SUMMARY
Sexually dimorphic traits play key roles in animal evolution and behavior. Little is known, however, about the mechanisms governing their development and evolution. One recently evolved dimorphic trait is the male-specific abdominal pigmentation of Drosophila melanogaster, which is repressed in females by the Bric-à-brac (Bab) proteins. To understand the regulation and origin of this trait, we have identified and traced the evolution of the genetic switch controlling dimorphic bab expression. We show that the HOX protein Abdominal-B (ABD-B) and the sex-specific isoforms of Doublesex (DSX) directly regulate a bab cis-regulatory element (CRE). In females, ABD-B and DSXF activate bab expression whereas in males DSXM directly represses bab, which allows for pigmentation. A new domain of dimorphic bab expression evolved through multiple fine-scale changes within this CRE, whose ancestral role was to regulate other dimorphic features. These findings reveal how new dimorphic characters can emerge from genetic networks regulating pre-existing dimorphic traits.
INTRODUCTION
Sexual dimorphism is widespread in the animal kingdom. Males and females differ in primary reproductive structures and are often distinguished by secondary sexual characteristics, some of which have evolved through sexual selection (Darwin, 1871). Some of the most conspicuous modifications of these secondary traits involve male morphology, such as the lion’s mane and the peacock’s elaborate tail. While the ecological roles of sexually dimorphic traits have been well-studied (Andersson, 1994), relatively little is known about how specific traits are produced or have evolved (Wilkins, 2004).
The development of a trait in one sex and not the other must be the result of differential gene expression. Correlations have been found between gene expression patterns and dimorphic trait production. For example, msxC expression is associated with the development of the male “sword” of the swordtail fish (Zauner et al., 2003) and the male-specific pattern of Sex combs reduced (Scr) correlates with the divergence in sex comb formation, size and morphology among Drosophila species (Barmina and Kopp, 2007). However, the regulatory mechanisms governing these gene expression patterns or the development of any male-limited morphological trait have not been elucidated.
While in vertebrates, sex-specific gene expression is often mediated by sex-specific hormones and their receptors (Robins, 2005; Verrijdt et al., 2003), in C. elegans and D. melanogaster, sex-specific gene expression is mediated by the transcriptional effectors of their sex-determination pathways. In these species, homologous regulatory proteins of their somatic sex-determination pathways bind to CREs for the yolk protein (yp) genes and regulate their female-specific expression (An and Wensink, 1995; Coschigano and Wensink, 1993; Yi et al., 2000; Yi and Zarkower, 1999).
Because of the prominent roles sexually dimorphic traits play in animal behavior and evolution, the origins of sex-limited traits has been of special interest in evolutionary biology. From the standpoint of developmental genetics, the central question has been whether new traits and gene expression patterns arise by the gain of expression of genes in one sex that were not previously expressed in either sex or, alternatively, whether dimorphism evolves by the repression of traits and genes in one sex that were formerly expressed in both sexes?
One promising model for understanding the development and evolution of dimorphic traits is the recently evolved male-specific pigmentation of the melanogaster species group in the genus Drosophila. Males of this clade typically have fully pigmented dorsal cuticular plates (tergites) on abdominal segments A5 and A6 (Figure 1A). In females (Figure 1B), A5 and A6 tergite pigmentation is restricted to a posterior stripe, similar to that present on the tergites of abdominal segments A2-A4 of both sexes. Much is known about the developmental genetics of this pattern in D. melanogaster, which requires the activities of several enzymes involved in pigment production as well as several transcription factors (Wittkopp et al., 2003). Two central regulators of posterior pigmentation are the proteins encoded by the tandemly duplicated genes bab1 and bab2 of the bab locus. Both genes encode DNA-binding proteins (Lours et al., 2003) that act as dominant repressors of pigmentation (Couderc et al., 2002; Kopp et al., 2000). While female pupae express bab in abdominal segments A2-A6, bab expression in males is limited to segments A2-A4, and the relative absence of bab expression in segments A5 and A6 is necessary for their greater pigmentation in males (Kopp et al., 2000). Genetic analyses have implicated the Hox gene Abd-B as a repressor of bab in these posterior segments and suggested that repression of bab is mitigated in females by the activity of dsxF, the sex-specific transcript derived from the doublesex (dsx) locus (Kopp et al., 2000). The male-specific repression of bab appears to have evolved from an ancestral monomorphic condition in which bab was expressed in the posterior of both sexes.
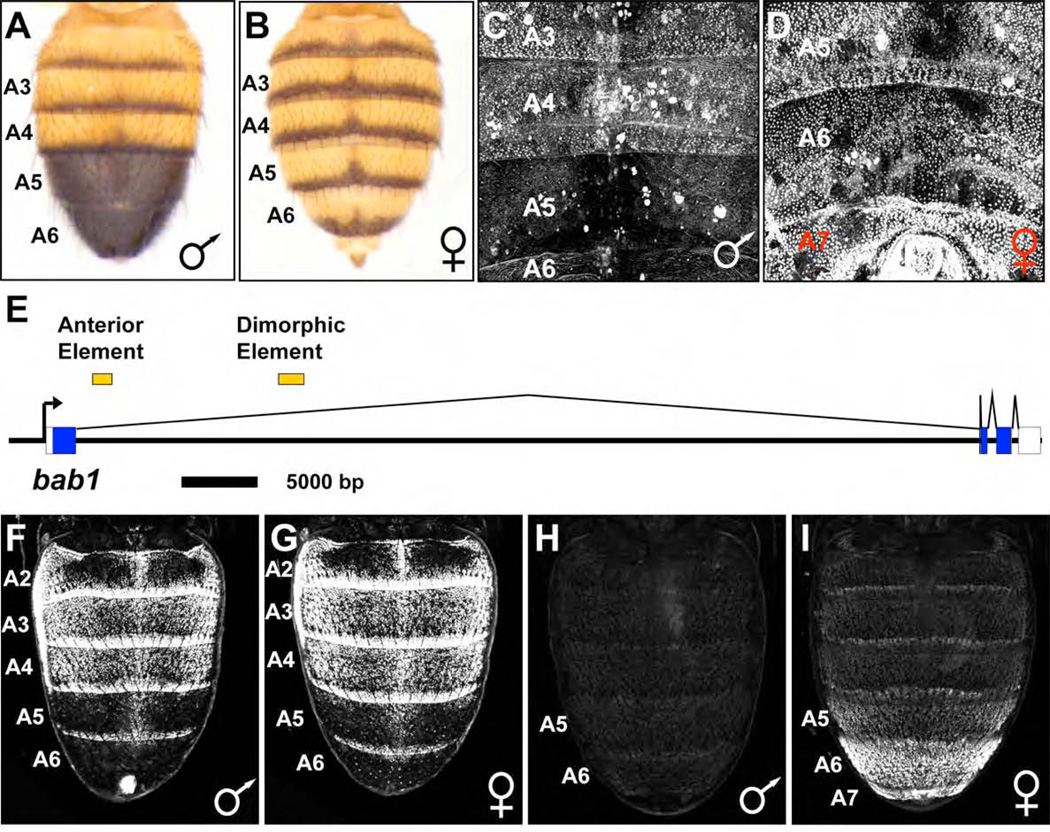
Figure 1. Bab1 expression in the abdomen is regulated by two CREs.
(A and B) Dorsal view of D. melanogaster adult abdomens. Male segments A5 and A6 of are fully pigmented (A). In females, pigmentation of these segments is limited to a posterior stripe (B)
(C and D) Expression of Bab1 in male and female pupae at 72 hours APF. Bab1 expression in males is limited to segments A2-A4 (C), but in females, Bab1 expression extends into segments A5 and A6, as well in the female-specific segment A7 (D).
(E) Two CREs, the anterior element and dimorphic element, reside in the large 1st intron of bab1 and govern Bab expression in the abdominal epidermis.
(F–I) GFP-reporter expression in dorsal pupal abdomens.
(F and G) The anterior element drove GFP-reporter gene activity in segments A2-A4 of both males (F) and females (G).
(H) The dimorphic element was inactive in males.
(I) The dimorphic element drove reporter expression in female segments A5-A7, with levels increasing from the anterior to posterior.
In order to understand the molecular mechanisms by which bab expression is regulated and has evolved, we sought to identify the CREs governing bab expression, to characterize the direct transcriptional regulators of their CREs, and to trace how functional changes in gene expression have occurred in Drosophila evolution. We found that two CREs govern bab expression in the pupal abdomen. These include one element that regulates bab expression in segments A2-A4 of both sexes and a second, dimorphic element that regulates expression in the posterior segments A5-A7 of females. We demonstrate that the dimorphic element is part of a genetic switch that, in combination with the HOX protein ABD-B and the sex-specific activities of the male and female isoforms of the DSX protein, directs female-specific activation and male-specific repression of bab in posterior segments. Surprisingly, we found that both the presence of this dimorphic CRE and its regulation by ABD-B and DSX predated the origin of dimorphic pigmentation. We discovered that the new domain of dimorphic CRE activity required for dimorphic pigmentation evolved from many fine-scale changes within the CRE. Our results show how new dimorphic characters can evolve from the genetic regulatory architecture governing other dimorphic traits.
RESULTS
Bab1 is expressed in a dimorphic pattern
Genetic studies that have shown that both bab1 and bab2 are required for the development of the wild-type D. mel. pigmentation pattern (Couderc et al., 2002; Kopp et al., 2000) and previous work has shown that Bab2 is expressed dimorphically in the pupal abdominal epidermis (Gompel and Carroll, 2003; Kopp et al., 2000). However, the expression of Bab1 has not been fully characterized and it was important to determine whether both proteins were expressed and therefore regulated in similar ways. In order to analyze the expression of Bab1, we developed an affinity-purified polyclonal antibody specific to Bab1 that did not cross-react with Bab2. We determined that during pupal development Bab1 was also expressed in segments A2-A4 of males with the highest levels occurring in A3 and A4 (Figure 1C), whereas in females Bab1 is expressed in segments A2 through A7 (Figure 1D). Therefore, both Bab1 and Bab2 are expressed dimorphically in the abdominal epidermis.
Two cis-regulatory elements direct Bab expression in the abdomen
In order to dissect the molecular mechanisms regulating dimorphic bab expression, we sought to identify the CREs within the bab locus that govern gene regulation in the abdomen. We conducted a systematic screen of the ~150 kb of non-coding sequence between the neighboring trio and CG13912 loci, excluding transposon-derived sequences, for regions with regulatory activity. Overlapping segments of DNA with a typical size of 7 kb were tested for their ability to direct reporter gene expression in the abdomens of transgenic D. mel. pupae (Figure S1A). Consistent with the pleiotropic roles of the Bab transcription factors (Couderc et al., 2002), we identified several separate CREs that drove reporter expression in specific tissues during pupal development, including the oenocytes, legs, bristles, and abdominal muscles (Figure S1).
Most importantly, two separate CREs were identified in the large first intron of bab1 that drove reporter expression in the pupal abdominal epidermis (Figure 1E and Figure S1). One CRE, which we refer to hereafter as the “anterior” element (1,357 base pairs (bp)), drove reporter expression in a monomorphic pattern in segments A2-A5 of both sexes with significantly lower levels of expression in segments A2 and A5 than in segments A3 and A4 (Figure 1F and 1G). The second CRE, referred to as the “dimorphic” element (663 bp), drove a female-specific pattern of reporter expression in segments A5-A7 (Figure 1H and 1I). A construct containing both elements drove reporter gene expression in a spatial pattern similar to the endogenous expression of Bab1 and Bab2 in the epidermis of the abdomen (Figure S1F–G). Since the regulatory elements are located in the first intron of bab1, we conclude that these CREs regulate bab1 in vivo. Furthermore, since no other CREs were found in the entire locus that directed gene expression in the abdomen, and bab2 is expressed in a similar pattern, we infer that the CREs also regulate bab2 expression.
Regulation of Bab expression by Abd-B and Dsx
The segment- and sex-specific activity of the bab CREs suggested that they are regulated by segment- and sex-specific transcription factors. In order to elucidate the factors regulating these CREs, we examined the activity of the anterior and dimorphic elements in mutant genetic backgrounds. The best candidate segment-specific regulator is the product of the Hox gene Abdominal-B (Abd-B), which is expressed at increasing levels from pupal abdominal segment A5 to A7 (Kopp and Duncan, 2002). Furthermore, Abd-B behaves genetically as a repressor of pigmentation and bab expression (Kopp et al., 2000). Consistent with these observations, compared to activity in a wild-type genetic background (Figure 2A), reporter gene expression driven by the anterior element was repressed in both males (Figure 2B and 2C) and females (data not shown) when Abd-B was ectopically expressed in segments anterior to A5. Conversely, the activity of the dimorphic element was expanded by ectopic expression of Abd-B in females (Figure 2D–2F). Ectopic expression of Abd-B had no effect on dimorphic element activity in males (data not shown). Thus, Abd-B behaves genetically as an activator of the dimorphic element and as a repressor of the anterior element.
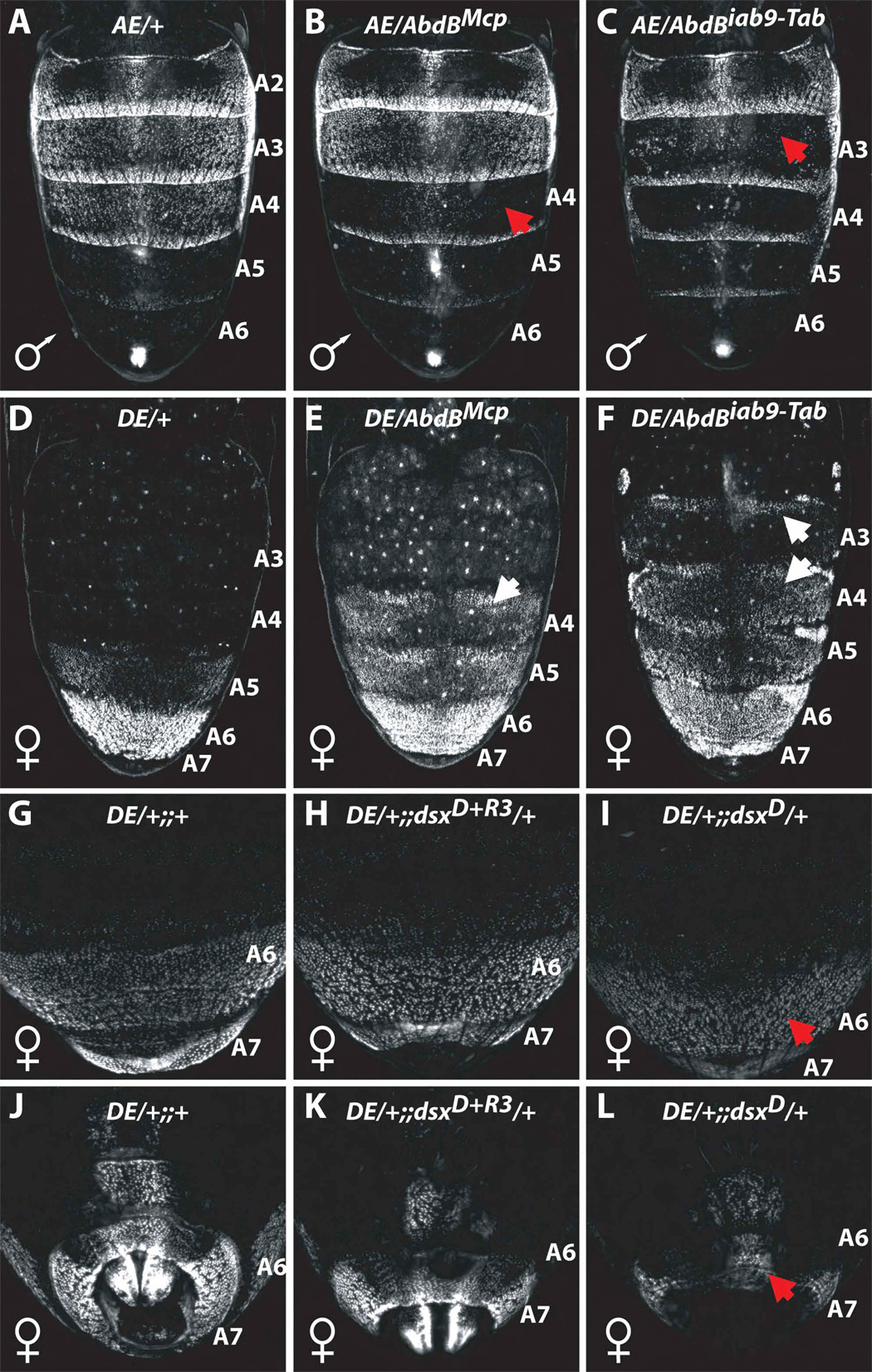
Figure 2. Genetic regulation of bab CRE activity by the Abd-B and Dsx loci.
Images are of dorsal (A–I) and ventral (J–L) abdomens of pupae heterozygous for GFP-reporter constructs. Genotypes are listed at the top of each image. The anterior element is denoted as ‘AE’ and the dimorphic element ‘DE’. Specimens are heterozygous for the AbdBMcp (B and E), AbdBiab9-Tab (C and F), dsxD+R3 (H and K), and dsxD (I and L) mutant allele. Red and white arrows respectively indicate regions where reporter activity in the mutant background is decreased or increased, respectively, compared to the wild-type control.
(A) The AE drove reporter expression in the anterior segments A2-A4.
(B and C) Ectopic expression of Abd-B in segments A4 (B) and A3-A4 (C) resulted in repression of AE activity in these segments.
(D, G and J) The DE drove reporter expression in the posterior segments A5-A7 of females.
(E and F) Ectopic expression of Abd-B in segments A4 (E) and A3-A4 (F) resulted in ectopic DE activity in these segments.
(H and K) In a dsx heterozygous null mutant genetic background, DE activity is indistinguishable from that in a wild-type background.
(I and L) In a chromosomal female intersex, where one dsx allele is producing the male transcript instead of the female, DE activity was reduced to 68±2% in A6 (I) and to 36% in A7 (L) of the activity of the DE in wild-type females.
However, genetic regulation by Abd-B cannot account for the absence of dimorphic element activity in males, as Abd-B expression is monomorphic. One candidate for controlling sex-specific gene expression is the doublesex (dsx) gene. dsx− flies exhibit an intermediate sexual identity (intersex) and their pigmentation on segments A5 and A6 is similar to that of a wild-type male (Jursnich and Burtis, 1993; Kopp et al., 2000). Transcripts from the dsx locus are alternatively spliced between males and females (Burtis and Baker, 1989), and these alternative transcripts encode sex-specific protein isoforms (DSXF and DSXM) which share a common N-terminal DNA-binding domain but have variant C-terminal domains that mediate different cofactor interactions (Garrett-Engele et al., 2002). To test whether dsxM genetically represses dimorphic element activity, we compared reporter activity in a chromosomal female genetic background (dsx>D/+) where the transcript produced by one allele of dsx is spliced as dsxM. In these intersexual pupae, reporter activity in segments A6 and A7 was reduced to 68% and 36% respectively (Figure 2I and 2L) of the activity in a wild-type background (Figure 2G and 2J). This reduction was due to the gain of dsxM activity rather than haplo-insufficieny for dsxF, because reporter activity in a female dsx+/− background (Figure 2H and 2K) was equivalent to that measured in the wild-type background (Figure 2G and 2J). These results demonstrate that sex-specific regulation of the dimorphic element is provided by dsxM acting as a repressor in males, and most likely by dsxF acting as an activator in females. In order to understand the molecular mechanisms of the dimorphic regulation of bab and how it evolved, we next sought to identify which of these regulatory interactions were direct.
ABD-B is a direct segment-specific activator of the dimorphic element
In order to determine if ABD-B is a direct regulator of bab CREs, we examined the anterior and dimorphic element for ABD-B binding sites. ABD-B and paralogous HOX proteins have been shown to preferentially bind to the five base pair motifs TTTAT or TTTAC, and with some affinity to the shorter motif TTAT (Ekker et al., 1994). Within the dimorphic element there were, remarkably, fourteen sites with the core sequence TTTAT or TTTAC (Figure 3A) and eight additional TTAT sites. DNaseI footprinting with purified ABD-B homeodomain (HD) protein identified that most of these sequences were bound (12 of the 14 core sites; Figure S2). To determine whether CRE activity required the full 663 bp of the dimorphic element containing all of these sites, truncation constructs that included subsets of the candidate ABD-B sites were tested for reporter activity. Compared to the wild-type element (Figure 3B), the truncated elements exhibited dramatic decreases in (Figure 3C and 3E) or the elimination (Figure 3D) of CRE activity. These results indicate that sequences required for full CRE activity are distributed throughout the entire dimorphic element.
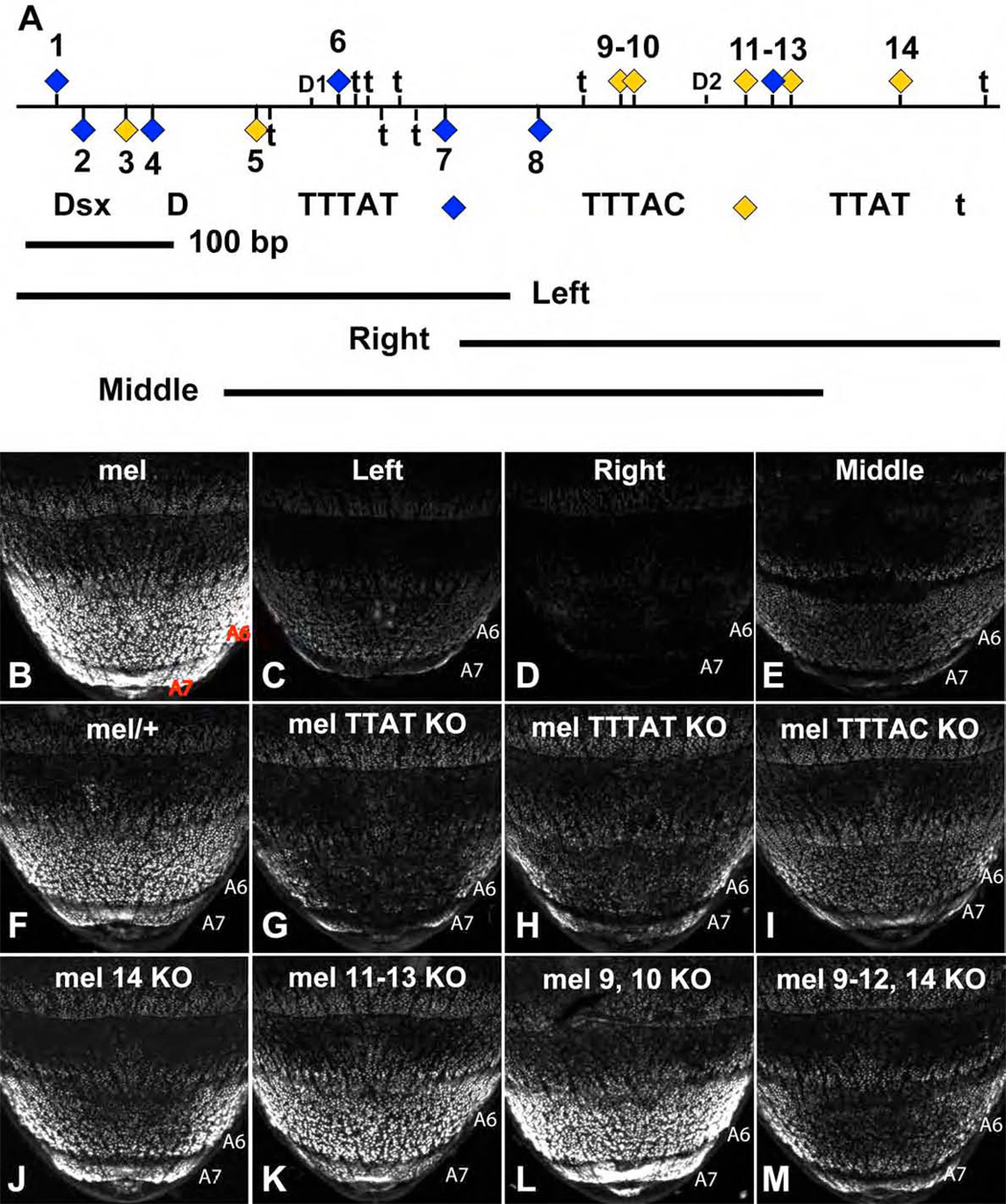
Figure 3. The dimorphic element is directly regulated by ABD-B through multiple binding sites.
(A) Schematic of the minimal D. mel. wild-type (mel) CRE sequence conferring robust female-specific activity, with the location of putative ABD-B (yellow and blue diamonds) and DSX (D1 and D2) binding sites indicated. Additional TTAT (non-TTTAT) motifs are indicated by “t”. The spans of the Left, Right, and Middle sub-constructs are indicated below the schematic.
(B–M) Comparison of GFP-reporter gene activity in transgenic female pupae at 75 hours APF. Activity measurements are represented as the % of the wild-type (mel) female A6 mean ±SEM.
(B) The wild-type dimorphic element drove reporter expression at high levels in A6 and A7.
(C–E) Truncation of the dimorphic element into Left (C), Right (D) and Middle (E) sub-fragments resulted in dramatically decreased reporter activity.
(F) Reporter activity is reduced to 59±2% in pupae heterozygous for the dimorphic element reporter transgene.
(G–L) Activity of dimorphic elements in which subsets of putative ABD-B binding sites have been mutated.
(G) Mutation of all fifteen TTAT sites reduced reporter activity to 9±0%.
(H) Mutation of all seven TTTAT sites reduced reporter activity to 19±2%.
(I) Mutation of all seven TTTAC sites reduced reporter activity to 26±3%.
(J) Mutation of ABD-B site 14 reduced reporter activity to 55±0%.
(K) Mutation of ABD-B sites 11–13 reduced reporter activity to 79±4%.
(L) Mutation of ABD-B sites 9 and 10 had no detectable affect on dimorphic activity.
(M) Mutation of ABD-B sites 9–12 and 14 reduced reporter activity to 26±6%.
In order to determine whether the candidate ABD-B sites were required for CRE activity in vivo, these sequences were mutated alone or in combination (from TTAT/TTAC to CGGC) within the context of the 663 bp dimorphic element, and the effects on reporter activity were analyzed in transgenic female pupae. In order to eliminate the potential variation introduced by the site of transgene insertion, comparisons between wild-type and mutant elements were performed with transgenes inserted into the same chromosomal position. We consistently observed low inter- and intra-transgenic line variation in reporter activity, which enabled us to detect modest quantitative changes in reporter activity.
Mutation of all TTAT sequences (a total of 15 sites) reduced reporter activity to 9±0% (%activity ±SEM) of the wild-type dimorphic element activity in segment A6 and substantially reduced reporter expression in segment A7 (Figure 3G). Mutation of the seven TTTAT sites reduced activity to 19±2% of the wild-type element (Figure 3H). This slightly lesser reduction in the TTTAT site mutant than in the TTAT site mutant indicated that there was some contribution of the TTAT sites to full CRE activity. Mutation of all seven TTTAC sites reduced activity to 26±3% of wild-type activity (Figure 3I), further indicating that regulation of the dimorphic element by ABD-B in vivo is mediated through many binding sites.
In order to better understand the contribution of individual or small groups of ABD-B binding sites to CRE activation, we mutated putative sites individually and in combination and analyzed their effects on reporter activity. Mutation of one or few sites ranged between having a large (Figure 3J), modest (Figure 3K and Figure 6B) or no measurable effect (Figure 3L and Figure 6C) on CRE activity. When more binding sites were mutated in combination, CRE activity was reduced further (Figure 3M). Collectively, these results demonstrate that ABD-B mediates the segment-specific activation of the dimorphic element by the cumulative effects of binding to many sites.
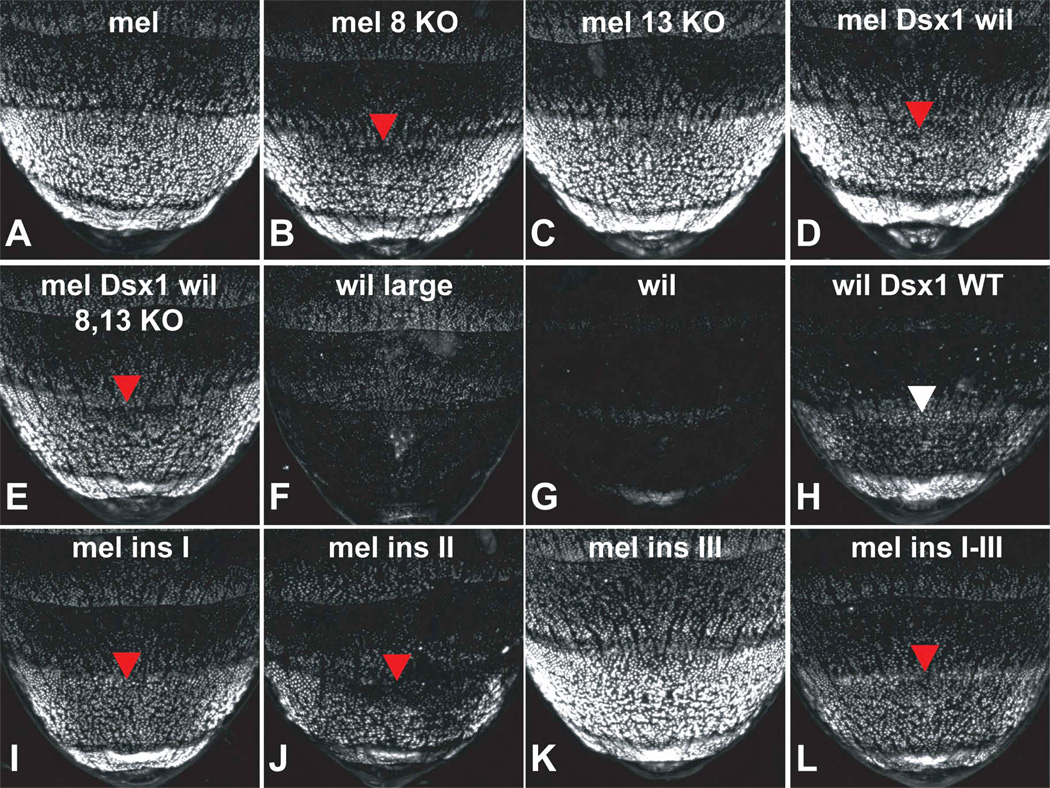
Figure 6. bab expression evolved via remodeling of the dimorphic element.
In all panels, GFP-reporter expression mediated by D. mel. (A–E and I–L) and D. wil. (F–H) dimorphic elements was assayed in transgenic female pupae at 75 hours APF Activity measurements are represented as the % of the wild-type (mel) female A6 mean ±SEM. Red arrow heads point to dorsal midline regions of A6 where reporter activity was reduced by modification of the D. mel. element. White arrow head points to area of segment A6 where reporter activity was increased by modification of the D. wil. element.
(A) Reporter expression driven by the wild-type D. mel. dimorphic element.
(B) Mutation of ABD-B site 8 reduced reporter activity to 78±5%.
(C) Mutation of ABD-B site 13 had no measurable affect on reporter activity.
(D) Reversal of the Dsx1 site polarity in the D. mel. element reduced reporter activity to 87±2%.
(E) Reversal of Dsx1 site polarity combined with mutation of ABD-B sites 8 and 13 reduced reporter activity to 66±3%.
(F) Reporter expression driven by the D. wil. dimorphic element that includes site 14.
(G) Reporter expression driven by the wild-type D. wil. dimorphic element.
(H) Reversal of Dsx1 site polarity in the D. wil. element resulted in a dramatic gain of reporter activity in segment A6. Activity increased from 1±1% to 34±3% of the wild-type D. mel element.
(I) Insertion of 58 base pairs between ABD-B site 5 and Dsx1 site (region I) reduced reporter expression to 62±3%.
(J) Insertion of 98 base pairs between ABD-B sites 8 and 9 (region II) reduced reporter expression to 41±3%.
(K) Insertion of 57 base pairs between the Dsx2 site and ABD-B site 11 (region III) increased the activity of the wild-type dimorphic element by 37±3%.
(L) Insertion of 58, 98, and 57 base pairs into regions I–III respectively, reduced dimorphic element activity to 44±4%.
In contrast to the dimorphic element, a similar mutational analysis of the bab anterior element provided no evidence for its direct regulation by ABD-B. We conclude that the repression of the anterior element in segments A5 and A6 is mediated indirectly by Abd-B through regulation of other genes. We have therefore focused our subsequent analyses almost exclusively on the regulation and evolution of the dimorphic element.
Sex-specific isoforms of DSX directly regulate female-specific activation and male-specific repression of the dimorphic element
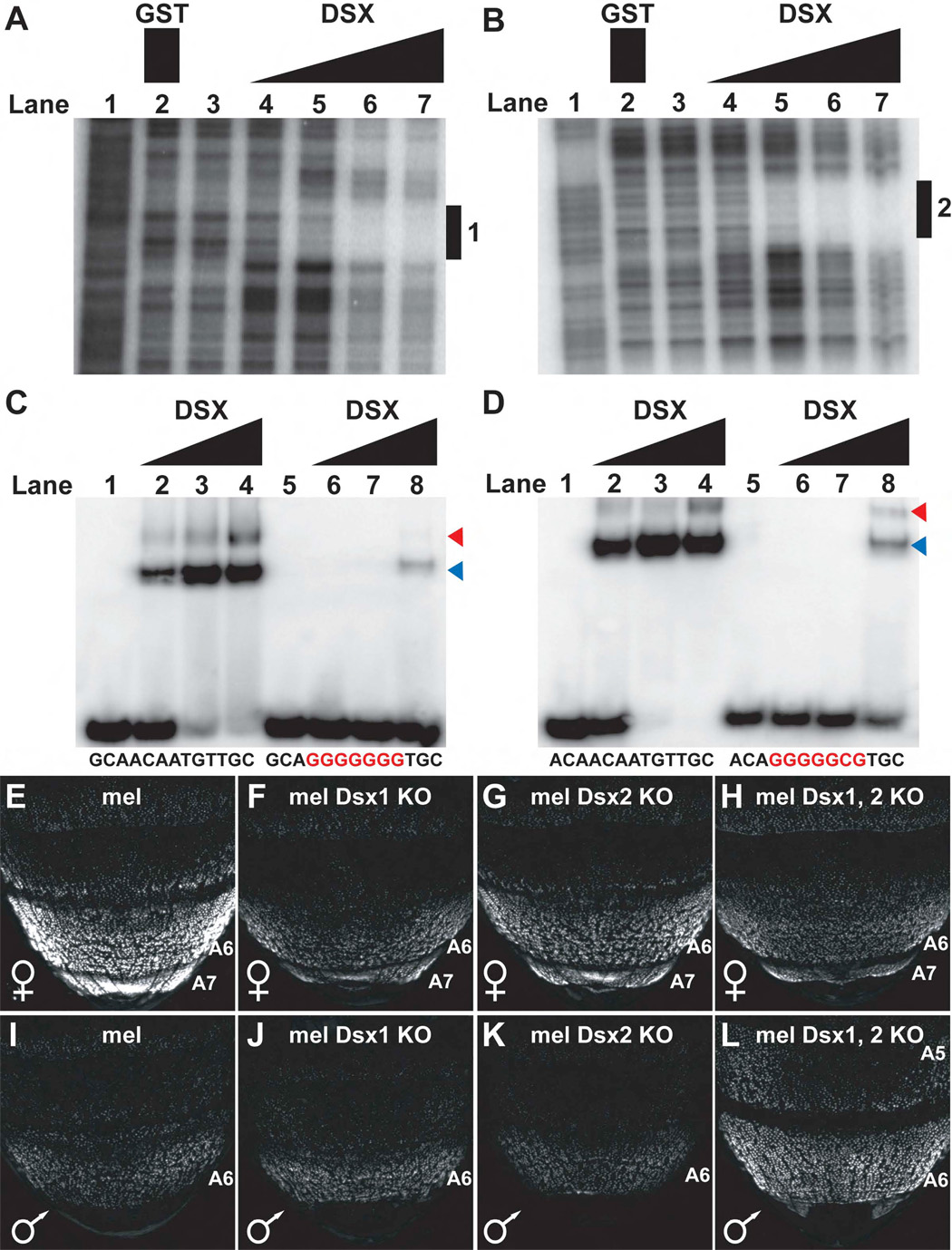
To determine whether the DSX proteins were direct sex-specific regulators of the dimorphic element, DNase I footprinting was performed with the DSX DNA-binding domain (DBD) on the minimal sequence sufficient for full dimorphic activity. Two sites were identified within the dimorphic element that were the only matches to an empirically determined consensus sequence for DSX binding (Erdman et al., 1996). The first site (Dsx1; Figure 4A), resides between ABD-B site 5 and 6 and the second site (Dsx2; Figure 4B), lies between ABD-B sites 10 and 11 (Figure 3A). DSX binding to these two sites was specific (Figure 4C and 4D, lanes 1–4), as mutations in them significantly decreased binding by DSX (Figure 4C and 4D, lanes 5–8).
Figure 4. The dimorphic element is directly regulated by sex-specific isoforms of DSX.
(A and B) DNaseI footprinting analysis of the dimorphic element with a GST-DSX DBD fusion protein identified two distinct sites bound by the DSX DNA-binding domain, referred to as Dsx1 (A) and Dsx2 (B). Amounts of each protein used were as follows: lane 2, 1,000 ng GST only; lane 3, no protein; lane 4, 64 ng DSX DBD; lane 5, 160 ng DSX DBD; lane 6, 400 ng DSX DBD; lane 7, 1,000 ng DSX DBD. A G+A sequencing ladder is included in lane 1. Footprinted regions are indicated by a black rectangle with an adjacent number.
(C and D) EMSAs on annealed oligonucleotide probes containing wild-type (lanes 1–4) and mutant (lanes 5–8) DSX binding sites. Below are the sequences of the wild-type and mutant Dsx1 (C) and Dsx2 (D) binding sites with mutated bases shown in red. For each probe, binding reactions were performed using increasing amounts of the DSX DBD protein (from left to right: 0 ng, 16 ng, 62 ng, 250 ng, and 1,000 ng). Blue and red arrowheads point to the respective locations on the gel of complexes containing a single or pair of DSX DBD monomers bound to the probe.
(E–L) GFP-reporter activity in pupae at 75 hours APF. Activity measurements are represented as the % of the wild-type (mel) female A6 mean ±SEM.
(E and I) Activity of the wild-type dimorphic element was much greater in the female (E) than the male (I).
(F and J) When the Dsx1 site was mutated, activity in the female was reduced to 23±2% (F), while activity in the male was unchanged (J).
(G and K) When the Dsx2 site was mutated, activity in females was reduced to 34±3% (G), while male activity was unchanged (K).
(H and L) When both the Dsx1 and Dsx2 sites were mutated, reporter activity in females was reduced to 24±1% (H), and increased to 53±3% in males (L).
To resolve whether DSX binding to sites Dsx1 and Dsx2 was required for dimorphic element function in vivo, GFP-reporter expression was monitored in transgenic pupae in which one or both of these sites were mutated. The wild-type CRE drove strong expression in segments A6 and A7 of female pupae (Figure 4E) and expression in males was limited to a low level in segment A6 (Figure 4I). When the Dsx1 site was mutated, reporter activity in females was reduced to 23±2% of the wild-type element (Figure 4F). Similarly, when the Dsx2 site was mutated, reporter activity was reduced to 34±3% (Figure 4G). However, neither mutation of the Dsx1 nor of the Dsx2 site alone altered the low level of reporter activity in males (Figure 4J and 4K). When both the Dsx1 and Dsx2 sites were mutated in combination, reporter activity in females was reduced to 24±1% of the wild-type element (Figure 4H), and the reporter activity in male pupae was increased to 53±3% to that of the wild-type dimorphic element in female pupae (Figure 4L).
These data demonstrate that the dimorphic element is directly regulated by the sex-specific isoforms of DSX and that, together with ABD-B, the two regulators and the CRE act as a segment- and sex-specific genetic switch to differentially regulate bab expression in the posterior segments of males and females. DSXF acts in conjunction with ABD-B to activate the CRE in females, whereas the DSXM isoform directly represses activity in males and overrides the positive regulation by ABD-B.
The recent evolution of dimorphic pigmentation and the operation of this genetic switch for controlling bab expression raises the question of how the switch evolved. Is the dimorphic element novel to D. melanogaster lineage? When did binding sites for and regulation by ABD-B and DSX evolve? In order to address these questions, we traced the evolutionary history of the bab CREs in the Drosophila genus.
The dimorphic element has a deep ancestry
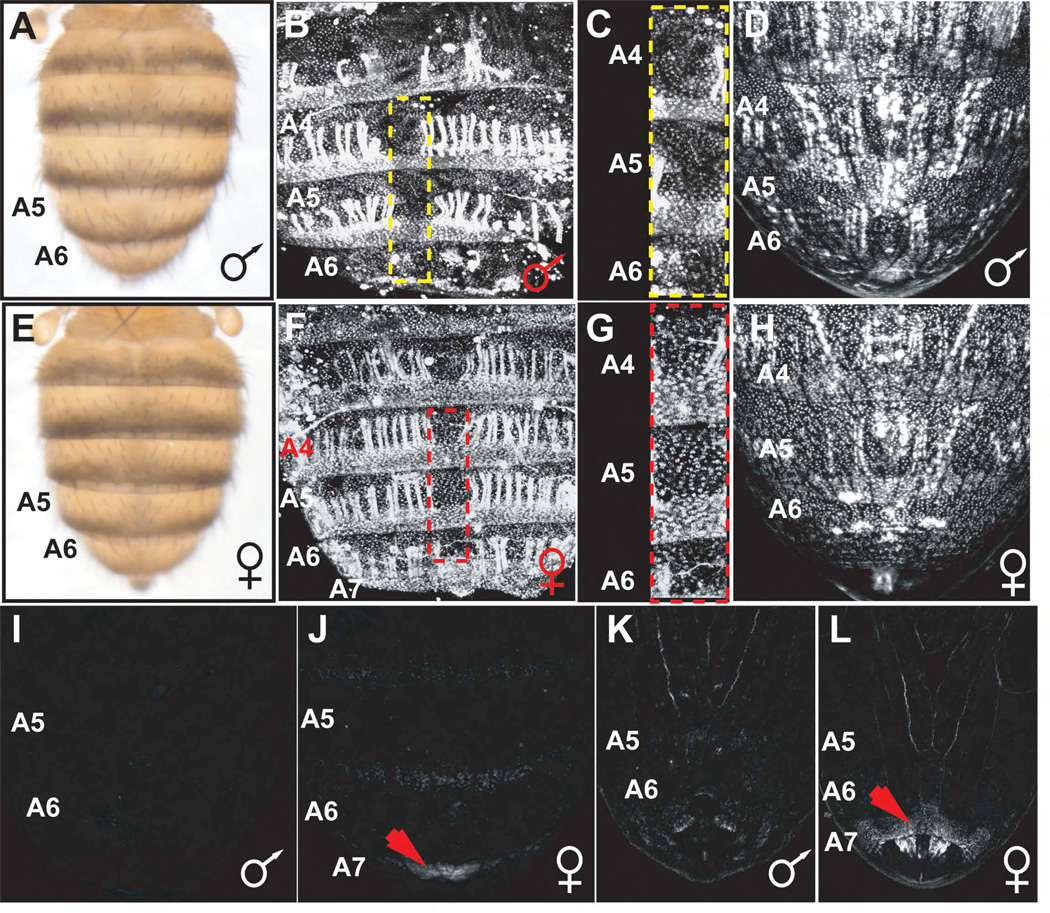
Monomorphic abdominal pigmentation (Jeong et al., 2006) and monomorphic Bab2 expression (Gompel and Carroll, 2003; Kopp et al., 2000) have been inferred to be the ancestral states within the subgenus Sophohora from which male-specific dimorphic pigmentation evolved in the D. mel. lineage. In order to trace the evolution of dimorphic bab expression, we selected D. willistoni (D. wil.) as a member of the monomorphic outgroup to the melanogaster species group and as a surrogate of the inferred ancestral monomorphic pigmentation state. In this species, pigmentation of segments A5 and A6 in males is limited to a posterior stripe, like that of females (Figure 5A and 5E). We found that Bab1 was expressed in a monomorphic pattern in segments A5 and A6 of D. wil. males (Figure 5B and 5C) and females (Figure 5F and 5G), similar to that shown for Bab2 (Kopp et al., 2000). Hence, the evolution of dimorphic pigmentation in the melanogaster species group involved the evolution of the repression of both Bab1 and Bab2 in the posterior segments of males.
Figure 5. The dimorphic element has a deep ancestry.
(A) Dorsal views of D. wil. male abdomen. Pigmentation of abdominal tergites on segments A2-A6 is limited to a posterior stripe.
(B, C, F, and G) Bab1 expression in pupal abdomens at 65 hours APF (a developmental time point equivalent to 72 hour APF assayed for D. mel.).
(B) Bab1 is expressed in segments A2-A6 (A2 not shown) of male pupae. Expression was also observed in longitudinal abdominal muscles.
(C) High magnification view of (yellow box; B) dorsal midline showing equivalent levels of Bab1 in segments A4-A6.
(D) The D. wil. anterior element drove GFP-reporter expression in segments A2-A6 of males (A2 not shown).
(E) Dorsal view of D. wil. female abdomen. The pigmentation pattern is identical to that of the male (compare with A).
(F) Bab1 is expressed in segments A2-A7 of female pupae.
(G) High magnification view of (red box; F) dorsal midline showing equivalent levels of Bab1 in segments A4-A6.
(H) The D. wil. anterior element drove reporter expression in segments A2-A7 of females (A2 and A3 not shown).
(I) The D. wil. dimorphic element does not activate reporter expression in posterior segments of males (dorsal view).
(J) The D. wil. dimorphic element drove expression in the A7 segment of females (red arrow; dorsal view).
(K) Ventral view showing the absence of D. wil. dimorphic element reporter activity in posterior segments of males.
(L) Ventral view showing reporter expression driven by the D. wil. dimorphic element in female segment A7 (red arrow), but not in the anterior A6 segment.
The evolutionary transition from monomorphic to dimorphic bab expression could have occurred through a variety of mechanisms including the evolution of new bab CREs, modifications to orthologous CREs that altered their regulatory function, or changes in the deployment or activity of transcription factors that regulate bab. Since dsx function and expression (Cho et al., 2007; Hediger et al., 2004) and Abd-B expression (Yoder and Carroll, 2006) are well-conserved in the family Drosophilidae, we focused on the evolution of bab CREs. It was necessary to first identify D. wil. bab CREs that drove gene expression in the abdomen. We analyzed the orthologous sequences in the first intron of the D. wil. bab1 gene for CREs with activity in transgenic D mel. pupae. We identified a large region (5.8 kb) that contained orthologous sequence to the anterior element of D. mel., and that drove GFP-reporter expression in the abdominal epidermis in segments A2 through the posterior-most segment in both sexes (Figure 5D and 5H), patterns similar to those of the endogenous Bab1 protein in D. wil. Thus, in the evolution of the D. mel. lineage the function of the anterior element was modified such that it no longer drove bab expression in segments A5 and A6 of either sex.
The activity of the D. wil. anterior element in males and females would appear sufficient to account for monomorphic bab expression and hence monomorphic pigmentation. This observation raised the possibility that the dimorphic element was novel to the D. mel. lineage. However, we also identified a D. wil. sequence orthologous to the dimorphic element, which drove reporter expression in D. mel. female segment A7, but not in segments A5 and A6 (Figure 5I–5L). These findings suggested that an active dimorphic element existed in a common ancestor of D. wil. and D. melanogaster. To test this idea further, we also isolated an orthologous dimorphic element from the more distantly related species D. virilis. The activity of this element was also limited to the female segment A7 (data not shown).
These results demonstrate that the dimorphic element of D. mel. has a deep ancestry and did not arise de novo in the D. melanogaster lineage. Rather, the CRE existed in a common ancestor of monomorphically and dimorphically pigmented species. In D. mel., bab is required to shape the development of particular features of the female-specific A7 segment (this segment is greatly reduced in males) (Kopp et al., 2000) and this is likely to be a deeply conserved role. Therefore, the D. mel. pattern of bab expression in segments A6 and A5 of females represents an expansion in the activity of the dimorphic element and the evolution of dimorphic pigmentation involved the functional modification of both ancestral bab CREs. We next sought to identify the sequence differences between the orthologous dimorphic elements that were responsible for their different activities.
Remodeling of the dimorphic element underlies the evolution of expanded dimorphic Bab expression
Since the D. mel. dimorphic element had fourteen putative ABD-B binding sites, one possible explanation for the expanded activity of the D. mel. dimorphic element would be the evolutionary gain of ABD-B binding sites and, hence, ABD-B regulation throughout segments A5 and A6. However, we were surprised to find that the D. wil. element contained fourteen ABD-B binding sites. Alignment of the orthologous dimorphic elements’ DNA sequences (Figure S5) revealed that the D. wil. element was collinear with the D. mel element and contained twelve of the fourteen D. mel. sites, and lacked only sites 8 and 13. The pair-wise differences between the two species could be due to either gains in the D. mel. lineage or losses in the D. wil. lineage, or both. To assess which was the case, we examined other taxa for the presence or absence of these ABD-B sites (Figure S5). Binding site 8 was identified in dimorphic elements from more distantly related non-Sophohoran species, indicating that this site was lost in the D. wil. lineage. The D. mel. site 13, however, was inferred to be a gain within the melanogaster species group, as this site did not occur in species outside of this clade, including representatives of the obscura and saltans groups. Conversely, the D. mel. element lacked two TTTAT sites that were adjacent to D. wil. site 12 that have been acquired since its divergence from the last common ancestor shared with D. melanogaster.
Although there is no net difference in the number of ABD-B binding sites, we analyzed whether the presence of sites 8 or 13 could account for some of the difference in CRE activity. To determine how much of the difference in activity was contributed by these sites in vivo, we mutated them and tested GFP-reporter activity of the dimorphic element in transgenic pupae. Dimorphic element activity was reduced to 78±5% of the wild-type CRE by mutation of site 8 (Figure 6B), while site 13 had no affect on activity (Figure 6C). Thus, differences in ABD-B sites account for a portion but not nearly all of the expanded activity of the D. mel. dimorphic element.
We ruled out that differences in DSX binding site number contributed to the activity difference between the dimorphic elements because the D. wil. element contained both the Dsx1 and Dsx2 sites. We noticed however, that while the two DSX sites were conserved, a two base pair change in the D. wil. Dsx1 site caused a reversal of the site’s polarity. To determine whether this polarity reversal affected CRE activity, we altered the D. mel. Dsx1 site to match that of the D. wil. site, and tested this modified element for GFP-reporter activity. This alteration reduced activity to 87±2% of the wild-type element (Figure 6D).
We also tested the effect of site polarity on the activity of the D. wil. element by making the reciprocal alteration to the D. wil. element, mutating the Dsx1 site to that of the D. mel., and analyzed this modified element for activity in transgenic pupae. Surprisingly, this alteration dramatically increased the activity of the D. wil. element from just 1±1% to 34±3% of the wild-type activity of the D. mel. element (compare Figures 6G and 6H). Importantly, the increased activity of this modified element was due to the polarity and not the affinity of DSXF for the site. The DSX protein bound both the D. mel. and D. wil. sites with equivalent affinity in EMSAs (Figure S4). These results indicated that the polarity of the DSX binding sites also contributed to the divergence of dimorphic CRE activity between D. wil. and D. melanogaster.
The contribution of both differences in ABD-B binding site number and Dsx1 site polarity to the activity difference between the D. mel. and D. wil. elements raised the possibility that cumulative mutational changes were responsible for CRE activity divergence. To test this possibility, we combined mutations of ABD-B sites 8 and 13 with mutations reversing the Dsx1 site polarity, and observed that dimorphic element activity was reduced to 66±3% of the wild-type element (Figure 6E), below the level caused by either of the mutations alone. However, additional changes beyond ABD-B binding site number and Dsx1 site polarity must also have contributed to the divergence of CRE activity in A5 and A6.
Other potential modifications to the divergence of dimorphic element activity could involve sites for other transcription factors and/or the spatial arrangement of binding sites. While we do not know yet the identity of any additional transcription factors that directly regulate the dimorphic element, we noted striking differences in the spacing of binding sites between the orthologous elements in the non-conserved regions between the ABD-B and DSX binding sites. The most prominent spacing differences resided between ABD-B site 5 and Dsx1 (58 base pairs), ABD-B sites 8 and 9 (98 base pairs), and Dsx2 and ABD-B site 11 (57 base pairs). We refer to these as regions I, II, and III respectively (Figure 7B), where in each region the D. wil. element possesses the greater spacing between binding sites. Compared to members of outgroups, the reduced spacing in regions I–III in D. mel is a general feature shared among the melanogaster species group (Figure S5). To test whether differences in the spatial topology of binding sites affected dimorphic element activity, we inserted the additional D. wil. sequences residing in regions I–III into the orthologous positions of the D. mel. element (Figure 6I–6L), making the distances between the adjacent binding sites in the D. mel element equal to those of the D. wil. dimorphic element. We found that the activity of the D. mel. dimorphic CRE was reduced to 62±3%, 41±3%, and 44±4% of the wild-type, respectively when the D. wil. sequences were inserted into regions I, II and I–III (Figure 6A, 6I, 6J and 6L). These effects appeared to be due to changes in spacing and not the insertion of sequences that were bound by repressors, because the reciprocal deletion of these sequences in the D. wil. element led to no net gain in CRE activity (data not shown). Interestingly, insertion of the D. wil. sequence into region III increased the activity of the wild-type dimorphic element by 37±3% (Figure 6K). This result demonstrated that the regulatory activity of the D. mel. element is not at a maximum and is further evidence of how changes in binding site topology can increase or decrease CRE activity.
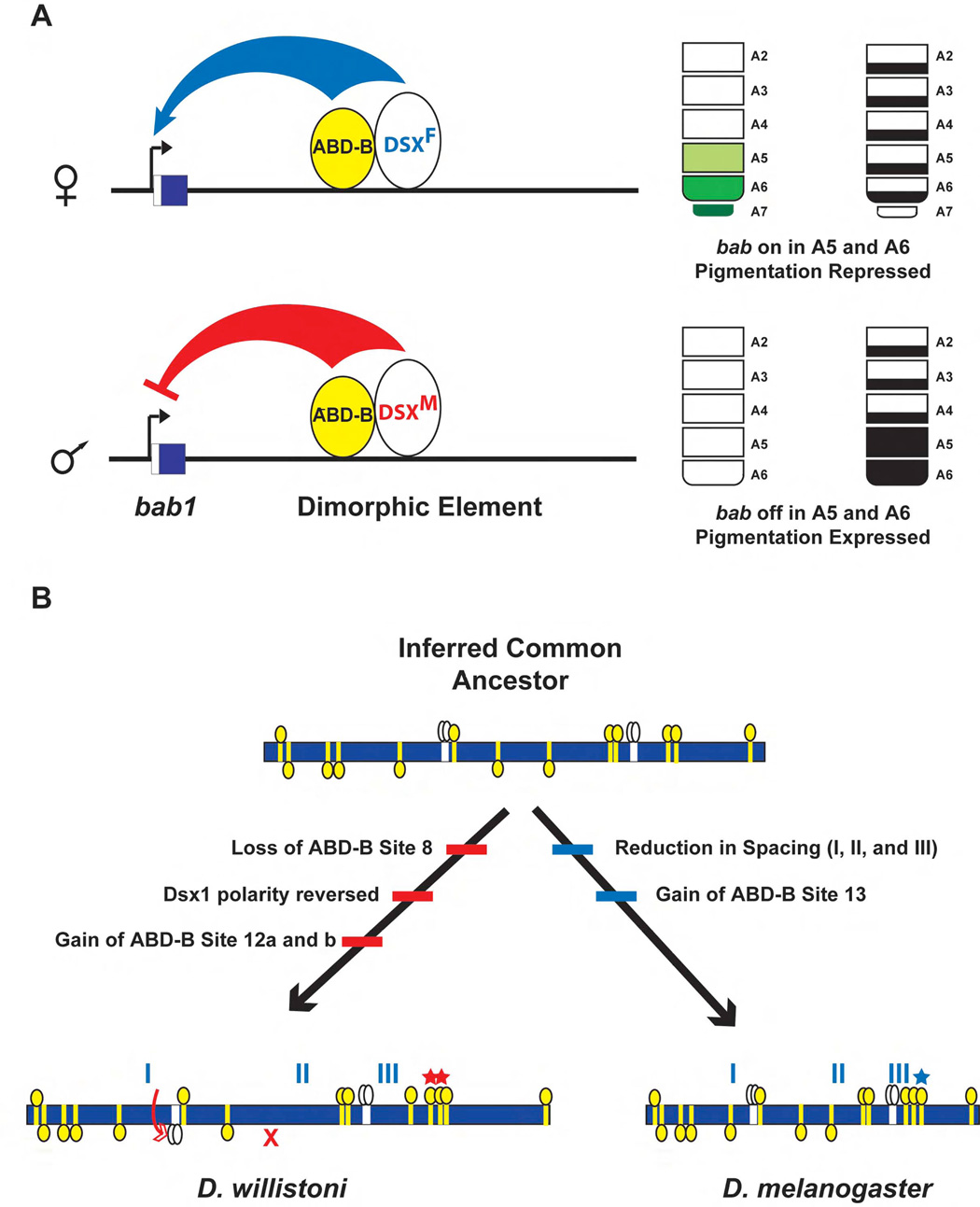
Figure 7. Model for the operation and evolution of the dimorphic genetic switch.
(A) The operation of the switch. Expression of bab in the posterior abdominal segments A5-A7 of females is mediated by the combined inputs of the segment-specific HOX protein ABD-B and the female-specific isoform DSXF. Expression of bab results in the repression of full tergite pigmentation in these segments. Expression of bab in male segments A5 and A6 is repressed by the male-specific isoform DSXM. The absence of bab expression in these segments allows for the development of fully-pigmented tergites.
(B) The evolution of the switch. Schematic depiction of the evolution of the dimorphic element from the inferred common ancestor of D. melanogaster and D. willistoni. Yellow boxes indicate binding sites for ABD-B and white boxes indicate DSX binding sites. Yellow and white ovals represent ABD-B and DSX protein monomers respectively. The common ancestral CRE contained two and thirteen orthologous binding sites for DSX and ABD-B, respectively. In the lineage leading to D. wil., ABD-B site 8 was lost, the polarity of Dsx1 was reversed (red arrow) and candidate ABD-B binding sites 12a and 12b were gained (red stars). In the lineage leading to D. mel., inter-binding site spacing was reduced in regions I, II, and III, and ABD-B site 13 was gained (blue star), which collectively contributed to the higher level of gene expression in female segments A5 and A6.
Together, these results reveal that the expansion of the activity of the D. mel. dimorphic element into anterior abdominal segments did not result from the gain of ABD-B or DSX binding sites. Rather this expanded activity resulted from an amalgam of changes in the CRE involving the number, polarity, and topology (spacing) of binding sites, what we describe as the molecular “remodeling” of a pre-existing DSX- and ABD-B-regulated CRE.
DISCUSSION
We have shown that bab expression in the abdominal epidermis is regulated by two separate CREs, one of which directs gene expression in the anterior abdomen of both sexes, and a second, dimorphic element that regulates female-specific gene expression in segments A5-A7. The dimorphic element, when bound by ABD-B and sex-specific isoforms of the DSX protein, acts as a genetic switch that allows pigmentation in males and represses pigmentation in females. We found that changes in the activities of both CREs have evolved in the course of the origin of the trait from a monomorphic ancestor. Furthermore, we demonstrated that dimorphic CRE function evolved by multiple fine-scale changes within the CRE. These results bear on our understanding of how sexually dimorphic traits develop, how new sex- and segment-restricted traits arise, and how CRE functions evolve.
A genetic switch controlling sexually dimorphic traits
Sex-restricted traits are the product of differences in gene expression between sexes, therefore, understanding how such traits develop requires the identification of those genes with sex-limited expression and elucidation of the genetic and molecular mechanisms governing their regulation. We showed that dimorphic bab expression is regulated by a discrete CRE whose activity is combinatorally regulated by the direct inputs of both region- (ABD-B) and sex-specific (DSX) transcription factors. In females, ABD-B acts in concert with the DSXF isoform through binding sites in the dimorphic element to activate bab expression in the posterior segments. Whereas in males, ABD-B activity is overridden by the repressive activity of the DSXM isoform which binds to the same sites as DSXF and hence, permits the formation of the male-specific posterior pigmentation (Figure 7A).
The genetic pathways that regulate sex-determination and sexual differentiation differ greatly across the animal kingdom, so this mode of male-specific trait regulation in Drosophila may not apply in detail to other animals. However, the integration of region- and sex-specific regulatory inputs must be a requirement for the production of dimorphic traits. We suggest that the integration of such combinatorial inputs by cis-regulatory elements, as we have demonstrated for bab, is a general feature of genetic switches within the pathways regulating the production of dimorphic traits.
The evolution of a new dimorphic trait
The origins of sexually dimorphic traits have long been of central interest in evolutionary biology. One of the key questions that Darwin (Darwin, 1871) grappled with, as have many others subsequently (Fisher, 1930), was whether dimorphic traits are limited to one sex at their origin, or whether these traits first appear in both sexes and then become restricted to one sex. This question has been particularly important and challenging in terms of genetics and evolutionary theory, as it has not been resolved previously how the effects of mutations could be restricted to one sex.
In the simplest genetic scenarios of sexual dimorphism, male-limited traits are the products of the male-limited expression of specific genes. The main evolutionary question then, as it has been phrased in classical genetic terms, is whether male-limited gene expression evolves via: i) “alleles” that are expressed only in males; or ii) alleles expressed in both sexes which are then suppressed in females or promoted in males (Coyne et al., 2008). The elucidation of the regulation and evolution of male-specific pigmentation provides a unique opportunity to reconstruct the genetic path of the evolution of a dimorphic trait.
Although posterior male-specific pigmentation is a relatively simple, two-dimensional morphological trait, it is clear that it did not originate via just one of the alternative genetic paths above. Rather, the evolution of this trait has involved three paths - the evolution of male-limited gene expression, of female-limited gene expression, and of non-sex-restricted gene expression. Specifically, we have shown here that in the course of the evolution from a monomorphically pigmented ancestor, the activity of the female-specific bab dimorphic CRE expanded into segments A6 and A5 and that the activity of the monomorphic bab anterior CRE retreated from segments A6 and A5 of both sexes. These two combined changes produced the sex-specific repression of bab expression in male segments A5 and A6. In addition, in previous work we showed that the yellow pigmentation gene gained high-level expression in segments A5 and A6 via the acquisition of ABD-B binding sites in a specific yellow gene CRE (Jeong et al., 2006), whose activity was male-limited due to repression by Bab (which is apparently indirect).
It is important to underscore that none of the genes in this newly-evolved regulatory circuit are globally restricted in their expression to one sex. Rather, the sex-specific features of their expression are controlled by modular CREs that are physically separate from those controlling gene expression in other developing body regions. The properties of these CREs resolve the question of how the effects of mutations can be restricted to one sex. Namely, mutations in a CRE that is under the direct (the female-specific bab dimorphic element) or indirect (the male-specific yellow CRE) control of an effector of sex determination will have sex-limited effects on gene expression. The findings here are a further demonstration of the general principle of how the modular CREs of pleiotropic genes enable the modification of gene expression in and morphology of one body part independent of other body parts, or in this case, the same body part in the opposite sex (Carroll, 2005; Carroll, 2008).
It is also notable that none of the CREs we have analyzed are new to the dimorphically pigmented melanogaster species group. It is clear, then, that the ancestral dimorphic CRE was active in segment A7 and modified to govern sexually dimorphic pigmentation in segments A6 and A5. Thus, in this example, we see that one path to evolving a new dimorphic trait is via the co-option of genetic components that regulate other pre-existing dimorphic traits.
Remodeling of CREs and the target size of functionally relevant mutations
One of the major questions concerning the evolution of gene expression is how new gene expression patterns arise. The two most obvious mechanisms would appear to be the gain of new regulatory elements or the gain of new transcription factor-CRE linkages. While the deep ancestry of the dimorphic element ruled out the former, we expected that the novel sex- and segment-specific regulation of this CRE by DSX and ABD-B in the D. mel. lineage would require the gain of binding sites for these two transcription factors. However, we found that the both DSX binding sites and most ABD-B sites were present in D. wil. and other monomorphic species and therefore were present in the last common ancestor of both monomorphic and dimorphic species. Thus, the expansion of the dimorphic CRE activity was not due to the wholesale gain of new DSX and ABD-B binding sites.
Rather, we discovered that the expanded, high level activity of the D. mel. dimorphic CRE in segments A6 and A5, relative to the A7-restricted activity of the D. wil. element, was due to an amalgam of changes involving the number, polarity, and topology of transcription factor binding sites. The evolution of dimorphic CRE activity demonstrates how changes beyond the simple gain or loss of binding sites shape CRE evolution. Similarly, we recently showed that changes in the topology and helical phasing of transcription factor binding sites shaped the evolution of a genetic switch controlling galactose utilization in yeast (Hittinger and Carroll, 2007). Our studies strongly support the view that the relationship between function and sequence variation in CREs is complex (Balhoff and Wray, 2005). A vast body of work on eukaryotic and prokaryotic transcriptional regulation has shown that binding site polarity and spacing influences the output of regulatory elements. Therefore, we suggest that one important, but generally unappreciated, class of functionally relevant mutations in CRE and trait evolution involves sequences outside of transcription factor binding sites. CREs thus present a very large target area for potential functionally relevant mutations that quantitatively modulate gene expression and trait development.
Hox genes and the evolution of axial traits
Finally, we suggest that our observations concerning the mechanisms underlying the expansion of dimorphic CRE activity help to shed light on another general aspect of the evolution of animal body plans - the evolution of segmental traits. A large number of studies have demonstrated that some of the major differences among arthropod and vertebrate body plans have involved evolutionary shifts in the spatial boundaries of gene expression along the main body axis (Carroll, 2005). However, the path by which such gene expression patterns are shifted has not been elucidated in any molecular detail. We submit that the expansion of the activity of the dimorphic element from the A7 segment into A6 and A5 is a model of this process. The remodeling of the dimorphic CRE in the course of evolution illustrates that one way such shifts can be accomplished is through numerous small, quantitative incremental changes in the activity of Hox-regulated CREs.
EXPERIMENTAL PROCEEDURES
Fly stocks
The CantonS strain of D. melanogaster (wild-type) and D. willistoni stocks were obtained from the Tucson Stock Center. Genetic analyses were performed using the following alleles: Abd-BMcp (Duncan, 1987) and Abd-Biab9-Tab (Celniker and Lewis, 1987), two gain-of-function alleles that drive ectopic Abd-B expression in segment A4 and A3-A4 respectively; dsxD+R3, a dsx null allele, and; dsxD, a mutant in which dsx RNA is altered from production of the female dsxF transcript to the production of male dsxM transcript, resulting in female intersexes (Duncan and Kaufman, 1975).
Reporter constructs and transgenic fly production
All DNA sequences used in GFP-reporter constructs were cloned into either p-element or site-specific transformation vectors. Additional information on the production of constructs, transgenic lines, and scoring of reporter expression phenotypes are provided in the Supplemental Experimental Procedures and Table S1–S3.
DNA-binding Analyses
DNaseI footprinting reactions and EMSAs were performed as previously described (Jeong et al., 2006) using a GST-DSX DNA Binding Domain (DBD) fusion protein (Supplementary Experimental Procedures). PAGE-purified oligos used in EMSAs are listed in Table S4.
Immunohistochemistry
Immunohistochemistry of pupal abdominal epidermis was performed as previously described (Gompel and Carroll, 2003) using an affinity purified anti-Bab1 antibody (Supplementary Experimental Procedures) on D. mel. and D. wil. specimens, that were dissected at 72 and 65 hours after puparium formation (APF) respectively. Detailed protocol is available at: http://www.molbio.wisc.edu/carroll/methods//methods.html.
Measurement of relative fluorescence intensity
The relative fluorescence intensities for A6 reporter expression was determined as previously described (Jeong et al., 2008) with modifications described in the Supplementary Experimental procedures.
Imaging of fly abdomens
Images of adult abdomens were taken using an Olympus SZX16 Zoom Stereo Microscope equipped with an Olympus DP71 microscope digital camera. Pupal immunohistochemistry and transgenic GFP-reporter line samples were imaged using an Olympus Fluoview FV 1000 confocal microscope and software. Pupae from transgenic reporter lines were dissected from pupal cases at 72–78 hours APF and mounted in Halocarbon 700 oil for confocal analysis.
Supplementary Material
Supplemental data includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at ………
ACKNOWLEDGMENTS
We thank V. Kassner, C. Walsh, M. Rebeiz, K. Vaccaro, and B. Prud’homme for technical assistance; J-L. Couderc for technical advice; K. Venken for providing genomic attP docking site lines; F. Karch for providing germline- specific □C31 integrase lines, and T. Shirangi for comments on the manuscript. This work was supported by NIH Postdoctoral fellowship (GM076935) to T.M.W., a Human Frontiers Science Program Fellowship (LT00640/2005-L) to T.W., and the Howard Hughes Medical Institute (S.B.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- An W, Wensink PC. Three protein binding sites form an enhancer that regulates sex- and fat body-specific transcription of Drosophila yolk protein genes. Embo J. 1995;14:1221–1230. doi: 10.1002/j.1460-2075.1995.tb07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. Sexual Selection. Princeton: Princeton University Press; 1994. [Google Scholar]
- Barmina O, Kopp A. Sex-specific expression of a HOX gene associated with rapid morphological evolution. Dev Biol. 2007;311:277–286. doi: 10.1016/j.ydbio.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo Devo and an Expanding Evolutionary Synthesis: A Genetic Theory of Morphological Evolution. Cell. 2008 doi: 10.1016/j.cell.2008.06.030. in press. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Lewis EB. Transabdominal, a dominant mutant of the Bithorax Complex, produces a sexually dimorphic segmental transformation in Drosophila. Genes Dev. 1987;1:111–123. doi: 10.1101/gad.1.2.111. [DOI] [PubMed] [Google Scholar]
- Cho S, Huang ZY, Zhang J. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics. 2007;177:1733–1741. doi: 10.1534/genetics.107.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 1993;7:42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- Couderc JL, Godt D, Zollman S, Chen J, Li M, Tiong S, Cramton SE, Sahut-Barnola I, Laski FA. The bric a brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila. Development. 2002;129:2419–2433. doi: 10.1242/dev.129.10.2419. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Kay EH, Pruett-Jones S. The genetic basis of sexual dimorphism in birds. Evolution Int J Org Evolution. 2008;62:214–219. doi: 10.1111/j.1558-5646.2007.00254.x. [DOI] [PubMed] [Google Scholar]
- Darwin C. The Descent of Man, and Selection in Relation to Sex. 1871 [Google Scholar]
- Duncan I. The bithorax complex. Annu Rev Genet. 1987;21:285–319. doi: 10.1146/annurev.ge.21.120187.001441. [DOI] [PubMed] [Google Scholar]
- Duncan IW, Kaufman TC. Cytogenic analysis of chromosome 3 in Drosophila melanogaster: mapping of the proximal portion of the right arm. Genetics. 1975;80:733–752. doi: 10.1093/genetics/80.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker SC, Jackson DG, von Kessler DP, Sun BI, Young KE, Beachy PA. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. Embo J. 1994;13:3551–3560. doi: 10.1002/j.1460-2075.1994.tb06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman SE, Chen HJ, Burtis KC. Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics. 1996;144:1639–1652. doi: 10.1093/genetics/144.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford: Clarendon Press; 1930. [Google Scholar]
- Garrett-Engele CM, Siegal ML, Manoli DS, Williams BC, Li H, Baker BS. Intersex, a gene required for female sexual development in Drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development. 2002;129:4661–4675. doi: 10.1242/dev.129.20.4661. [DOI] [PubMed] [Google Scholar]
- Gompel N, Carroll SB. Genetic mechanisms and constraints governing the evolution of correlated traits in drosophilid flies. Nature. 2003;424:931–935. doi: 10.1038/nature01787. [DOI] [PubMed] [Google Scholar]
- Hediger M, Burghardt G, Siegenthaler C, Buser N, Hilfiker-Kleiner D, Dubendorfer A, Bopp D. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev Genes Evol. 2004;214:29–42. doi: 10.1007/s00427-003-0372-2. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rokas A, Carroll SB. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125:1387–1399. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- Jursnich VA, Burtis KC. A positive role in differentiation for the male doublesex protein of Drosophila. Dev Biol. 1993;155:235–249. doi: 10.1006/dbio.1993.1021. [DOI] [PubMed] [Google Scholar]
- Kopp A, Duncan I. Anteroposterior patterning in adult abdominal segments of Drosophila. Dev Biol. 2002;242:15–30. doi: 10.1006/dbio.2001.0529. [DOI] [PubMed] [Google Scholar]
- Kopp A, Duncan I, Godt D, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–559. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- Lours C, Bardot O, Godt D, Laski FA, Couderc JL. The Drosophila melanogaster BTB proteins bric a brac bind DNA through a composite DNA binding domain containing a pipsqueak and an AT-Hook motif. Nucleic Acids Res. 2003;31:5389–5398. doi: 10.1093/nar/gkg724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins DM. Androgen receptor and molecular mechanisms of male-specific gene expression. Novartis Found Symp. 2005;268:42–52. discussion 53-46, 96-49. [PubMed] [Google Scholar]
- Verrijdt G, Haelens A, Claessens F. Selective DNA recognition by the androgen receptor as a mechanism for hormone-specific regulation of gene expression. Mol Genet Metab. 2003;78:175–185. doi: 10.1016/s1096-7192(03)00003-9. [DOI] [PubMed] [Google Scholar]
- Wilkins AS. Deciphering the swordtail's tale: a molecular and evolutionary quest. Bioessays. 2004;26:116–119. doi: 10.1002/bies.10414. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Carroll SB, Kopp A. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 2003;19:495–504. doi: 10.1016/S0168-9525(03)00194-X. [DOI] [PubMed] [Google Scholar]
- Yi W, Ross JM, Zarkower D. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000;127:4469–4480. doi: 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- Yi W, Zarkower D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development. 1999;126:873–881. doi: 10.1242/dev.126.5.873. [DOI] [PubMed] [Google Scholar]
- Yoder JH, Carroll SB. The evolution of abdominal reduction and the recent origin of distinct Abdominal-B transcript classes in Diptera. Evol Dev. 2006;8:241–251. doi: 10.1111/j.1525-142X.2006.00095.x. [DOI] [PubMed] [Google Scholar]
- Zauner H, Begemann G, Mari-Beffa M, Meyer A. Differential regulation of msx genes in the development of the gonopodium, an intromittent organ, and of the "sword," a sexually selected trait of swordtail fishes (Xiphophorus) Evol Dev. 2003;5:466–477. doi: 10.1046/j.1525-142x.2003.03053.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at ………