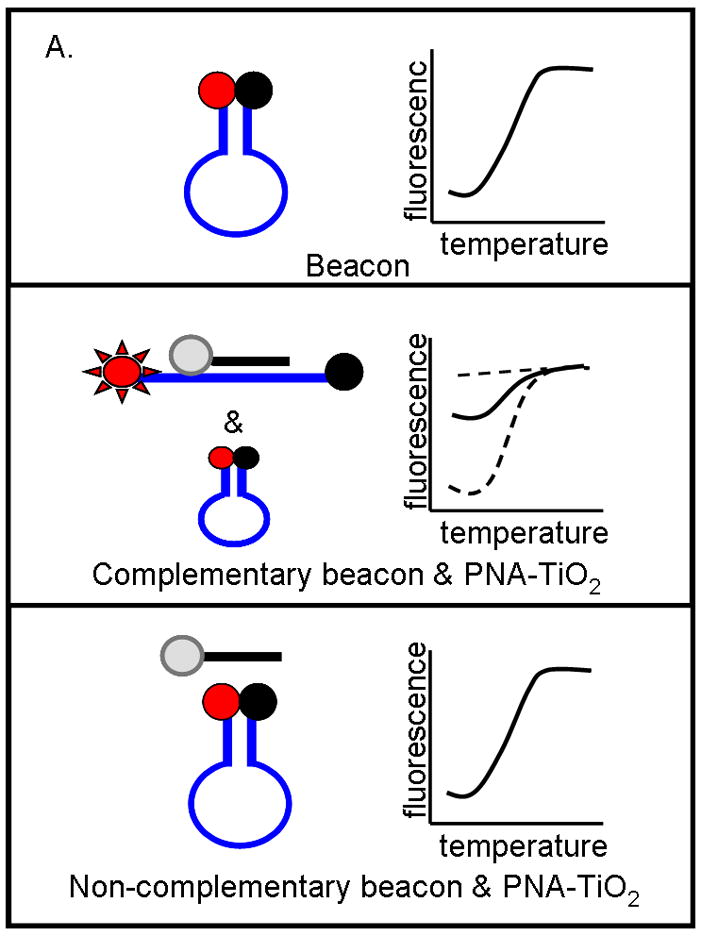
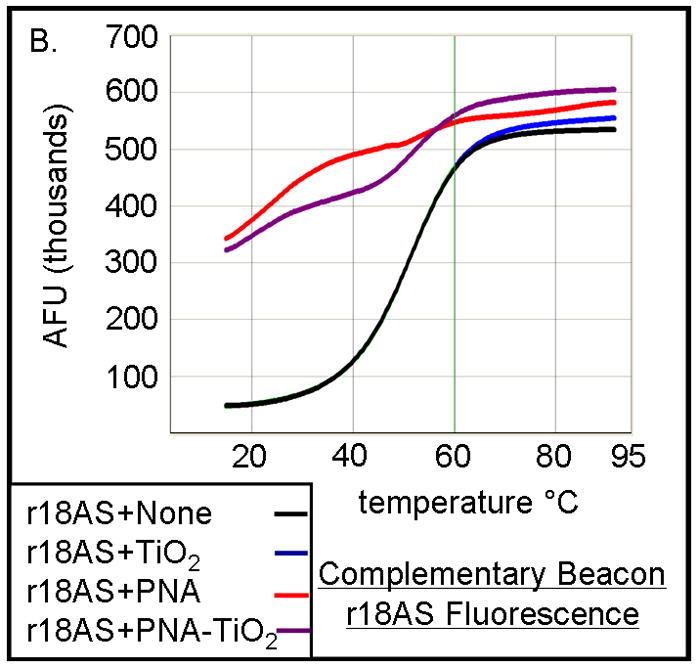
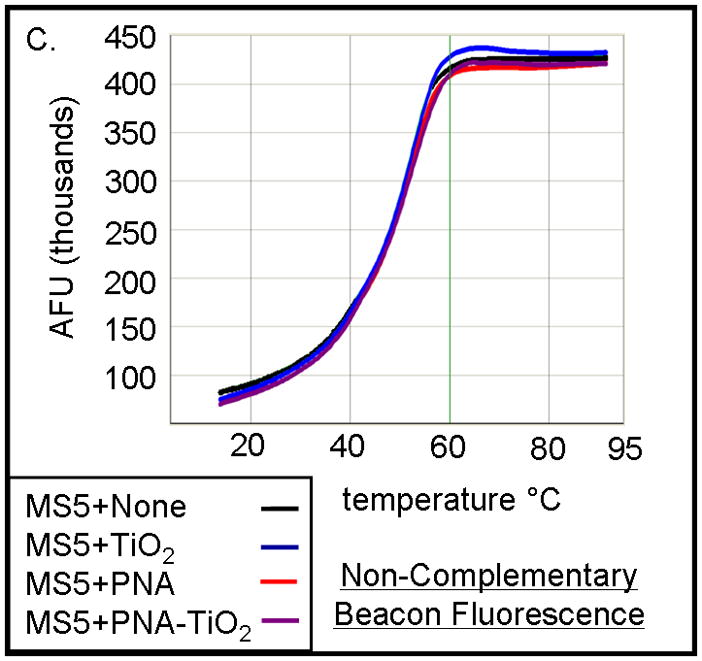
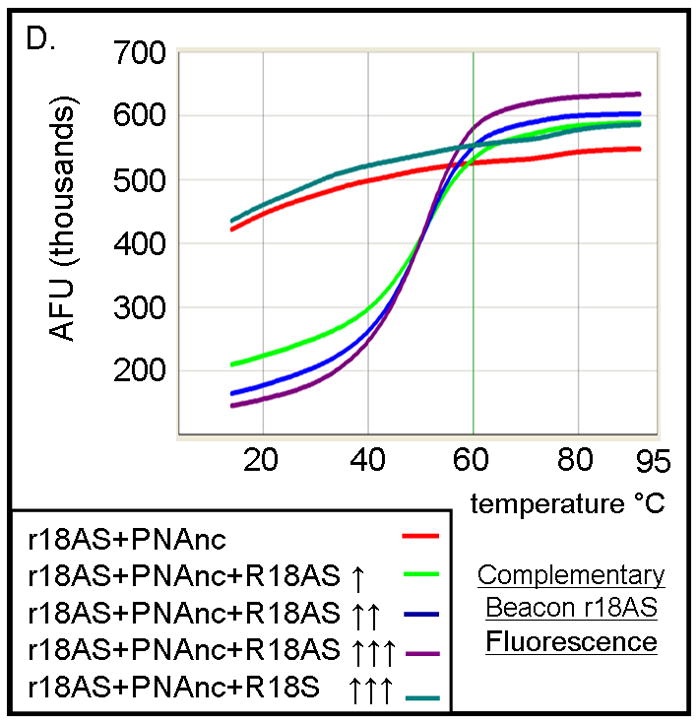
TiO2-PNA nanoconjugates can hybridize to complementary DNA. (A) Schematic representation depicts changes in fluorescence resulting from self hybridization and melting of molecular beacons; changes in fluorescence caused by hybridization between molecular beacons and PNA-TiO2 and self hybridization and melting of molecular beacon; changes in fluorescence when non-complementary PNA-TiO2 and molecular beacon are mixed. Representative dissociation curves resulting from hybridization reactions containing test samples (TiO2 = nanoparticle; PNA = PNA; PNA-TiO2 = PNA-TiO2 nanoconjugates) and (B) complementary molecular beacon or (C) non-complementary molecular beacon are shown. Hybridization of either PNA-TiO2 or PNA to molecular beacons containing a complementary target results in alterations in fluorescence curves, while no such alterations are observed when a non-complementary molecular beacon is used. (D) The addition of various amounts of excess unlabeled PNA-complementary DNA oligonucleotide (r18AS), as molecular beacon competitor, to samples containing PNA-TiO2 or PNA and complementary molecular beacons gradually returns the shape of the fluorescence curve to that obtained with samples containing molecular beacons alone. The addition of excess oligonucleotide identical to the sequence of PNA (r18S) shows a slight increase of the fluorescence (which can be expected because of additional hybridization of these DNA oligonucleotides with the beacon). In this and all other molecular beacon experiments concentrations of PNA-TiO2 and/or PNAs were 0.66 μM, and the concentration of beacon was 0.33 μM. Concentrations of competitor in this experiment were ↑ = 0.66 μM; ↑↑ = 1.0 μM; ↑↑↑ = 1.25 μM. AFU = arbitrary fluorescent units. The results shown are representative examples from five independent experiments.
The hybridization abilities of PNA-TiO2 nanoconjugates were further characterized by determining their capability to engage in strand exchange with dsDNA. We developed a new method to differentiate between DNA:DNA and PNA:DNA complexes (Figure 3) based on a knowledge that DNA intercalating dyes show little or no binding to PNA:DNA complexes [24]. Testing of the DNA-binding dye Power Sybr Green (Applied Biosystems) for monitoring DNA dissociation revealed that this DNA dye yields a fluorescent signal that is strong in the presence of dsDNA, but greatly reduced in the presence of PNA:DNA complexes (Figure 3a). We then tested the ability of PNA-TiO2 nanoconjugates to outcompete a homologous ssDNA sequence for a complementary ssDNA target, as determined by a decrease in the derivative of fluorescence intensity peak at the melting temperature of the DNA duplex. Samples containing oligonucleotide dsDNA yielded a characteristically sharp peak indicative of the increased rate of fluorescence loss at the melting temperature (Figure 3b, black curve). The addition of free glycidyl isopropyl ether coated nanoparticles had little effect on the fluorescence curve, although a small increase in the peak was observed perhaps due to nanoparticle-concentration-induced stabilization of the duplex or background fluorescence from the nanoparticle (Figure 3b, blue curve). However, the addition of PNA-TiO2 nanoconjugates resulted in over a 50% decrease in the fluorescence derivative peak at the melting temperature of the dsDNA, indicative of reduced dsDNA at this stage (Figures 3b, purple curve). The addition of free PNAs to dsDNA oligonucleotides yielded similar results, affirming that PNA also competes during hybridization (Figure 3b, red curve). The magnitude of this signal dampening effect was dependent upon the quantity of PNA-TiO2 nanoconjugates added (Figure 3c), and such reductions in fluorescence and the rate of fluorescence loss were not obtained when PNA-TiO2 nanoconjugates were added to samples of heterologous dsDNA oligonucleotides (Figures 3d). This indicates that the changes in fluorescence were sequence-specific and caused by the PNA-TiO2 nanoconjugate outcompeting the homologous DNA strand for hybridization with the complementary DNA strand.
Figure 3.
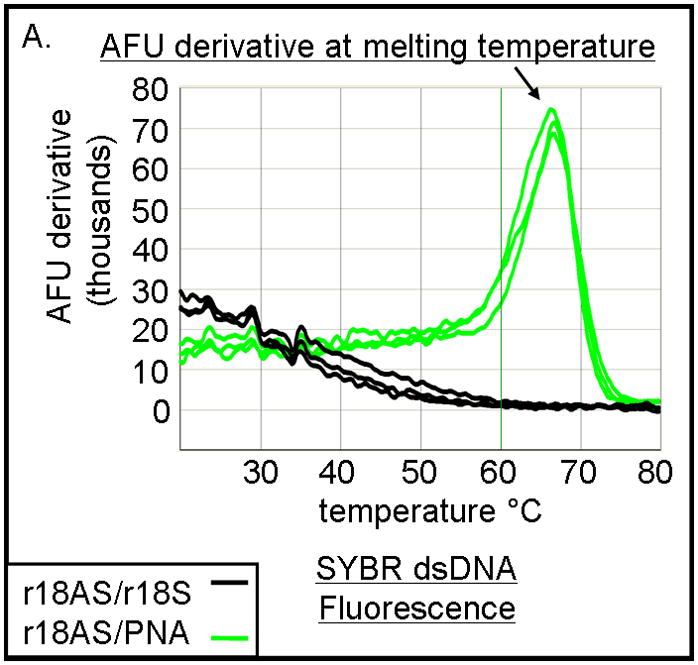
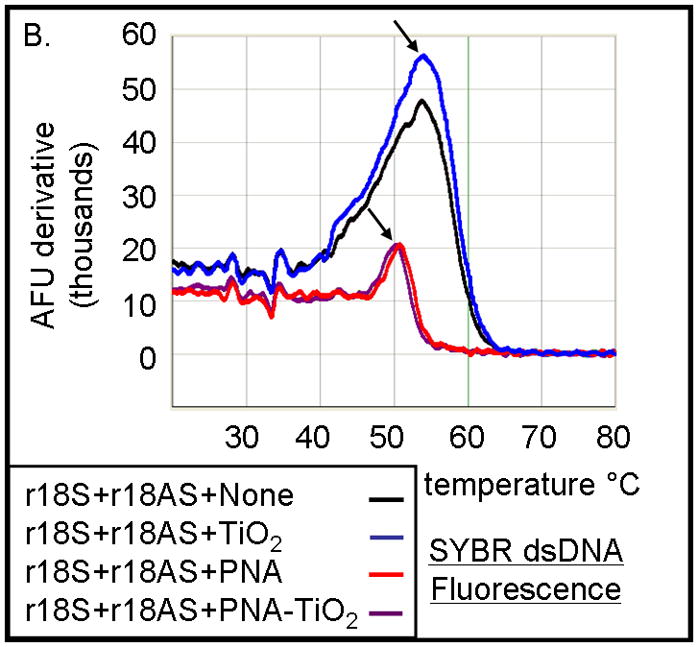
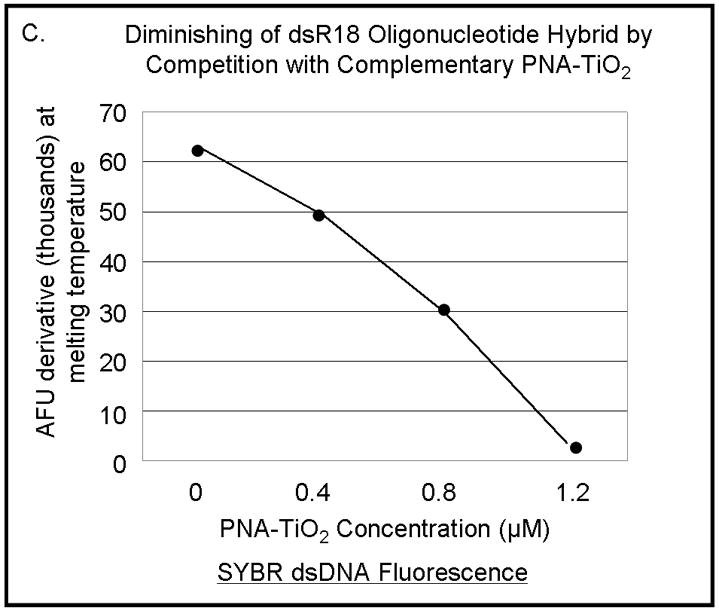
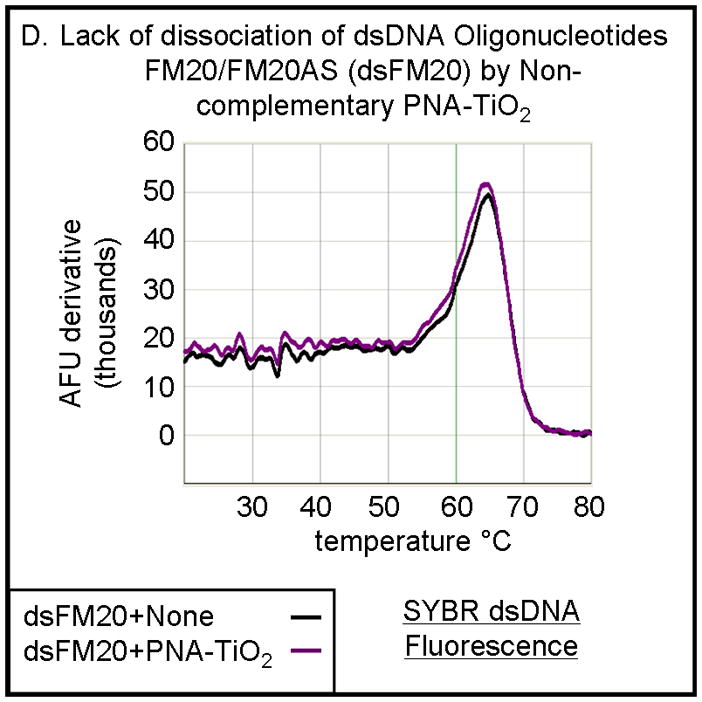
PNA-TiO2 nanoconjugates outcompete (replace) homologous DNA oligonucleotides in formation of the double strand hybrid (labeled R18 when made of two DNA oligonucleotides). (A) Derivative dissociation curves of fluorescence intensities show a well defined Tm peak (indicated by an arrow) for dsDNA hybrids (green), but not PNA:DNA complexes (black); three replicates each. (B) Derivative dissociation curves show a lowering of Tm peak height associated with competition between the homologous DNA strand of dsR18 and PNA or PNA-TiO2 for the same target (complementary DNA oligonucleotide). Black arrows point to the AFU derivative at melting temperature. (C) A graph showing inverse dependence of the peak height for the value of AFU derivative at Tm peak, related to the concentration of TiO2-PNA nanoconjugates added to oligonucleotides creating dsR18 molecules is shown. (D) Representative derivative dissociation curves show no change in the Tm peak intensity when PNA-TiO2 is combined with the non-complementary dsDNA (hybrid of FM20 and FM20AS oligonucleotides, labeled dsFM20). In these and other experiments where Power Sybr Green signal was used for measurements, concentrations of complementary DNA oligonucleotides (r18S and r18AS) were 0.5 μM each, while the nanoconjugate and PNA concentrations were 1 μM. AFU = arbitrary fluorescence units; TiO2 = nanoparticle; TiO2-PNA= TiO2-PNA nanoconjugates
Modifications of the nanoparticle surface
TiO2 nanoconjugates are capable of binding bidentate ligands enabling attachment of additional intracellular targeting agents and/or therapeutic payloads; moreover, carboxyl groups of peptides bind weakly to the nanoparticle surface. Therefore, it is essential to determine the effect of adding such a peptide on the ability of PNA-TiO2 nanoconjugates to hybridize to target DNA. Hybridization-dissociation studies conducted with PNA-TiO2 nanoconjugates coated with an epidermal growth factor peptide segment (pEGF), containing a nuclear localization sequence (NLS), indicate that peptide-modified nanoconjugates maintain the ability to hybridize to complementary molecular beacons (Figure 4a, purple curve). Peptide-coated nanoconjugates appear to have slightly enhanced hybridization capabilities compared to their naked counterparts (compare fluorescence values at 14°C in Figures 2b, purple curve and 4a, purple curve), which may be due to a locally increased DNA concentration induced by the positively charged peptides. Such relative comparisons between figures were possible since all experimental conditions were set against the same intra-experimental molecular beacon controls. This suggests that peptide coating is not only a useful tool to increase cellular uptake [reviewed in (11)], but it may also be improving interactions between the nanoconjugate and its molecular beacon target. Interestingly, when the PNA alone in the presence of pEGF peptide was used, the molecular beacon showed an aberrant fluorescence curve, perhaps due to interference between the free peptides and reopening of the molecular beacon (Figure 4a, blue curve). Since no such inhibition was observed with the addition of peptide-coated nanoconjugates or nanoparticles (Figure 4a, red curve), the ability of TiO2 to readily bind surface modifiers and alter their function is further accentuated.
Figure 4.
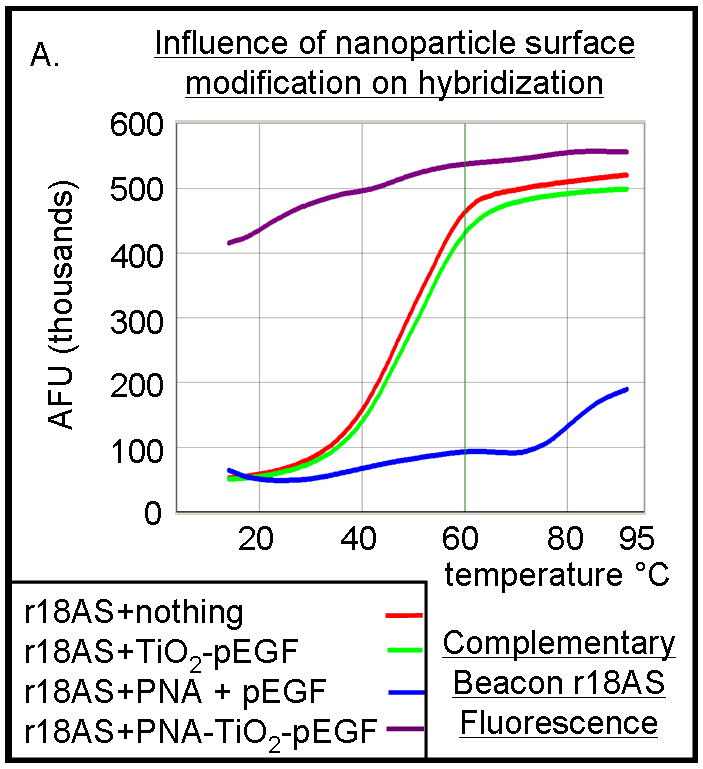
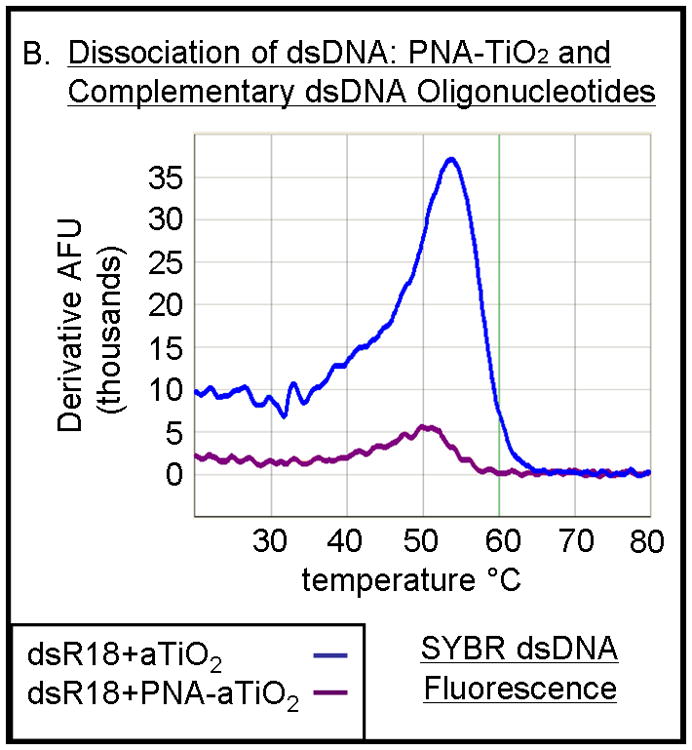
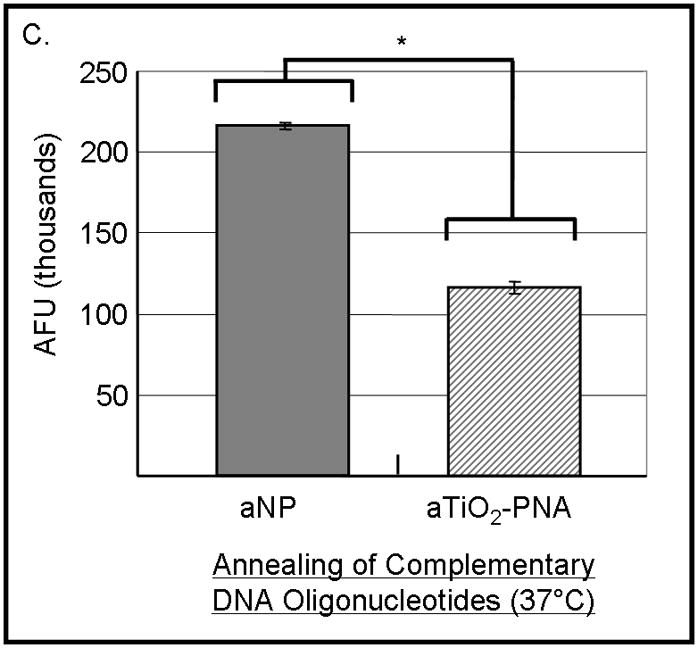
PNA-TiO2 nanoconjugates (PNA-TiO2) retain their hybridization abilities after additional modification of the nanoparticle. (A) PNA-TiO2 nanoconjugates conjugated with a peptide that represents a segment of epidermal growth factor (pEGF) maintain the ability to hybridize to target molecular beacon. Representative dissociation curves resulting from hybridization-dissociation reactions containing various test samples and a complementary molecular beacon. Hybridization of either PNA-TiO2 or PNA to molecular beacon r18AS results in alterations in fluorescence curves. (B) Alizarin red s-coated PNA-TiO2 nanoconjugates (PNA-aTiO2) maintain the ability to outcompete homologous DNA oligonucleotide from a dsR18 dsDNA oligonucleotide. Derivative dissociation curves show a lowering of Tm peak height associated with addition of PNA-aTiO2 to complementary DNA hybridization mixture. (C) Alizarin red s-coated PNA-TiO2 nanoconjugates (PNA-aTiO2) are able to engage in strand exchange with dsDNA at 37° C under 137mM sodium conditions. AFU = arbitrary fluorescent units; TiO2 = nanoparticle; PNA = PNA; pEGF = a peptide that represents a segment of epidermal growth factor; PNA-TiO2-pEGF = PNA-TiO2 nanoconjugates additionally coated with a peptide that represents a segment of epidermal growth factor; dsR18 = r18S/r18AS dsDNA hybrid; aTiO2 = Alizarin red s coated nanoparticle; PNA-aTiO2 = PNA and Alizarin red s nanoconjugates. * = p <0.05
Previous studies have noted that Alizarin red s bound to the surface of the nanoparticle permits visualization either in cells [11] or in vitro (Figure 1). Hybridization-competition assays were done to determine whether Alizarin red s, when bound to the nanoparticle, affects hybridization of PNA bound to the same nanoparticle. Alizarin red s-coated PNA-TiO2 nanoconjugates demonstrated an ability to outcompete homologous DNA in binding with the complementary DNA target as indicated by seven-fold decrease in the fluorescence derivative peak (Figure 4b, compare blue and purple curves). Therefore, Alizarin red s coating did not adversely affect the hybridization behavior of nanoconjugates; in fact, coating the nanoconjugate enhanced the hybridization abilities by a factor of 2 ½ compared to naked nanoconjugates and naked PNAs (compare Figures 4b and 3b). Furthermore, alizarin-coated PNA-TiO2 nanoconjugates proved able to engage in strand exchange in 137mM conditions at 37°C as indicated by a reduction in Sybr Power Green fluorescence in reactions containing dsDNA and nanoconjugates compared to only dsDNA (Figure 4c, p < 0.05).
Ability of PNA-TiO2 nanoconjugates to invade plasmid DNA containing a mixed base target under physiological-like temperature and ionic conditions
The use of PNA-TiO2 nanoconjugates in cellular and whole animals systems depends on their ability to hybridize to complementary DNA under physiological-like conditions, and the sodium concentration affects hybridization of DNA oligonucleotides [25] and PNA strand invasion of duplex DNA [26,27]. Additionally, targeting of PNAs to previously inaccessible mixed base sequences will allow for increased diversification of potential therapeutic targets. Testing of PNA nanoconjugate behavior under physiological-like temperature and ionic conditions showed that PNA-TiO2 nanoconjugates hybridized well with molecular beacons under such varied conditions (Supplemental Figure 1a–b). To affirm that PNA nanoconjugates are able to invade a supercoiled plasmid DNA containing a mixed base target under physiological-like salt and temperature conditions (137 mM sodium, 37.5 °C), we conducted the experiments presented in Figure 5. For these studies we developed an assay based on the precipitation of TiO2 nanoparticles from aqueous solutions of neutral pH (while plasmid DNA does not precipitate in 100% aqueous solutions) when centrifuged at 0.2 g. pKaede-MN1 (MLB) and pKaede-MN1-R18 plasmids were used, and the only difference between these plasmids was that the latter contains a mixed base sequence that is complementary to the sequence of PNA-TiO2 nanoconjugate (schematically depicted in Figure 5a). All reactions were incubated at 37.5° C for two hours with periodic mixing followed by centrifugations at 0.2 g to pellet nanoparticles or nanoconjugates. Presence of plasmid DNA in the pellet was then analyzed on an agarose gel (Figure 5b). Three independent experiments are seen on this gel.
Figure 5.
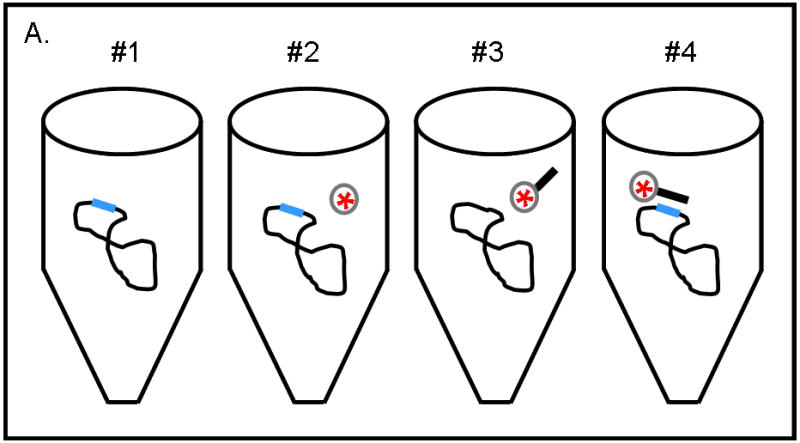
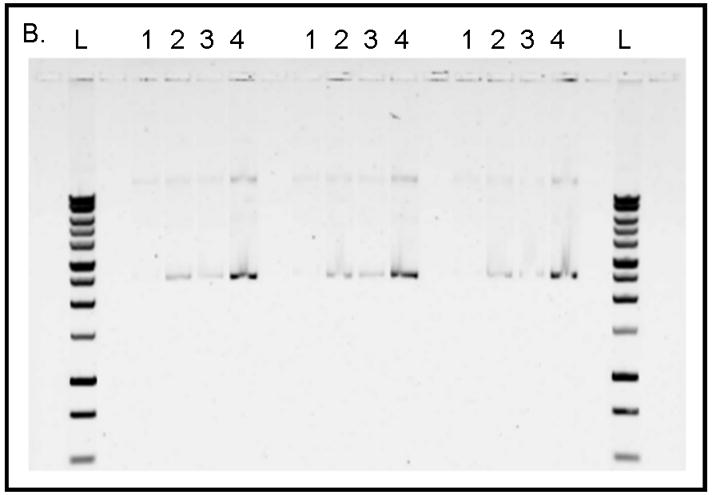
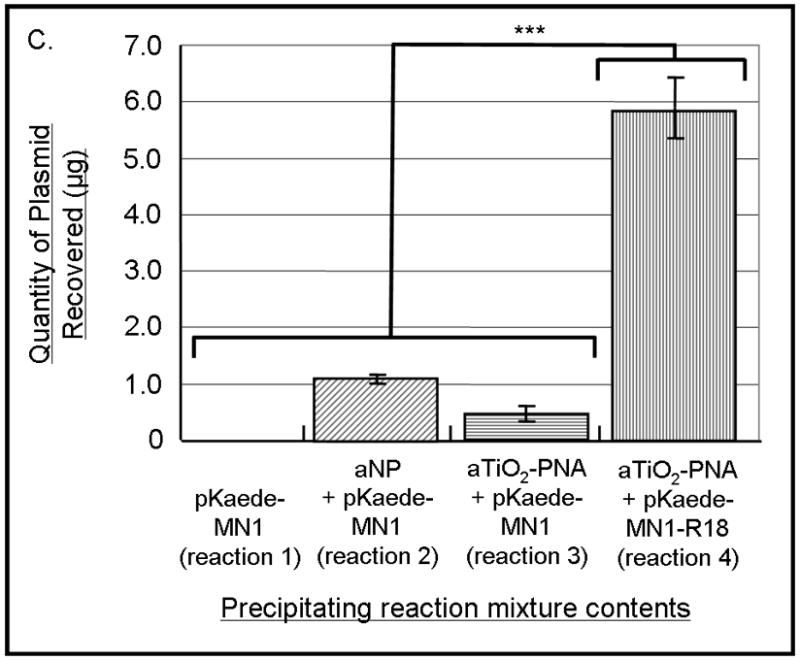
(A) Schematic of the experimental design for sample preparation for each well type on the gel is shown. Reactions of types 1, 2, and 4 contained plasmid pKaede-MN1-R18 (containing the insert that is complementary to the PNA sequence), in addition, reaction type 2 contained TiO2 nanoparticles and reaction type 4 also contained PNA-TiO2 nanoconjugates. Reaction type 3 contained plasmid pKaede-MN1 without PNA target sequence and PNA-TiO2 nanoconjugates. (B) Gel demonstrates the ability of PNA-TiO2 nanoconjugates to invade target-containing plasmid DNA pKaede-MN1-R18 (wells containing reaction type 4), at 37° C in 137mM sodium, much more avidly than the “empty” pKaede-MN1 plasmid (wells containing reaction type 3), thus allowing plasmid precipitation from a 100% aqueous solution. L = DNA ladder. In lanes 1 through 4, top bands are relaxed plasmids and bottom bands are supercoiled plasmids. Three separate experiments are shown on the same gel. (C) Semi-quantitative comparison of plasmid precipitation resulting from reaction types 2–4 is shown. *** = p < 0.001
As expected, virtually no plasmid was recovered when the plasmid was incubated alone in the aqueous solution (Figure 5b, lanes labeled 1). When Alizarin red s-coated nanoparticles were incubated with the plasmid (Figure 5b, lanes labeled 2) or when Alizarin red s-coated nanoconjugates were incubated with plasmid pKaede-MN1 devoid of a complementary insert R18 (Figure 5b, lanes labeled 3) a small amount of plasmid precipitated. However, a significant quantity of plasmid material was recovered when PNA-aTiO2 nanoconjugates were incubated with the plasmid pKaede-MN1-R18 containing a complementary sequence R18 (Figure 5b, lanes labeled 4). The limited precipitation of the plasmid found in samples 2 and 3 can be explained by non-sequence specific interaction between surface of nanoparticles and polyphosphate of the DNA backbone; the affinity of TiO2 surface sites for polyphosphates was previously established in the literature [28]. Comparisons of these results indicate that recovery of the plasmid in reaction mixture containing TiO2-PNA and supercoiled plasmid with the target sequence was due to the invasion of the supercoiled plasmid DNA (reaction mixture 4) by nanoconjugates in a sequence-specific manner. Rate of plasmid recovery (Figure 5c) suggests that 5.9 μg of the 19.5 μg plasmid sample (30% recovery rate) was achieved by precipitation of plasmid containing the mixed base DNA target using TiO2-PNA.
Discussion
Advances in nanoparticle technology have provided biologists with a set of new tools with unique physico-chemical properties [4–6,11,29]. Previous research from our laboratory was focused on the properties of conjugates of DNA and TiO2 nanoparticles, their synthesis, use in enzymatic reactions (e.g. PCR), cleavage of DNA upon excitation, and imaging by X-ray fluorescence microscopy [7,8]. Optically fluorescent agents that are ortho-substituted enediol ligands [5,6,11], may be added to the nanoconjugate to provide a new dimension to intracellular studies—imaging by confocal fluorescent microscopy [11]. As previously discussed, the nanoparticle can be further modified and detected with multiple other imaging modalities, including those that are of interest to medicine [9,10].
The main benefits of using TiO2 nanoparticles are that they can serve as a platform/scaffold to which numerous molecules can be conjugated, and therefore serve as a multi-modal imaging agent and therapeutic agent; moreover, when excited by white light [7], these nanoconjugates have the ability to cleave DNA. Benefits of these nanoconjugates will, however, largely depend on their targeting and retention specificity, and these can be provided by conjugating DNA analogs to the nanoparticle surface, in addition to any other targeting, diagnostic, and therapeutic modifications. Studies from our lab have shown that conjugating DNA to the nanoparticle surface greatly enhances cellular localization and retention of our nanoparticles [7,8]. Replacing DNA with PNA will lead to increased stability of the nucleic acid component of the nanoconjugate and will, therefore, prolong targeting life of the nanoconjugates and may even increase their sequence selectivity. Previously, fluorescently tagged homopyrimidine PNAs had been used to invade plasmid DNA for tracking without disturbing transgene expression [30]. However, to our knowledge, the work herein is the first study demonstrating that modified, mixed based PNAs are capable of invading and labeling supercoiled plasmid DNA. Development of PNA-TiO2 nanoconjugates and their ability to invade mixed base sequences will undoubtedly enhance the targeting of deleterious cellular DNA in manners previously not possible. On the other hand, PNA/PNA binding affinity is extremely strong and often results in unwanted interactions. For this reason, it was essential to fully characterize PNA-TiO2 nanoconjugates containing a low surface density of PNAs in this study, before introducing additional variables in the form of potential PNA/PNA interactions.
To our knowledge no studies currently exist that have characterized factors affecting hybridization behaviors of PNAs conjugated to TiO2 nanoparticles, so direct comparisons between the results herein with others is not possible. In a different nanoparticle regime, PNAs conjugated to gold nanoparticles and immobilized on glass had the ability to discriminate between DNA targets with a single base mismatch [31]. Numerous other studies involving gold nanoparticles have demonstrated single base hybridization specificity of nucleic acid-nanoparticle conjugates by utilizing various methods such as gel electrophoresis, DNA arrays combined with a traditional flatbed scanner, single-strand-specific nucleases, and high fidelity DNA ligases [32–35]. Additionally, DNA oligonucleotides conjugated to gold nanoparticles showed higher binding affinities and a more rapid melting transition compared to free oligonucleotides; this system was sensitive to many factors, including DNA surface density, nanoparticle size, salt concentration, and interparticle distances [36–38]. Using TiO2 nanoparticles linked to ssDNA, it has recently been demonstrated that a single base mismatch between the nanoconjugate and its target significantly reduces photocatalytic cleavage due to disruption of the of the π stack, indicating that nucleic acid-TiO2 nanoconjugates can also exhibit a high level of sequence specificity [39]. Under these circumstances, there is no reason to suspect that PNA-TiO2 nanoconjugates would exhibit less sequence specificity. Relative differences in melting temperatures between Au-DNA nanoconjugates and free DNAs (compared to PNA-TiO2 nanoconjugates and free PNAs) may be explained by many factors, including the increased nucleic acid surface density of Au-DNA nanoconjugates compared to PNA-TiO2 nanoconjugates, as well as the dissimilar nucleic acid backbone. Au-DNA nanoconjugates are able to migrate through gels during electrophoresis [40], while DNA-TiO2 [7] or PNA-TiO2 nanoconjugates do not (Figure 1); this suggests that Au and TiO2 nanoparticles differentially affect the nucleic acids to which they are conjugated. Considering that Au is a noble metal whereas TiO2 is a semiconductor with very reactive surface chemistry, these differences should be expected. These studies highlight the complexity of hybridization dynamics involving nucleic acid-conjugated nanoparticles and emphasize the need for further studies of the effect of factors modulating hybridization between PNA-TiO2 nanoconjugates and target DNA.
Conjugating peptides or imaging agents to the TiO2 nanoparticles will aid in cellular uptake and visualization of PNA-TiO2 nanoconjugates during intracellular studies. Additionally, the imaging agent Alizarin red s can also be conjugated to TiO2 nanoparticles and serve as a fluorescent marker. This study demonstrates that PNA-TiO2 nanoconjugates coated with peptide segment of EGF or Alizarin red s have an improved ability to hybridize to complementary DNA; these modifications will lead to enhanced cellular targeting and visualization of PNA-TiO2 nanoconjugates in cells, without sacrificing the hybridization proficiency of the nanoconjugates. Moreover, it is possible that the presence of peptides may improve hybridization performance of nanoconjugates, as shown by a comparison of the Figures 2b and 4a. Studies have demonstrated that such additional modifications of the nucleic acid-TiO2 nanoconjugates with various ligands do not impede intracellular hybridization of nanoconjugates. DNA-TiO2 nanoconjugates, coated with bulky gadolinium contrast agent for detection with MRI, retained their intracellular hybridization specificity and retention [9, 10].
Research investigating charge transfer in PNA/DNA complexes is limited, but one study has shown that a charge injected into the PNA strand can be transferred to guanine doublets on the complementary strand DNA via a hopping mechanism [41]. Titanium dioxide nanoparticles larger than 2 nm have been shown capable of creation of electropositive holes [6]. Viewing these finding in light of our current study, indicates that like their DNA counterparts, PNA-TiO2 nanoconjugates larger than 2 nm could be used to induce DNA cleavage when excited.
It is well established in the literature that PNAs have a longer half-life in vivo and intracellularly compared to DNAs [42]. However, the same body of research established that PNAs are not as soluble as DNAs and that some PNA sequences have proved difficult to synthesize in the past. The later issues have since been circumvented with the development of automated synthesis protocols [43]; while challenges of solubility and delivery of PNAs into cells were solved by new solubilization strategies For example, PNAs can be annealed to a negatively charged DNA oligonucleotide and complexed with cationic lipids for intracellular delivery [44,45]. Alternative methods of intracellular delivery of PNAs include synthesis of a PNA with a nuclear localization signal (NLS) [46], electroporation [47,48], and possibly microinjection. The studies shown here, however, suggest that in the context of PNA-TiO2 nanoconjugates many different molecules can be attached to the nanoparticle part of the nanoconjugate (and in multiple copies) to increase cellular uptake of nanoparticle conjugated PNAs.
Conclusion
This study has shown that PNA-TiO2 nanoconjugates are capable of hybridizing with target DNA in a sequence-specific manner, regardless of conjugation of peptides or imaging agents to the nanoparticle surface, and invasion supercoiled plasmid DNA containing a mixed base target. PNA stability (established in the literature) should make PNA-TiO2 nanoconjugates very useful in other intracellular and in vivo studies, improving targeting specificity and increasing sequence-specific intracellular retention. Also, the new methods for investigation of nanoparticle-nucleic acid conjugates hybridization parameters described here will facilitate study of other DNA and DNA analog-nanoparticle systems. The evidence shown in this manuscript indicates that PNA-TiO2 nanoconjugates are a viable alternative to their DNA counterparts and warrant further in-depth study.
Supplementary Material
Supplemental Figure 1. Ability of TiO2-PNA nanoconjugates to hybridize to target DNA (complementary molecular beacon) is affected by sodium concentration. (A) Increasing the sodium concentration of buffer increases the melting temperature of the molecular beacon stem. (B) Increasing the sodium concentration of buffer reduces the ability of both PNA-TiO2 nanoconjugates and PNA to hybridize to the molecular beacon loop. Representative dissociation curves resulting from the hybridization-dissociation reaction between each test sample and molecular beacon are shown. AFU = arbitrary fluorescent units; TiO2 = nanoparticle; PNA = PNA; PNA-TiO2 = PNA-TiO2 nanoconjugates.
Acknowledgments
The authors would like to acknowledge, T. Chew, D. Dean, Y. Fukui, J. Maser, C. Cruz, A. Babbo, and D. Jakubczak for their scientific advice and G. Brown for her support. National Institutes of Health (CA107467, EB002100, P50 CA89018, U54CA119341).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gonzalez-Alegre P. Therapeutic RNA interference for neurodegenerative diseases: From promise to progress. Pharmacol Ther. 2007;114:34–55. doi: 10.1016/j.pharmthera.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Natt F. siRNAs in drug discovery: target validation and beyond. Curr Opin Mol Ther. 2007;9:242–247. [PubMed] [Google Scholar]
- 3.Scherer L, Rossi JJ, Weinberg MS. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Ther. 2007;14:1057–1064. doi: 10.1038/sj.gt.3302977. [DOI] [PubMed] [Google Scholar]
- 4.Roco MC, TAW, Henkart MP, Kalil TA, Trew R, Murday JS, Yoshida PG, Casassa MP, Shull RD, Thomas IL. National nanotechnology initiative: the initiative and its implementation plan. The Office of Science and Technology. 2007 [Google Scholar]
- 5.Rajh T, Chen LX, Lukas K, Liu T, Thurnauer MC, Tiede DM. Surface restructuring of nanoparticles: An efficient route for ligand-metal oxide crosstalk. J Phys Chem B. 2002;106:10543–10552. [Google Scholar]
- 6.Rajh T, Poluektov O, Dubinski AA, Wiederrecht G, Thurnauer MC, Trifunac AD. Spin polarization mechanisms in early stages of photoinduced charge separation in surface-modified TiO2 nanoparticles. Chemical Physics Letters. 2001;344:31–39. [Google Scholar]
- 7.Paunesku T, Rajh T, Wiederrecht G, Maser J, Vogt S, Stojicevic N, Protic M, Lai B, Oryhon J, Thurnauer M, Woloschak G. Biology of TiO2-oligonucleotide nanocomposites. Nat Mater. 2003;2:343–346. doi: 10.1038/nmat875. [DOI] [PubMed] [Google Scholar]
- 8.Paunesku T, Vogt S, Lai B, Maser J, Stojicevic N, Thurn KT, Osipo C, Liu H, Legnini D, Wang Z, Lee C, Woloschak GE. Intracellular distribution of TiO2-DNA oligonucleotide nanoconjugates directed to nucleolus and mitochondria indicates sequence specificity. Nano Lett. 2007;7:596–601. doi: 10.1021/nl0624723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endres PJ, Paunesku T, Vogt S, Meade TJ, Woloschak GE. DNA-TiO2 nanoconjugates labeled with magnetic resonance contrast agents. J Am Chem Soc. 2007;129:15760–15761. doi: 10.1021/ja0772389. [DOI] [PubMed] [Google Scholar]
- 10.Paunesku T, Ke T, Dharmakumar R, Mascheri N, Wu A, Lai B, Vogt S, Maser J, Thurn K, Szolc-Kowalska B, Larson A, Bergan RC, Omary R, Li D, Lu ZR, Woloschak GE. Gadolinium-conjugated TiO(2)-DNA oligonucleotide nanoconjugates show prolonged intracellular retention period and T1-weighted contrast enhancement in magnetic resonance images. Nanomedicine. 2008;20 doi: 10.1016/j.nano.2008.04.004. e-published June 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thurn KT, Brown EMB, Wu A, Vogt S, Lai B, Maser J, Paunesku T, Woloschak GE. Nanoparticles for Applications in Cellular Imaging. Nanoscale Research Letters. 2007 doi: 10.1007/s11671-007-9081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 13.Summerton JE. Morpholinos and PNAs compared. In: During CGJaMJ., editor. Peptide Nucleic Acids, Morpholinos, and Related Antisense Biomolecules. Springer US; 2006. [Google Scholar]
- 14.Demidov VV, Potaman VN, Frankkamenetskii MD, Egholm M, Buchard O, Sonnichsen SH, Nielsen PE. Stability of Peptide Nucleic-Acids in Human Serum and Cellular-Extracts. Biochem Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton SE, Iyer M, Norton JC, Corey DR. Specific and nonspecific inhibition of transcription by DNA, PNA, and phosphorothioate promoter analog duplexes. Bioorg Med Chem Lett. 1996;6:2897–2900. [Google Scholar]
- 16.Kaihatsu K, Janowski BA, Corey DR. Recognition of chromosomal DNA by PNAs. Chem Biol. 2004;11:749–758. doi: 10.1016/j.chembiol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Brown SC, Thomson SA, Veal JM, Davis DG. NMR solution structure of a peptide nucleic acid complexed with RNA. Science. 1994;265:777–780. doi: 10.1126/science.7519361. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson M, Nielsen PE. Solution structure of a peptide nucleic acid-DNA duplex. Nat Struct Biol. 1996;3:410–413. doi: 10.1038/nsb0596-410. [DOI] [PubMed] [Google Scholar]
- 19.Betts L, Josey JA, Veal JM, Jordan SR. A nucleic acid triple helix formed by a peptide nucleic acid-DNA complex. Science. 1995;270:1838–1841. doi: 10.1126/science.270.5243.1838. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen H, Kastrup JS, Nielsen JN, Nielsen JM, Nielsen PE. Crystal structure of a peptide nucleic acid (PNA) duplex at 1.7 A resolution. Nat Struct Biol. 1997;4:98–101. doi: 10.1038/nsb0297-98. [DOI] [PubMed] [Google Scholar]
- 21.Armitage B, Koch T, Frydenlund H, Orum H, Batz HG, Schuster GB. Peptide nucleic acid-anthraquinone conjugates: strand invasion and photoinduced cleavage of duplex DNA. Nucleic Acids Res. 1997;25:4674–4678. doi: 10.1093/nar/25.22.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaihatsu K, Shah RH, Zhao X, Corey DR. Extending recognition by peptide nucleic acids (PNAs): binding to duplex DNA and inhibition of transcription by tail-clamp PNA-peptide conjugates. Biochemistry (Mosc) 2003;42:13996–14003. doi: 10.1021/bi035194k. [DOI] [PubMed] [Google Scholar]
- 23.Smolina IV, Demidov VV, Soldatenkov VA, Chasovskikh SG, Frank-Kamenetskii MD. End invasion of peptide nucleic acids (PNAs) with mixed-base composition into linear DNA duplexes. Nucleic Acids Res. 2005;33:e146. doi: 10.1093/nar/gni151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittung P, Kim SK, Buchardt O, Nielsen P, Norden B. Interactions of DNA binding ligands with PNA-DNA hybrids. Nucleic Acids Res. 1994;22:5371–5377. doi: 10.1093/nar/22.24.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salzberg S, Levi Z, Aboud M, Goldberger A. Isolation and characterization of DNA-DNA and DNA-RNA. Biochemistry (Mosc) 1977;16:25–29. doi: 10.1021/bi00620a004. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence specific inhibition of DNA restriction enzyme cleavage by PNA. Nucleic Acids Res. 1993;21:197–200. doi: 10.1093/nar/21.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peffer NJ, Hanvey JC, Bisi JE, Thomson SA, Hassman CF, Noble SA, Babiss LE. Strand-invasion of duplex DNA by peptide nucleic acid oligomers. Proc Natl Acad Sci U S A. 1993;90:10648–10652. doi: 10.1073/pnas.90.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michelmore A, Gong WQ, Jenkins P, Ralston J. The interaction of linear polyphosphates with titanium dioxide surfaces. Phys Chem Chem Phys. 2000;2:2985–2992. [Google Scholar]
- 29.Rajh T, Nedeljkovic JM, Chen LX, Poluektov O, Thurnauer MC. Improving optical and charge separation properties of nanocrystalline TiO2 by surface modification with vitamin C. J Phys Chem B. 1999;103:3515–3519. [Google Scholar]
- 30.Hillery E, Munkonge FM, Xenariou S, Dean DA, Alton FW. Nondisruptive, sequence-specific coupling of fluorochromes to plasmid DNA. Anal Biochem. 2006;352:169–175. doi: 10.1016/j.ab.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 31.Endo T, Kerman K, Nagatani N, Takamura Y, Tamiya E. Label-free detection of peptide nucleic acid-DNA hybridization using localized surface plasmon resonance based optical biosensor. Anal Chem. 2005;77:6976–6984. doi: 10.1021/ac0513459. [DOI] [PubMed] [Google Scholar]
- 32.Qin W, Yung L. Nanoparticle-based detection and quantification of DNA with single nucleotide polymorphism (SNP) discrimination selectivity. Nucleic Acids Res. 2007;35(17):e111. doi: 10.1093/nar/gkm602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taton T, Mirkin C, Letsinger R. Scanometric DNA array detection with nanoparticle probes. Science. 2000 Sep 8;289(5485):1757–60. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Hsu C, Hou S. Detection of single-nucleotide polymorphisms using gold nanoparticles and single-strand-specific nucleases. Anal Biochem. 2008 Apr 15;375(2):299–305. doi: 10.1016/j.ab.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Chu X, Liu Y, Jiang J, He Z, Zhang Z, Shen G, Yu R. A colorimetric method for point mutation detection using high-fidelity DNA ligase. Nucleic Acids Res. 2005 Oct 27;33(19):e168. doi: 10.1093/nar/gni163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Jin R, Mirkin CA, Letsinger RL. Multiple thiol-anchor capped DNA-gold nanoparticle conjugates. Nucleic Acids Res. 2002;30:1558–1562. doi: 10.1093/nar/30.7.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin R, Wu G, Li Z, Mirkin CA, Schatz GC. What controls the melting properties of DNA-linked gold nanoparticle assemblies? J Am Chem Soc. 2003;125:1643–1654. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- 38.Lytton-Jean AK, Mirkin CA. A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J Am Chem Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 39.Tachikawa T, Asanoi Y, Kawai K, Tojo S, Sugimoto A, Fujitsuka M, Majima T. Photocatalytic cleavage of single TiO2/DNA nanoconjugates. Chemistry. 2008;14(5):1492–8. doi: 10.1002/chem.200701030. [DOI] [PubMed] [Google Scholar]
- 40.Fu A, Micheel CM, Cha J, Chang H, Yang H, Alivisatos AP. Discrete nanostructures of quantum dots/Au with DNA. J Am Chem Soc. 2004;126:10832– 10833. doi: 10.1021/ja046747x. [DOI] [PubMed] [Google Scholar]
- 41.Armitage B, Ly D, Koch T, Frydenlund H, Orum H, Batz HG, Schuster GB. Peptide nucleic acid-DNA duplexes: long range hole migration from an internally linked anthraquinone. Proc Natl Acad Sci U S A. 1997;94:12320–12325. doi: 10.1073/pnas.94.23.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMahon BM, Mays D, Lipsky J, Stewart JA, Fauq A, Richelson E. Pharmacokinetics and tissue distribution of a peptide nucleic acid after intravenous administration. Antisense Nucleic Acid Drug Dev. 2002;12:65–70. doi: 10.1089/108729002760070803. [DOI] [PubMed] [Google Scholar]
- 43.Mayfield LD, Corey DR. Automated synthesis of peptide nucleic acids and peptide nucleic acid peptide conjugates. Anal Biochem. 1999;268:401–404. doi: 10.1006/abio.1998.3052. [DOI] [PubMed] [Google Scholar]
- 44.Braasch DA, Corey DR. Synthesis, analysis, purification, and intracellular delivery of peptide nucleic acids. Methods. 2001;23:97–107. doi: 10.1006/meth.2000.1111. [DOI] [PubMed] [Google Scholar]
- 45.Herbert B, Pitts AE, Baker SI, Hamilton SE, Wright WE, Shay JW, Corey DR. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc Natl Acad Sci U S A. 1999;96:14276–14281. doi: 10.1073/pnas.96.25.14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braun K, Peschke P, Pipkorn R, Lampel S, Wachsmuth M, Waldeck W, Friedrich E, Debus J. A biological transporter for the delivery of peptide nucleic acids (PNAs) to the nuclear compartment of living cells. J Mol Biol. 2002;318:237–243. doi: 10.1016/S0022-2836(02)00031-1. [DOI] [PubMed] [Google Scholar]
- 47.Karras JG, Maier MA, Lu T, Watt A, Manoharan M. Peptide nucleic acids are potent modulators of endogenous pre-mRNA splicing of the murine interleukin-5 receptor-alpha chain. Biochemistry (Mosc) 2001;40:7853–7859. doi: 10.1021/bi010263l. [DOI] [PubMed] [Google Scholar]
- 48.Shammas MA, Simmons CG, Corey DR, Shmookler-Reis RJ. Telomerase inhibition by peptide nucleic acids reverses ‘immortality’ of transformed human cells. Oncogene. 1999;18:6191–6200. doi: 10.1038/sj.onc.1203069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Ability of TiO2-PNA nanoconjugates to hybridize to target DNA (complementary molecular beacon) is affected by sodium concentration. (A) Increasing the sodium concentration of buffer increases the melting temperature of the molecular beacon stem. (B) Increasing the sodium concentration of buffer reduces the ability of both PNA-TiO2 nanoconjugates and PNA to hybridize to the molecular beacon loop. Representative dissociation curves resulting from the hybridization-dissociation reaction between each test sample and molecular beacon are shown. AFU = arbitrary fluorescent units; TiO2 = nanoparticle; PNA = PNA; PNA-TiO2 = PNA-TiO2 nanoconjugates.


