Abstract
Human TRIM5α restricts N-tropic murine leukemia virus (N-MLV) but not B-tropic MLV (B-MLV) infection. Here we study B30.2/SPRY domain mutants of human TRIM5α that acquire the ability to inhibit B-MLV infection prior to reverse transcription without losing the ability to restrict N-MLV infection. Remarkably, these mutants gain the ability to decrease the amount of particulate B-MLV capsids in the cytosol of infected cells. In addition, these mutants gain the ability to restrict SIVmac and HIV-2 infection. B-MLV and SIVmac infections were blocked by the mutant TRIM5α proteins prior to reverse transcription. Thus, the range of retroviruses restricted by human TRIM5α can be increased by changes in the B30.2/SPRY domain, which also result in the ability to cause premature uncoating of the restricted retroviral capsid.
Keywords: Retrovirus, restriction factor, uncoating, tripartite motif, mechanism, capsid, reverse transcription
Introduction
Following entry into cells, some retroviruses encounter dominant blocks to infection that act prior to reverse transcription (Cowan et al., 2002; Munk et al., 2002). For example, human immunodeficiency virus (HIV-1) is blocked in the cells of most Old World monkeys. Infection by the simian immunodeficiency virus of macaques (SIVmac) encounters similar restrictions in New World monkeys (Hofmann et al., 1999). These restrictions are mediated by TRIM5α, a tripartite motif protein with a RING, B-box 2 and coiled-coil (CC) domain, as well as a carboxy-terminal B30.2/SPRY domain (Stremlau et al., 2004). The TRIM5α B30.2/SPRY domain is essential for retroviral restriction, and differences in the antiretroviral potency of TRIM5α from different species map to this domain (Ohkura et al., 2006; Perron, Stremlau, and Sodroski, 2006; Sawyer et al., 2005; Song et al., 2005a; Stremlau et al., 2005; Yap, Nisole, and Stoye, 2005).
Human TRIM5α (TRIM5αhu) potently restricts N-tropic murine leukemia virus (NMLV) but not B-MLV infection (Hatziioannou et al., 2004; Keckesova, Ylinen, and Towers, 2004; Perron et al., 2004). TRIM5αhu partially inhibits equine infectious anemia virus (EIAV) and feline immunodeficiency virus (FIV) infections (Diaz-Griffero et al., 2007b). TRIM5 αhu does not efficiently restrict HIV-1 infection; however, a single amino acid change in its B30.2/SPRY domain (TRIM5αhu R332P) converts TRIM5αhu into a potent restrictor of HIV-1 and SIVmac infections (Li et al., 2006; Stremlau et al., 2005; Yap, Nisole, and Stoye, 2005). In contrast to the wild-type protein, TRIM5αhu R332P efficiently binds HIV-1 capsid-nucleocapsid (CA-NC) complexes assembled in vitro (Li et al., 2006). Thus, variation in the surface of the B30.2/SPRY domain can modulate the specificity of retroviral restriction.
Changes in arginine 110 of the N-MLV capsid (CA) protein determine susceptibility to TRIM5αhu-mediated restriction. Changing this arginine residue to a glutamic acid residue, the corresponding amino acid from B-MLV, generates a virus that can partially overcome TRIM5αhu restriction (Perron et al., 2004; Towers et al., 2000). Conversely, replacement of glutamic acid 110 of the B-MLV capsid with an arginine residue generates a virus that is susceptible to TRIM5αhu restriction. Particulate N-MLV capsids in the cytoplasm of TRIM5αhu-expressing cells undergo a premature conversion to soluble capsid proteins (Perron et al., 2007). Decreases in the amounts of particulate cytosolic capsids have been observed for other retroviruses, including HIV-1, in cells expressing a restricting TRIM5α protein (Diaz-Griffero et al., 2007a; Stremlau et al., 2006). Thus, there is a good correlation between the restriction of a retrovirus and the premature uncoating of the viral capsid in the cytoplasm of the infected cell. However, other mechanistic models for TRIM5α-mediated restriction, including those that involve blocks after reverse transcription, have been proposed (Anderson et al., 2006; Wu et al., 2006).
In this report, we study changes in the B30.2/SPRY domain of TRIM5αhu that result in an increase in the ability to restrict B-MLV infection prior to reverse transcription. Remarkably, these mutants gain the ability to decrease the amount of cytosolic particulate B-MLV capsid during infection. These mutants also acquire the ability to restrict SIVmac potently by interfering with a step in the virus life cycle prior to reverse transcription.
Results
TRIM5αhu B30.2/SPRY domain mutants that gain the ability to inhibit B-MLV infection
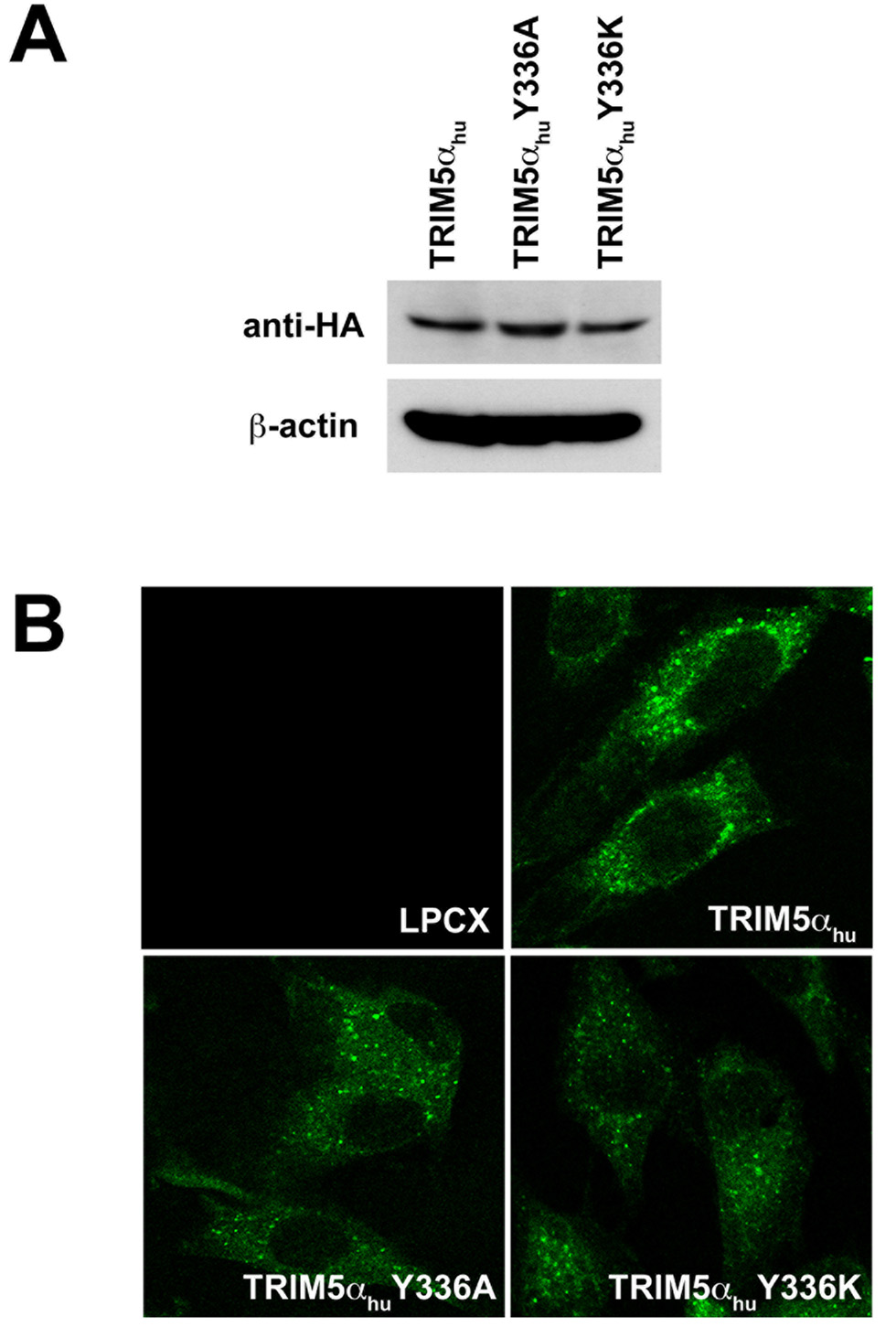
Changes in tyrosine 336 of TRIM5αhu have been reported to allow this protein to inhibit B-MLV as well as N-MLV infection (Maillard et al., 2007). We took advantage of this gain of function to investigate the mechanism of B-MLV restriction by these mutants. We stably expressed the wild-type TRIM5αhu protein and two mutants, TRIM5αhu Y336A and Y336K, in canine Cf2Th cells. These cells were used to assess the level of expression and localization of the TRIM5αhu variants; viral restriction, viral cDNA formation and the fate of retroviral capsids following virus entry into the cell were evaluated.
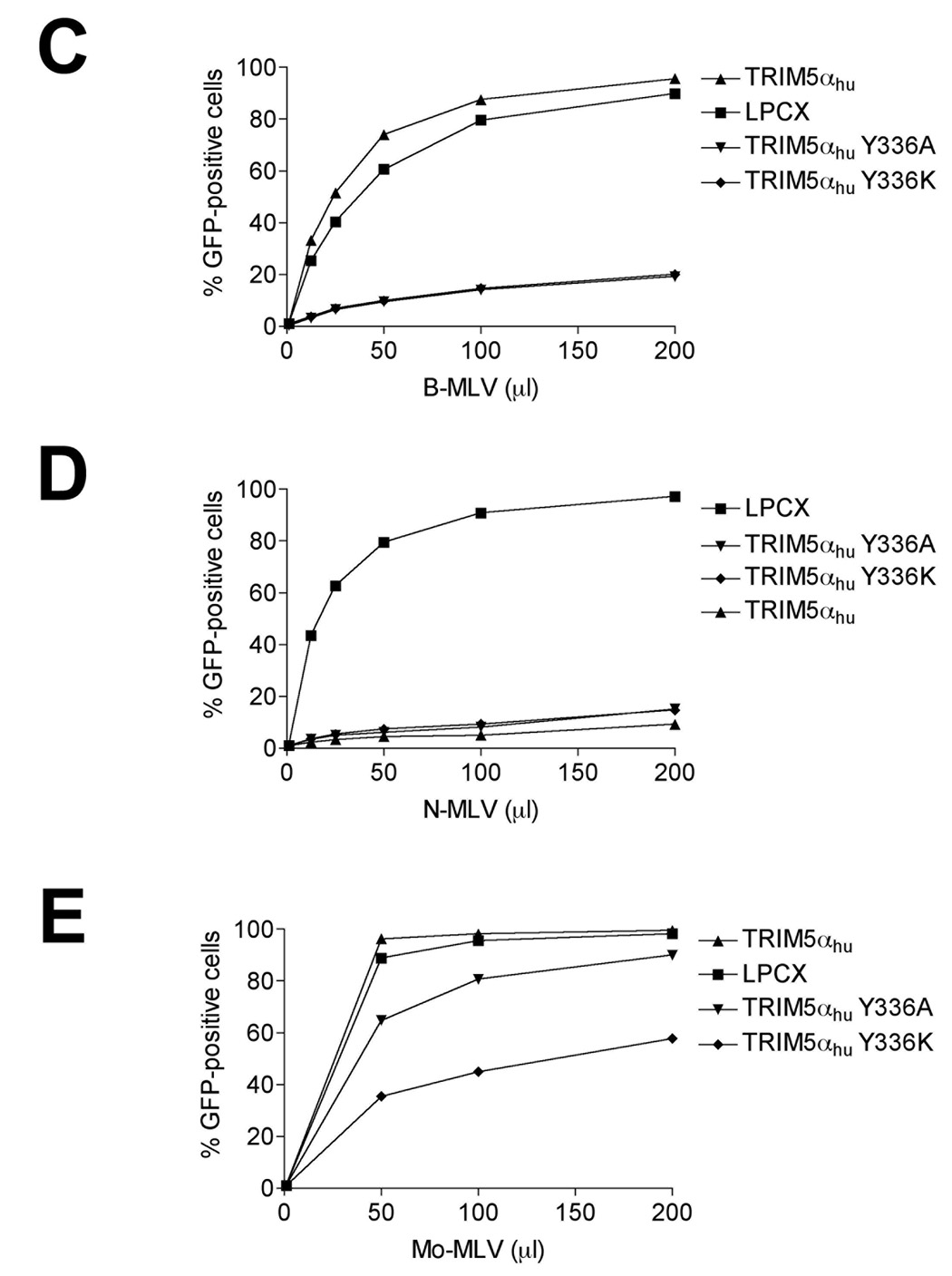
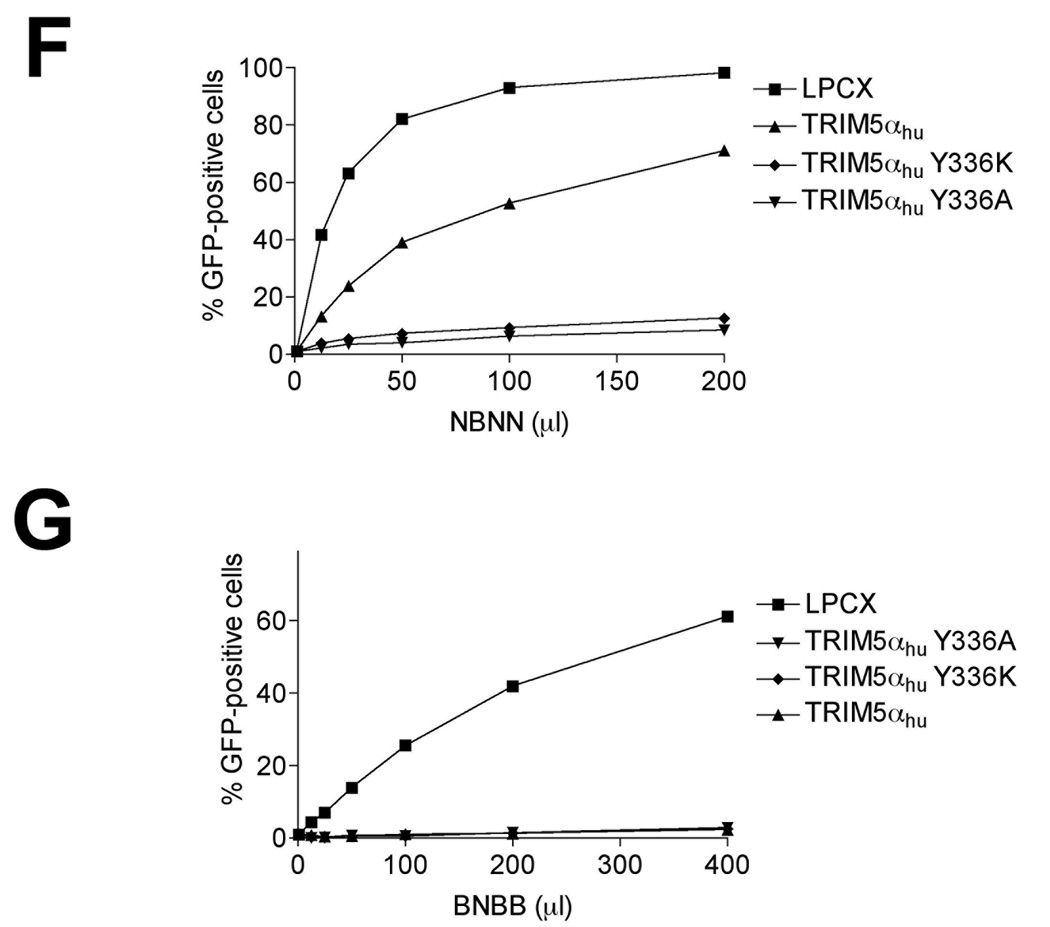
The wild-type, Y336A and Y336K TRIM5αhu proteins were expressed at similar steady-state levels in the Cf2Th cells (Figure 1A). Wild-type and mutant TRIM5αhu proteins localized in the cytoplasm, concentrating in speckles (Figure 1B). As previously reported (Maillard et al., 2007), both TRIM5αhu Y336A and Y336K gain the ability to restrict B-MLV potently (Figure 1C) without losing the ability to restrict N-MLV (Figure 1D). In addition, TRIM5αhu Y336K can partially block the infection of the NB-tropic Moloney MLV (Mo-MLV) (Figure 1E). Replacing arginine at position 110 in the N-MLV capsid with glutamic acid, the corresponding residue from the B-MLV capsid, rendered this virus (NBNN) partially resistant to wild-type TRIM5αhu; however, the NBNN virus remained sensitive to the TRIM5αhu Y336A and Y336K mutants (Figure 1F). The converse capsid mutant, BNBB, which is identical to the B-MLV capsid except for an arginine residue at position 110, was restricted by the wild-type and mutant TRIM5αhu proteins (Figure 1G). Thus, the TRIM5αhu Y336A and Y336K mutants can potently restrict both N-MLV and B-MLV; in contrast, the wild-type TRIM5αhu protein restricts only N-MLV or a modified form of B-MLV that has an arginine residue at position 110 of the capsid protein.
Figure 1. Expression, localization and MLV-restricting ability of TRIM5αhu Y336A and Y336K.
(A) Wild-type, Y336A and Y336K TRIM5αhu proteins were stably expressed in canine Cf2Th cells. Cell lysates were Western blotted and probed with antibodies directed against the HA epitope tag (top panel) or β-actin (bottom panel). (B) Cf2Th cells transduced with the empty LPCX vector or stably expressing the wild-type and mutant TRIM5αhu proteins were fixed and stained using a fluorescein isothiocyanate-conjugated anti-HA antibody, as described in Materials and Methods. Representative confocal microscope images are shown. (C–G) Cf2Th cells transduced with the empty LPCX vector or expressing the indicated wild-type or mutant TRIM5αhu proteins were challenged with B-MLV-GFP (C), N-MLV-GFP (D), Mo-MLV-GFP (E), NBNN-GFP (F) and BNBB-GFP (G). GFP-positive cells were counted. Similar results were obtained in three independent experiments.
Restriction of B-MLV infection prior to the completion of reverse transcription by TRIM5αhu Y336A and Y336K
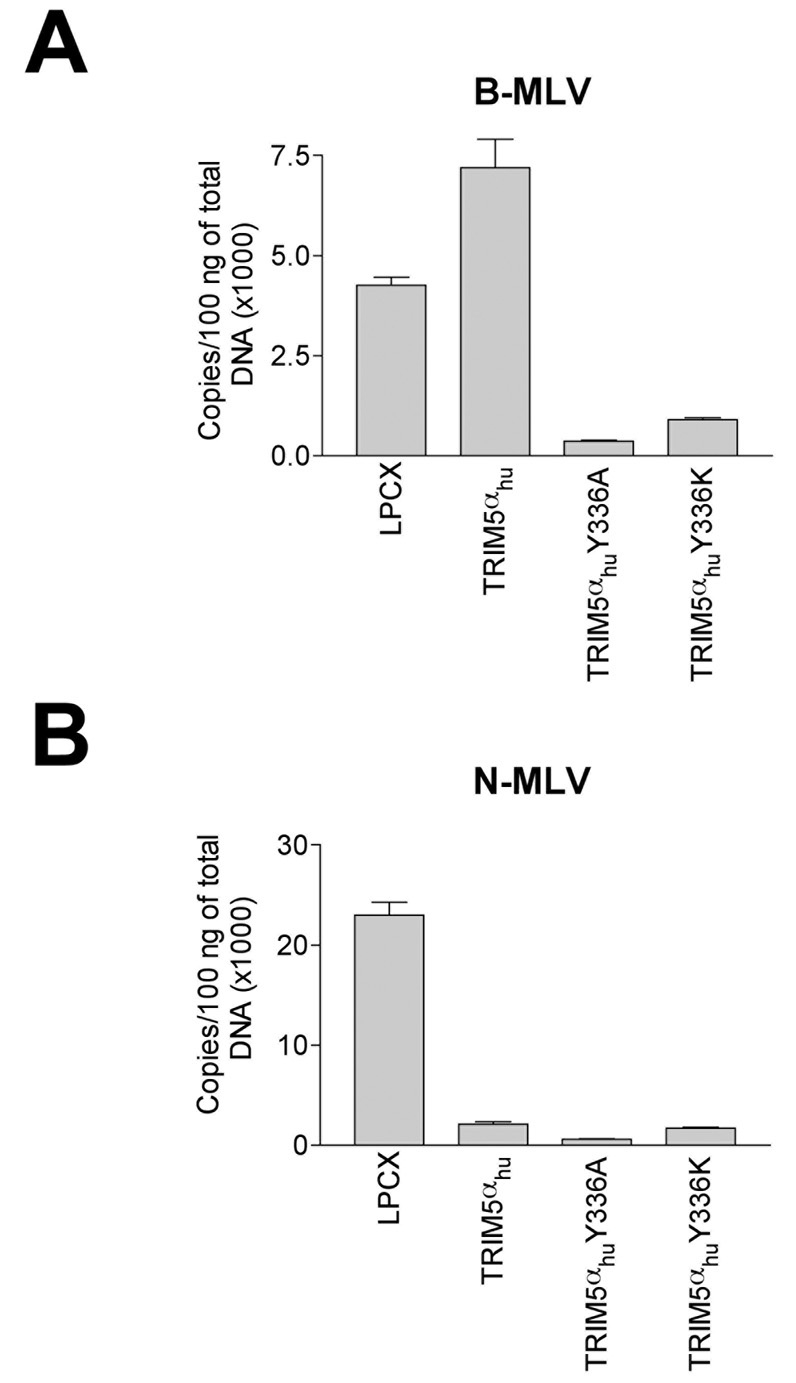
The above results indicate that TRIM5αhu Y336A and Y336K potently restrict B-MLV infection. To investigate the mechanism of this gain of virus-restricting ability, we measured the synthesis of late reverse transcripts (LRTs) during B-MLV infection of cells expressing the wild-type and mutant TRIM5αhu proteins. Compared with the levels of LRTs in control LPCX cells or in cells expressing wild-type TRIM5αhu, the levels of LRTs in cells expressing TRIM5αhu Y336A and Y336K were significantly lower (Figure 2A). When these cells were incubated with N-MLV, the levels of LRTs were lower in cells expressing the wild-type and mutant TRIM5αhu proteins, compared with those in the control LPCX cells (Figure 2B). We conclude that the TRIM5αhu Y336A and Y336K mutants block B-MLV and N-MLV infection prior to the completion of reverse transcription.
Figure 2. Levels of MLV reverse transcripts in infected cells expressing TRIM5αhu Y336A and Y336K.
Cf2Th cells expressing the indicated wild-type and mutant TRIM5αhu proteins or containing the empty vector LPCX were challenged at an MOI of 0.2 with DNAse-pretreated B-MLV-GFP (A) or N-MLV-GFP (B). After 7 hours, cells were lysed and total DNA extracted. The levels of viral DNA were measured by quantitative real- time PCR, using a probe against GFP as described in Materials and Methods. Similar results were obtained in three independent experiments.
Effects of TRIM5αhu Y336A and Y336K on the fate of cytosolic MLV capsids
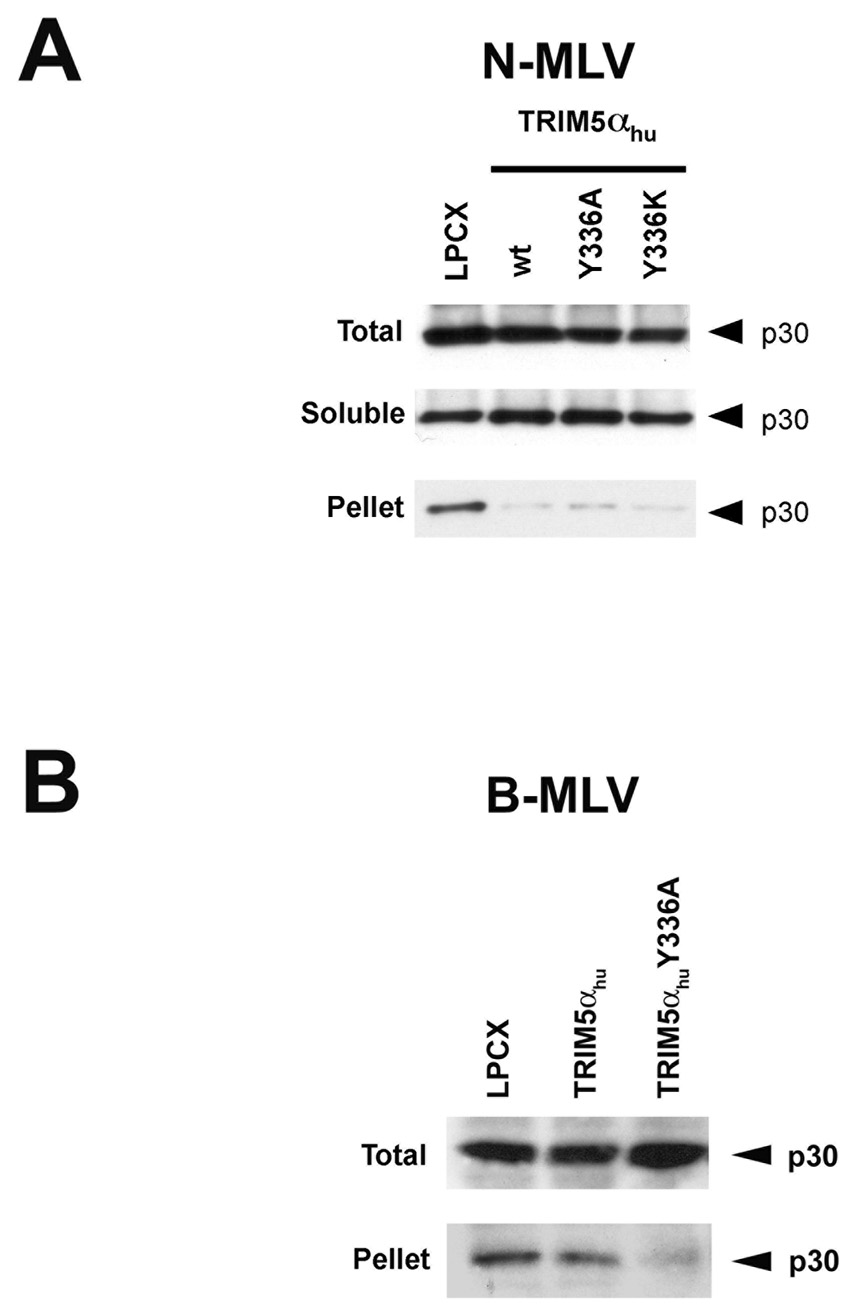
To investigate further the mechanism underlying the ability of the TRIM5αhu tyrosine 336 mutants to block B-MLV infection, we examined the effect of expression of wild-type and mutant TRIM5αhu proteins on the state of the viral capsid in the cytosol of infected cells. As previously demonstrated (Perron et al., 2007), the amount of particulate N-MLV capsid in the cytosol of cells expressing the wild-type TRIM5αhu protein was significantly reduced compared with that seen in the control LPCX-transduced cells (Figure 3A). The relative increase in soluble N-MLV capsid protein indicates that the particulate NMLV capsid was converted into soluble forms in the TRIM5αhu-expressing cells. This premature uncoating of the N-MLV capsid was also seen in cells expressing the TRIM5αhu Y336A and Y336K mutants (Figure 3A).
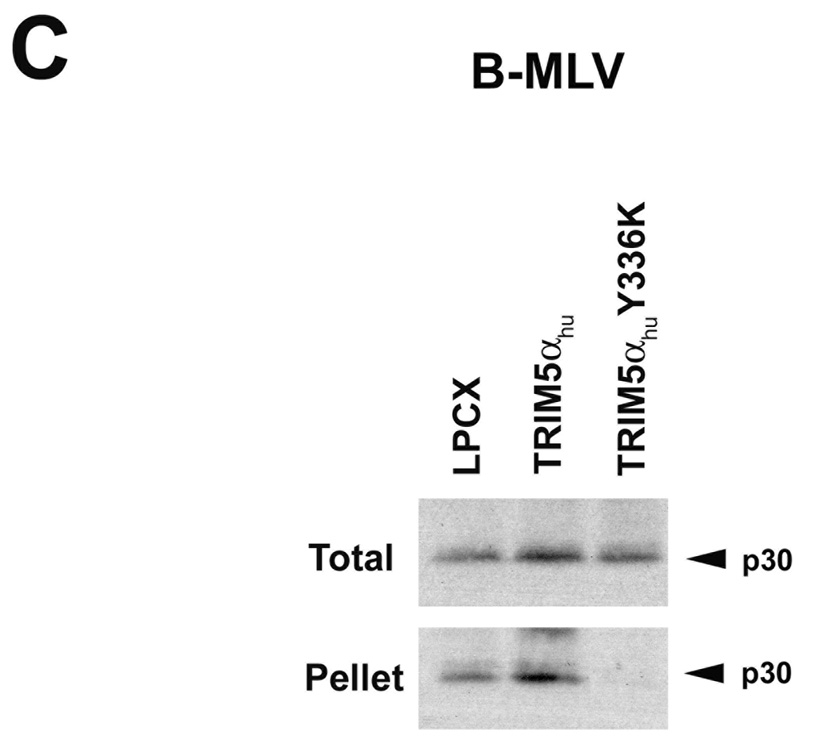
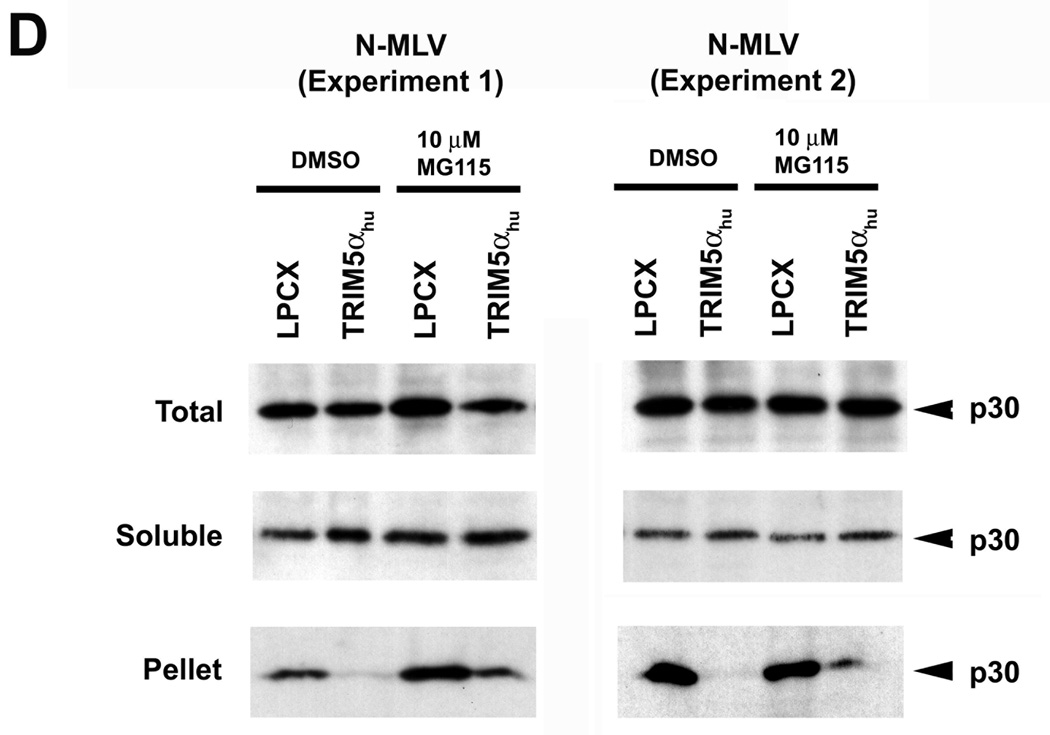
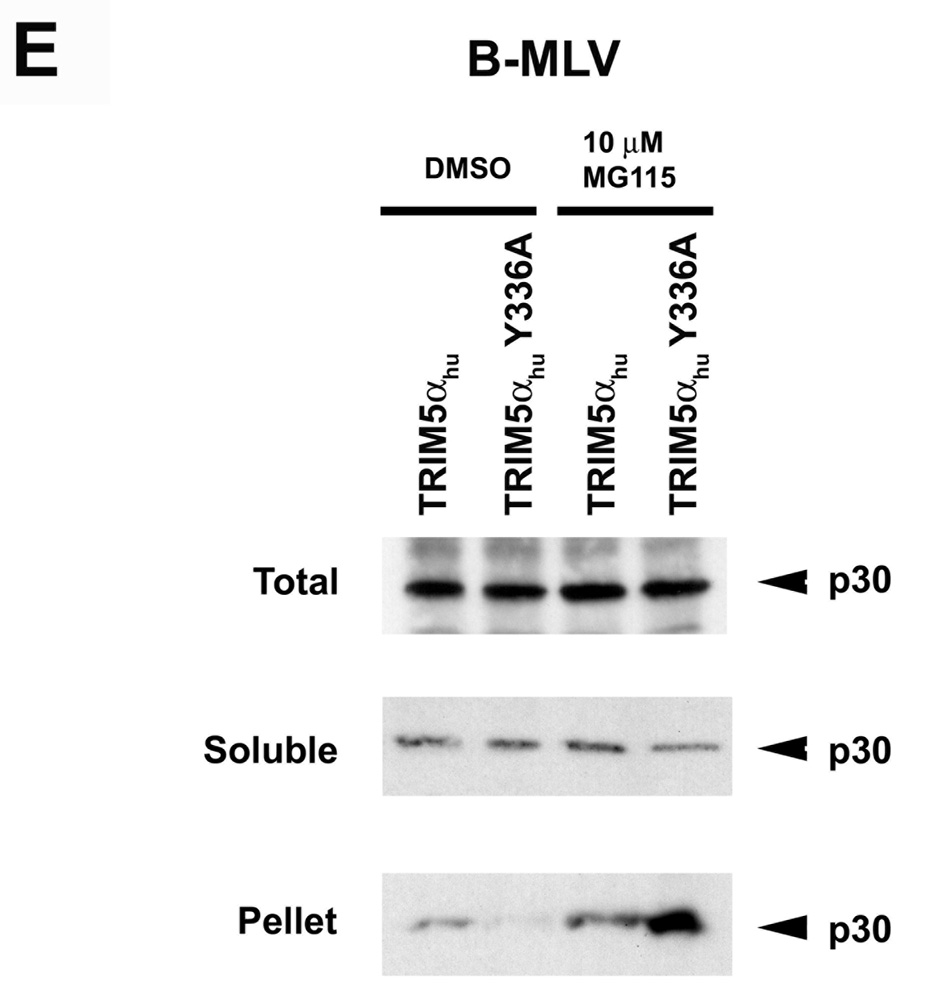
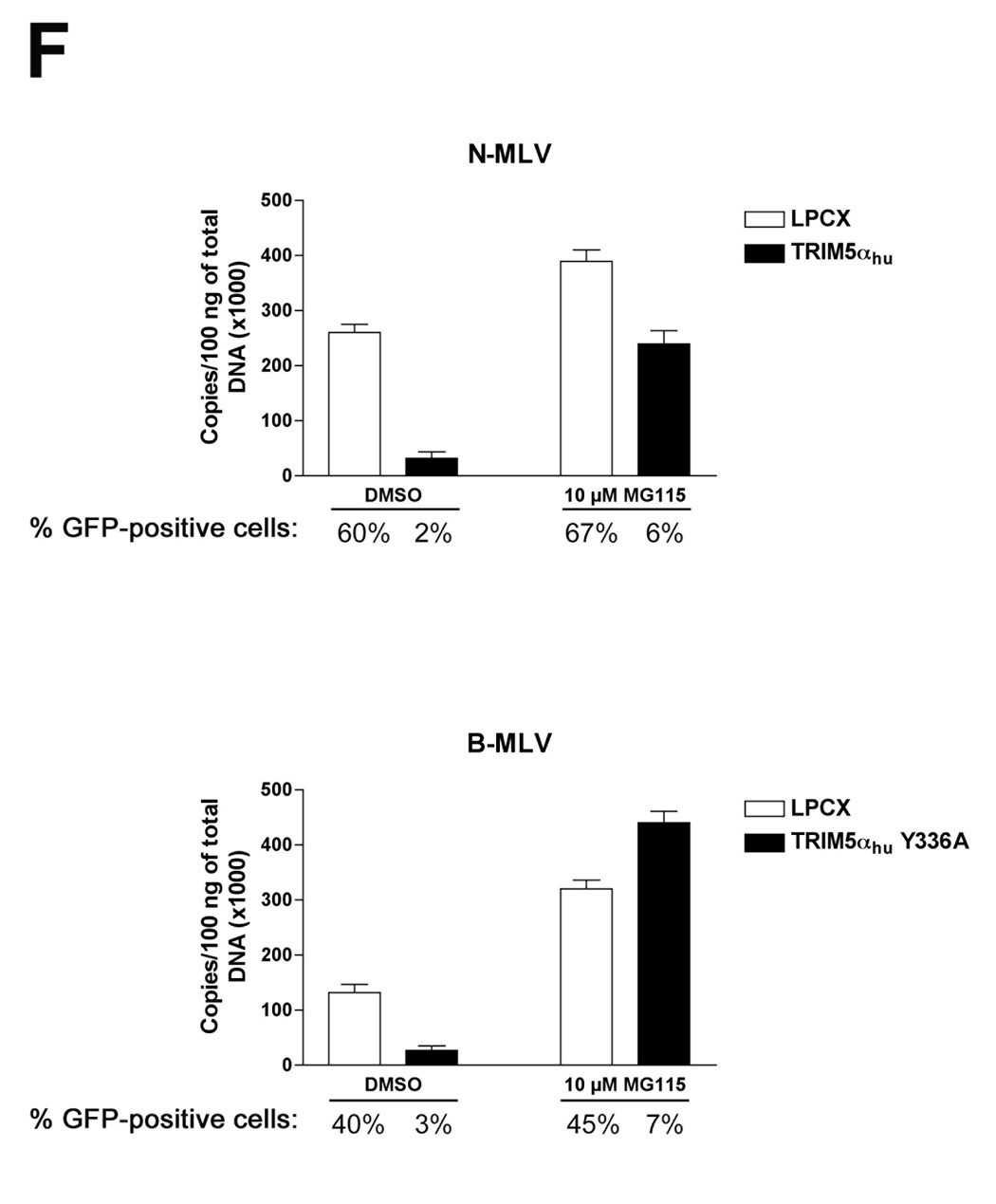
Figure 3. Fate of the cytosolic MLV capsid and levels of late reverse transcripts following infection of cells expressing TRIM5αhu Y336A and Y336K.
Cf2Th cells expressing wild-type or mutant TRIM5αhu proteins, or transduced with the empty LPCX control vector, were incubated with VSV G-pseudotyped N-MLV (A, D) or B-MLV (B, C, E) VLPs for 4 hours. In D and E, target cells were preincubated with 10 µM MG115, a proteasomal inhibitor, or the control DMSO solvent for 1 hour at 37°C. N-MLV and B-MLV VLPs were then added and the incubation at 37°C was continued for another 4 hours. Following lysi s of the target cells, cytoplasmic lysates were analyzed directly (“Total”) or were separated on sucrose gradients into endosomal, soluble and particulate (“Pellet”) fractions, as described in Materials and Methods. The fractions were analyzed by Western blot, using an antibody directed against the MLV p30 capsid protein. (F) Cf2Th cells were preincubated for one hour with DMSO or 10 µM MG115 and then exposed to VSV G-pseudotyped N-MLV-GFP or B-MLV-GFP. After 4 hours, an aliquot of the cells was preserved for assessment of the percentage of GFP-positive cells at 48 hours after infection. The remaining cells were lysed and cytoplasmic lysates were used for DNA extraction and quantitative real-time PCR, utilizing a probe against GFP.
The effect of expression of the TRIM5αhu Y336A and Y336K proteins on the state of the cytosolic B-MLV capsid was examined. Remarkably, both mutants caused a decrease in the amount of cytosolic B-MLV capsid, relative to the levels observed in cells transduced with the empty LPCX vector or expressing the wild-type TRIM5αhu protein (Figures 3B and 3C). Thus, concomitant with a gain of B-MLV-restricting ability, the TRIM5αhu tyrosine 336 mutants acquired the ability to decrease the amount of particulate B-MLV capsid in the cytosol of infected cells.
Effects of proteasome inhibition on viral cDNA and cytosolic capsids
We tested the ability of TRIM5αhu and TRIM5αhu Y336A to decrease the amount of particulate capsid in the presence of MG115, a proteasomal inhibitor, during infection with N-MLV and B-MLV, respectively (Figures 3D and 3E). Proteasome inhibition resulted in a general increase in the amount of pelletable N-MLV and B-MLV capsids in the cytosol of infected cells, regardless of the expression of a restricting TRIM5α protein in the cells. This increase in the background level of particulate capsid obscured any effect of TRIM5αhu Y336A on B-MLV capsids (Figure 3E); however, the TRIM5αhu-induced uncoating of N-MLV capsids, which are more efficiently recovered from sucrose gradients (Perron et al., 2007), was still evident after treatment with the proteasome inhibitor (Figure 3D).
We next examined the effects of proteasome inhibition on the levels of late reverse transcripts (LRTs) in control Cf2Th cells or Cf2Th cells expressing TRIM5αhu or TRIM5αhu Y336A, following exposure to N-MLV or B-MLV. The cells were treated with the proteasome inhibitor, MG115, or with the control solvent, DMSO. As was seen for the levels of cytosolic particulate capsids above, proteasome inhibition resulted in a general increase in the levels of LRTs, even in cells not expressing TRIM5αhu variants (Figure 3F). Relative to the level of N-MLV LRTs in the control cells, LRT levels in the TRIM5αhu-expressing cells were reduced in both DMSO-treated and MG115-treated cells. The reduction in the level of B-MLV LRTs due to expression of TRIM5αhu Y336A was observed in the Cf2Th cells treated with the DMSO solvent, but not in the MG115-treated cells. Thus, the levels of LRTs (Figure 3F) closely paralleled the levels of cytosolic particulate N-MLV and B-MLV capsids (Figures 3D and 3E).
The effects of proteasome inhibition on the efficiency of N-MLV and B-MLV infection of control Cf2Th cells or Cf2Th cells expressing TRIM5αhu or TRIM5αhu Y336A were examined. Slight increases in the percentages of N-MLV- and B-MLV-infected cells were observed following proteasome inhibition (Figure 3F), as has been previously observed for HIV-1 infection (Stremlau et al., 2006; Wu et al., 2006; Schwartz et al., 1998; Fredericksen et al., 2002; Dueck and Guatelli, 2007). However, TRIM5αhu and TRIM5αhu Y336A efficiently inhibited infection by N-MLV and B-MLV, respectively, of cells treated with the MG115 proteasome inhibitor. Thus, proteasome inhibition resulted in general increases in the levels of particulate cytosolic capsids and LRTs, but minimally affected the efficiency of MLV infection and restriction by TRIM5αhu.
TRIM5αhu Y336A and Y336K potently restrict SIVmac and HIV-2 infection
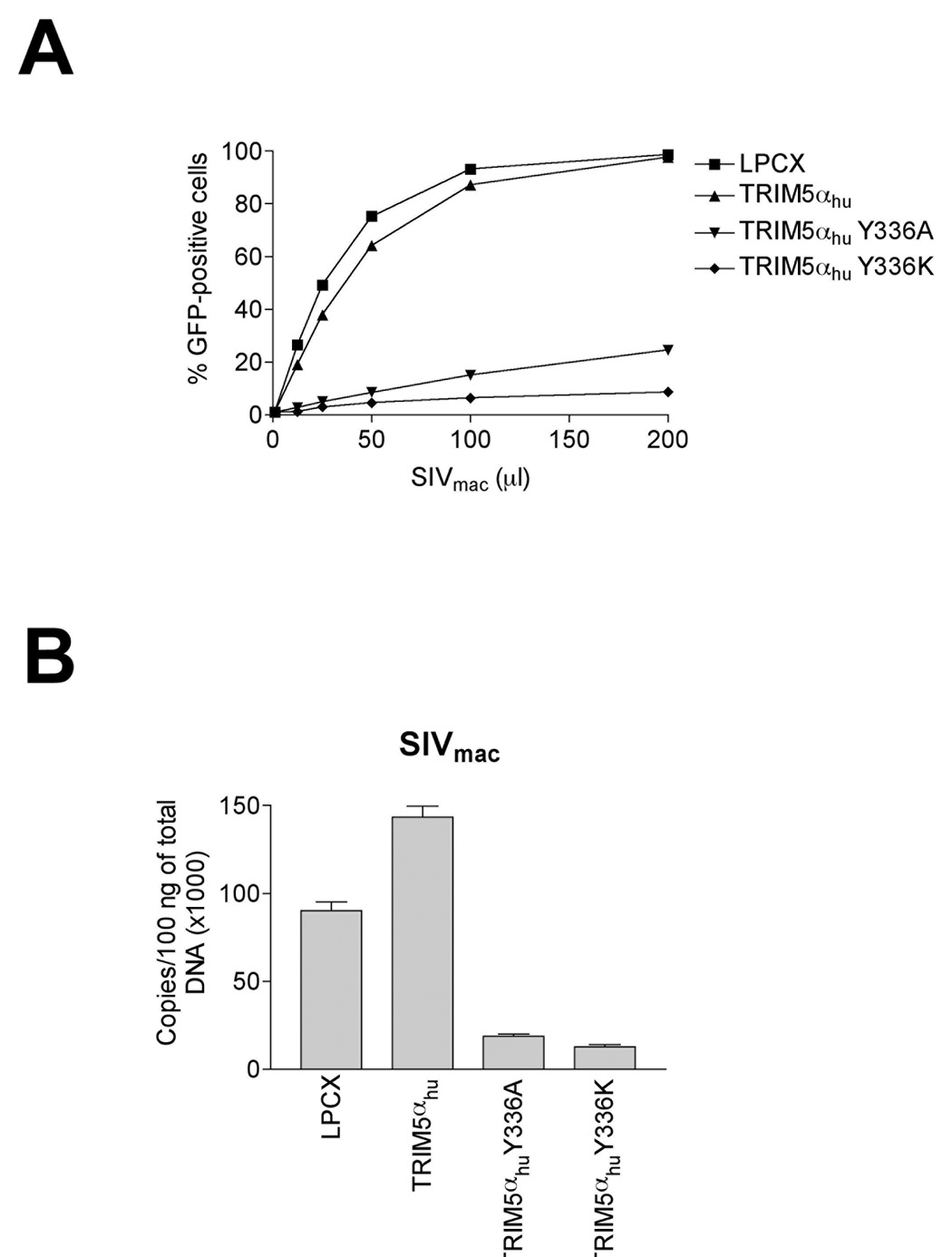
The wild-type TRIM5αhu protein does not restrict infection by SIVmac (Song et al., 2005b). However, TRIM5αhu Y336A and Y336K potently restricted SIVmac infection (Figure 4A). The Y336K mutant was more potent in this regard than the Y336A variant (Figure 4A). As shown in Figure 4B, the expression of TRIM5αhu Y336A and Y336K resulted in a marked decrease in the synthesis of LRTs following infection. Thus, changes in tyrosine 336 allow TRIM5αhu to restrict infection of SIVmac prior to reverse transcription.
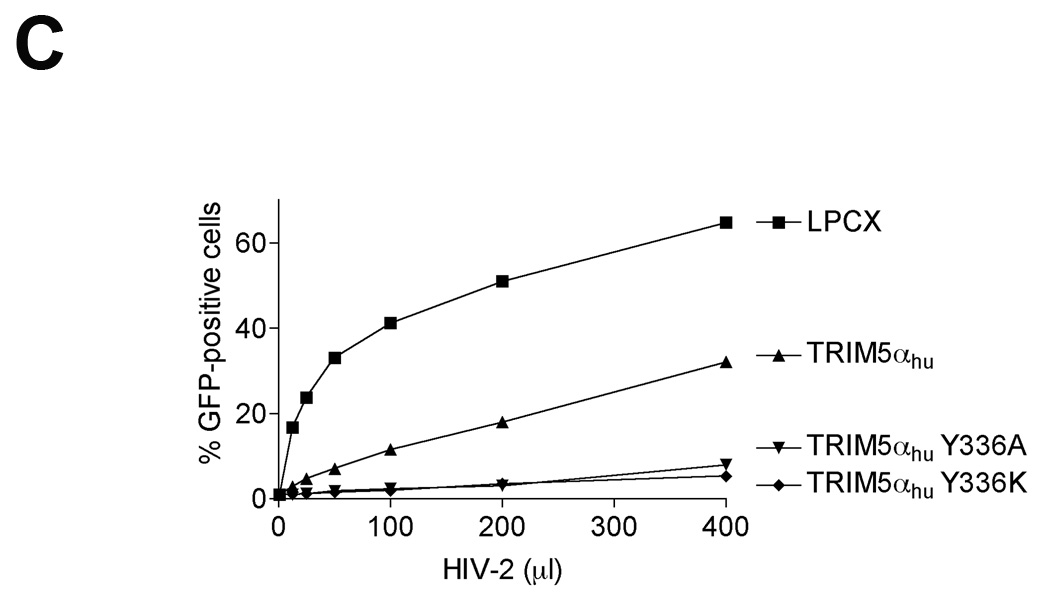
Figure 4. Inhibition of SIVmac and HIV-2 infection in cells expressing TRIM5αhu Y336A and Y336K.
(A) Cf2Th cells expressing the indicated wild-type and mutant TRIM5αhu proteins or containing the empty vector LPCX were challenged with SIVmac-GFP. GFP-positive cells were counted. (B) Cf2Th cells transduced with the empty LPCX vector or stably expressing wild-type and mutant TRIM5αhu proteins were challenged with SIVmac-GFP at an MOI of 0.4. After 7 hours, cells were lysed and total DNA extracted. The levels of viral DNA were measured by quantitative real-time PCR, using a probe against GFP as described in Materials and Methods. Similar results were obtained in three independent experiments. (C) Cf2Th cells expressing the indicated wild-type and mutant TRIM5αhu proteins or containing the empty vector LPCX were challenged with HIV-2-GFP. GFP-positive cells were counted. Similar results were obtained in two independent experiments.
The wild-type TRIM5αhu protein partially restricts infection by HIV-2 (Ylinen et al., 2005). The potency of HIV-2 restriction by TRIM5αhu was increased by the Y336A and Y336K changes (Figure 4C).
Effect of TRIM5αhu Y336A and Y336K on other retroviruses
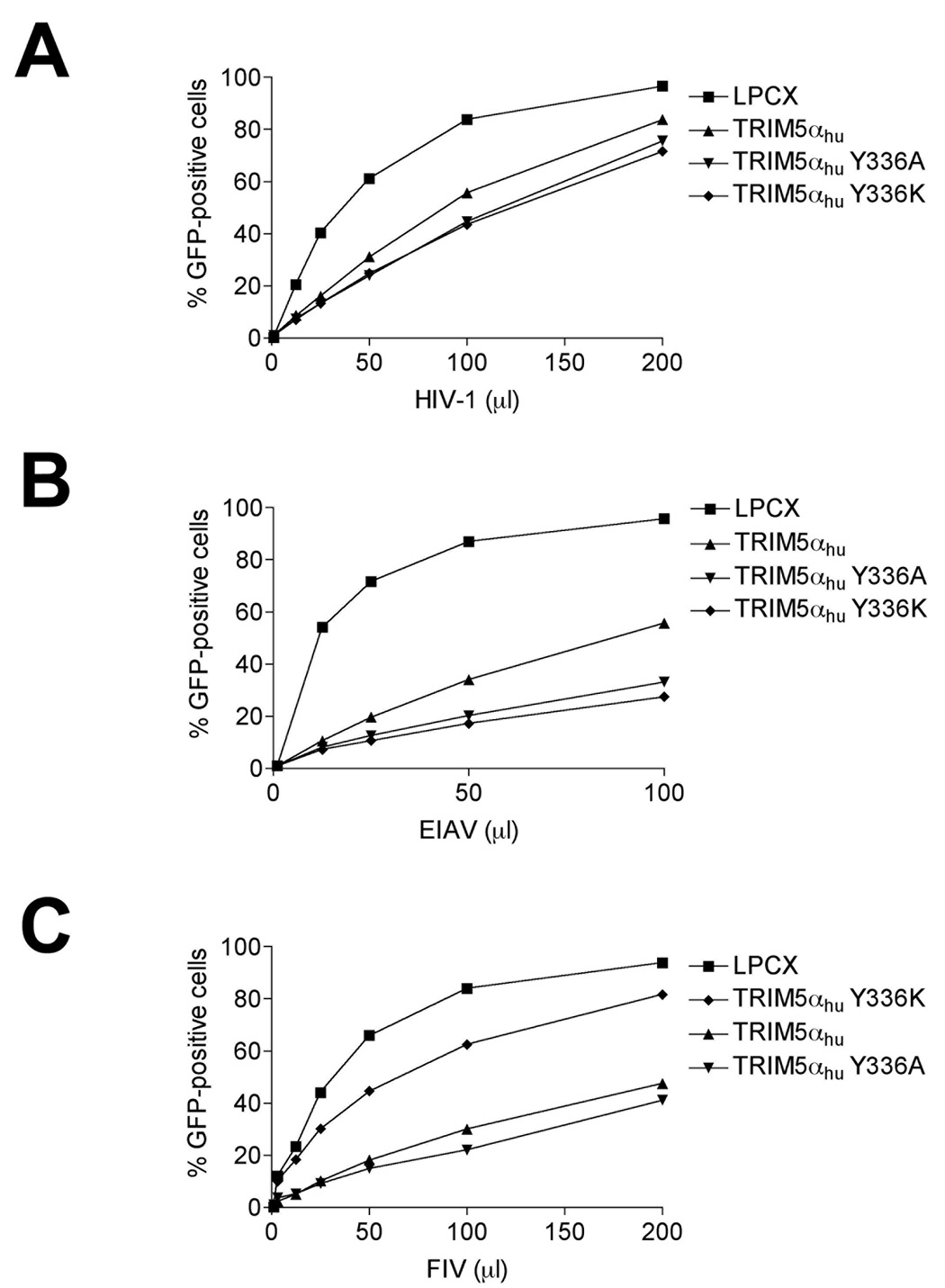
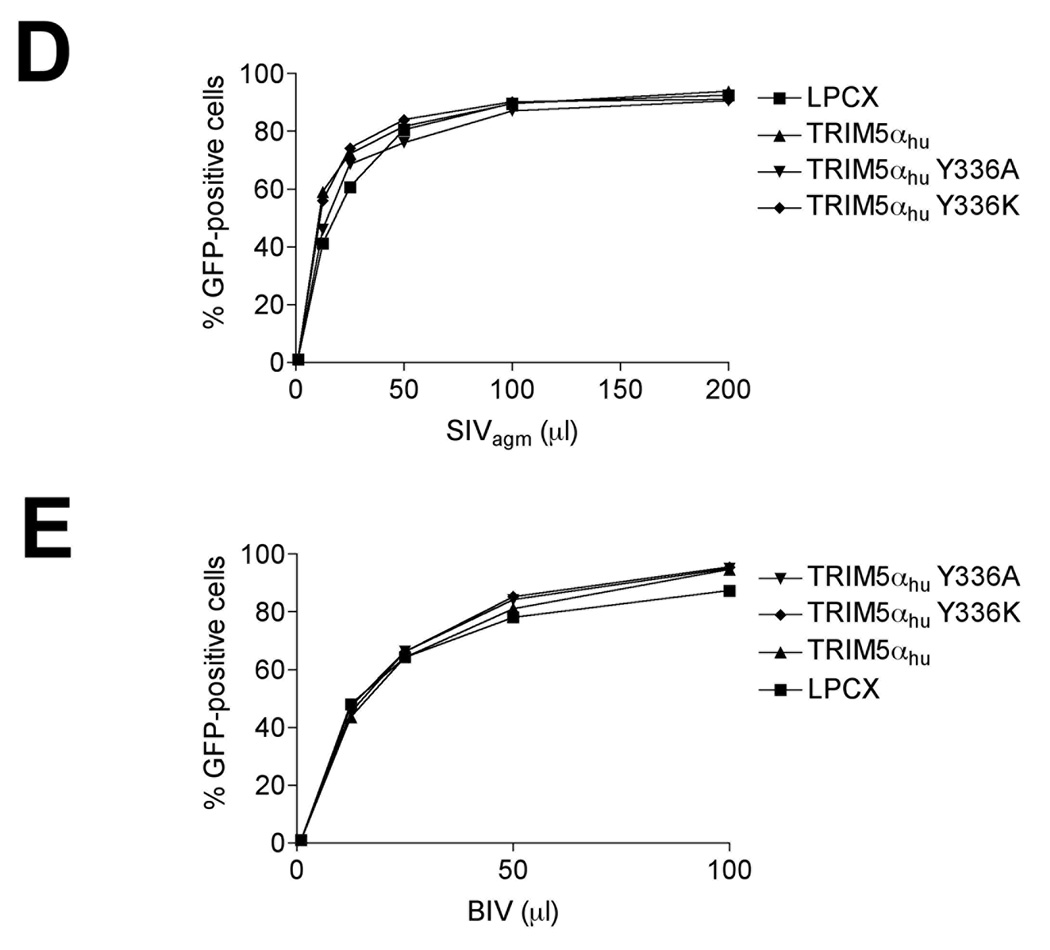
To test if infection by other retroviruses can be restricted by the TRIM5αhu Y336A and Y336K proteins, we challenged Cf2Th cells expressing these mutants with different retroviruses, including HIV-1, SIVagm, FIV, EIAV and BIV (Figure 5A–E). As expected, expression of the wild-type TRIM5αhu protein resulted in modest inhibition of HIV-1, EIAV and FIV infections (Figure 5A–C). The Y336A and Y336K mutants were slightly more effective than the wild-type TRIM5αhu protein in restricting HIV-1 and EIAV infection (Figure 5A and B). By contrast, the Y336K, but not the Y336A, mutant exhibited diminished ability to restrict FIV infection, compared with the wild-type TRIM5αhu protein (Figure 5C). The wild-type and mutant TRIM5αhu proteins did not affect SIVagm or BIV infection (Figure 5D and E). Thus, changes in tyrosine 336 in the B30.2/SPRY domain selectively increase or decrease the ability of TRIM5αhu to restrict infection by various retroviruses.
Figure 5. Inhibition of other retroviruses by TRIM5αhu Y336A and Y336K.
Cf2Th cells expressing the indicated wild-type and mutant TRIM5αhu proteins or the empty LPCX vector were challenged with HIV-1-GFP (A), EIAV-GFP (B), FIV-GFP (C), SIVagm-GFP (D) and BIV-GFP (E). GFP-positive cells were counted. Similar results were obtained in three independent experiments.
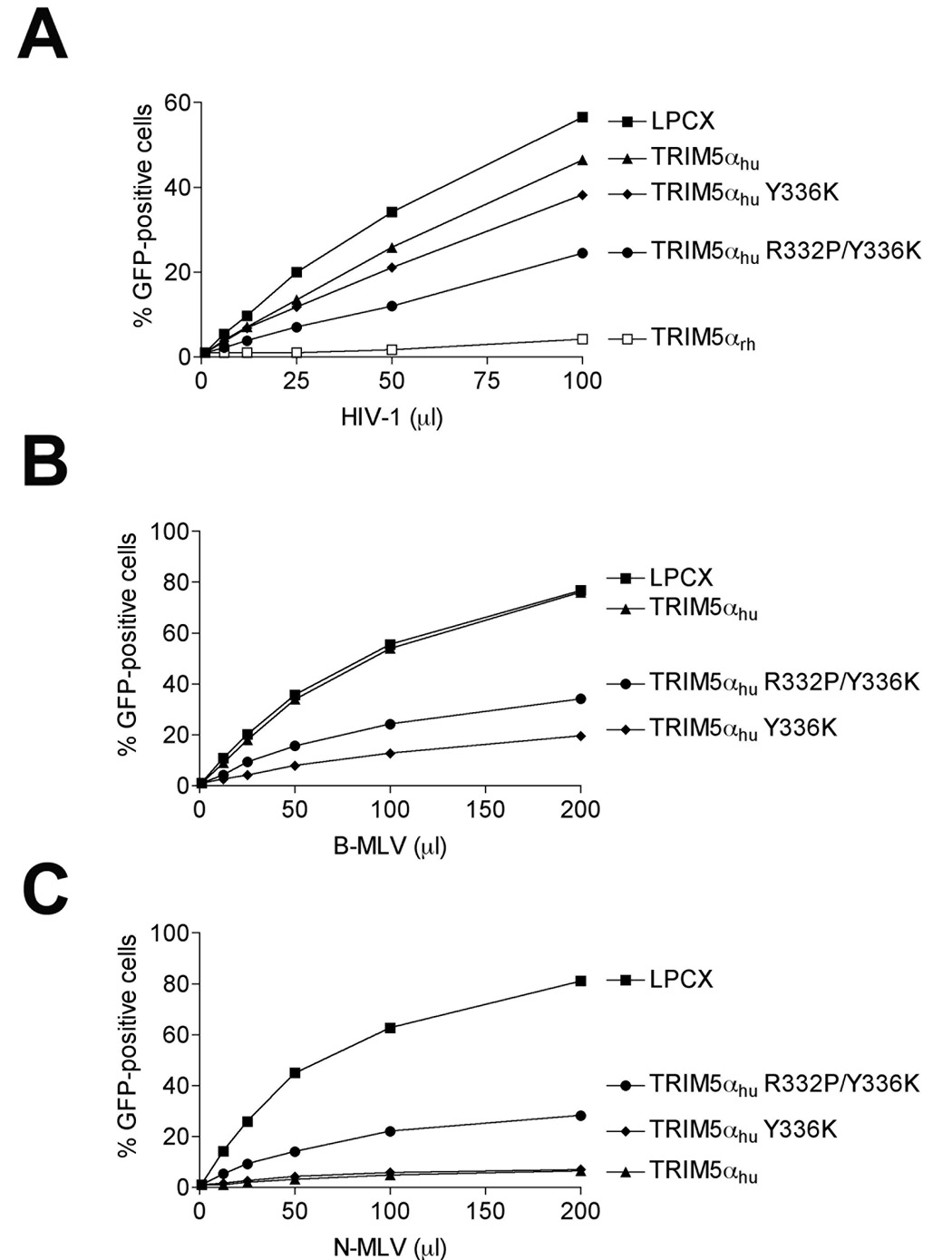
The combination of R332P and Y336K in TRIM5αhu only partially increased its anti-HIV-1 activity
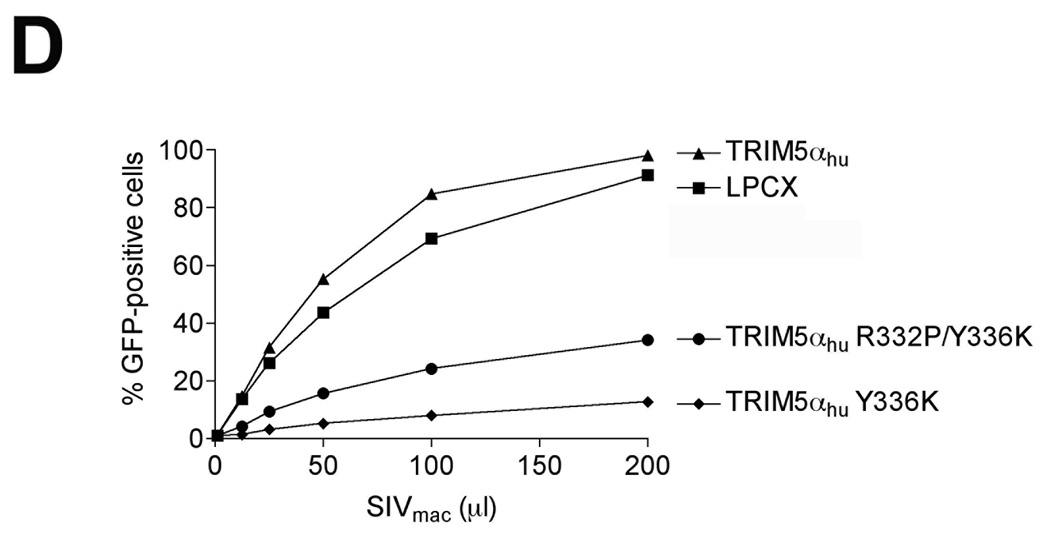
The R332P mutant of TRIM5αhu potently blocks HIV-1 and SIVmac infection (Li et al., 2006; Yap, Nisole, and Stoye, 2005). To determine if the R332P change would enhance the antiretroviral potency of TRIM5αhu Y336K, we constructed TRIM5αhu R332P/Y336K. The TRIM5αhu R332P/Y336K protein restricted HIV-1 infection slightly more efficiently than TRIM5αhu Y336K (Figure 6A). By contrast, the addition of the R332P change slightly decreased the restriction activity of TRIM5αhu Y336K against B-MLV, N-MLV and SIVmac (Figure 6B–D). Thus, in most cases, the phenotypes of the TRIM5αhu R332P and Y336K changes are not additive.
Figure 6. Effect of the R332P change on the anti-retroviral activity of TRIM5αhu Y336K.
Cf2Th cells expressing the indicated TRIM5α protein or the empty LPCX vector were challenged with HIV-1-GFP (A), B-MLV-GFP (B), N-MLV-GFP (C), and SIVmac-GFP (D). GFP-positive cells were counted. Similar results were obtained in three independent experiments.
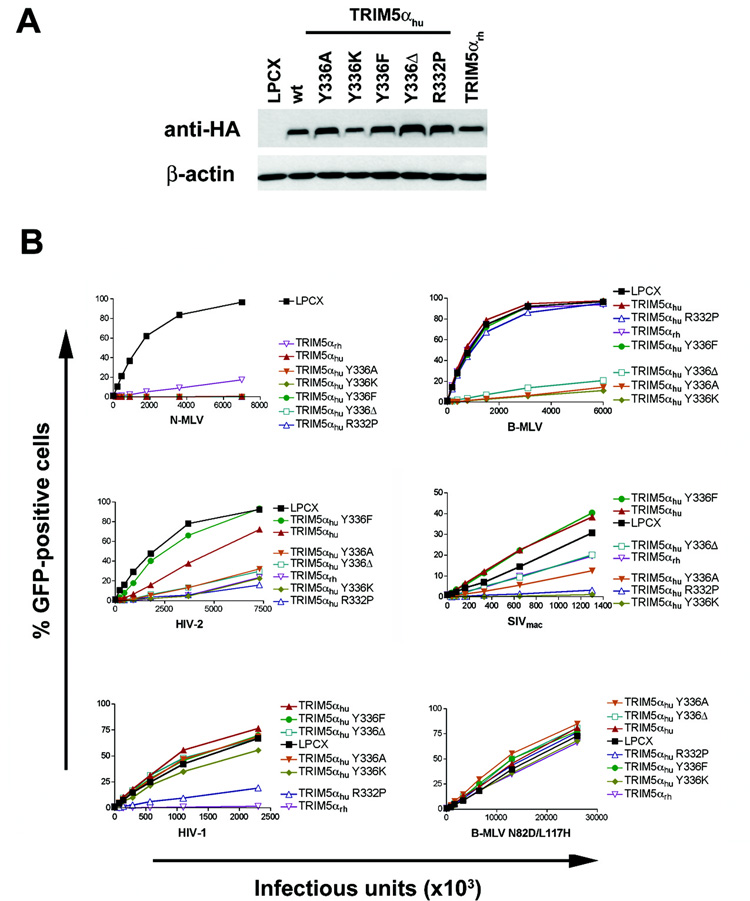
Ability of additional TRIM5αhu tyrosine 336 variants to restrict different retroviruses
The retrovirus-restricting abilities of additional TRIM5αhu mutants altered in tyrosine 336 were examined. TRIM5αhu Y336F has the more conservative substitution of a phenylalanine for tyrosine 336. TRIM5αhu Y336α has tyrosine 336 deleted. The mutants were efficiently expressed in Mus dunni tail fibroblasts cells (Figure 7A) and tested for the ability to inhibit infection by recombinant N-MLV, B-MLV, HIV-1, HIV-2 and SIVmac expressing GFP (Figure 7B). A B-MLV mutant with two amino acid changes in the capsid protein, B-MLV N82D L117H, was also tested for susceptibility to restriction by these TRIM5αhu variants. None of the tyrosine 336 changes affected the ability of TRIM5αhu to inhibit N-MLV infection potently. TRIM5αhu Y336K, Y336A and Y336Δ inhibited B-MLV and HIV-2 infection, but TRIM5αhu Y336F did not. Likewise, TRIM5αhu Y336F did not inhibit SIVmac infection, and TRIM5αhu Y336Δ exhibited only mild inhibition of SIVmac infection. Infection by B-MLV N82D L117H was not affected by any of the TRIM5αhu variants tested. HIV-1 infection was only mildly inhibited by TRIM5αhu Y336K and not the other TRIM5αhu variants with changes in tyrosine 336. Thus, TRIM5αhu and TRIM5αhu Y336F exhibited similar abilities to restrict infection by various retroviruses, whereas the TRIM5αhu Y336α protein exhibited restriction activity closer to that of the TRIM5αhu Y336A mutant.
Figure 7. Expression and retrovirus-restricting ability of TRIM5αhu variants with other changes in tyrosine 336.
(A) Wild-type (wt) TRIM5αhu or the indicated TRIM5αhu variants were stably expressed in Mus dunni tail fibroblasts. Cell lysates were Western blotted and probed with antibodies directed against the HA epitope tag (top panel) or α-actin (bottom panel). (B) Mus dunni tail fibroblasts transduced with the empty LPCX vector or stably expressing TRIM5αrh or the wild-type and mutant TRIM5αhu proteins were challenged with the indicated amounts of recombinant GFP-expressing viruses. GFP-positive cells were counted. Similar results were obtained in three independent experiments.
Discussion
The results presented herein underscore previous observations that alteration of the TRIM5α B30.2/SPRY v1 variable region can dramatically alter the range of retroviruses restricted (Li et al., 2006; Maillard et al., 2007; Sawyer et al., 2005; Yap, Nisole, and Stoye, 2005). Some alterations of tyrosine 336 in TRIM5αhu allow a broadening of the range of retroviruses potently restricted to include B-MLV, SIVmac and HIV-2, in addition to N-MLV. This result is reminiscent of the effects of changes in the nearby residue, arginine 332, in human TRIM5α; alteration or removal of this arginine creates a protein that acquires more potent restricting activity against HIV-1 and SIVmac (Yap et al., 2005; Stremlau et al., 2005; Li et al., 2006). In the case of HIV-1, this increased restricting ability results from improved binding of the TRIM5αhu derivative to viral capsid complexes (Li et al., 2006). Given the proximity of arginine 332 and tyrosine 336, the changes in the latter residue studied herein likely improve TRIM5αhu binding to the B-MLV capsid.
Most of the phenotypes observed for the TRIM5αhu Y336A and Y336K mutants were similar, despite the different nature of the substituted residues. As was seen for arginine 332 (Li et al., 2006; Yap, Nisole, and Stoye, 2005), deletion of tyrosine 336 results in a gain of ability to restrict some retroviruses. The TRIM5αhu mutant lacking tyrosine 336 was similar in phenotype to TRIM5αhu Y336A. By contrast, TRIM5αhu Y336F exhibited restriction phenotypes similar to those of wild-type TRIM5αhu. Apparently, an aromatic residue at position 336 of TRIM5αhu is an impediment to the restriction of some retroviruses (Maillard et al., 2007).
Remarkably, the gain in restricting activity against B-MLV that results from the tyrosine 336 changes also results in the acquisition of two other activities: 1) the ability to inhibit the synthesis of viral cDNA; and 2) the ability to promote the disappearance of particulate capsids from the cytosol. These observations support a growing correlation between TRIM5-mediated restriction of infection and premature conversion of particulate capsids to soluble proteins (Diaz-Griffero et al., 2007a; Perron et al., 2007; Stremlau et al., 2006). The results imply a likely mechanistic relationship among these processes, and support a model in which premature uncoating of the incoming retroviral capsid by a restricting TRIM5α protein leads to the disruption of the reverse transcription process.
Inhibition of the proteasome does not relieve TRIM5α-mediated restriction of retrovirus infection (Stremlau et al., 2006; Anderson et al., 2006; Wu et al., 2006), but increases the level of viral reverse transcripts in the target cell (Anderson et al., 2006; Wu et al., 2006). MLV-infected cells treated with proteasome inhibitors exhibited large increases in the amount of particulate, cytosolic capsids, regardless of the expression of TRIM5α in the target cells. Similar increases were previously observed in the levels of particulate capsids in the cytosol of HIV-1-infected control cells and cells expressing TRIM5α or TRIMCyp as a consequence of proteasome inhibition (Diaz-Griffero et al., 2007a). The levels of late reverse transcripts changed in parallel with the levels of particulate cytosolic capsids, consistent with the notion that loss of capsid integrity leads to disruption of reverse transcription. In many cases, the generalized increases in the levels of cytosolic capsids and late reverse transcripts resulting from proteasome inhibition obscured the TRIM5α- or TRIMCyp-mediated decrease in these parameters (Diaz-Griffero et all, 2007). However, the efficiency with which N-MLV capsids can be recovered from sucrose gradients allowed detection of human TRIM5α-associated decreases in pelletable capsid and late reverse transcripts even in the presence of a proteasome inhibitor. Therefore, an active proteasome is not absolutely required for the accelerated uncoating of N-MLV capsids by human TRIM5α, or for its subsequent impact on the efficiency of reverse transcription.
Future studies should further clarify the structural requirements for TRIM5α interaction with the retroviral capsid and the mechanistic consequences of this recognition event.
Materials and Methods
Creation of cells stably expressing TRIM5α variants
Retroviral vectors encoding wild-type or mutant TRIM5αhu proteins were created using the pLPCX vector. Recombinant viruses were produced in 293T cells by cotransfecting the pLPCX plasmids with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus (VSV) G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells. Cf2Th canine thymocytes or Mus dunni tail fibroblasts were transduced and selected in 1–5 µg/ml puromycin (Sigma).
Infection with viruses expressing green fluorescent protein (GFP)
Recombinant HIV-1, simian immunodeficiency viruses from macaques and African green monkeys (SIVmac and SIVagm, respectively), N-tropic murine leukemia virus (N-MLV) and B-tropic murine leukemia virus (B-MLV) expressing GFP were prepared as described (Diaz-Griffero et al., 2006; Stremlau et al., 2004). The feline immunodeficiency virus (FIV) vector expressing GFP was obtained from System Biosciences (Mountain View, CA) and was prepared following the manufacturer’s instructions. The equine infectious anemia virus (EIAV) vector expressing GFP was a gift from Dr. John Olsen (Olsen, 1998). Bovine immunodeficiency virus (BIV) vectors expressing GFP were prepared as described (Berkowitz et al., 2001). All recombinant viruses were pseudotyped with the VSV G glycoprotein. For infections, 3 × 104 Cf2Th cells seeded in 24-well plates were incubated with virus for 24 hours. Cells were washed and returned to culture for 48 hours, and then subjected to FACS analysis with a FACScan (Becton Dickinson). HIV-1, N-MLV, B-MLV, FIV, BIV, SIVmac, SIVagm and EIAV viral stocks were titrated by serial dilution on Cf2Th cells to determine the concentration of infectious viruses. Similarly, viruses used in the experiments shown in Figure 7 were titrated in Mus dunni tail fibroblasts.
Protein analysis
Cellular proteins were extracted with radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris, pH 7.4; 100 mM NaCl; 1% sodium deoxycholate; 0.1% sodium dodecyl sulfate [SDS]; 1% NP-40; 2 mg/ml aprotinin; 2 mg/ml leupeptin; 1 mg/ml pepstatin A; 100 mg/ml phenylmethylsulfonyl fluoride). The cell lysates were analyzed by SDS-PAGE (10% acrylamide), followed by blotting onto nitrocellulose membranes (Amersham Pharmacia Biotech). Detection of protein by Western blotting utilized monoclonal antibodies directed against the HA epitope tags (Roche) and monoclonal antibodies to β-actin (Sigma). Detection of proteins was performed by enhanced chemiluminescence (NEN Life Sciences Products), using anti-mouse immunoglobulin (for β-actin) and anti-rat immunoglobulin (for HA) secondary antibodies (Amersham Pharmacia Biotech).
Localization of wild-type and mutant TRIM5α proteins
The localization of TRIM5αhu and variants was investigated as previously described (Diaz-Griffero, Hoschander, and Brojatsch, 2002). Briefly, cells were grown overnight on 12-mm-diameter coverslips and fixed in 3.9% paraformaldehyde (Sigma) in phosphate-buffered saline (PBS; Cellgro) for 30 minutes. Cells were washed in PBS, incubated in 0.1 M glycine (Sigma) for 10 minutes, washed in PBS, and permeabilized with 0.05% saponin (Sigma) for 30 minutes. Samples were blocked with 10% donkey serum (Dako, Carpinteria, CA) for 30 minutes, and incubated for 1 hour with antibodies. The anti-HA FITC-conjugated 3F10 antibody (Roche) was used to stain HA-tagged proteins. Subsequently, samples were mounted for fluorescence microscopy by using the ProLong Antifade Kit (Molecular Probes, Eugene, OR). Images were obtained with a BioRad Radiance 2000 laser scanning confocal microscope with Nikon 60X N.A.1.4 optics.
Fate-of-capsid assay
Recombinant N-MLV and B-MLV virus-like particles (VLPs) were produced by calcium phosphate cotransfection of 293T cells with the pCIG-N or pCIG-B vectors (Bock et al., 2000), respectively, along with pVPack-VSV-G at a weight ratio of 15:4, respectively. Stably transduced Cf2Th (1.5 × 106) cells expressing the different TRIM5αhu variants were seeded in 80 cm2 flasks. The following day, the cells were incubated with 5–10 ml (approximately 2.5– 5.0 × 105 reverse transcriptase units) of N-MLV or B-MLV VLPs at 4°C for 30 minutes to allow vira l attachment to the cells. The cells were then shifted to 37°C until they were harvest ed at 4 hours post-infection. Cells were washed three times using ice-cold PBS and detached by treatment with 1.0 ml of pronase (7.0 mg/ml in DMEM) for 5 minutes at 25°C. The cells were then washed three times with PBS. The cells were resuspended in 2.5 ml hypotonic lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM KCl, 1 mM EDTA and one complete protease inhibitor tablet) and incubated on ice for 15 minutes. The cells were lysed using 15 strokes in a 7.0 ml Dounce homogenizer with pestle B. Cellular debris was cleared by centrifugation for 3 minutes at 3000 rpm. To allow assessment of the total input (Total or Input) MLV p30 capsid protein, 100 µl of the cleared lysate were collected, made 1× in SDS sample buffer, and analyzed by SDS-PAGE and Western blotting. Then 2.0 ml of the cleared lysate were layered onto a 50% sucrose (weight:volume) cushion in 1× PBS and centrifuged at 125,000 × g for 2 hours at 4°C in a Be ckman SW41 rotor. Following centrifugation, 100 µl of the top-most portion of the supernatant were collected and made 1× in SDS sample buffer; this sample is referred to as the soluble p30 fraction. The pellet was resuspended in 50 µl 1× SDS sample buffer and is referred to as the particulate p30 fraction. All samples were then subjected to SDS-PAGE and Western blotting. The MLV p30 protein was detected using a rat monoclonal antibody purified from the R187 hybridoma cell line (American Type Culture Collection).
To examine the effects of proteasome inhibition, the Cf2Th target cells were incubated in the presence of 10 µM MG115, a proteasome inhibitor, for 1 hour at 37°C prior to the addition of N-MLV or B-MLV VLPs. In some experiments, this 1-hour preincubation was not performed, with no effect on the outcome (data not shown). Following the preincubation step, viruses and either DMSO or MG115 were incubated with the target cells at 37°C for 4 hours. Cells were washed, harvested and processed for the fate-of-capsid assay (see above) or for measurement of LRTs (see below).
Quantitative real-time PCR
Cf2Th cells expressing wild-type and mutant TRIM5αhu proteins were challenged at a multiplicity of infection (MOI) of 0.2 for N-MLV-GFP and B-MLV-GFP, or 0.4 for SIVmac-GFP. Viruses were pretreated with DNAse to prevent contamination from carry-over plasmid DNA. As a control to establish background, viruses were heat-inactivated (30 minutes at 65°C). In some cases, the Cf2Th cells wer e treated with MG115 (10 µg/ml) or DMSO, as described above. After 4 or 7 hours of incubation with the virus (and compound, if included), cells were lysed and DNA was extracted using a Qiagen Blood Tissue DNA extraction kit. PCR reactions were prepared using the QuantiTect probe PCR kit. Each sample contained 100 ng of total cellular DNA. PCR was carried out using two primers that amplify a 263-bp fragment of GFP:
GFP-fwd: 5’-GAC GTA AAC GGC CAC AAG-3’
GFP-rev: 5’-GGT CTT GTA GTT GCC GTC GT-3’
GFP-Probe: 5’-56-FAM-CCT ACG GCA AGC TGA CCC TGA-36-TAMRA-3’
The calibration curve was prepared using an MLV-GFP plasmid. The background observed for the heat-inactivated viruses was subtracted from all values.
Acknowledgments
We thank Ms. Yvette McLaughlin and Ms. Elizabeth Carpelan for manuscript preparation, Drs. Gregory Towers and Andrew Lever for providing recombinant HIV-2, and the National Institutes of Health (AI063987, AI076094 and a Center for AIDS Research Award AI60354), the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, and the late William F. McCarty-Cooper for research funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JL, Campbell EM, Wu X, Vandegraaff N, Engelman A, Hope TJ. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J Virol. 2006;80(19):9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz R, Ilves H, Lin WY, Eckert K, Coward A, Tamaki S, Veres G, Plavec I. Construction and molecular analysis of gene transfer systems derived from bovine immunodeficiency virus. J Virol. 2001;75(7):3371–3382. doi: 10.1128/JVI.75.7.3371-3382.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock M, Bishop KN, Towers G, Stoye JP. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J Virol. 2000;74(16):7422–7430. doi: 10.1128/jvi.74.16.7422-7430.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan S, Hatziioannou T, Cunningham T, Muesing MA, Gottlinger HG, Bieniasz PD. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci U S A. 2002;99(18):11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Hoschander SA, Brojatsch J. Endocytosis is a critical step in entry of subgroup B avian leukosis viruses. J Virol. 2002;76(24):12866–12876. doi: 10.1128/JVI.76.24.12866-12876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Kar A, Lee M, Stremlau M, Poeschla E, Sodroski J. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology. 2007a;369(2):400–410. doi: 10.1016/j.virol.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Kar A, Perron M, Xiang SH, Javanbakht H, Li X, Sodroski J. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5alpha B-box 2 domain. J Virol. 2007b;81(19):10362–10378. doi: 10.1128/JVI.00703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Vandegraaff N, Li Y, McGee-Estrada K, Stremlau M, Welikala S, Si Z, Engelman A, Sodroski J. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology. 2006;351(2):404–419. doi: 10.1016/j.virol.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Dueck M, Guatelli J. Evidence against a direct antiviral activity of the proteasome during the early steps of HIV-1 replication. Virology. 2007;361:1–8. doi: 10.1016/j.virol.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Wei BL, Yao J, Luo T, Garcia JV. Inhibition of endosomal/lysosomal degradation increases the infectivity of human immunodeficiency virus. J Virol. 2002;76:11440–11446. doi: 10.1128/JVI.76.22.11440-11446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc Natl Acad Sci U S A. 2004;101(29):10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. Species-specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999;73(12):10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LM, Towers GJ. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci U S A. 2004;101(29):10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li X, Stremlau M, Lee M, Sodroski J. Removal of arginine 332 allows human TRIM5alpha to bind human immunodeficiency virus capsids and to restrict infection. J Virol. 2006;80(14):6738–6744. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, Reynard S, Serhan F, Turelli P, Trono D. Interfering residues narrow the spectrum of MLV restriction by human TRIM5alpha. PLoS Pathog. 2007;3(12):e200. doi: 10.1371/journal.ppat.0030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk C, Brandt SM, Lucero G, Landau NR. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc Natl Acad Sci U S A. 2002;99(21):13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura S, Yap MW, Sheldon T, Stoye JP. All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol. 2006;80(17):8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JC. Gene transfer vectors derived from equine infectious anemia virus. Gene Ther. 1998;5(11):1481–1487. doi: 10.1038/sj.gt.3300768. [DOI] [PubMed] [Google Scholar]
- Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J. The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol. 2007;81(5):2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MJ, Stremlau M, Sodroski J. Two surface-exposed elements of the B30.2/SPRY domain as potency determinants of N-tropic murine leukemia virus restriction by human TRIM5alpha. J Virol. 2006;80(11):5631–5636. doi: 10.1128/JVI.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MJ, Stremlau M, Song B, Ulm W, Mulligan RC, Sodroski J. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci U S A. 2004;101(32):11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 2005;102(8):2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard JM. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J Virol. 1998;73:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Gold B, O'Huigin C, Javanbakht H, Li X, Stremlau M, Winkler C, Dean M, Sodroski J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J Virol. 2005a;79(10):6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Javanbakht H, Perron M, Park DH, Stremlau M, Sodroski J. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J Virol. 2005b;79(7):3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79(5):3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers G, Bock M, Martin S, Takeuchi Y, Stoye JP, Danos O. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci U S A. 2000;97(22):12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci U S A. 2006;103(19):7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15(1):73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- Ylinen LM, Keckesova Z, Wilson SJ, Ranasinghe S, Towers GJ. Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5alpha alleles. J Virol. 2005;79(18):11580–11587. doi: 10.1128/JVI.79.18.11580-11587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]