Abstract
Coupling of autonomous cellular oscillators is an essential aspect of circadian clock function but little is known about its circuit requirements. Functional ablation of the pigment-dispersing factor-expressing lateral ventral subset (LNV) of Drosophila clock neurons abolishes circadian rhythms of locomotor activity. The hypothesis that LNVs synchronize oscillations in downstream clock neurons was tested by rendering the LNVs hyperexcitable via transgenic expression of a low activation threshold voltage-gated sodium channel. When the LNVs are made hyperexcitable, free-running behavioral rhythms decompose into multiple independent superimposed oscillations and the clock protein oscillations in the dorsal neuron 1 and 2 subgroups of clock neurons are phase-shifted. Thus, regulated electrical activity of the LNVs synchronize multiple oscillators in the fly circadian pacemaker circuit.
Keywords: arrhythmia, behavior, circadian rhythms, desynchronization, Drosophila, sodium channel
Introduction
All animals exhibit physiological and/or behavioral rhythms with periods of ∼24 h, even in the absence of environmental cues to time of day. This includes cave dwellers that have existed for generations without any daily variation in light, temperature, or other environmental parameters (Pradhan et al., 1989; Koilraj et al., 2000). These free-running circadian rhythms indicate that animals possess an intrinsic timekeeping mechanism. Although we now know a great deal about how oscillators work at the molecular genetic level, relatively little is known about how individual molecular oscillators affect the functional properties of neurons or how they are coupled within pacemaker circuits.
A number of studies suggest that coordinated physiological and behavioral rhythmicity requires synchronization of multiple oscillators at the circuit level (de la Iglesia et al., 2000, 2004; Low-Zeddies and Takahashi, 2001; Nagano et al., 2003). Individual clock neurons isolated from the mammalian suprachiasmatic nucleus (SCN) and placed in dispersed culture or brain slices prepared from SCN continue to oscillate, but asynchronously with their neighbors (Welsh et al., 1995; Yamaguchi et al., 2003). The variation in the intrinsic period of individual dispersed mammalian clock neurons is substantially greater than that exhibited by either the intact animal or a semi-intact brain explant (Herzog et al., 2004). Coupling is clearly important in vivo because certain unusual environmental light/dark (LD) conditions that induce complex behavioral rhythms with multiple periods and/or phases also cause dissociation of cellular cycling in discrete anatomical compartments of the SCN (de la Iglesia et al., 2000, 2004; Nagano et al., 2003; Schaap et al., 2003).
The pigment-dispersing factor neuropeptide (PDF)-expressing lateral ventral subset (LNV) of clock neurons has been proposed to be functionally critical within the fly circadian control circuit, because Pdf01-null mutant flies and flies in which the LNV subset of clock neurons have been genetically ablated or electrically silenced exhibit severe deficits in free-running rhythms (Renn et al., 1999; Nitabach et al., 2002; Lin et al., 2004). Circadian regulation, along with PDF function, has also been studied intensively in other insects, notably in cockroaches but also in locusts, moths, beetles, and ants. Much of this comparative work is covered comprehensively in a previous review of insect circadian function (Helfrich-Forster, 2004). Cyclic release of PDF from the terminals of the LNV pacemaker neurons has been proposed to provide an important circadian signal to PDF-sensitive downstream targets (Renn et al., 1999; Helfrich-Forster et al., 2000; Park et al., 2000; Peng et al., 2003; Lin et al., 2004). Consistent with this hypothesis, levels of anti-PDF immunoreactivity in the dorsomedial terminals of the small LNV (sLNV) pacemaker neurons have been observed to oscillate both in diurnal cycling light/dark and free-running constant-darkness conditions, with a much greater amplitude of cycling in diurnal conditions (Helfrich-Forster et al., 2000; Park et al., 2000). Furthermore, pacemaker neurons overexpressing PDF still exhibit cycling in the terminals in contrast to nonclock neurons overexpressing PDF, where no terminal cycling is observed (Helfrich-Forster et al., 2000). These results suggest that PDF cycling in the terminals may be clock regulated. In contrast to cyclic levels of anti-PDF immunoreactivity in the dorsomedial terminals, PDF RNA synthesis appears to be noncyclic (Park and Hall, 1998). Thus, given the complexity of factors that appear to contribute to PDF cycling in nerve terminals, it should be noted with caution that levels of anti-PDF immunoreactivity in the dorsomedial terminals may be influenced by many cellular processes, including protein synthesis, transport, and release.
Additional studies indicate a more complex role for PDF in the insect circadian system: PDF injected into the cockroach brain resets the phase of free-running behavioral rhythms (Petri and Stengl, 1997), and ectopic release of PDF from nonclock neuron terminals in the region of the dorsal protocerebrum where the DNs reside results in complex behavioral rhythms (Helfrich-Forster et al., 2000). It may be the case that as long as some PDF is present in the dorsal protocerebrum with little spatial or temporal constraints, rhythms may persist. Alternatively, rhythms and oscillator synchronization may require precisely timed activity of the LNVs at their normal targets. This cannot be addressed by ectopic PDF administration or release in flies or other insects, because such approaches do not account for the spatial or temporal aspects of neuronal LNV-based signaling to their normal targets. These approaches, although having provided valuable information, do not account for possible effects of focal versus nonfocal release of PDF or other unknown neurotransmitters that may be released by the LNVs. It is an open question as to whether it is regulated LNV neural activity and temporally controlled PDF release that synchronizes the pacemaker circuit, or rather, whether the requirement for PDF revealed by the phenotypes of Pdf01-null flies is permissive instead. To address this question, we have developed a physiological approach in which we induce electrical hyperexcitability of the LNVs by targeted expression of a low-threshold slowly inactivating voltage-gated sodium channel in transgenic flies. Mutations in human and mammalian voltage-gated sodium channels that slow inactivation lead to hyperexcitability in nerve and muscle (for review, see Lehmann-Horn and Jurkat-Rott, 1999). The expression of NaChBac, a bacterial depolarization-activated sodium channel, in Drosophila cells can thus be expected to increase membrane excitability; depolarization of the membrane will induce inward currents through NaChBac, which will further depolarize the membrane, thus, triggering a low-threshold positive-feedback loop.
Materials and Methods
DNA constructs, P-element transformation, electrophysiology, and fly strains. The NaChBac coding cDNA (Ren et al., 2001) was truncated 16 nucleotides 5′ of the stop codon by cleavage with HindIII and fused in frame to enhanced green fluorescent protein (eGFP; Clontech, Mountain View, CA) to generate NaChBac-eGFP. Untagged NaChBac constructs included the entire coding sequence of NaChBac. All cDNAs were subcloned into pCS2+ and pUAST plasmids for Xenopus oocyte expression and Drosophila P-element transformation, respectively. Drosophila embryos were microinjected with pUAST-NaChBac or pUAST-NaChBac-eGFP constructs as described previously (Brand and Perrimon, 1993) to obtain multiple independent chromosomal insertions of UAS-NaChBac and NaChBac-eGFP transgenes. Oocyte expression and two-electrode voltage-clamp recording was performed in a standard low-potassium extracellular bath solution with 0.3–1.2 MΩ electrodes filled with 3 m potassium chloride (KCl) according to standard procedures as described previously (Nitabach et al., 2001). The voltage step protocols to measure NaChBac currents were adapted from Ren et al., 2001. NaChBac currents in larval muscle fiber 6 (third instar) of animals homozygous for the mesodermal 24B-GAL4 driver and containing one copy of the UAS-NaChBac4 transgene were measured by two-electrode voltage clamp. Dissections and recordings were performed at room temperature (22 ± 2°C) in modified hemolymph-like saline (Stewart et al., 1994) containing 0 mm calcium (Ca2+) to eliminate Ca2+ currents. Voltage electrodes were filled with 3 m KCl and had resistances of 7–9 MΩ. The shafts of the current electrodes were filled with 3 m K-acetate and the tips were backfilled with 2 m cesium chloride (CsCl). Final resistances were 3–5 MΩ. Before recording, CsCl was introduced intracellularly through iontophoresis (1 Hz; 500 ms; +5 nA pulses for 5 min) to block K+ currents. pdf-GAL4, UAS-dORKΔ-NC, and 24B-Gal4 fly lines are as described previously (Brand and Perrimon, 1993; Blau and Young, 1999; Renn et al., 1999; Nitabach et al., 2002).
Circadian behavioral analysis. Locomotor activity of individual flies was measured using the TriKinetics infrared beam-crossing system recording total crosses in 15 min bins as described previously (Nitabach et al., 2002). Raw activity histograms were analyzed for circadian rhythms using Actimetrics (Wilmette, IL) Clocklab software. Lomb–Scargle periodograms were constructed according to Van Dongen et al. (1999), and significant circadian rhythmicity was defined as presence of a peak in periodogram power that extends above the p = 0.05 significance line.
Immunocytochemistry. Adult brains were processed for anti-period (PER) immunocytochemistry using the same antibody and as described previously (Price et al., 1998; Nitabach et al., 2002). Adult brains were processed for anti-PDF immunocytochemistry as described previously (Price et al., 1998; Nitabach et al., 2002) using an anti-PDF antibody raised in rabbit against amidated chemically synthesized PDF (CNSE-LINSLLSLPKNMNDA), and for anti-par domain protein 1 (PDP1) immunocytochemistry using the same antibody and as described previously (Cyran et al., 2003). PER was visualized using an HRP-conjugated secondary antibody followed by diaminobenzene-based colorimetric staining. PDF and PDP1 were visualized using a Texas Red-conjugated secondary antibody. Staining intensity of the most darkly anti-PER-stained cell in the sLNV, lateral dorsal subset of pacemaker neurons (LND), and DN1 subgroups in each hemisphere was quantified on a scale from 0 to 4 by a scorer blind to the experimental group to which any particular brain belonged. A value of 0 denotes undetectable staining, 1 denotes just-barely detectable, and 4 denotes maximal staining. This is a standard method for quantifying clock protein expression in Drosophila clock neurons (Kaneko et al., 1997; Stanewsky et al., 1998; Helfrich-Forster et al., 2001; Nitabach et al., 2002, 2005). Because the DN2 subgroup is not invariably distinguishable anatomically from the nearby DN1s, DN2 staining was scored for each brain hemisphere only as the presence or absence of a distinct group of two neurons ventral to the DN1s, and analyzed by comparing the percentage of brain hemispheres in which stained DN2s could be unambiguously identified.
Anti-PDF and anti-PDP1 immunofluorescence images were collected using a charge-coupled device (CCD) camera mounted on a Zeiss (Oberkochen, Germany) Axioskop microscope. An average pixel value was computed for a 30 × 30 pixel region selected from each image by eye to best represent the background staining intensity in the region of tissue adjacent to the dorsomedial processes of the LNVs or the relevant subgroup of clock neuron cell bodies. A pixel-value threshold was chosen for each image individually by eye to include pixels in the dorsomedial processes of the LNVs or the relevant subgroup of clock neuron cell bodies, and to exclude background pixels. The average background pixel value for each image was then subtracted from the threshold-selected pixels of that image to yield the final threshold-selected background-subtracted images, which were pseudocolored (with hotter colors representing greater pixel values) and used for quantitative analysis. Statistical analysis was performed on the integrated pixel values of the threshold-selected background-subtracted images, except that DN2s were scored solely by their detectable presence or absence, as described above for anti-PER-stained specimens. For statistical analysis of anti-PDP1 staining, integrated pixel values were normalized within each cell group and day in constant darkness (DD) to the average absolute integrated pixel value for the time point and genotype with the greatest average.
Results
Functional expression of NaChBac, a bacterial depolarization-activated sodium channel in transgenic Drosophila
In previous studies, we used targeted potassium channel expression as a tool for decreasing membrane excitability in vivo (White et al., 2001; Nitabach et al., 2002, 2005). We have now developed a tool for performing the reciprocal manipulation for increasing membrane excitability. NaChBac is a voltage-gated sodium channel cloned from a halophilic bacterium (Ren et al., 2001), which differs considerably in structure from eukaryotic sodium channels and is, thus, less likely to be sensitive to intracellular signaling pathways that animal cells use to modulate ion channel activity and membrane excitability. When expressed in Xenopus laevis oocytes, NaChBac conducts inward currents that begin to activate near –60 mV and reverse near 65 mV, with slower kinetics of activation and inactivation relative to most animal voltage-gated sodium channels (Fig. 1A,B). In contrast, Drosophila voltagegated sodium channels exhibit a higher threshold for activation of approximately –40 mV and much faster inactivation rates (for review, see Wicher et al., 2001). To examine the effects of increased membrane excitability in vivo in specific cells, we exploited the GAL4-upstream activation sequence (UAS) system to target expression of NaChBac (Brand and Perrimon, 1993). We generated transgenic Drosophila melanogaster containing transgenes with cDNAs encoding either NaChBac or a NaChBac-eGFP fusion protein downstream of five binding sites for the yeast GAL4 transcriptional activator (Fig. 1C). To allow an electrophysiological test for functional expression of NaChBac in transgenic Drosophila, we crossed UAS-NaChBac-GFP flies with 24B-GAL4 enhancer-trap flies, which express GAL4 in all muscle fibers (Brand and Perrimon, 1993). NaChBac-eGFP expression in third-instar larval progeny was verified by fluorescence microscopy (Fig. 1F). NaChBac currents in larval muscle fiber 6 were measured by two-electrode voltage clamp under conditions that isolate sodium currents (see Materials and Methods). We observed robust voltage-gated inward currents with slow kinetics of activation and inactivation that began to activate at approximately –60 mV, peak at approximately –30 mV, and fall off as the transmembrane voltage approaches the reversal potential for sodium, similarly to the currents measured from NaChBac-expressing oocytes (Fig. 1D,E). This confirms that NaChBac expresses functional currents in excitable cells of transgenic Drosophila melanogaster, and indicates that NaChBac expression can be used to increase the electrical excitability of Drosophila neurons.
Figure 1.
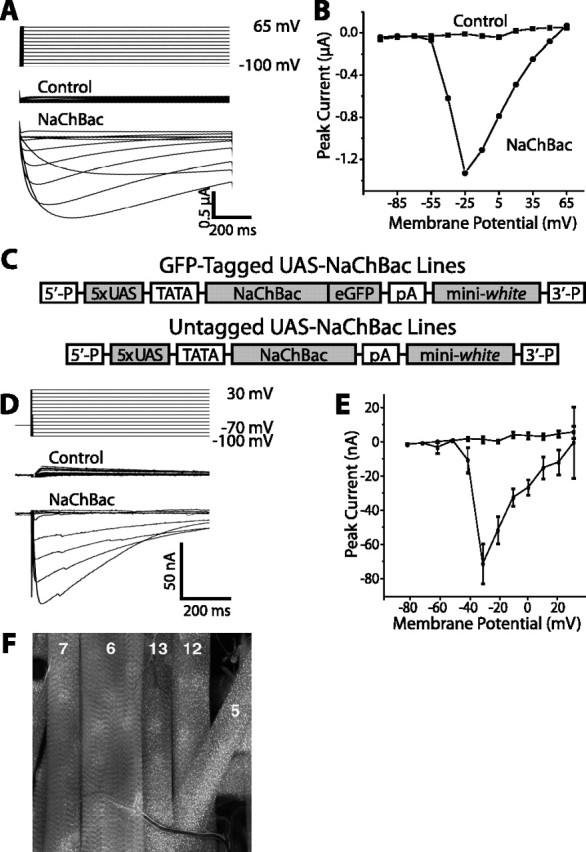
Functional expression of voltage-gated bacterial sodium channel NaChBac in Xenopus laevis oocytes and transgenic Drosophila melanogaster. A, Two-electrode voltage-clamp measurements of transmembrane current in an uninjected Xenopus oocyte (control) or an oocyte injected with cRNA encoding NaChBac (NaChBac). Oocytes were held at –100 mV and stepped in increments of 15 mV to a maximum of 65 mV. Inward NaChBac currents exhibit slower activation and inactivation kinetics than those of Drosophila sodium channels that underlie neuronal action potentials. B, Current–voltage relationships for the currents measured in A. NaChBac begins to activate at approximately –60 mV. After reaching a peak at approximately –25 mV, the current begins to fall off as the transmembrane voltage approaches the reversal potential for sodium. The activation threshold of NaChBac is 20–25 mV lower than that of Drosophila neuronal sodium currents (Wicher et al., 2001). C, Constructs used for P-element transformation of the Drosophila germline. Multiple independent insertion lines were generated containing either NaChBac alone or NaChBac fused to eGFP downstream of the UAS promoter, thus allowing cell-specific expression driven by GAL4. D, E, GFP-tagged NaChBac was expressed in third instar Drosophila larval muscles using the 24B-GAL4 enhancer-trap line and the NaChBac4 insertion line, and currents were measured by two-electrode voltage clamp. Voltage-clamp recordings performed under conditions that isolate Na+ currents show robust voltage-gated inward currents with slow kinetics of activation and inactivation that begin to activate at approximately –50 mV, peak at approximately –30 mV, and fall off as the transmembrane voltage approaches the reversal potential for sodium, similar to the currents measured from NaChBac-expressing oocytes. Control muscle fibers, with the 24B-Gal4 driver alone, lack inward currents, consistent with the absence of Na+ channels in Drosophila muscle. F, Muscle fibers (as numbered) expressing GFP-tagged NaChBac channel are brightly fluorescent. The voltage-clamp measurements depicted were made in muscle fiber number 6. Error bars indicate SE.
NaChBac expression in LNV pacemaker neurons abolishes PDF cycling in dorsomedial sLNV nerve terminals
To determine whether LNV membrane excitability alters phasic anti-PDF immunoreactivity in the dorsomedial terminals and whether manipulation of LNV membrane excitability can be used to interfere with the amplitude and cycling of anti-PDF immunoreactivity in the dorsomedial terminals, we compared PDF oscillations in the dorsomedial terminals of sLNVs in flies expressing either NaChBac or a nonconducting point-mutant form of a truncated Drosophila open-rectifier potassium channel (dORKΔ-NC) (Nitabach et al., 2002) as a negative control.
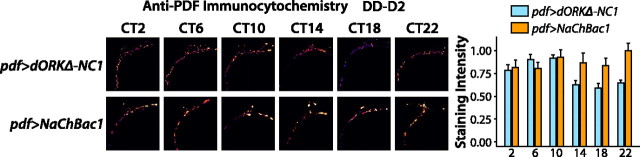
Flies expressing either NaChBac or dORKΔ-NC specifically in the PDF-expressing LNV pacemaker neurons were generated by crossing UAS-NaChBac or UAS-dORKΔ-NC flies with pdf-GAL4 flies (which express GAL4 solely in the PDF-expressing LNVs). Double imaging of NaChBac-eGFP or dORKΔ-NC-GFP fusion protein fluorescence and anti-PDF immunofluorescence in the brains of progeny from these crosses demonstrates colocalization of NaChBac or dORKΔ-NC and PDF in the cell bodies and processes of the small and large LNVs, but not in any other neurons in the fly brain (data not shown). Flies were kept in constant darkness after entrainment in diurnal 12 h light/dark conditions, and processed for single-label imaging of anti-PDF immunofluorescence in the dorsal medial terminals of the LNvs at 4 h intervals [circadian time (CT) 2, CT6, CT10, CT14, CT18, and CT22] on circadian day 2. The time points for data collection were chosen to capture potential differences in phase cycling of PDF accumulation in the dorsomedial terminals between the two experimental groups. Control flies expressing dORKΔ-NC in the LNVs exhibit significantly greater anti-PDF immunofluorescence in the dorsomedial terminals of the sLNVs at CT2, CT6, and CT10 during subjective day, as compared with subjective night at CT14, CT18, and CT22 of circadian day 2 (p < 0.05; ANOVA with Tukey–Kramer paired comparison) (Fig. 2). Thus, anti-PDF immunofluorescence in the dorsomedial terminals of the sLNVs exhibits cycling. In contrast, anti-PDF immunofluorescence in the dorsomedial terminals of the sLNVs of experimental flies expressing NaChBac in the LNVs does not vary significantly from subjective day to night of circadian day 2, and is maintained at constitutively higher levels statistically indistinguishable (p > 0.20; ANOVA with Tukey–Kramer paired comparison) from that exhibited by control flies during the subjective day of circadian day 2 (CT2, CT6, and CT10).
Figure 2.
NaChBac expression in LNV pacemaker neurons abolishes cyclic accumulation of PDF in dorsomedial LNV nerve terminals. pdf-GAL4 virgin female flies were crossed with UAS-NaChBac or UAS-dORKΔ-NC males. Progeny of the indicated genotypes were entrained in 12 h LD conditions followed by DD and processed for anti-PDF immunofluorescence at the indicated times on circadian day 2 (genotypes are indicated with “UAS” and “GAL4” omitted for simplicity; numbers denote specified independent insertion lines for the NaChBac and dORKΔ-NC transgenes). After collection with a CCD camera on a Zeiss Axioskop microscope, images were thresholded to exclude pixels not in the LNV processes, and then the background-subtracted intensities of remaining pixels were integrated over the image, resulting in the plotted integrated pixel intensities (mean±SE). Pseudocolor images depict the background-subtracted pixels remaining after thresholding, with hotter colors representing greater pixel intensity. The dorsomedial LNV terminals of control pdf>dORKΔ-NC1 flies expressing the nonconducting point-mutant potassium channel dORKΔ-NC in the LNVs exhibit a significantly greater anti-PDF immunofluorescence at CT2, CT6, and CT10 than at CT14, CT18, and CT22 (p < 0.05; ANOVA with Tukey–Kramer multiple-comparison test). In contrast, anti-PDF immunofluorescence in the LNV terminals of experimental pdf>NaChBac1 flies expressing NaChBac in the LN Vs did not significantly vary from day to night and was maintained for both time points at a high level statistically indistinguishable from that exhibited by control LNVs at the time points corresponding to subjective day (p > 0.20; ANOVA with Tukey–Kramer multiple-comparison test). n > 12 hemispheres for each experimental group. Error bars indicate SE.
NaChBac expression in LNV pacemaker neurons induces complex free-running behavioral rhythms
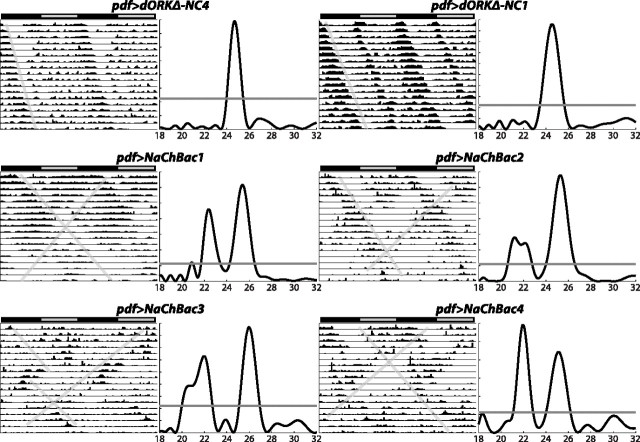
To assess the functional role of regulated membrane activity by the LNV pacemaker neurons, we examined the free-running locomotor activity patterns of flies expressing either NaChBac or control dORKΔ-NC specifically in the PDF-expressing LNV pacemaker neurons. Flies were entrained for 5 d in 12 h light/dark conditions and then released into constant darkness for automated monitoring of the locomotor activity of individual flies for 17 d. Control flies expressing dORKΔ-NC in the LNVs exhibit steady free-running circadian rhythms of locomotor activity with a single statistically significant period of ∼24.5 h (Figs. 3, 4A,B). Actograms of representative flies expressing dORKΔ-NC from two independent transgene chromosomal insertions are shown in the top row of Figure 3, with the angled lines across the actogram providing a visual cue to the free-running rhythms. Lomb–Scargle periodograms computed for these same flies show single statistically significant peaks, with the horizontal lines across the periodograms indicating the p = 0.05 significance threshold (Fig. 3). Of 31 dORKΔ-NC-expressing flies examined, all of them exhibited a single statistically significant periodogram peak (Fig. 4A,B).
Figure 3.
NaChBac expression in LNV pacemaker neurons induces complex free-running behavioral rhythms with multiple superimposed periods. Double-plotted locomotor actograms and Lomb–Scargle periodograms are shown, spanning 17 d in DD of representative male progeny of the indicated genotypes after release from diurnal 12 h light/dark entraining conditions. The bar above each actogram indicates subjective day (gray) and subjective night (black), and the angled lines across the actograms indicate free-running rhythms. Periodograms determined by Lomb–Scargle analysis (Van Dongen et al., 1999) corresponding to each actogram show activity power in arbitrary units (y-axis) as a function of period in hours (x-axis), with the horizontal line across the periodograms indicating the p=0.05 statistical significance level. pdf>dORKΔ-NC control flies expressing dORKΔ-NC in the LNVs exhibit a single statistically significant free-running rhythm of locomotor activity. In contrast, many pdf>NaChBacflies expressing NaChBac in the LNVs exhibit multiple statistically significant superimposed free-running rhythms of locomotor activity, mathematically defined by periodogram peaks extending above the p = 0.05 threshold and separated by intervening regions that dip below the p = 0.05 threshold.
Figure 4.
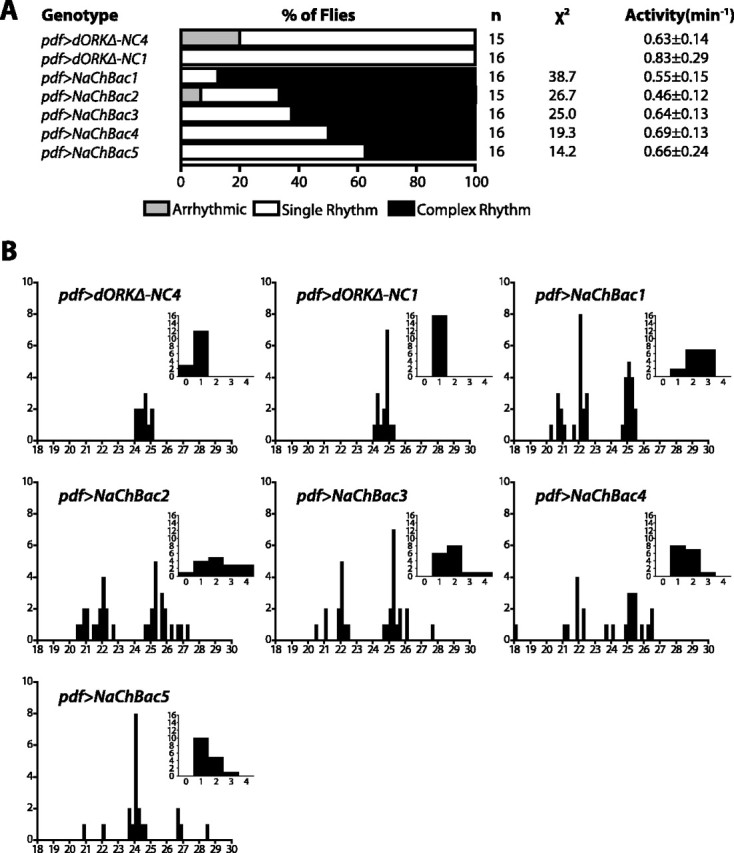
Summary of behavioral analysis of flies expressing either NaChBac or dORKΔ-NC in the LNV pacemaker neurons. A, Control flies expressing dORKΔ-NC never exhibit more than one statistically significant periodogram peak for p = 0.05 (single rhythm; white bar), and a small proportion are arrhythmic (light gray bar). In contrast, experimental flies expressing NaChBac frequently exhibit multiple significant peaks (complex rhythm; black bar), with the exact frequency depending on the particular UAS-NaChBac transgene chromosomal insertion. The difference in proportion of behavioral phenotypes between each of the NaChBac-expressing genotypes and the pooled dORKΔ-NC-expressing genotypes were all highly statistically significant; for each of the listed χ2 values, p < 0.001. Average activity levels in units of beam-crossings per minute are shown ± SD. There were no statistically significant differences in average activity among the groups (one-way ANOVA; p > 0.05). B, Histograms of the aggregate number of statistically significant (p < 0.05) periodogram peaks for flies of the indicated genotypes as a function of period in hours. The distribution of periods of control flies expressing dORKΔ-NC show a single peak centered between 24 and 25 h. In contrast, the distribution of periods of pdf>NaChBac flies show at least two peaks: one between 25 and 26 h and one near 22 h. In the case of the NaChBac1 and NaChBac2 insertions lines, which exhibit the greatest frequency of complex rhythmicity, there may be a third peak between 20 and 21 h. The NaChBac5 insertion line, which exhibits the lowest frequency of multiple rhythmicity, shows a single major peak between 24 and 25 h similar to that of the control flies, but with a number of short-period and long-period outliers. The distribution of periods for each of the experimental groups was significantly different from the distribution of periods for the pooled control groups (χ2; p < 0.001; χ2 > 29). The insets show histograms of the number of flies as a function of the number of statistically significant periodogram peaks. The distribution of the number of periodogram peaks for each of the experimental groups was significantly different from the distribution of periods for the pooled control groups (χ2;p< 0.0025; t >14).
In contrast, experimental flies expressing NaChBac in the LNVs frequently exhibit multiple superimposed free-running circadian rhythms of locomotor activity with different periods (Figs. 3, 4A,B). Actograms of representative flies expressing NaChBac from four independent transgene insertions are shown in the bottom two rows of Figure 3. The angled lines on the actograms point out the presence of multiple free-running rhythms in single flies, some with periods shorter than 24 h and some longer. The corresponding periodograms exhibit multiple significant peaks for each fly, with one longer-than-normal period of ∼25.5 h, and one shorter-than-normal period of ∼22 h. The percentage of flies that exhibit multiple free-running rhythms of locomotor activity varies depending on the particular independent UAS-NaChBac transgene insertion, from 87.5% of pdf>NaChBac1 flies to 37.5% of pdf>NaChBac5 flies (Fig. 4A, multiple rhythms depicted as black bars). This range of penetrance is most likely attributable to chromosomal position effects on the level of transcription of the transgene, and provides an “allelic series” with NaChBac1 strongest (i.e., of probable highest expression) and NaCh-Bac5 weakest (i.e., of probable lowest expression). In contrast, multiple free-running rhythms were not observed in any of the control pdf>dORKΔ-NC flies, which most frequently exhibited single free-running rhythms (Fig. 4A, single rhythms depicted as white bars). The differences in proportion of behavioral phenotypes between each of the NaChBac-expressing genotypes and the pooled dORKΔ-NC-expressing genotypes were all highly statistically significant (χ2 >14; p < 0.001).
Some of the NaChBac-expressing flies exhibit three significant rhythms, with periods of ∼21, ∼22.5, and ∼25 h, as can be seen from histograms plotting the number of significant periodogram peaks as a function of period and the number of flies as a function of number of significant periods (Fig. 4B). The distribution of periods for each of the NaChBac-expressing genotypes was significantly different from the distribution of periods for the pooled dORKΔ-NC-expressing genotypes (χ2; p < 0.001; χ2 > 29), and the distribution of the number of significant periods for each of the NaChBac-expressing genotypes was significantly different from the distribution of the number of significant periods for the pooled dORKΔ-NC-expressing genotypes (χ2; p < 0.0025; χ2 > 14). Interestingly, there are no significant differences among any of the control and pdf>NaChBac flies in overall locomotor activity over the 17 d test period (one-way ANOVA; p > 0.05) (Fig. 4A), suggesting the existence of an intrinsic regulator of the amount of daily activity that is independent of the circadian system.
Coexpression of NaChBac rescues behavioral arrhythmicity induced by expression of inward-rectifier potassium channel
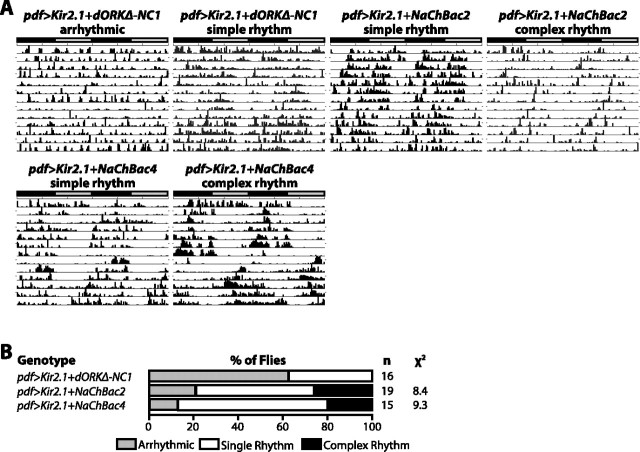
To confirm that expression of NaChBac in Drosophila pacemaker neurons increases membrane excitability, we examined the effect of coexpression of NaChBac and Kir2.1, a mammalian inward-rectifier potassium channel known to hyperpolarize the plasma membrane and decrease neuronal excitability when expressed in transgenic Drosophila (Baines et al., 2001; Nitabach et al., 2002, 2005). We have previously shown that decreasing the excitability of the LNVs by Kir2.1 expression induces behavioral arrhythmicity (Nitabach et al., 2002). When coexpressed in the LNVs with dORKΔ-NC, Kir2.1 induces behavioral arrhythmicity in >60% of flies (Fig. 5B, arrhythmicity depicted as gray bars). When coexpressed with NaChBac from either the NaChBac2 or NaChBac4 insertion, the arrhythmicity induced by Kir2.1 is significantly suppressed, with only 12–20% of flies exhibiting behavioral arrhythmicity and most flies exhibiting single rhythms (p < 0.05; χ2) (Fig. 5B, single rhythms depicted as white bars). Representative actograms of Kir2.1-expressing flies in which rhythmicity was rescued by NaChBac coexpression shows that the rescued rhythms tend to be weaker than those exhibited by normal flies (Fig. 5A). Coexpression of Kir2.1 with NaChBac decreases the proportion of flies exhibiting complex rhythmicity induced by NaChBac expression (compare Figs. 4A, 5B, multiple rhythms depicted as black bars).
Figure 5.
Coexpression of NaChBac in the LNV pacemaker neurons rescues the arrhythmic locomotor activity induced by expression of Kir2.1 inward-rectifier potassium channel. A, B, Representative locomotor actograms of male flies of the indicated genotypes and a summary of the percentages of flies exhibiting arrhythmic (light gray bar), single rhythmic (white bar), and complex rhythmic (black bar) locomotor activity. Expression of Kir2.1 in the LNVs decreases the excitability of the LNV plasma membrane (Nitabach et al., 2002) and induces arrhythmic locomotor activity in >60% of flies when coexpressed with dORKΔ-NC. Coexpression of NaChBac from the NaChBac2 or NaChBac4 transgene insertions significantly suppressed the arrhythmicity induced by Kir2.1 (p < 0.02; χ2). Note that coexpression of Kir2.1 with NaChBac decreases the proportion of flies with complex behavioral rhythms induced by NaChBac expression alone (compare with Fig. 4).
NaChBac expression in LNV pacemaker neurons transsynaptically alters the spatiotemporal phase relationships of both par domain protein 1 and period clock proteins in two subsets of downstream dorsal neurons
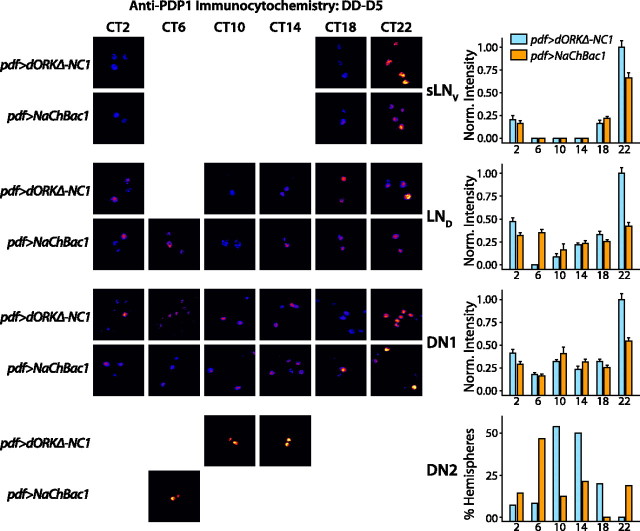
The results described above show that electrical hyperexcitation of the LNVs results in multiple-period behavioral rhythms in constant darkness. To test the hypothesis that electrical hyperexcitation alters the molecular and anatomical substrates of independent oscillators throughout the pacemaker circuit, PER and PDP1 (Cyran et al., 2003) clock protein levels were assayed in the sLNV,LND, DN1, and DN2 neurons of flies expressing NaChBac in the LNVs. Two other cell groups, the large LNVs (lLNVs) and the DN3 neurons, which express both PER and TIM (timeless), were excluded from the analysis. Previous reports suggest that the lLNV neurons in normal control animals do not exhibit molecular clock oscillations under free-running conditions (Yang et al., 1998; Veleri et al., 2003; Grima et al., 2004). The DN3 neurons are small cells that are diffusely distributed and close to the brain surface. The DN3 neurons did not stain as reliably as the other groups, and they were excluded from further analysis. Flies were entrained in diurnal 12 h light/dark conditions and then released into constant darkness for 2 or 5 d before fixation at different circadian times, followed by anti-PER or anti-PDP1 immunocytochemistry (Kaneko et al., 1997; Price et al., 1998; Stanewsky et al., 1998; Blau and Young, 1999; Helfrich-Forster et al., 2001; Nitabach et al., 2002, 2005; Cyran et al., 2003). The use of the 2 d time point for analysis insures measurement of the circadian clock, whereas the 5 d time point was chosen because this was the earliest time that behavioral “splitting” could be discerned reliably (Figs. 3, 4).
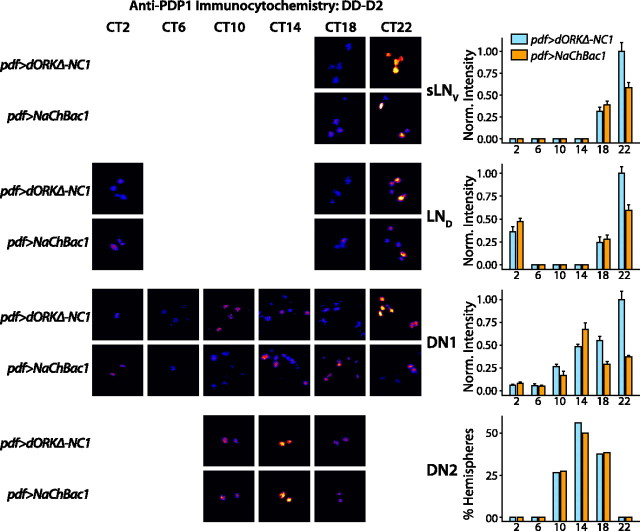
Detection of anti-PDP1 labeling used a fluorescent-conjugated secondary antibody and fluorescence levels were quantified objectively by integrating background subtracted pixel intensities of the clock neuron cell bodies in each anatomical subgroup of each brain hemisphere (see Materials and Methods). Control pdf>dORKΔ-NC1 and experimental pdf>NaChBac1 flies expressing NaChBac in the LNVs exhibit a similar temporal pattern of PDP1 accumulation in the sLNV and LND cell groups, with peak levels at CT22 (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test), late in subjective night, on both day 2 and day 5 in constant darkness (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test) (Figs. 6, 7). However, PDP1 oscillation in pdf>NaChBac1 flies is damped in the sLNV and LND cell groups in comparison with pdf>dORKΔ-NC1 flies (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test). Thus, cell autonomous effects of NaChBac expression are limited to clock-cycling amplitude but not phase shift of the molecular oscillator in sLNV neurons.
Figure 6.
NaChBac expression in LNV pacemaker neurons alters the phase of PDP1 clock protein accumulation in clock neurons on circadian day 2 in constant darkness. pdf-GAL4 virgin female flies were crossed to UAS-NaChBac1 or UAS-dORK Δ-NC1 flies. After entrainment in diurnal 12 h light/dark conditions, pdf>dORKΔ-NC1 and pdf>NaChBac1 progeny were released into constant darkness and then processed for anti-PDP1 immunofluorescence at the indicated CT on the second day in constant darkness. Bar graphs show mean ±SEM normalized integrated anti-PDP1-staining intensities, except for the DN2s, in which case the bar graph shows the percentage of hemispheres in which an anatomically distinguishable pair of stained DN2s was detectable above background (see Materials and Methods). Representative pseudocolored photomicrographs of clock neurons of the indicated cell groups and genotypes at the indicated circadian times are shown for those time points and genotypes where staining was detectable above background or, in the case of the DN2s, where >25% of hemispheres exhibited detectable DN2 staining. Whereas control pdf>dORKΔ-NC1 flies exhibit a similar temporal pattern of PDP1 accumulation in the sLNV, LND, and DN1 groups, with peak levels at CT22 (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test), late in subjective night, the DN2s exhibit a peak of accumulation centered at CT14 (p < 0.05;χ2), 8 h earlier. pdf>NaChBac1 flies expressing NaChBac in the LNVs exhibit temporal patterns of PDP1 accumulation in the sLNV,LND, and DN2 cell groups that are similar to control, except that peak accumulation is significantly less than in controls for the sLNV and LND cell groups (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test). In contrast, pdf>NaChBac1 flies exhibit a significantly different temporal profile of PDP1accumulation from controls in the DN1 group of clock neurons, with peak accumulation 8 h earlier at CT14 (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test). In addition, the amplitude of PDP1 oscillation in the sLNVs, LNDs, and DN1s is damped in the pdf>NaChBac1 flies, with peak accumulation at CT22 significantly less than in controls (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test). The value n > 12 hemispheres for each experimental group.
Figure 7.
NaChBac expression in LNV pacemaker neurons alters the phase of PDP1 clock protein accumulation in clock neurons on circadian day 5 in constant darkness. Control pdf>dORKΔ-NC1 flies exhibit temporal patterns of PDP1 accumulation on day 5 in constant darkness very similar to those on day 2 (Fig. 6), with peak accumulation at CT22 in the sLNV, LND, and DN1 groups (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test), and with peak accumulation centered between CT10 and CT14 in the DN2s (p < 0.05; χ2). Unlike the control flies, pdf>NaChBac1 flies exhibit temporal patterns of PDP1 accumulation in the DN1s and DN2s that are phase shifted relative to day 2 in constant darkness, with DN1 peak accumulation at CT22 (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test) and DN2 peak accumulation at CT6 (p < 0.05; χ2). As on day 2, the amplitude of PDP1 oscillation in the sLNVs, LNDs, and DN1s is damped in the pdf>NaChBac1 flies, with peak accumulation at CT22 significantly less than in controls (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test). The value n > 12 hemispheres for each experimental group. Error bars indicate SE.
In control flies, the DN1s also exhibit a peak at CT22 both on day 2 and day 5 in constant darkness (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test;) (Figs. 6, 7). In marked contrast, the DN1s of pdf>NaChBac1 flies exhibit a peak of PDP1 accumulation on day 2 in constant darkness at CT14, 8 h earlier than in control flies, whereas by day 5, this peak has damped in comparison to control flies and a significant additional peak has appeared at CT22 (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test) (Figs. 6, 7).
The DN2s of control flies exhibit peak PDP1 accumulation at CT14 on day 2 in constant darkness and at CT10–CT14 on day 5 in constant darkness (p < 0.05; χ2) (Figs. 6, 7). Although the DN2s of pdf>NaChBac1 flies exhibit a peak of PDP1 accumulation at CT14 on day 2 in constant darkness, in phase with control flies, by day 5 in constant darkness, PDP1 accumulation peaks in the DN2 neurons of pdf>NaChBac flies at CT6, 4–8 h earlier than in controls (p < 0.05; χ2) (Figs. 6, 7), indicating that the DN2 molecular oscillator of pdf>NaChBac1 flies is running faster than that of control flies.
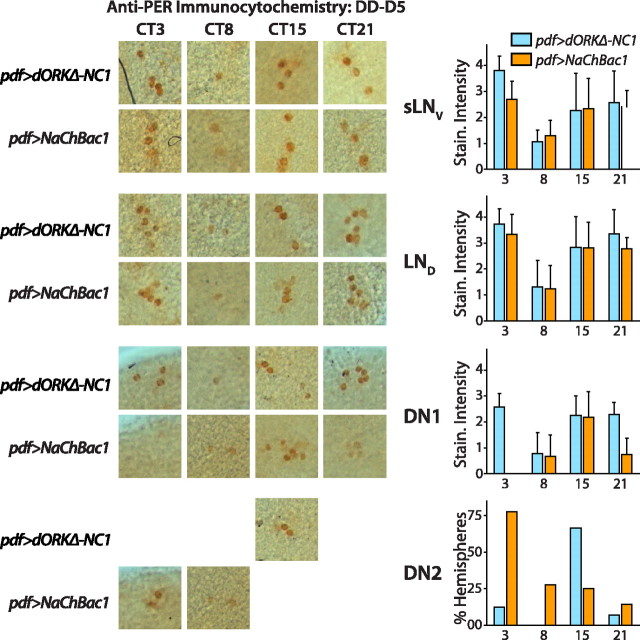
Anti-PER labeling was detected using an HRP-conjugated secondary antibody and levels were quantified using a scorer-blind subjective scale of colorimetric staining intensity applied to the most intensely stained cell in each subgroup of sLNV, LND, DN1, and DN2 neurons of each brain hemisphere (see Materials and Methods). All of the subgroups in control pdf>TM3 flies exhibit robust free-running PER oscillation after 5 d in constant darkness, with trough levels of PER in the sLNV, LND, and DN1 neurons in the second half of the subjective day (Fig. 8). There were no significant differences in the temporal pattern of PER accumulation in the sLNV and LND subgroups between control flies and experimental flies expressing NaChBac in the LNVs (p > 0.05; ANOVA with Tukey–Kramer multiple comparison test) (Fig. 8). Although PER levels in the DN1s of pdf>NaChBac1 flies exhibit a peak during subjective night, as in control flies, this peak is shorter in duration than in controls and occurs earlier (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test) (Fig. 8). The DN2 group of clock neurons in experimental flies expressing NaChBac in the LNVs exhibits a quite different pattern of PER accumulation from both controls and DN1s, with a peak at CT3, 12 h out-of-phase from controls (p < 0.05; χ2) (Fig. 8).
Figure 8.
NaChBac expression in LNV pacemaker neurons alters the phase of PER clock protein accumulation in clock neurons on circadian day 5 in constant darkness. pdf-GAL4 virgin female flies were crossed to UAS-NaChBac1/TM3 flies. pdf>TM3 siblings served as negative controls for pdf>NaChBac1 experimental flies. pdf>TM3 negative controls do not exhibit any complex behavioral rhythms (data not shown), in contrast to complex behavioral rhythms seen in ∼85% of pdf>NaChBac1 flies. After entrainment in diurnal 12 h light/dark conditions, flies were released into constant darkness and then processed for anti-PER immunocytochemistry at the indicated CT on the fifth day in constant darkness. Representative photomicrographs are shown for those time points and genotypes for which staining was detectable above background or, in the case of the DN2s, where >25% of hemispheres exhibited detectable DN2 staining. Bar graphs show mean ± SEM anti-PER staining intensity of the most intensely stained neuron in each of the indicated cell groups as assayed on a subjective scorer-blind arbitrary scale, or the percentage of hemispheres in which an anatomically distinguishable pair of stained DN2s was detectable above background (see Materials and Methods). Control pdf>TM3 flies exhibit a similar temporal pattern of PER accumulation in the sLNV,LND, and DN1 cell groups, with the lowest levels at CT8 (just after the middle of subjective day) and higher levels at other circadian times, with a peak centered around late subjective night/early subjective day (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test). PER accumulation in the DN2s of control flies oscillates out of phase with that of the other cell groups, with peak levels at CT15, early in subjective night (p < 0.05;χ2). pdf>NaChBac1 flies expressing NaChBac in the LNVs exhibit a different pattern of PER accumulation in the DN1s and DN2s from controls, with DN1 PER accumulation peaking at CT15 (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test), and DN2 PER accumulation peaking at CT3 (p < 0.05; χ2). Patterns of PER accumulation in the sLNV and LND cell groups are similar to controls, with lowest levels at CT8 (just after the middle of subjective day) and higher levels at other circadian times, with a peak centered around late subjective night/early subjective day (p < 0.05; ANOVA with Tukey–Kramer multiple comparison test). The value n > 12 hemispheres for each experimental group.
Discussion
Understanding the mechanisms for synchronizing multiple independent neural oscillators in circadian circuits is a key issue in circadian biology. In this study we have provided evidence that the excitability state of the LNV subset of clock neurons plays a critical role in coordinating multiple oscillators in the fly brain. When the LNVs are made electrically hyperexcitable by genetically targeted expression of NaChBac (Fig. 1), transgenic flies exhibit complex free-running behavioral rhythms with multiple periods along with desynchronization of clock protein cycling throughout the pacemaker circuit and disrupted cycling of PDF levels in the dorsomedial terminal projections of the sLNVs (Figs. 2, 3, 4, 6, 7, 8).
We observe similar anti-PDF immunofluorescence in the dorsomedial terminals of the sLNVs, as observed previously, in control flies of the same genetic background (Park et al., 2000). However, anti-PDF immunofluorescence in the dorsomedial terminals of the sLNVs of experimental flies expressing NaChBac in the LNVs is maintained at constitutively higher levels (Fig. 2). This result is unexpected if PDF release at nerve terminals is the only cellular function influenced by alterations in cellular electrical excitability. Although there remains a formal possibility that NaChBac expression does not cause increased electrical excitability in pacemaker neurons, we consider this highly unlikely because of the robust and opposite effects of NaChBac expression compared with open-rectifier potassium-channel expression on behavior, reciprocal rescue of behavior by coexpression, clock oscillation, and direct electrophysiological recordings of muscle and photoreceptor neurons expressing NaChBac (Luan et al., 2006) (Nitabach et al., 2002, 2005). Furthermore, hyperpolarization of LNv membrane potential after the targeted expression of open-rectifier potassium channels to these cells causes accumulations of PDF in the cell bodies of the LNVs (Nitabach et al., 2005), providing further evidence that membrane potential regulates the rates of synthesis and/or trafficking of PDF as well as release. These results together suggest that regulated electrical excitability of the sLNV plasma membrane underlies cycling PDF levels in the dorsomedial terminals, and that rendering the sLNVs hyperexcitable through NaChBac expression disrupts one or more of the cellular processes (synthesis, trafficking, or release) that determine PDF accumulation in the dorsomedial terminals. It remains unclear whether changes in neuronal membrane excitability directly influences PDF accumulation or whether this is caused by indirect effects via the molecular clock, because PDF accumulation appears to be restricted to pacemaker neurons (Helfrich-Forster et al., 2000).
The behavioral and circuit alterations caused by NaChBac expression in the LNVs may be attributable in part to an altered pattern of PDF release or a yet-unidentified neurotransmitter released by the LNVs, or to complex circuit properties of the pacemaker circuit. Regulated membrane electrical excitability of other neuropeptide-secreting neurons of the insect nervous system is known to be essential for appropriate control of the temporal patterns of peptide release (Ewer et al., 1997; Hewes, 1999). PDF may act as an intrinsic coupling signal within the circadian clock circuit that synchronizes multiple oscillators that otherwise free-run independently. This interpretation is consistent with a synchronizing role for PDF proposed on the basis of gradual phase dispersal within the sLNV subgroup of Pdf01-null mutant flies in constant darkness (Lin et al., 2004). In addition, our results are consistent with the idea that temporally regulated PDF release by the LNVs synchronizes the circuit, and are inconsistent with the hypothesis that PDF plays a purely permissive role.
Recent electrophysiological evidence in another insect suggests a mechanism for PDF- and GABA-mediated synchronization of multiple oscillators of pacemaker circuits (Schneider and Stengl, 2005). Extracellular multiunit recordings of the candidate circadian neurons in excised preparations of the cockroach accessory medulla exhibit ultradian oscillatory action potential firing that is synchronized by local application of pressure ejected PDF and GABA through glass micropipettes or bath applied GABA (Schneider and Stengl, 2005). Similarly, circadian neurons in the fly may fire in PDF-regulated assemblies. Although there is as yet insufficient electrophysiological evidence to allow direct comparison of our results in Drosophila with this recent finding in the cockroach. This raises the interesting possibility that NaChBac expression in the Drosophila LNVs may result in desynchronized firing of pacemaker neurons throughout the circuit, starting with the LNVs themselves. This would be consistent with the biophysical property of NaChBac of low-threshold voltage activation (Fig. 1). Interestingly, similar mechanisms for oscillator coupling at the circuit level may also be important in mammals. GABA also modulates phase coupling between the ventral and dorsal oscillators in brain slices prepared from the rat SCN (Albus et al., 2005).
The behavioral results shown in Figures 3 and 4 confirm that the Drosophila circadian control circuit contains multiple clocks capable of oscillating independently and capable of independently controlling the pattern, but not the amount, of locomotor activity (Helfrich, 1986; Yoshii et al., 2004). They further indicate that properly regulated electrical excitability of the LNVs (and perhaps of particular importance, the LNVs) is required to synchronize these multiple clocks throughout the pacemaker neuronal circuit. The synchronization of multiple oscillators appears to be necessary to generate coherent single-period behavioral rhythms.
The reciprocal suppression by NaChBac of the arrhythmicity induced by Kir2.1, and by Kir2.1 of the complex rhythmicity induced by NaChBac (Fig. 5), strongly supports the interpretation that NaChBac and Kir2.1 have opposite effects on the electrical excitability of the LNVs, with Kir2.1 decreasing excitability and NaChBac increasing excitability. When expressed individually in the LNVs, K+ channels and Na+ channels have opposite behavioral effects: hyperpolarizing K+-channel expression results in arrhythmic behavior, whereas depolarizing Na+-channel expression results in hyper-rhythmic behavior. The coexpression of these two channel types together results in functional reciprocal compensation, yielding nearly normal behavior.
In our previous studies, we manipulated LNV membrane potential to be hypoexcitable through the targeted expression of modified open-rectifier or inward-rectifier potassium channels (Nitabach et al., 2002, 2005). This caused behavioral arrhythmicity and cell autonomous dampening of the free-running molecular clock in LNV neurons in constant darkness, along with no discernable changes in the cycling of the molecular clock in downstream pacemaker neuronal subgroups at circadian day 2. Our results are consistent with the findings that desynchrony of downstream cell groups does not become apparent in pdf01-null mutant flies until 2 d in constant darkness (Lin et al., 2004). In the present study, LNV hyperexcitability induces trans-synaptic changes in the free-running temporal pattern of clock protein accumulation in the DN1s and DN2s (Figs. 6, 7, 8), thus, the DN neuronal groups appear to be functionally downstream of the LNV neurons in the pacemaker circuit. In negative control flies, the DN1s oscillate in phase with the sLNVs and LNDs, maintaining synchrony on both days 2 and 5 after release into constant darkness from a diurnal 12 h light/dark entraining regime, whereas the DN2s gradually advance from synchrony in 12 h light/dark to a 12 h phase difference by circadian day 5 (Figs. 6, 7, 8). The DN2s of control flies exhibit peak PDP1 accumulation at CT14 on day 2 in constant darkness and at CT10-CT14 on day 5 in constant darkness (Figs. 6, 7). This gradual shift of DN2 PDP1 oscillation from synchrony with the other cell groups in LD (data not shown) to a 12 h phase advance after 5 d in constant darkness is consistent with other published observations of DN2 PER cycling (Veleri et al., 2003). In pdf>NaChBac1 flies expressing NaChBac in the LNVs, the DN1s exhibit a PDP1 molecular peak 8 h earlier than control flies on day 2 in constant darkness, and by circadian day 5 this peak has significantly damped and an additional significant peak has appeared at CT22 (Figs. 6, 7). The DN2s of pdf>NaChBac1 flies exhibit a peak of PDP1 accumulation at CT14 on day 2 in constant darkness, in phase with control flies; by day 5 in constant darkness they peak at CT6, 4–8 h earlier than in controls (Figs. 6, 7). This phase shift suggests that the DN2 molecular oscillator of pdf>NaChBac1 flies is running faster than that of control flies. These differences in the temporal pattern of PDP1 accumulation in the DN1s and DN2s induced by NaChBac expression in the LNVs indicate that properly regulated electrical activity is required for normal patterns of molecular oscillation in these dorsal cell groups.
The DN2s may be capable of independently driving behavioral outputs, and are possibly the cellular substrate for the ∼22 h short-period component of the complex behavioral rhythmicity exhibited by flies expressing NaChBac in the LNVs. The cellular substrates for the ∼25.5 h long-period component are likely to reside in other cells within the circuit. In control pdf>TM3 flies, robust free-running PER oscillation is observed in the sLNV,LND, and DN1 neurons after 5 d in constant darkness, with trough levels of PER in the second half of subjective day (Fig. 8), consistent with previous observations (Yang and Sehgal, 2001; Shafer et al., 2002; Peng et al., 2003; Veleri et al., 2003; Lin et al., 2004). The differences in the spatiotemporal pattern of PER accumulation induced by NaCh-Bac expression in the LNVs shown in Figure 8 confirm, as indicated by the effects on PDP1 accumulation (Figs. 6, 7), that hyperexcitation of electrical activity in the LNVs causes desynchronization of the coupling and phase of molecular oscillation in dorsal clock neurons.
Recent work shows the existence of multiple oscillators distributed throughout the pacemaker circuit in Drosophila (Grima et al., 2004; Stoleru et al., 2004). The present study confirms and extends evidence for multiple oscillators in the pacemaker circuit in Drosophila. The independent oscillators driving the multiple period components of the behavioral rhythms that we observe do not appear to correspond directly to the “morning” and “evening” oscillators, which have been localized to the LNVs and LNDs, respectively (Grima et al., 2004; Stoleru et al., 2004). Our results emphasize that the activity of the LNVs controls the synchronization of independent oscillators throughout the pacemaker circuit. The normal pattern of DN1 and DN2 clock oscillation requires properly regulated electrical excitability of the LNVs. Further, our results suggest that the DN2s, and at least some other cell groups, possess independent output pathways to the downstream locomotor circuitry.
We have introduced a novel method for inducing electrical hyperexcitability in neurons of interest by the expression of the low-threshold voltage-gated sodium channel NaChBac. This method is likely to be useful for the analysis of other neural circuits. In a previous study (Luan et al., 2006), we further demonstrate the utility of the NaChBac channel for enhancing excitability in other neurons. Targeted expression of ion channel subunits in vivo provides a powerful means for precisely perturbing neuronal membrane excitability to probe the role of activity on neuronal development and function (Baines et al., 2001; White et al., 2001; Nitabach et al., 2002; Borodinsky et al., 2004). The need for more precise in vivo“circuit-breaking” methods was noted first by Francis Crick (1988). This goal began to be realized shortly thereafter by investigators who have used the exogenous expression of ion channels in vertebrate and invertebrate neurons (Jones and Ribera, 1994; Jones et al., 1995; Baines et al., 2001; White et al., 2001; Nitabach et al., 2002; MacLean et al., 2003). The initial methods to exogenously regulate electrical excitability in neurons in vivo have used potassium channel expression to electrically silence neurons. More recently, new methods have been developed to electrically hyperexcite neurons in vivo (Borodinsky et al., 2004; Lima and Miesenbock, 2005). Exogenous manipulation of electrical excitability within specific Drosophila neurons can be combined with finer parsing of neural circuits using GAL80 and other genetic approaches.
Expression of ion channel subunits from exogenous sources has been used in vivo as a method for rescuing cardiomyocytes from disorders of electrical excitability (Brunner et al., 2003; Kodirov et al., 2003). However, we are not aware of any studies demonstrating the feasibility of such in vivo approaches to disorders of electrical excitability in neurons and behavioral rescue. We have shown that aberrations of electrical excitability in Drosophila neurons, either hyperexcitability induced by NaChBac or hypoexcitability induced by Kir2.1, can be rescued by coexpression of an ion channel with an opposite effect on excitability. This provides reason to believe that such an approach to neurological disorders of aberrant electrical activity such as epilepsy might indeed be feasible.
Footnotes
This work was supported by National Science Foundation Grants IBN-0323466 and IBN-0092753, National Institutes of Health (NIH) Grant R01-NS046750 (T.C.H.), an Individual Postdoctoral NIH National Research Service Award, Yale University School of Medicine start-up funds (M.N.N.), and the Intramural Research Program of the NIH National Institute of Mental Health (B.H.W.). We extend special thanks to Justin Blau for his insightful suggestions throughout this project, provision of reagents, and comments on this manuscript. We also thank Vijay Sharma, Joel Levine, Hugh Piggins, Patrick Emery, and Ravi Allada for comments on experiments and this manuscript, Michael Schwartz for use of optical equipment, Dejian Ren and David Clapham for providing the NaChBac clone, and the Bloomington Stock Center.
Correspondence should be addressed to Todd C.Holmes, Department of Biology, NewYork University, 1009 Silver Center, 100 Washington Square East, New York, NY 10003. E-mail: todd.holmes@nyu.edu.
DOI:10.1523/JNEUROSCI.3915-05.2006
Copyright © 2006 Society for Neuroscience 0270-6474/06/260479-11$15.00/0
References
- Albus H, Vansteensel MJ, Michel S, Block GD, Meijer JH (2005) A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Curr Biol 15: 886–893. [DOI] [PubMed] [Google Scholar]
- Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M (2001) Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci 21: 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Young MW (1999) Cycling vrille expression is required for a functional Drosophila clock. Cell 99: 661–671. [DOI] [PubMed] [Google Scholar]
- Borodinsky LN, Root CM, Cronin JA, Sann SB, Gu X, Spitzer NC (2004) Activity-dependent homeostatic specification of transmitter expression in embryonic neurons. Nature 429: 523–530. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brunner M, Kodirov SA, Mitchell GF, Buckett PD, Shibata K, Folco EJ, Baker L, Salama G, Chan DP, Zhou J, Koren G (2003) In vivo gene transfer of Kv1.5 normalizes action potential duration and shortens QT interval in mice with long QT phenotype. Am J Physiol 285: H194–H203. [DOI] [PubMed] [Google Scholar]
- Crick F (1988) What mad pursuit: a personal view of scientific discovery. New York: Basic Books.
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J (2003) vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112: 329–341. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Meyer J, Carpino Jr A, Schwartz WJ (2000) Antiphase oscillation of the left and right suprachiasmatic nuclei. Science 290: 799–801. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Cambras T, Schwartz WJ, Diez-Noguera A (2004) Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus. Curr Biol 14: 796–800. [DOI] [PubMed] [Google Scholar]
- Ewer J, Gammie SC, Truman JW (1997) Control of insect ecdysis by a positive-feedback endocrine system: roles of eclosion hormone and ecdysis triggering hormone. J Exp Biol 200: 869–881. [DOI] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F (2004) Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431: 869–873. [DOI] [PubMed] [Google Scholar]
- Helfrich C (1986) Role of the optic lobes in the regulation of the locomotor activity rhythm of Drosophila melanogaster: behavioral analysis of neural mutants. J Neurogenet 3: 321–343. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C (2004) The circadian clock in the brain: a structural and functional comparison between mammals and insects. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 190: 601–613. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Tauber M, Park JH, Muhlig-Versen M, Schneuwly S, Hofbauer A (2000) Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J Neurosci 20: 3339–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C, Winter C, Hofbauer A, Hall JC, Stanewsky R (2001) The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30: 249–261. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H (2004) Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J Biol Rhythms 19: 35–46. [DOI] [PubMed] [Google Scholar]
- Hewes RS (1999) Voltage-dependent ionic currents in the ventromedial eclosion hormone neurons of Manduca sexta J Exp Biol 202: 2371–2383. [DOI] [PubMed] [Google Scholar]
- Jones SM, Ribera AB (1994) Overexpression of a potassium channel gene perturbs neural differentiation. J Neurosci 14: 2789–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SM, Hofmann AD, Lieber JL, Ribera AB (1995) Overexpression of potassium channel RNA: in vivo development rescues neurons from suppression of morphological differentiation in vitro J Neurosci 15: 2867–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Helfrich-Forster C, Hall JC (1997) Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J Neurosci 17: 6745–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodirov SA, Brunner M, Busconi L, Koren G (2003) Long-term restitution of 4-aminopyridine-sensitive currents in Kv1DN ventricular myocytes using adeno-associated virus-mediated delivery of Kv1.5. FEBS Lett 550: 74–78. [DOI] [PubMed] [Google Scholar]
- Koilraj AJ, Sharma VK, Marimuthu G, Chandrashekaran MK (2000) Presence of circadian rhythms in the locomotor activity of a cave-dwelling millipede Glyphiulus cavernicolus sulu (Cambalidae, Spirostreptida). Chronobiol Int 17: 757–765. [DOI] [PubMed] [Google Scholar]
- Lehmann-Horn F, Jurkat-Rott K (1999) Voltage-gated ion channels and hereditary disease. Physiol Rev 79: 1317–1372. [DOI] [PubMed] [Google Scholar]
- Lima SQ, Miesenbock G (2005) Remote control of behavior through genetically targeted photostimulation of neurons. Cell 121: 141–152. [DOI] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH (2004) The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci 24: 7951–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Zeddies SS, Takahashi JS (2001) Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell 105: 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Lemon WC, Penbody NC, Pohl JB, Zolensky PK, Wang D, Nitabach MN, Holmes TC, White BH (2006) Functional dissection of a neuronal network required for cuticle tanning and wing expansion in DrosophilaJ Neurosci 26: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean JN, Zhang Y, Johnson BR, Harris-Warrick RM (2003) Activity independent homeostasis in rhythmically active neurons. Neuron 37: 109–120. [DOI] [PubMed] [Google Scholar]
- Nagano M, Adachi A, Nakahama K, Nakamura T, Tamada M, Meyer-Bernstein E, Sehgal A, Shigeyoshi Y (2003) An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J Neurosci 23: 6141–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Llamas DA, Araneda RC, Intile JL, Thompson IJ, Zhou YI, Holmes TC (2001) A mechanism for combinatorial regulation of electrical activity: potassium channel subunits capable of functioning as Src homology 3-dependent adaptors. Proc Natl Acad Sci USA 98: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC (2002) Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109: 485–495. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Sheeba V, Vera DA, Blau J, Holmes TC (2005) Membrane electrical excitability is necessary for the free-running larval Drosophila circadian clock. J Neurobiol 62: 1–13. [DOI] [PubMed] [Google Scholar]
- Park JH, Hall JC (1998) Isolation and chronobiological anaylsis of a neuropeptide pigment-dispersing factor gene in Drosophila melanogaster. J Biol Rhythms 13: 219–228. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA 97: 3608–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M (2003) Drosophila free-running rhythms require intercellular communication. PLoS Biol 1: E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri B, Stengl M (1997) Pigment-dispersing hormone shifts the phase of the circadian pacemaker of the cockroach Leucophaea maderae J Neurosci 17: 4087–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan RK, Pati AK, Agarwal SM (1989) Meal scheduling modulation of circadian rhythm of phototactic behaviour in cave dwelling fish. Chronobiol Int 6: 245–249. [DOI] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW (1998) Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83–95. [DOI] [PubMed] [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE (2001) A prokaryotic voltage-gated sodium channel. Science 294: 2372–2375. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791–802. [DOI] [PubMed] [Google Scholar]
- Schaap J, Albus H, VanderLeest HT, Eilers PH, Detari L, Meijer JH (2003) Heterogeneity of rhythmic suprachiasmatic nucleus neurons: implications for circadian waveform and photoperiodic encoding. Proc Natl Acad Sci USA 100: 15994–15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider NL, Stengl M (2005) Pigment-dispersing factor and GABA synchronize cells of the isolated circadian clock of the cockroach Leucophaea maderae. J Neurosci 25: 5138–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW (2002) Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci 22: 5946–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC (1998) The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95: 681–692. [DOI] [PubMed] [Google Scholar]
- Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF (1994) Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 175: 179–191. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M (2004) Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431: 862–868. [DOI] [PubMed] [Google Scholar]
- Van Dongen HP, Olofsen E, VanHartevelt JH, Kruyt EW (1999) A procedure of multiple period searching in unequally spaced time-series with the Lomb-Scargle method. Biol Rhythm Res 30: 149–177. [DOI] [PubMed] [Google Scholar]
- Veleri S, Brandes C, Helfrich-Forster C, Hall JC, Stanewsky R (2003) A self-sustaining, light-entrainable circadian oscillator in the Drosophila brain. Curr Biol 13: 1758–1767. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM (1995) Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14: 697–706. [DOI] [PubMed] [Google Scholar]
- White BH, Osterwalder TP, Yoon KS, Joiner WJ, Whim MD, Kaczmarek LK, Keshishian H (2001) Targeted attenuation of electrical activity in Drosophila using a genetically modified K+ channel. Neuron 31: 699–711. [DOI] [PubMed] [Google Scholar]
- Wicher D, Walther C, Wicher C (2001) Non-synaptic ion channels in insects: basic properties of currents and their modulation in neurons and skeletal muscles. Prog Neurobiol 64: 431–525. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H (2003) Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302: 1408–1412. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sehgal A (2001) Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron 29: 453–467. [DOI] [PubMed] [Google Scholar]
- Yang Z, Emerson M, Su HS, Sehgal A (1998) Response of the timeless protein to light correlates with behavioral entrainment and suggests a nonvisual pathway for circadian photoreception. Neuron 21: 215–223. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Funada Y, Ibuki-Ishibashi T, Matsumoto A, Tanimura T, Tomioka K (2004) Drosophila cryb mutation reveals two circadian clocks that drive locomotor rhythm and have different responsiveness to light. J Insect Physiol 50: 479–488. [DOI] [PubMed] [Google Scholar]