SUMMARY
FoxO transcription factors regulate the transcription of genes that control metabolism, cellular proliferation, stress tolerance and possibly lifespan. A number of post-translational modifications within the forkhead DNA binding domain regulate FoxO mediated transcription. We report the crystal structures of FoxO1 bound to three different DNA elements and measure the change in FoxO1-DNA affinity with acetylation and phosphorylation. The structures reveal additional contacts and increased DNA distortion for the highest affinity DNA site. The flexible wing 2 region of the forkhead domain was not observed in the structures but is necessary for DNA-binding, and we show that p300 acetylation in wing 2 reduces DNA affinity. We also show that MST1 phosphorylation of FoxO1 prevents high affinity DNA binding. The observation that FoxO-DNA affinity varies between response elements and with post-translational modifications suggests that modulation of FoxO-DNA affinity is an important component of FoxO regulation in health and misregulation in disease.
INTRODUCTION
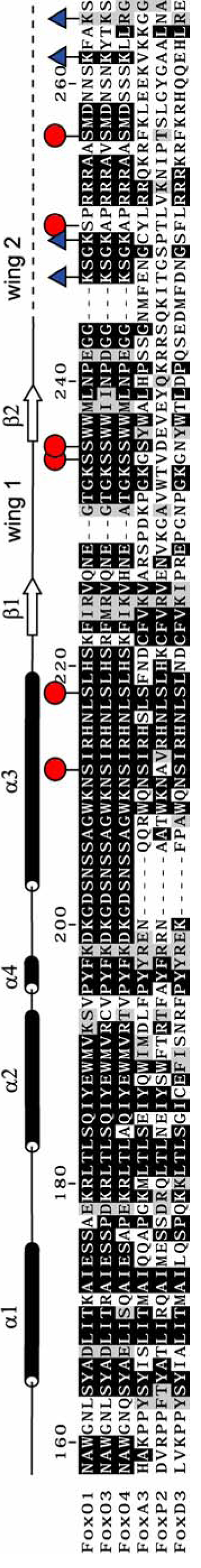
FoxO1 belongs to a family of transcription factors that share a conserved 100 amino acid forkhead box or winged helix DNA-binding domain. The nomenclature for the more than 100 members of this gene family has been standardized such that all members start with Fox (Forkhead box), followed by a letter to distinguish 17 subfamilies, and a number to distinguish individual members (Kaestner et al., 2000) (Figure 1). The many members of this family are emerging as critical regulators of development, immunity, metabolism and cancer. The FoxO subfamily has been shown to have roles in apoptosis, stress resistance, cell cycle arrest, DNA damage repair response and glucose metabolism in mammalian cells (Greer and Brunet, 2005). In addition, recent mouse knockout studies have shown that FoxOs are tumor suppressors (Paik et al., 2007). FoxO also represents the closest human homologues of the C. elegans longevity gene daf-16, the downstream target of daf-2 insulin/IGF-I receptor and C. elegans sir-2.1 (Kenyon et al., 1993; Mazet et al., 2003; Tissenbaum and Guarente, 2001). In humans, FoxO activity is regulated by a number of post-translational modifications (Huang and Tindall, 2007) (Figure 1), and misregulation of FoxO has been shown to play a role in diseases of aging (Anderson et al., 1998; Borkhardt et al., 1997; Cheong et al., 2003; Cohen et al., 2006; Davis et al., 1994; Galili et al., 1993; Hillion et al., 1997; Hu et al., 2004; Modur et al., 2002; Paik et al., 2007; Parry et al., 1994; Seoane et al., 2004; Sunters et al., 2006; Tothova et al., 2007).
Figure 1.
FoxO1 secondary structure and sequence alignment of human FoxO1 DBD with other forkhead domains. Residues that undergo phosphorylation or acetylation are marked with a red circle or a blue triangle respectively.
Previous structures of the FoxA3/HNF-3γ, FoxD3/Genesis, FoxP2, and FoxK1a/ILF-1 forkhead domains bound to their DNA consensus sequences have revealed a compact three helix fold with the third helix sitting in the major groove of B-form DNA, and C-terminal β–strands projecting along the axis of the DNA to contact one or both of the adjacent minor grooves (Clark et al., 1993; Jin et al., 1999; Stroud et al., 2006; Tsai et al., 2006). Solution structures have also been reported for FoxO4/AFX and FoxC2/FREAC11 (van Dongen et al., 2000; Weigelt et al., 2001). The FoxA3/HNF-3γ structure, the namesake of the winged helix domain, was the first reported and it revealed two well ordered loops or wings contacting each of the adjacent minor grooves (Clark et al., 1993). Later structures revealed significant diversity in DNA binding, particularly in the C-terminal wing 2 region. The FoxD3/Genesis, FoxK1a/ILF-1 and FoxP2 structures all contain a combination of α-helix and loop in the wing 2 region and mediate divergent interactions with DNA (Jin et al., 1999; Liu et al., 2002; Stroud et al., 2006; Tsai et al., 2006). Outside of the conserved DNA binding motif, forkhead transcription factors also show important differences in their domain structure, expression, regulation and disease association (Myatt and Lam, 2007).
FoxO1 was originally named FKHR or forkhead in rhabdomyosarcomas due to its association with a chromosomal translocation in alveolar rhabdomyosarcoma (Galili et al., 1993). Later, daf-16, the C. elegans homolog of FoxO1/FKHR, was characterized as a key downstream target in the insulin/IGF-1 signaling pathway that is required for mutations in the insulin/IGF-1-like receptor daf-2 to confer increased lifespan in a C. elegans longevity model (Gottlieb and Ruvkun, 1994; Kenyon et al., 1993; Larsen et al., 1995; Ogg et al., 1997). This pathway is well conserved in humans where nutrient abundance triggers insulin/IGF-1 receptor signaling that leads to Akt phosphorylation of FoxO1 at Thr24, Ser256, and Ser319 (Biggs et al., 1999; Brunet et al., 1999; Rena et al., 1999; Tang et al., 1999). This phosphorylation causes an interaction with 14-3-3 proteins that localizes FoxO1 to the cytoplasm to block transcriptional activation by FoxO1 (Brunet et al., 1999). Both CDK1 and CDK2 have been reported to phosphorylate FoxO1 at Ser249 to regulate subcellular localization of FoxO1 (Huang et al., 2006; Yuan et al., 2008). The effect of this modification on FoxO1 localization and activity is controversial with CDK1 reported to cause nuclear localization and transcriptional activation (Yuan et al., 2008), and CDK2 reported to cause cytoplasmic localization and inhibition of FoxO1 (Huang et al., 2006). MST1 kinase has been shown to phosphorylate four sites on FoxO3, Ser207, Ser213, Ser229, and Ser230, in response to oxidative stress in neurons (Lehtinen et al., 2006). This phosphorylation is reported to disrupt FoxO’s interaction with 14-3-3 proteins, and allow nuclear translocation of FoxO where it initiates apoptotic cell death (Lehtinen et al., 2006). The MST1 sites are conserved in FoxO1 and are also likely targets of MST1 regulation.
In addition to phosphorylation, the DNA-binding domain of FoxO can be acetylated by CBP/p300, and SIRT1 and SIRT2, human homologs of the yeast Sir2 longevity protein, can deacetylate FoxO (Brunet et al., 2004; Daitoku et al., 2004; Fukuoka et al., 2003; Jing et al., 2007; Motta et al., 2004; van der Horst et al., 2004). This provides an additional level of control over the FoxO transcription factors and a connection to the Sir2 model for longevity by calorie restriction. It has been shown that acetylation of three lysines, all within or near the DNA-binding domain of FoxO1 reduces the affinity of FoxO1 for target DNA and increases Akt phosphorylation of FoxO1 (Jing et al., 2007; Matsuzaki et al., 2005). The biological effects of the acetylation state of FoxO are diverse and dependent on both the family member (FoxO1, FoxO3, FoxO4 and FoxO6) and on the cellular context. Deacetylation of FoxO3 by SIRT1 is associated with repressed FoxO induced apoptosis (Motta et al., 2004); however, in response to oxidative stress, SIRT1 deacetylation of FoxO3 promotes cell cycle arrest and resistance to oxidative stress (Brunet et al., 2004). In adipocytes, SIRT2 deacetylation of FoxO1 causes FoxO1 to suppress adipogenesis through its transcriptional regulation of cell cycle inhibitors (Jing et al., 2007; Nakae et al., 2003).
The post-translational modifications discussed here only represent the phosphorylation and acetylation sites within or near the DNA-binding domain (DBD) of FoxO1, or amino acids 158–248 in the case of FoxO1. These sites are interesting from a structural perspective because they may regulate FoxO1 activity in different ways. They may disrupt or enhance DNA binding directly or they may mediate protein-protein interactions that regulate FoxO1. Yet another factor that is reported to be important for regulation of target gene expression by forkhead transcription factors is variations in affinity for different DNA response elements. For FoxA transcription factor in C. elegans, it is reported that high affinity DNA elements cause FoxA target genes to be expressed earlier in embryonic development and low affinity DNA elements cause delayed onset of target gene expression(Gaudet and Mango, 2002).
Here we report structures of the FoxO1 DBD bound to closely related recognition elements: the insulin response element (IRE) with the consensus sequence of TT(G/A)TTTTG was the first recognition element reported for FoxO1 (Brunet et al., 1999; Guo et al., 1999; Kops et al., 1999; Tang et al., 1999) and the Daf-16 family binding element (DBE) with the consensus sequence of TT(G/A)TTTAC, which is bound by FoxO1 more strongly (Furuyama et al., 2000). We also report a third structure of FoxO1 DBD bound to a higher affinity DBE sequence, called DBE2 here, which reveals that FoxO1 bending of the flanking bases outside of the 8-base consensus sequence may be important for optimal DNA binding by FoxO1. We then measure the effect of acetylation and phosphorylation on DNA binding affinity of FoxO1 to provide a structural basis for DNA recognition and post-translational regulation of FoxO1.
RESULTS
Structures of FoxO1 DBD bound to DBE and IRE DNA
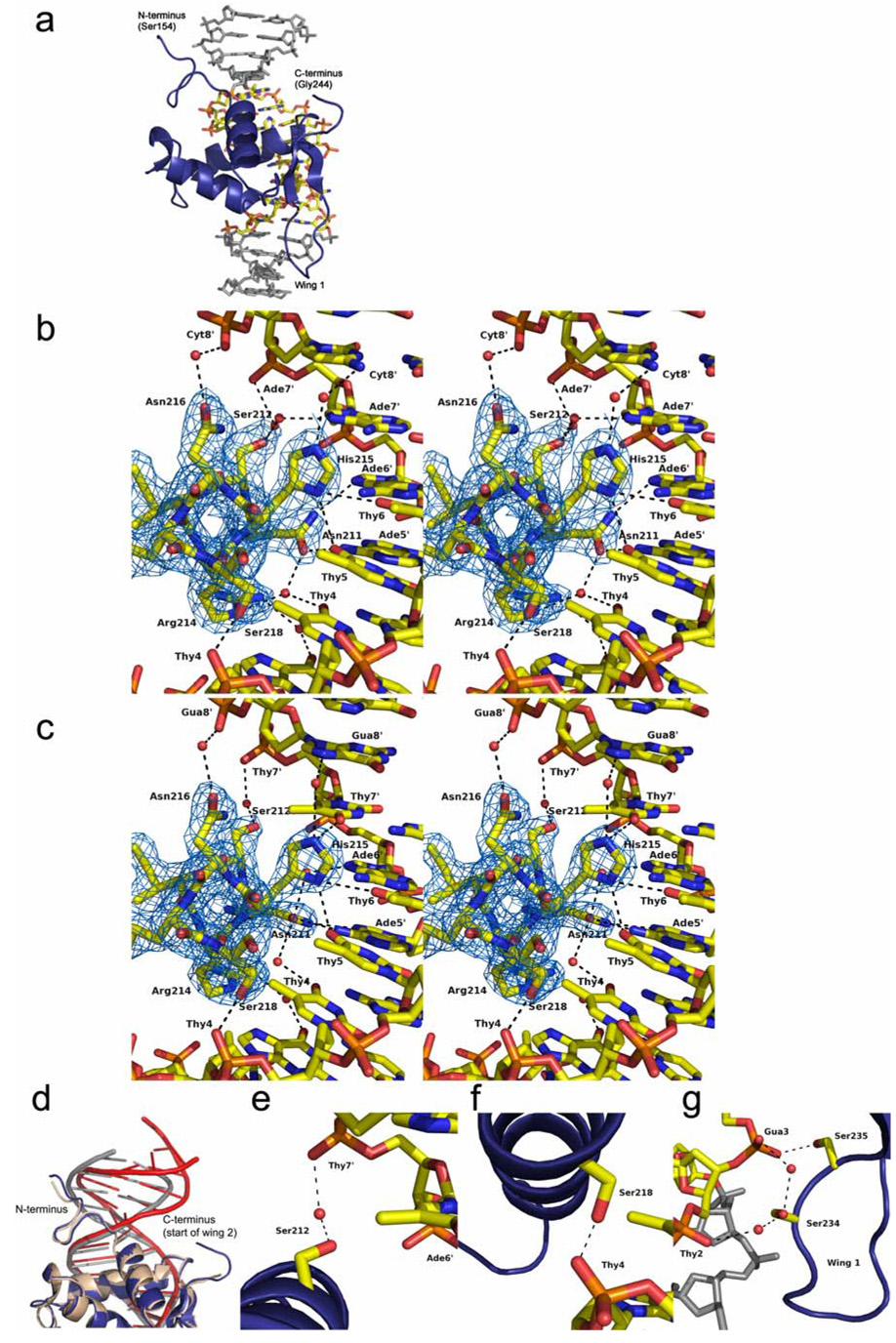
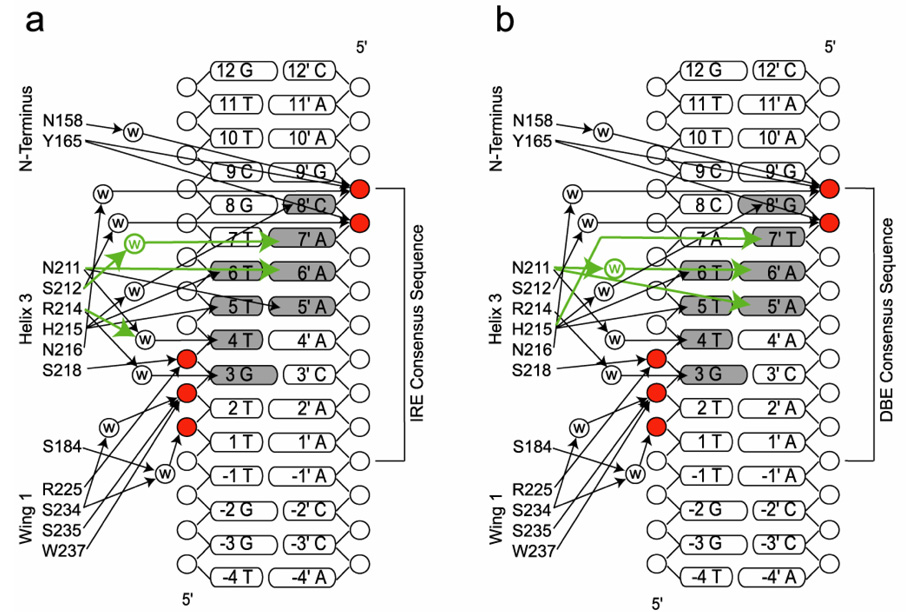
Structures of the FoxO1 DBD bound to 16-base-pair DNA containing the DBE recognition sequence of 5’-TTGTTTAC-3’ or the IRE recognition sequence of 5’-TTGTTTTG-3’ were determined to 2.1 Å and 2.2 Å respectively (Table 1 and Figure 2). The structures reveal the expected forkhead or winged helix DNA binding motif with helix 3 binding in the major groove of the DNA and the side chains of Asn211 and His215 making all of the direct base specific contacts (Figure 2b, 2c and Figure 3). Side chains of Asn158 and Tyr165 from the N-terminus of the DBD and Arg225, Ser234, Ser235 and Trp237 from wing 1 make phosphate contacts to adjacent minor grooves of the DNA (Figure 3). The 2.9 Å structure of FoxO1 bound to the high affinity DBE2 DNA sequence shows the same interactions around helix 3; however, it also shows a greater amount of bend in the 6 base pairs flanking the DBE consensus sequence, where wing 2 is predicted to make interactions (Figure 2d). Interestingly, unlike previously reported forkhead structures, electron density was not observed for the C-terminal wing 2 region of the FoxO1 DBD suggesting that it is flexible and disordered in the crystals. Given the lack of sequence homology to other forkhead subfamilies in this region (Figure 1) and the previously observed structural diversity at the C-terminus of forkhead domains this finding was not surprising, but required further investigation as described below.
Table 1.
Data collection and refinement statistics for FoxO1/DNA complexes.
| FoxO1/IRE DNA | FoxO1/DBE1 DNA | FoxO1/DBE2 DNA | |||
|---|---|---|---|---|---|
| Native | Hg S184C | Hg A207C | Native | Native | |
| Data Collection | |||||
| Wavelength (Å) | 0.88560 | 1.54178 | 1.54178 | 1.00000 | 0.97949 |
| Space Group | P21212 | I222 | I222 | I222 | P32 |
| a, b, c (Å) | 76.06 | 65.40 | 65.30 | 65.52 | 99.64 |
| 102.43 | 76.17 | 76.66 | 76.44 | 99.64 | |
| 65.45 | 102.34 | 102.36 | 102.14 | 98.47 | |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 30-2.2 | 30-3.1 | 30-3.2 | 30-2.1 | 30-2.9 |
| Redundancy | 11.0(9.9) | 5.1(5.0) | 6.2(5.8) | 4.8(4.9) | 4.3(4.3) |
| Completeness (%) | 99.4(98.1) | 99.3(98.9) | 99.8(99.5) | 99.9(100.0) | 99.6(99.8) |
| Rsym (%) | 6.4(32.7) | 11.8(31.1) | 16.0(37.3) | 7.2(34.0) | 8.8(32.4) |
| I/σ (I) | 39.4(6.9) | 12.8(6.9) | 11.9(6.3) | 20.2(4.5) | 15.7(4.9) |
| Phasing Analysis | |||||
| Resolution (Å) | 30-3.20 | ||||
| Number of sites | 2 | ||||
| Figure of merit (FOM) | 0.40 | ||||
| Refinement | |||||
| Resolution (Å) | 30-2.21 | 30-2.10 | 30-2.91 | ||
| Number of reflections | 26167 | 15307 | 23754 | ||
| Rwork/Rfree | 21.5 / 24.9 | 21.1 / 25.5 | 26.7 / 27.6 | ||
| B factors (Å2) | |||||
| All atoms | 30.1 | 39.9 | 69.0 | ||
| Protein | 26.0 | 35.9 | 69.1 | ||
| DNA | 34.1 | 43.2 | 68.9 | ||
| Water | 31.7 | 44.3 | 54.1 | ||
| Ions | 53.1 | 60.5 | N/A | ||
| Ramachandran plot | |||||
| Most favored | 94.9% | 92.6% | 80.4% | ||
| Allowed | 5.1% | 4.9% | 19.6% | ||
| Generously allowed | 0.0% | 2.5% | 0.0% | ||
| Disallowed | 0.0% | 0.0% | 0.0% | ||
| Rms deviations | |||||
| Bond lengths (Å) | 0.008 | 0.008 | 0.007 | ||
| Bond angles (°) | 1.3 | 1.3 | 1.2 | ||
Datasets were collected from a single crystal. Values in parentheses are for the highest resolution shell. Rfree was calculated using 10% of the reflection data chosen randomly and omitted at the start of refinement.
Figure 2.
Structures of FoxO1/DNA complexes (a) Representative FoxO1 DBD/DNA complex - 2.1 Å structure of FoxO1 bound to the DBE1 sequence. The eight base DBE sequence is in CDK coloring and flanking bases are gray. (b and c) Stereo view of helix 3 interactions with DNA showing the different interactions with IRE (b) and DBE (c) DNA mediated by Asn211 and His215. Electron density shown is from a simulated annealing omit map contoured at 1.0 σ around the DNA recognition helix. (d) Superposition of FoxO1 DBD bound to DBE1 and DBE2 DNAs to illustrate the larger amount of bend in the DNA for the DBE2 structure. FoxO1 DBD bound to DBE1 is colored in blue and gray. FoxO1 DBD bound to DBE2 is colored in tan and red. (e–g) The targets of MST1 phosphorylation and their interactions with the phosphate backbone of DNA. Structure shown is FoxO1 DBD/DBE1 DNA. The interactions are conserved in the FoxO1/IRE DNA structure. (e) Ser212. (f) Ser218. (g) Ser234 and Ser235.
Figure 3.
Schematic showing FoxO1 DBD interactions with IRE (a) and DBE1 (b) DNA sequences. Differences in hydrogen bonding between the DBE and IRE sequences are highlighted in green. Bases that are contacted directly or through water mediated interactions are shaded. Phosphates that are contacted directly or through water mediated interactions are highlighted in red.
Wing 2 is flexible but essential for DNA binding
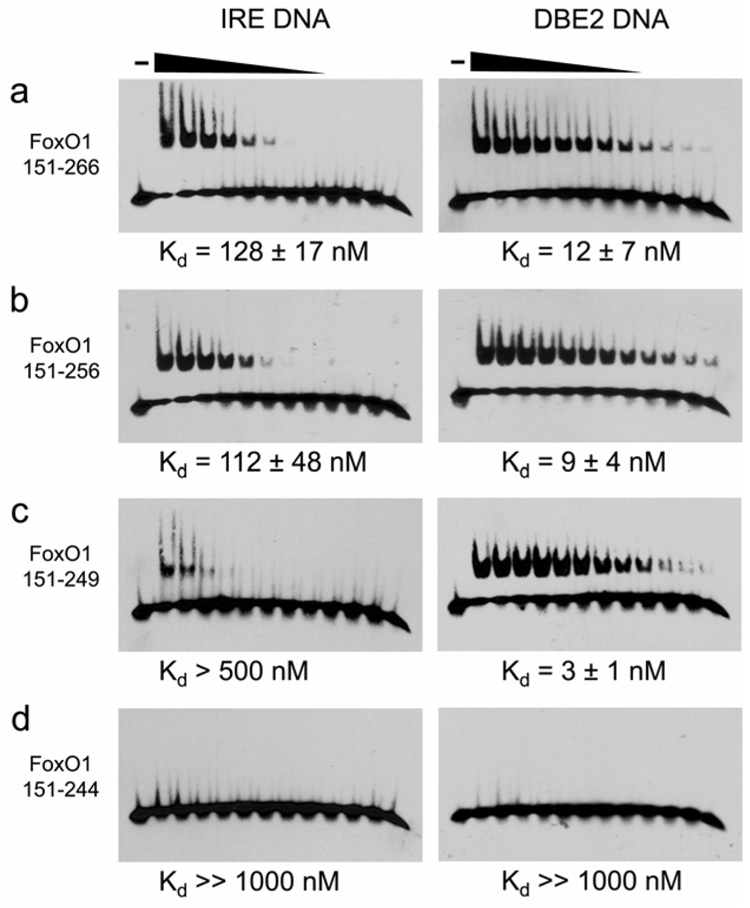
The extent of FoxO1 DBD wing 2 interactions with DNA was not known at the start of this study. By sequence alignment, wing 2 was predicted to start at Lys245 and possibly extend as far as Ser256, the target of Akt phosphorylation. To be inclusive of all post-translational modification sites we crystallized and determined the structure of FoxO1 151–266 bound to the IRE and DBE2 sequences, but electron density was not observed for residues 242–266. To rule out proteolysis during crystal preparation, FoxO1/IRE crystals were redissolved in water and TOF mass spectrometry was carried out to confirm the presence of full length protein in the crystals (Supplemental Figure 1a). We then mapped the extent of wing 2 contributions to DNA binding by preparing four FoxO1 DBD constructs with varying wing 2 lengths and measuring DNA binding using EMSA (Figure 4a–d). FoxO1 constructs 151–266 and 151–256 both bound to the IRE and DBE2 sequences with roughly equal affinity. FoxO1 151–249 also bound to the DBE2 sequence with roughly equal affinity, while binding of the IRE sequence was reduced several fold. Surprisingly, the FoxO1 151–244 construct showed no detectable binding for IRE or DBE2 DNA indicating that some or all of wing 2 residues 244–249 are required for FoxO1 to bind DNA. To rule out an artifact of the EMSA assay, we also performed the same assay on 151–244 by fluorescence polarization using 151–266 as a positive control, and we observed the same result (Supplemental Figure 1b).
Figure 4.
(a–d) Measurement of binding affinity to IRE and DBE2 DNA for C-terminally truncated FoxO1 DBD constructs.
These findings suggest that amino acids 244–249 are flexible but nonetheless important for DNA binding, most likely through phosphate contacts by Lys245 and Lys248. Previous reports have suggested that the wing 2 region of FoxO may be more flexible than the rest of the protein. Specifically, wing 2 was found to be disordered in a FoxO4 DBD solution structure (without DNA) by NMR (Weigelt et al., 2001). Molecular Dynamics simulations of a theoretical FoxO/DNA complex also suggested that wing2 may be more flexible than the rest of the DBD (Boura et al., 2007).
To lend further support to the hypothesis that wing 2 is the most flexible part of the FoxO1 DBD even in the presence of DNA, we performed limited proteolysis of FoxO1 151–266 in the presence of a 3-fold excess of DBE2 DNA and sequenced the proteolysis products by N-terminal Edman degradation (Supplemental Figure 2a–e). The results confirmed that papain, proteinase K and trypsin all remove C-terminal amino acids preferentially even in the presence of DNA. Trypsin also removed N-terminal amino acids 151–157. TOF mass spectrometry of the papain digestion product confirmed that papain removes amino acids 245–266, but the remaining FoxO1 151–244 is stable towards papain. These results indicate that residues 245–266 are flexible and accessible to protease digestion even in the presence of a 3-fold molar excess DBE2 DNA. These data, and S249E mutagenesis (see CDK phosphorylation), indicate a minimal FoxO1 DBD of residues 158–248 and a short but essential wing 2 comprised of residues 245–248. There also seems to be some DNA binding contribution from arginines 250–252 under certain circumstances.
Structural basis for high affinity binding of DBE sequences
When the bases flanking the FoxO consensus sequence are kept the same, the FoxO1 DBD binds to the DBE1 DNA with 2-fold greater affinity compared with the IRE DNA (Figure 5a). High resolution structures reveal that the DBE and IRE consensus sequences are bound with different hydrogen bonding and water mediated interactions through the versatile side chains of Asn211 and His215 (Figure 2b, 2c and Figure 3). The overall structural differences between the two FoxO1/DNA complexes are quite small with an alignment rmsd less than 0.2 Å. However, the network of hydrogen bonds is different at the protein/DNA interface due to the different arrangement of hydrogen bond donors and acceptors in the major groove of the DNA around bases 7’ and 8’.
Figure 5.
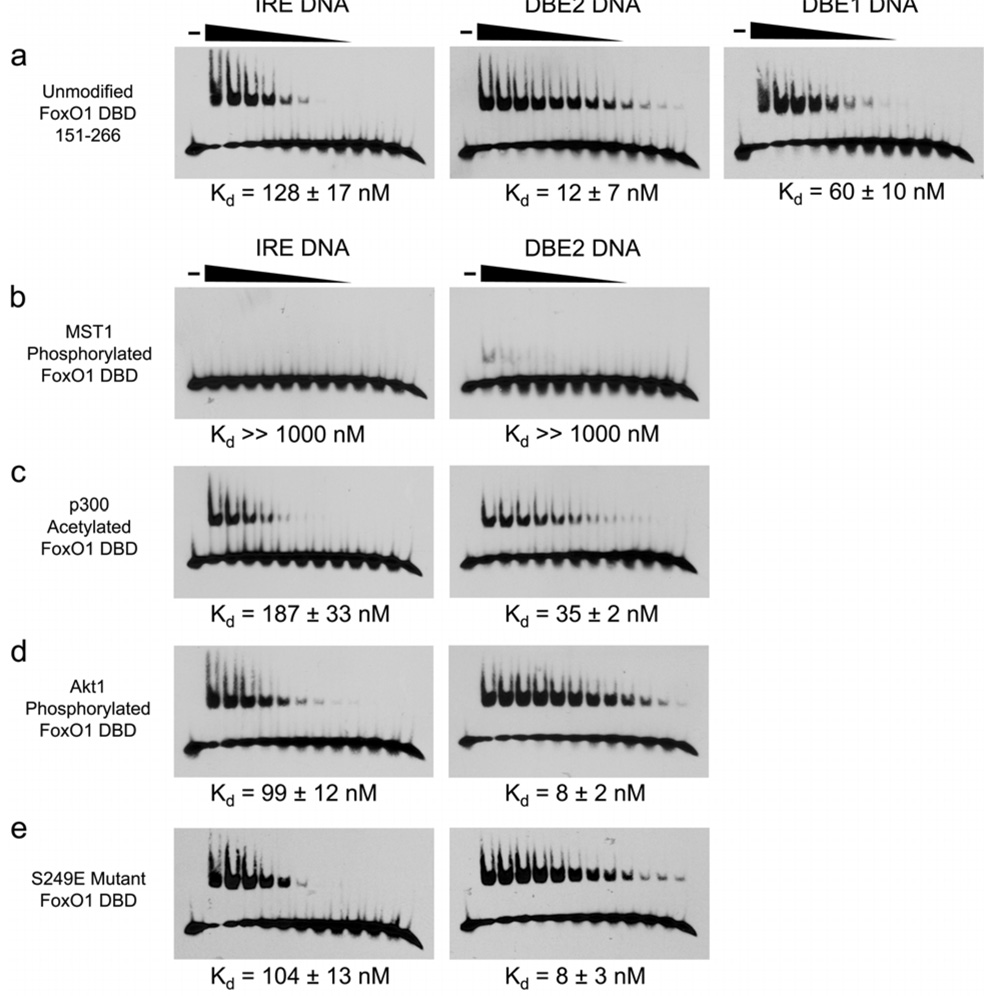
(a) EMSA results showing the difference in affinity for FoxO1 151–266 binding to IRE, DBE2 and DBE1 DNA sequences. (b–e) EMSA results showing the effects of FoxO1 151–266 modifications on binding affinity for IRE and DBE2 DNA sequences.
In the FoxO1/DBE structure Asn211 forms bidentate hydrogen bonds with Ade5’ (Figure 2c). His215 in the FoxO1/DBE structure is protonated with delocalized positive charge between ND1 and NE2 mediating direct interactions with Thy7’, Thy5 and Thy6, which are within 3.07, 2.90, and 3.07 Å respectively, and forming a water mediated hydrogen bond to N7 of Gua8’. In the FoxO1/IRE structure, the Asn211 side chain is oriented parallel with the DNA axis (Figure 2b) and is hydrogen bonding with Ade5’ and Ade6’. His215 in the FoxO1/IRE structure is still interacting with Thy5 and Thy6, and makes a water mediated hydrogen bond to Cyt8’ in place of Gua8’ of the DBE sequence; however, Ade7’, replacing Thy7’ of the DBE sequence, lacks a hydrogen bond acceptor for His215. Instead, the hydrogen bond donating amino group of Ade7’ is 3.47 Å away from His215 and is unlikely to be contributing to protein-DNA affinity. We believe this difference at base 7’ accounts for the modest 2-fold increased affinity of FoxO1 for the DBE1 sequence versus the IRE sequence (Figure 5a).
The DBE2 DNA sequence contains the same 8 base consensus sequence as DBE1 DNA, but the 3’ flanking sequence is two bases longer and has been replaced with the 5’-ATTTTG-3’ sequence. The 5’-NTTT-3’ sequence is a high affinity consensus sequence for the 3’ flanking region enriched by random oligonucleotide selection using mouse FoxO1 DBD which shares an identical sequence to human FoxO1 within the DBD (Furuyama et al., 2000). An analysis of 1387 putative human DBE sequences in the Database of Transcriptional Start Sites (http://dbtss.hgc.jp) suggests that thymine rich 3’ flanking sequences are also present in the human genome. We observed a 5-fold increase in affinity of FoxO1 DBD for DBE2 DNA versus DBE1 DNA (Figure 5a). The structure of the FoxO1 DBD/DBE2 DNA complex exhibits the same binding motif as the DBE1 structure, but in contrast to the DBE1 structure, the DBE2 structure reveals bent and distorted DNA in the 3’ flanking region where wing 2 of FoxO1 is predicted to make interactions with the DNA minor groove (Figure 2d).
MST1 phosphorylation blocks DNA binding
MST1 kinase is reported to phosphorylate four serines within the forkhead domains of FoxO1 and FoxO3 to promote FoxO translocation to the nucleus and expression of cell death target genes (Lehtinen et al., 2006). The FoxO1/DNA structures reported here however reveal that these four serines (Ser212, Ser218, Ser234 and Ser235) all make either direct or water mediated hydrogen bond interactions with the phosphate backbone of the DNA (Figure 2e–g). Based on this observation, we hypothesized that MST1 phosphorylated FoxO1 would be unable to bind DNA due to steric and electrostatic repulsion of the negatively charged phosphoserines and the negatively charged DNA backbone. To directly measure the effect of MST1 phosphorylation on DNA affinity, we phosphorylated the FoxO1 DBD with MST1 and purified the reaction product by gel filtration. TOF mass spectrometry confirmed the addition of four phosphates (Supplemental Figure 3a). EMSA was done on the MST1 phosphorylated sample and an unmodified control to measure affinity for the DBE2 and IRE sequences (Figure 5b). The assay reveals that MST1 phosphorylated FoxO1 shows almost no detectable binding to either DBE2 or IRE DNA for the 1 µM to 0.5 nM FoxO1 concentration range tested. To rule out an artifact of the EMSA assay, we also measured affinity for DBE2 DNA by fluorescence polarization using MST1 phosphorylated FoxO1 and unmodified FoxO1 as a positive control, and we observed the same result (Supplemental Figure 3c). This finding is not necessarily inconsistent with the observation that MST1 activates FoxO; however, it suggests that additional steps, probably involving phosphatase activity in the pathway for MST1 activation of FoxO.
CBP/p300 acetylation reduces DNA binding
Previous studies have investigated the in vivo effect of FoxO acetylation on DNA binding (Daitoku et al., 2004). In vitro DNA binding assays have also been done in which CBP/p300 target lysines are mutated to alanine, glutamine, or arginine (Matsuzaki et al., 2005). These studies show that removal of the positive charge at Lys245, 248, and 262 reduces DNA binding and transcription of FoxO target genes. To more directly measure the effect of CBP/p300 acetylation on DNA affinity in vitro, we acetylated the FoxO1 DBD with p300 and purified the reaction product by gel filtration. We used a combination of proteolysis and TOF mass spectrometry to confirm that four acetyl groups were added to lysines 245, 248, 262, and 265 (Supplemental Figures 2b and 4a–c). The fourth acetyl group at Lys265 was not previously reported, but is consistent with preference of CBP/p300 for acetylating KXXK sequences. EMSA was carried out on the p300 acetylated sample and an unmodified control to measure affinity for the DBE2 and IRE sequences (Figure 5c). The assay reveals that the DNA binding affinity of p300 acetylated FoxO1 is reduced approximately three fold to the DBE2 sequence and close to two fold to the IRE sequence. These results reveal that CBP/p300 acetylation of the FoxO1 DBD does indeed cause a modest reduction in DNA binding in vitro.
DNA binding is unchanged by Akt or CDK phosphorylation
The sites of Akt and CDK1/2 phosphorylation, serines 256 and 249 respectively, are reported to regulate FoxO1 through mechanisms unrelated to DNA binding. Phosphorylation of Ser256 by Akt has been shown to regulate FoxO1 by causing an inhibitory interaction with 14-3-3 proteins. Phosphorylation of Ser249 is proposed to either disrupt the FoxO1/14-3-3 interaction in the cytoplasm or interfere with the adjacent nuclear localization signal (NLS, arginines 251–253).To test the possibility that phosphorylation of these two sites may also reduce FoxO1 activity by interfering with DNA binding, we used Akt kinase to phosphorylate Ser256, and we prepared a S249E mutant to mimic CDK phosphorylated FoxO1. For the Akt phosphorylated sample, TOF mass spectrometry was done to confirm the addition of one phosphate (Supplemental Figure 5). CDK2 was inactive towards the FoxO1 DBD due to the absence of the C-terminal regions of FoxO1, amino acids 267–655, as previously reported (Huang et al., 2006), so the S249E mutant was made to mimic the modification. EMSA was done on both samples and on an unmodified control to measure affinity for the DBE2 and IRE sequences (Figure 5d and 5e). DNA affinity for both the Akt phosphorylated FoxO1 and the S249E mutant were found to be unchanged compared with unmodified FoxO1, confirming that these post-translational modifications do not affect the DNA binding activity of FoxO1 under the conditions tested.
DISCUSSION
DNA recognition by members of the 43 human forkhead domain containing proteins is diverse in terms of sequence recognition and structural mode of binding. The C-terminal wing 2 region of forkhead domains is especially diverse in terms of sequence, structure and extent of DNA interactions (Clark et al., 1993; Jin et al., 1999; Liu et al., 2002; Stroud et al., 2006; Tsai et al., 2006). Notably, none of the FoxO1/DNA complexes reported here show ordered electron density for wing 2, suggesting that wing 2 is highly flexible and disordered in the crystal lattice. Further investigation revealed that wing 2 is necessary for high affinity DNA binding by FoxO1. This observation suggests that wing 2 enhances affinity through forming transient and/or multiple nonspecific electrostatic interactions with the phosphate backbone of the DNA. This hypothesis is supported by the observation that acetylation of Lys245 and Lys248 in wing 2 by CBP/p300 reduces DNA binding affinity (Figure 5c).
Comparison of the FoxO1/DBE and /IRE DNA structures also reveals new details about how forkhead domains can preferentially bind one sequence over another slightly different sequence. The more favorable interaction of His215 with the carbonyl oxygen of Thy7’ of the DBE DNA sequence likely accounts for most of the 2-fold difference in affinity between the DBE1 and IRE DNA (Figure 5a). The DBE2 DNA, with thymine rich 3’ flanking bases, exhibited a 5-fold increase in FoxO1 DBD affinity over DBE1, suggesting that the 3’ flanking sequence may be a factor for optimal DNA binding by FoxO1. The FoxO1/DBE2 structure reveals that the 3’ flanking region of the DNA is more bent and distorted compared with the FoxO1/DBE1 structure (Figure 2d). In all three structures reported here the DNA forms a pseudo-continuous helix in the crystal lattice, so it is possible that crystal packing has influenced the DNA geometry for the structures; however, the difference in affinity combined with the evidence that wing 2 contributes to DNA binding in this region suggests that the increased A–T content in the 3’ flanking region of the DBE2 DNA may allow for more DNA bending and more optimal interaction with wing 2 of FoxO1. It has been reported for FoxA transcription factors that DNA response element affinity can determine the timing of gene expression in embryonic development (Gaudet and Mango, 2002). It would be interesting to investigate whether this holds true for FoxO transcription factors. Genes regulated by high affinity DBE sequences may have a different timing of expression or level of expression compared with genes regulated by lower affinity IRE sequences.
Acetylation of the wing 2 region of FoxO by CBP/p300 has been previously shown to alter transcriptional activity of the protein (Brunet et al., 2004; Daitoku et al., 2004; Fukuoka et al., 2003; Jing et al., 2007; Matsuzaki et al., 2005; Motta et al., 2004; Nakae et al., 2003; van der Horst et al., 2004). Our studies show that this change in activity can work, at least in part, via a direct reduction in FoxO1/DNA affinity. However, this decrease in DNA binding by FoxO1 as a function of wing 2 acetylation is not nearly as dramatic as the decrease in DNA binding that is caused by MST1 mediated phosphorylation. In light of this, it is possible that CBP/p300-mediated acetylation of FoxO1 my also change the transcriptional activity of FoxO through altering protein-protein interactions that are necessary for FoxO-mediated transcriptional regulation.
Regulation of FoxO1 activity by Akt, CDK1/2, and MST1 kinases is reported to work by three different mechanisms. Akt kinase phosphorylation of Ser256 was found to have no direct effect on DNA binding (Figure 5d). This finding is consistent with regulation through an inhibitory interaction with 14-3-3 proteins, and is also consistent with data that shows wing 2 DNA interactions do not extend to Ser256 (Figure 4). CDK2 phosphorylation of Ser249 was mimicked with a S249E mutant. This mutant was found to have similar DNA binding affinity to unmodified FoxO1 DBD (Figure 5e). This finding is also consistent with the proposed mechanism by which phosphorylation of Ser249 regulates FoxO1 activity indirectly by disrupting the FoxO1 interaction with 14-3-3 proteins or by blocking the adjacent nuclear localization signal (Huang et al., 2006; Yuan et al., 2008). Interestingly, MST1 phosphorylation of Ser212, 218, 234, and 235 was found to almost completely block DNA binding by FoxO1. This is consistent with our observation that the target serines are making direct or water mediated contacts to the phosphate backbone (Figure 2e–g); however, this suggests that the proposed mechanism by which MST1 phosphorylation promotes nuclear translocation and transcription of FoxO1 target genes requires an additional step. We propose that dephosphorylation of serines 212, 218, 234 and 235 occurs before DNA binding and transcriptional regulation by MST1 restored FoxO1. Alternatively, MST1 phosphorylated FoxO1 may participate in transcriptional regulation through association with other regulatory proteins rather than binding DNA directly.
The diverse biological roles of the FoxO family of transcription factors and the mechanisms by which FoxO activity is regulated in the cell remains incompletely understood. Previous reports have indicated that FoxOs are tumor suppressors and mediators of longevity as both downstream targets of insulin signaling and as substrates for Sir2 deacetylase (Biggs et al., 1999; Brunet et al., 1999; Brunet et al., 2004; Daitoku et al., 2004; Fukuoka et al., 2003; Kenyon et al., 1993; Motta et al., 2004; Paik et al., 2007; Rena et al., 1999; Tang et al., 1999; van der Horst et al., 2004). The three crystal structures reported herein help to explain the structural basis for DNA recognition by FoxO1, and the factors that contribute to optimal DNA binding by FoxO1. Our rigorous evaluation of DNA binding affinity of directly acetylated and phosphorylated FoxO1 DBD also provides a more complete understanding of FoxO1 regulation by post-translational modification, and how misregulation of FoxO may lead to disease.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The gene encoding human FoxO1, amino acids 151–266, was cloned into a pETDuet-1 vector containing the gene for yeast SMT3, a ubiquitin-like protein of the SUMO family, in the first multiple cloning site. Overnight expression at 15 °C in BL21(DE3)LysS (Novagen) yields a 6x histidine-SUMO-FoxO1 fusion protein. Following lysis and Ni-NTA purification, the his-SUMO tag was removed by incubation with ULP1 (Ubiquitin-like-specific protease 1) at 4 °C overnight. The FoxO1 DBD was further purified by cation exchange, ammonium sulfate precipitation, and Superdex-75 size exclusion chromatography in 20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 2 mM DTT buffer. The protein was finally concentrated to 8 mg/ml and stored at 4 °C until use. Protein mutants were generated by site-directed mutagenesis based on the QuickChange protocol from Stratagene (Papworth, 1996). The FoxO1 DBD truncation constructs and point mutants were purified as described above.
Crystallization
DNA for binding assays and crystallization was purchased from Integrated DNA Technologies (Coralville, IA). The IRE sequence contains strands 5’-CAAGCAAAACAAACCA-3’ and 5’-TGGTTTGTTTTGCTTG-3’. The DBE1 sequence contains strands 5’-CAAGGTAAACAAACCA-3’ and 5’-TGGTTTGTTTACCTTG-3’. The DBE2 sequence contains strands 5’-CAAAATGTAAACAAGA-3’ and 5’-TCTTGTTTACATTTTG-3’. Complementary strands were annealed in 20 mM HEPES pH 7.5, 50 mM NaCl buffer by heating to 80 °C for 10 minutes and slowly cooling to room temperature over a period of three hours. FoxO1-DNA complexes for crystallization were prepared by mixing concentrated FoxO1 DBD, amino acids 151–266 for the IRE and DBE2 structures and 151–249 for the DBE1 structure, with concentrated, annealed DNA in a ratio of 1:1.2. The final concentration of FoxO1 for crystallization was 5 mg/ml. Crystals of FoxO1 DBD/IRE DNA were grown by hanging drop vapor diffusion in 4 days at 4 °C using a well solution containing 50 mM sodium cacodylate pH 6.8, 0.2 M NH4Cl, 0.01 M CaCl2, and 30% PEG 4,000. Crystals of FoxO1 DBD/DBE1 DNA were grown under the same conditions except with 50 mM HEPES pH 7.9 and 22% PEG 4,000. Crystals of FoxO1 DBD/DBE2 DNA were also grown under the same conditions except with 50 mM sodium cacodylate pH 6.4 and 21% PEG 4,000. All crystals were frozen in well solution plus 25% glycerol for cryoprotection. The IRE crystals formed in the space group P21212, the DBE1 crystals formed in the space group I222, and the DBE2 crystals formed in the space group P32. Mercury chloride soaking changed cysteine mutant FoxO1 DBD/IRE crystals from the P21212 space group to the I222 space group with approximately the same unit cell dimensions.
Data Collection and Structure Determination
Native datasets were collected at the NSLS X6A and APS 23ID-B beamlines. Mercury soaked FoxO1 S184C and A207C bound to IRE DNA datasets were collected on a Rigaku Raxis-IV home source. Data were processed using HKL2000 (Otwinowski, 1997). Single Isomorphous Replacement (SIR) was performed with Solve and Resolve (Terwilliger, 2000; Terwilliger and Berendzen, 1999) to find two mercury sites in a FoxO1 DBD S184C mutant/IRE DNA crystal. A mercury soaked A207C mutant dataset was initially set as the native dataset since mercury soaking changed the space group to I222, and the true native dataset was in the P21212 space group. Initial model building with Coot (Emsley and Cowtan, 2004) and refinement with CNS (Brunger et al., 1998) yielded a low resolution structure that was used for molecular replacement into the higher resolution native P21212 (IRE DNA) and P32 (DBE2 DNA) space groups using Phaser (Read, 2001; Storoni et al., 2004). The DBE1 DNA structure, also in the I222 space group, was solved later using Phaser and the FoxO1 DBD/IRE DNA complex as a search model. Models were initially refined with simulated annealing, energy minimization and group B-factor refinement. For later stages of refinement ions and solvent molecules were added to the model and individual atomic B-factors were refined. The final model was checked for errors against a simulated annealing omit map. Refinement of all three structures resulted in models with excellent statistics and geometries (Table 1). Figures were prepared using PyMOL (DeLano Scientific, Palo Alto, CA).
MST1 Phosphorylation
Human full length MST1 kinase was amplified and cloned into pachis-Tev baculoviral transfer vector. Recombinant viruses were selected, amplified and harvested in Sf9 cells as described (Hutchison et al., 1998). Cells were lysed in 50 mM TRIS pH 8.0, 400 mM NaCl, 2 mM imidazole, and 1 mM β-mercaptoethanol supplemented with protease inhibitors. The clarified supernatant was loaded onto Ni-NTA agarose, washed, and eluted with 25 mM HEPES pH 7.5, 100 mM NaCl, and 200 mM imidazole. The eluted protein was further purified by Superdex-200 size exclusion chromatography in 25 mM HEPES pH 7.5, 100 mM NaCl. The eluted fractions were pooled, concentrated to 2 mg/ml, and flash frozen until use. MST1 was incubated with the FoxO1 DBD (151–266) in a 1/10 ratio (MST/FoxO1) at 30 °C for 4 hours in buffer containing 20 mM TRIS pH 7.5, 5 mM MgCl2, and 2 mM ATP. A control sample was also incubated at 30 °C for 4 hours in the absence of MST1 or ATP. After 4 hours, the sample was put on ice for 10 minutes and then injected onto a Superdex-75 size exclusion column to purify the FoxO1 DBD from the MST1 and the ATP. Purified FoxO1 was then concentrated and analyzed by MALDI-TOF mass spectrometry to confirm phosphorylation. The mass of the largest peak corresponded to addition of 4 phosphates. Species corresponding to 3 and 5 phosphates were also observed (Supplemental Figure 3a).
p300 Acetylation
p300 (a gift from X.L, L.W., P.C. and R.M.) was incubated with the FoxO1 DBD (151–266) in a 1/20 ratio (p300/FoxO1) at 30 °C for 3 hours in buffer containing 50 mM HEPES pH 7.5, 0.1 mM EDTA, 1 mM DTT, 0.05 mg/mL BSA, and 3 mM Acetyl-CoA. A control sample was also incubated at 30 °C for 3 hours with the same buffer conditions but with p300 absent. After 3 hours, the sample was put on ice for 10 minutes and then injected onto a Superdex-75 size exclusion column to purify the FoxO1 DBD from the p300 and the Acetyl CoA. Purified FoxO1 was then concentrated and analyzed by MALDI-TOF mass spectrometry to confirm acetylation. Results indicated one distinct species with an increased mass of 172 Da or approximately 4 acetyl groups (Supplemental Figure 4a). A combination of papain digestion, MALDI-TOF mass spectrometry and N-terminal sequencing was used to confirm that the acetyl groups were added to the C-terminus of FoxO1 (amino acids 245–266) (Supplemental Figures 2b and 4c).
Akt Phosphorylation
Akt1 (PKB α) was purchased (Biomol International, SE-416) and incubated with the FoxO1 DBD (151–266) at 30 °C for 3 hours in buffer containing 25 mM MOPS pH 7.2, 12.5 mM β-glycerophosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, 0.4 mM DTT and 2 mM ATP. A control sample was also incubated at 30 °C for 3 hours with the same buffer conditions but with Akt absent. After 3 hours, the sample was put on ice for 10 minutes and then injected onto a Superdex-75 size exclusion column to purify the FoxO1 DBD from the Akt and the ATP. Purified FoxO1 was then concentrated and analyzed by MALDI-TOF mass spectrometry to confirm phosphorylation. Results showed one distinct species with an increased mass of 90 Da or approximately 1 phosphate (Supplemental Figure 5a).
DNA Binding Assays
Electrophoretic mobility shift assays (EMSAs) were done with biotinylated DNA duplexes (Integrated DNA Technologies, Coralville, IA) and developed with a chemiluminescent nucleic acid detection kit (Pierce 89880). Briefly, a serial dilution of FoxO1 protein was prepared to give a final concentration range from 1 µM–0.5 nM FoxO1. FoxO1 serial dilutions were each equilibrated at room temperature with 1 nM DNA for 30 minutes in binding buffer containing 20 mM TRIS pH 8.0, 50 mM KCl, 5% glycerol, 2 mM DTT, 0.2 mM EDTA, 2 mM MgCl2, 0.1 mg/mL BSA, and 1 ng/uL poly dI·dC. The equilibrated mixture was loaded onto a 6% DNA retardation gel (Invitrogen) in 0.5X TRIS-borate-EDTA (TBE) and run at 100 V for 1 hour at 4 °C. The gel was blotted onto a Biodyne B (Invitrogen) membrane at 380 mA for 1 hour in 0.5X TBE at 4 °C. The blotted DNA was crosslinked to the membrane using a Stratagene crosslinker. The membrane was developed according to the nucleic acid detection protocol from Pierce, films were then exposed, developed and scanned. All assays were done in duplicate. Apparent Kd values were determined by measuring shifted band intensity with ImageJ (from the NIH, http://rsb.info.nih.gov/ij/) and fitting a plot of intensity versus Log[FoxO1] to one-site competitive binding in GraphPad Prism software.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Mary Fitzgerald, Manqing Hong, Santosh Hodawadekar, Troy Messick, Brandi Sanders and other members of the Marmorstein Lab for helpful discussions. We thank Xin Liu, Ling Wang, and Philip Cole for the contribution of purified p300. We also thank Nicole DiFlorio and Matt Hart of Wistar Proteomics, and Marc Allaire, Fabiano Yokaichiya and Jean Jakoncic of NSLS beamline X6A for help with data collection. This work was supported by an NIH grant to R.M. (GM 052880) and institutional grants to the Wistar Institute from the Commonwealth Universal Research Enhancement Program, Pennsylvania and the NCI (CA01015).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers - Coordinates for the FoxO1 DBD/IRE, /DBE1, and /DBE2 DNA complexes have been deposited in the Protein Data Bank with accession codes 3COA, 3CO6, and 3CO7.
REFERENCES
- Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- Biggs WH, 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkhardt A, Repp R, Haas OA, Leis T, Harbott J, Kreuder J, Hammermann J, Henn T, Lampert F. Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23) Oncogene. 1997;14:195–202. doi: 10.1038/sj.onc.1200814. [DOI] [PubMed] [Google Scholar]
- Boura E, Silhan J, Herman P, Vecer J, Sulc M, Teisinger J, Obsilova V, Obsil T. Both the N-terminal loop and wing W2 of the forkhead domain of transcription factor Foxo4 are important for DNA binding. J Biol Chem. 2007;282:8265–8275. doi: 10.1074/jbc.M605682200. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cheong JW, Eom JI, Maeng HY, Lee ST, Hahn JS, Ko YW, Min YH. Constitutive phosphorylation of FKHR transcription factor as a prognostic variable in acute myeloid leukemia. Leuk Res. 2003;27:1159–1162. doi: 10.1016/s0145-2126(03)00102-4. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, D'Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Daitoku H, Hatta M, Matsuzaki H, Umemura S, Fukamizu A. Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. Int J Mol Med. 2003;12:503–508. [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, 3rd, Emanuel BS, Rovera G, Barr FG. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- Hillion J, Le Coniat M, Jonveaux P, Berger R, Bernard OA. AF6q21, a novel partner of the MLL gene in t(6;11)(q21;q23), defines a forkhead transcriptional factor subfamily. Blood. 1997;90:3714–3719. [PubMed] [Google Scholar]
- Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–297. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- Hutchison M, Berman KS, Cobb MH. Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J Biol Chem. 1998;273:28625–28632. doi: 10.1074/jbc.273.44.28625. [DOI] [PubMed] [Google Scholar]
- Jin C, Marsden I, Chen X, Liao X. Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J Mol Biol. 1999;289:683–690. doi: 10.1006/jmbi.1999.2819. [DOI] [PubMed] [Google Scholar]
- Jing E, Gesta S, Kahn CR. SIRT2 Regulates Adipocyte Differentiation through FoxO1 Acetylation/Deacetylation. Cell Metab. 2007;6:105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Liu PP, Chen YC, Li C, Hsieh YH, Chen SW, Chen SH, Jeng WY, Chuang WJ. Solution structure of the DNA-binding domain of interleukin enhancer binding factor 1 (FOXK1a) Proteins. 2002;49:543–553. doi: 10.1002/prot.10227. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazet F, Yu JK, Liberles DA, Holland LZ, Shimeld SM. Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene. 2003;316:79–89. doi: 10.1016/s0378-1119(03)00741-8. [DOI] [PubMed] [Google Scholar]
- Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J Biol Chem. 2002;277:47928–47937. doi: 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- Nakae J, Kitamura T, Kitamura Y, Biggs WH, 3rd, Arden KC, Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papworth C, Bauer J, Braman J, Wright DA. Site-directed mutagenesis in one day with >80% efficiency. Strategies. 1996;9:3–4. [Google Scholar]
- Parry P, Wei Y, Evans G. Cloning and characterization of the t(X;11) breakpoint from a leukemic cell line identify a new member of the forkhead gene family. Genes Chromosomes Cancer. 1994;11:79–84. doi: 10.1002/gcc.2870110203. [DOI] [PubMed] [Google Scholar]
- Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Rena G, Guo S, Cichy SC, Unterman TG, Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J Biol Chem. 1999;274:17179–17183. doi: 10.1074/jbc.274.24.17179. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006;14:159–166. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Sunters A, Madureira PA, Pomeranz KM, Aubert M, Brosens JJ, Cook SJ, Burgering BM, Coombes RC, Lam EW. Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res. 2006;66:212–220. doi: 10.1158/0008-5472.CAN-05-1997. [DOI] [PubMed] [Google Scholar]
- Tang ED, Nunez G, Barr FG, Guan KL. Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem. 1999;274:16741–16746. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Tsai KL, Huang CY, Chang CH, Sun YJ, Chuang WJ, Hsiao CD. Crystal structure of the human FOXK1a-DNA complex and its implications on the diverse binding specificity of winged helix/forkhead proteins. J Biol Chem. 2006;281:17400–17409. doi: 10.1074/jbc.M600478200. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- van Dongen MJ, Cederberg A, Carlsson P, Enerback S, Wikstrom M. Solution structure and dynamics of the DNA-binding domain of the adipocyte-transcription factor FREAC-11. J Mol Biol. 2000;296:351–359. doi: 10.1006/jmbi.1999.3476. [DOI] [PubMed] [Google Scholar]
- Weigelt J, Climent I, Dahlman-Wright K, Wikstrom M. Solution structure of the DNA binding domain of the human forkhead transcription factor AFX (FOXO4) Biochemistry. 2001;40:5861–5869. doi: 10.1021/bi001663w. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Becker EB, Merlo P, Yamada T, DiBacco S, Konishi Y, Schaefer EM, Bonni A. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science. 2008;319:1665–1668. doi: 10.1126/science.1152337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.