Abstract
Hypomorphic RAG mutants with severely reduced V(D)J recombination activity cause Omenn Syndrome (OS), an immunodeficiency with features of immune dysregulation and a restricted T cell receptor repertoire. Precisely how RAG mutants produce autoimmune and allergic symptoms has been unclear. Current models posit that the severe recombination defect restricts the number of lymphocyte clones, a few of which are selected upon antigen exposure. We show that murine RAG1 R972Q, corresponding to an OS mutation renders the recombinase hypersensitive to select coding sequences at the hairpin formation step. Other RAG1 OS mutants tested do not manifest this sequence sensitivity. These new data support a novel mechanism for OS: by selectively impairing recombination at certain coding flanks, a RAG mutant can cause primary repertoire restriction, as opposed to a more random, limited repertoire that develops secondary to severely diminished recombination activity.
Keywords: Omenn Syndrome, RAG, V(D)J recombination, antigen receptor repertoire, SCID
Introduction
Omenn Syndrome (OS) is a severe immune deficiency that is, peculiarly, accompanied by both autoimmune and allergic symptoms (1, 2). Patients usually present by the age of six months, suffering recurrent and opportunistic infections, as well as hepatosplenomegaly, lymphadenopathy, erythrodermia, eosinophilia, and elevated serum IgE. T cells from OS patients have a highly restricted, oligoclonal repertoire, and B cells are generally absent from peripheral blood (3–7). The disease is fatal without bone marrow transplant.
OS is typically caused by mutations in the RAG1 and RAG2 genes, which encode the lymphoid-specific components of the V(D)J recombinase responsible for generating T cell receptor (TCR) and immunoglobulin (Ig) diversity in developing T and B lymphocytes (8–11). Most OS-causing mutant RAG proteins display severely decreased recombination activity in vitro, and it is thought that this creates a “bottleneck” in lymphocyte differentiation, resulting in production of the restricted T cell receptor repertoire characteristic of OS (8).
It is easy to see how defective RAG mutants could cause immunodeficiency, but it has been more difficult to elucidate the connection between reduced V(D)J recombination and the immunoregulatory defects characteristic of OS. Recent work in mouse models bearing RAG mutations offers some suggestions. First, the constrained repertoire may cause a relative deficiency in regulatory T cells, leading to impaired self tolerance (12, 13). Second, the diminished production of T cells likely fosters homeostatic proliferation in the periphery, which could lead to TH2 skewing (14–16).
The reduced V(D)J recombinase activity underlying most cases of OS may stem from defects affecting any of several distinct steps in the recombination reaction. The RAG complex first recognizes antigen receptor V, D, and J coding segments by binding to recombination signal sequences (RSSs) that border each segment and synapsing a pair of RSSs. The recombinase then generates single-strand nicks precisely between each RSS and the adjacent coding flank (the sequence of the coding segment directly 5’ of the RSS). These nicks liberate 3’-hydroxyl groups that are used by the RAG proteins to attack the other strand, forming a covalently sealed DNA hairpin structure at each coding flank and a blunt double-strand break at each RSS (17). The hairpins are then opened and joined together to form a coding joint, while the signal ends form a signal joint (18). OS-causing RAG mutants can be defective in the binding, nicking, hairpin formation, or joining steps of V(D)J recombination (8, 9, 19, 20).
The prevailing pathogenetic model for OS holds that hypomorphic RAG mutations severely reduce recombinase activity uniformly, across all antigen receptor gene segments. According to this model, the observed repertoire restriction is secondary to the extremely low throughput of productive antigen receptor rearrangements and selective expansion of those few mature cells that encounter their cognate antigens. We considered an alternative hypothesis: certain RAG mutations might directly affect the ability of particular coding segments to undergo recombination, resulting in a primary defect in repertoire generation. Two lines of evidence make such a hypothesis worth considering. First, the coding flank sequence is well known to affect the efficiency of recombination by the wild-type RAG proteins (21–25). Second, a certain group of RAG1 mutants shows dramatic hypersensitivity to certain coding flank sequences: these mutations cluster near a known catalytic amino acid (D600), mapping to the region between amino acids 606–611 and include a deletion-insertion mutation (known as “D32”) and two point mutations, H609L and K608A (19, 26–28). All of these mutant proteins are severely defective for hairpin formation at certain coding flank sequences, leading to the deduction that the region of RAG1 between amino acids 606 and 611 contacts the coding flank and is directly involved in hairpin formation (19, 26–28). No known OS mutations, however, map to this region, nor have coding flank-sensitive mutants been described elsewhere in the RAG coding sequences.
Given the close proximity of all known coding flank-sensitive mutants to a catalytic amino acid (D600), we reasoned that coding flank hypersensitivity might also result from mutations in other regions of RAG1 that are directly involved in catalysis. We now report that the RAG1 R972Q mutation, located in the primary sequence near the catalytic amino acid, E962, is hypersensitive to certain coding flank sequences. This mutation is found at the orthologous human RAG1 residue, R975Q, in a patient with OS (9) and was serendipitously discovered in mice that display an OS-like phenotype (14). Our results provide the first evidence of an OS mutation that has a primary (rather than secondary) effect on the repertoire and support an alternative model for the molecular pathogenesis of OS and perhaps other disease states involving lymphopenia-associated autoimmunity (29, 30).
Materials and Methods
RAG Protein Purification
Experiments were performed with recombinant glutathione S-transferase (GST)-tagged mouse core RAG1 and core RAG2. RAG1 and RAG2 proteins are co-purified from CHO cells, as described in (27), using GST affinity resin beads (GE). Three different protein purifications for each mutant were made, and mutant phenotypes were consistent between different preparations.
In vitro cleavage assay
Oligonucleotides and cleavage reactions were described in (31, 32). Reaction products were detected and quantified using a PhosphorImager and ImageQuant software.
Cellular V(D)J recombination assays
Wild-type and mutant murine full-length RAG1 and wild-type full-length murine RAG2 were expressed from a modified pEBG vector (lacking GST tag) (33). Reporter substrates were transfected into RMP41 CHO cells, as described (33). The GFP V(D)J recombination reporter substrates have been described (33). Briefly, a poly(A) sequence between 12-RSS and 23-RSS elements were cloned into pEGFP-N1 (Clontech) between the CMV promoter and GFP gene. RAG-mediated recombination results in deletion of the poly(A) sequence and expression of GFP. In the coding joint substrate, RSSs are oriented such that GFP is expressed after coding joint formation, while in the signal joint substrate, inversion of both RSSs allows expression after signal joint formation. Cells were harvested 48h after transfection, trypsinized, spun at 1200 rpm for 5 min, and resuspended in 50µl PBS with 0.5% BSA and 5mM EDTA. Cells were analyzed by flow cytometry using LSR II flow cytometer (BD Biosciences) and FlowJo software.
Altered coding flank substrates
Coding flank sequences at both 12-RSS and 23-RSS of fluorescent reporter coding joint and signal joint substrates, as described above, were changed to all sixteen permutations by site-directed mutagenesis kit (Stratagene).
Substrate-specific integrated cell lines
RMP41 CHO cell lines bearing integrated recombination substrates were developed as previously described (33). 1 × 105 stable substrate-integrated cells were plated per well in 24-well plates and transfected 20 hours later with 100ng each of RAG-1 and RAG-2 expression plasmids using Fugene6 reagent (3:2 ratio of Fugene6:DNA). Cells were harvested and analyzed as described above.
Results
R972Q displays coding flank hypersensitivity in vitro
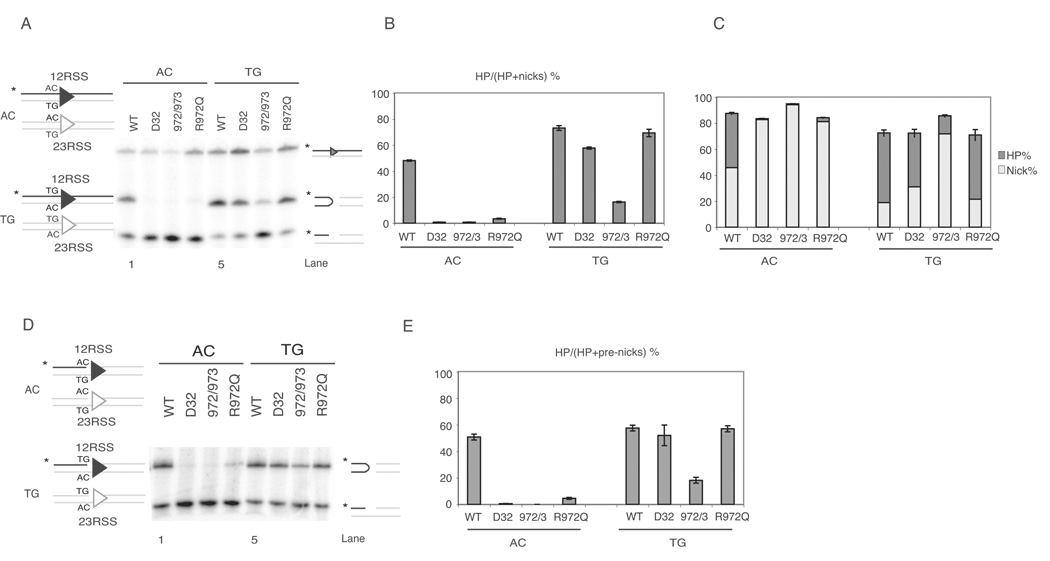
Human RAG1 R975, the ortholog of murine RAG1 R972, is mutated to glutamine in a patient with OS (9). This mutation shares two features with mutations in the 606–611 region of RAG1: it lies near a catalytic amino acid (E962) (34), and the R972A/K973A mutant exhibits a specific defect in hairpin formation (19). We asked whether R972Q might also show hypersensitivity to certain coding flank sequences. We purified the R972Q mutant as well as wild-type RAG1, D32, and R972A/K973A proteins as controls, and examined the ability of these proteins, in conjunction with wild-type RAG2, to cleave oligonucleotide substrates containing either AC or TG coding flanks (Figure 1A). As expected, wild-type RAG proteins catalyzed robust nicking and hairpin formation with both substrates (lanes 1 and 5). D32 was specifically defective for hairpin formation with AC, but not TG, coding flanks (lanes 2 and 6) (26), and R972A/K973A was severely defective for hairpin formation on both substrates, with a slight increase in activity at a TG flank (lanes 3,7). Interestingly, R972Q was severely impaired at the AC flank but formed hairpins at wild-type levels on the TG flank (lanes 4, 8). We define this sequence-dependent effect on hairpin formation as coding flank hypersensitivity.
Figure 1. R972Q RAG1 mutant shows preferences for coding flank sequence in vitro.
(A) Cleavage assay using oligonucleotide substrates as depicted on the left, with either AC (non-preferred) or TG (preferred) flank. Only one strand of each oligo pair was 5’-radiolabled (*, strand in black), and thus detectable after electrophoresis.
(B) Quantitative analysis of gel in (A), showing the percentage of hairpin formation. Error bars represent standard error of the mean of three experiments.
(C) Quantitative analysis of gel in (A). The percentage of hairpins and nicks are shown in dark grey and light grey, respectively, and the length of each bar represents total nicking activity, as nicks were converted into hairpins.
(D) Cleavage assay using oligonucleotide substrates with same sequence as in (A), except these contain a preformed nick.
(E) Quantitative analysis of gel in (D), showing the percentage of pre-nicks converted to hairpins.
Quantitative analyses of the efficiency of hairpin formation from multiple experiments confirmed these observations (Figure 1B). Regardless of the coding flank sequence, total nicking activity (the sum of nicking and hairpin formation) was not affected by the mutations (Figure 1C). To confirm that the coding flank sequence specifically affects hairpinning, we turned to pre-nicked substrates (Figure 1D). Wild-type RAG proteins performed similar levels of hairpin formation at the pre-nicked AC and TG substrates (lanes 1 and 5), whereas the mutants formed little or no hairpins at a pre-nicked AC flank (lane 2–4), though they efficiently convert a pre-nicked TG flank to hairpin form, as expected (lane 6–8). Data from three experiments are quantified in Figure 1E, confirming that hairpin formation by the mutants is not enhanced by providing a pre-nicked substrate and is essentially the same as with uncleaved substrates (Figure 1B). These results localize the coding flank sensitivity of these RAG1 mutants to the hairpin formation step.
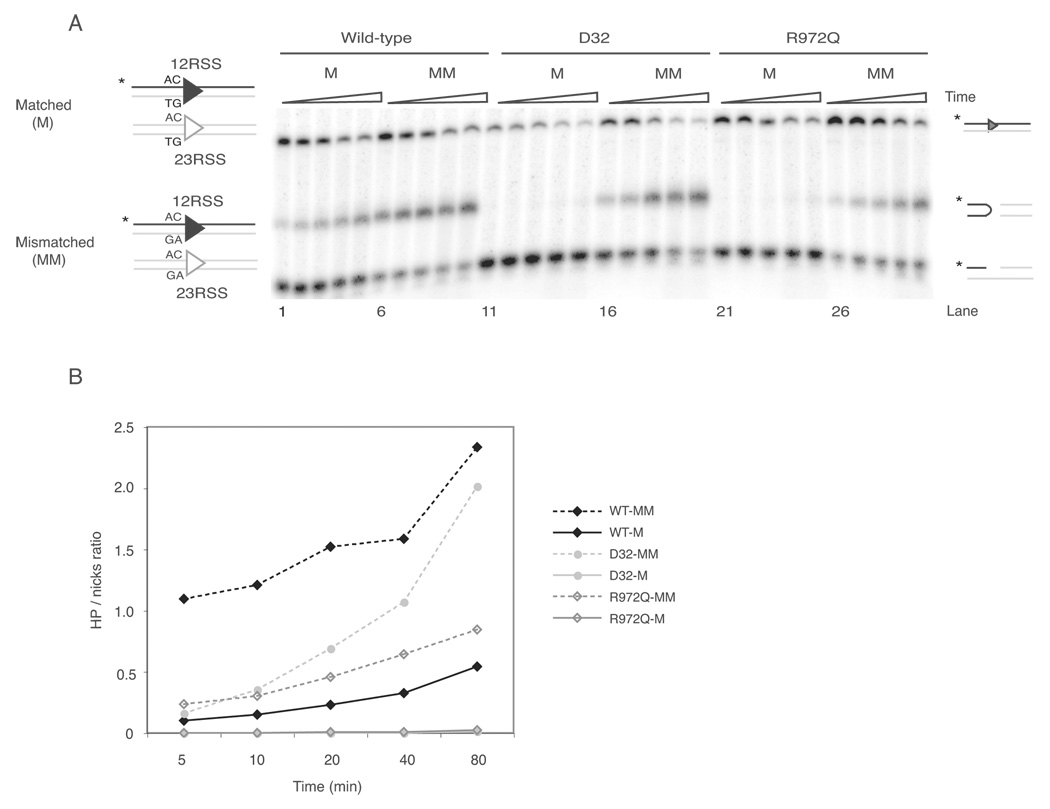
Furthermore, as previously found for the D32 mutant (27), hairpin formation by R972Q was rescued by oligonucleotide substrates containing two mismatched bases at both coding flanks (Figure 2A, 2B, and data not shown), indicating that, at certain coding flanks, R972Q renders the RAG recombinase unable to generate the DNA distortion required for hairpin formation, as suggested previously for mutants in the 606–611 region (26, 27).
Figure 2. Hairpin formation defect of coding flank sensitive RAG1 mutant can be rescued by oligonucleotide substrate that contains mismatched base pairs.
(A) Oligonucleotide substrates containing matched (M) or mismatched (MM) base pairs as depicted on the left. Triangles above gel indicate increasing incubation times: 5, 10, 20, 40, and 80 min. Reaction products are depicted on the right (from top to bottom): uncleaved substrates, hairpins, and nicks.
(B) Quantitative analysis of gel shown in (A). Reactions performed with matched substrates are shown in solid lines and mismatched substrates in dashed lines.
R972Q is hypersensitive to coding flank sequence in cells
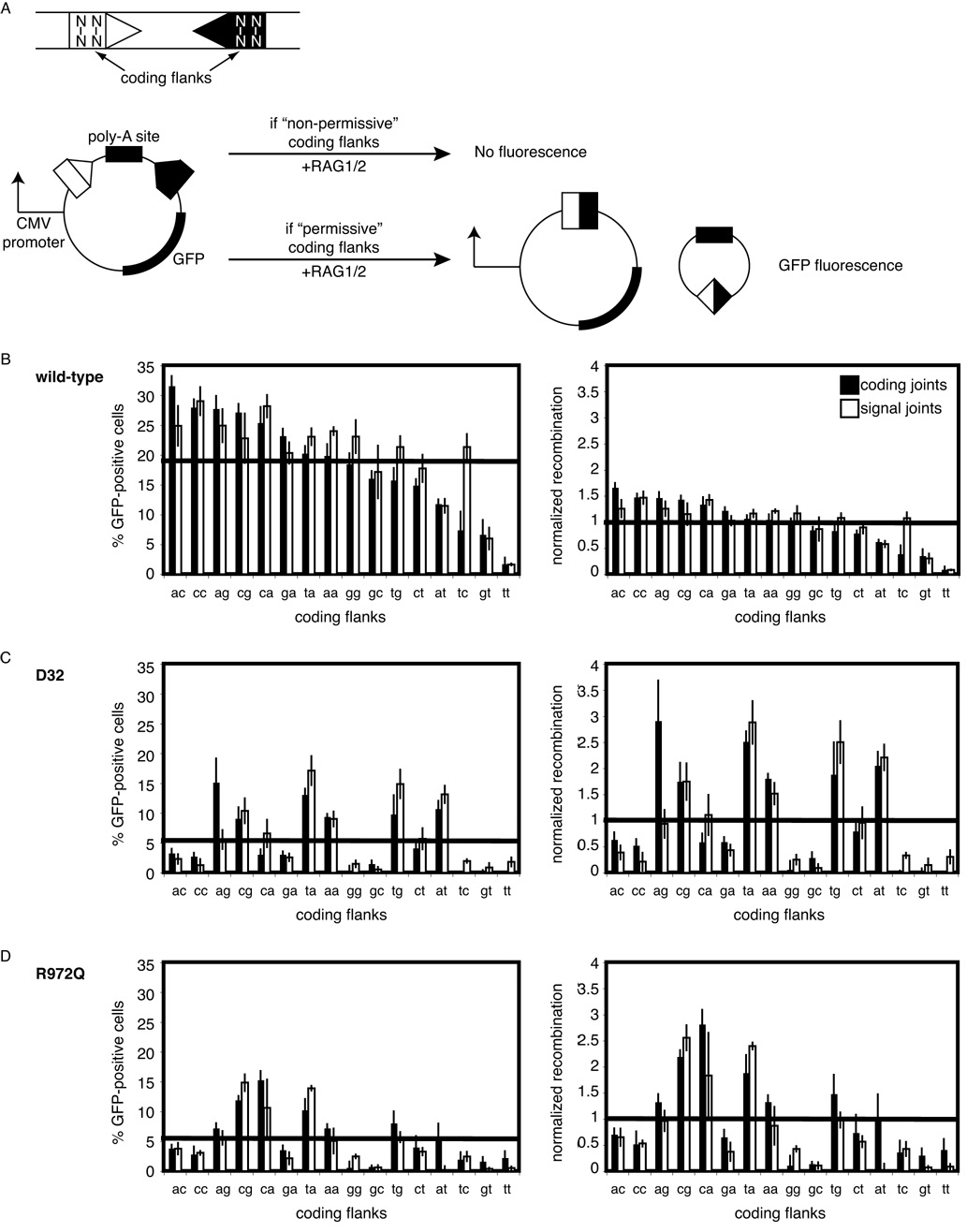
We next examined the behavior of the wild-type and mutant RAG1 proteins in cells. Previous work has shown that the 2 bp proximal to the cleavage site are the major determinant of sequence hypersensitivity of D32 (26). We therefore constructed a set of extrachromosomal fluorescent reporter substrates (33) bearing each of the sixteen possible permutations of the two coding flank base pairs (Figure 3A) and used them to examine coding flank sensitivity of the mutants in transiently transfected Chinese hamster ovary (CHO) cells. As measured by the proportion of GFP-positive cells, wild-type RAG proteins catalyzed recombination over a 30-fold range that varied according to the sequence of the last two bases of the coding flank (Figure 3B, left panel). D32 shows a distinct profile of coding flank preferences, in agreement with previous reports (26). In comparison with wild-type RAG1, R972Q displays strikingly different coding flank preferences, with similar effects on both signal and coding joints. These data are in agreement with our in vitro results, which demonstrate that coding sequence affects the efficiency of cleavage.
Figure 3. Wild-type and R972Q RAG1 display distinct coding flank preferences during V(D)J recombination in a cell culture assay.
(A) Schematic of V(D)J recombination assay for coding joint formation. Rearrangement of plasmid results in GFP fluorescence. Coding flank sequences marked N. A total of sixteen coding flank sequence permutations were generated and tested. Substrates contained the same coding flank adjacent to both 12-RSS (open triangle) and 23RSS (closed triangle).
(B, C, D) Plasmids containing RAG1 (wild-type, D32, or R972Q, repectively), wild-type RAG2, and each of sixteen coding joint (filled bars) substrates or sixteen signal joint (open bars) substrates were transfected into CHO cells and analyzed by FACS for GFP expression. Percentage of GFP-positive cells is graphed in the left panel. The graph on the right depicts the same data normalized over the mean. Error bars represent standard error of the mean of ≥3 experiments.
To facilitate comparisons between wild-type and mutant RAG proteins, we normalized the data obtained from the reporter substrates to the mean levels of coding and signal joints observed over all coding flanks (~20% GFP positive cells for both with wild-type RAG proteins). This analysis revealed that the efficiency of recombination by wild-type RAG1 over most coding flanks is close to the mean. In contrast, both D32 and R972Q display reduced levels of V(D)J recombination overall (~5% for coding joints and ~6% for signal joints), and the individual substrates show a much wider variance from the mean (right column of Figures 3B, 3C, and 3D). In sharp contrast with wild-type RAG1, R972Q strongly prefers a few coding flanks, especially CA, CG, and TA, but shows very poor recombination at GG and GC flanks, (Figure 3, bottom right). The distribution of sequence preferences shown by R972Q over all 16 coding flanks differs significantly from wild-type RAG1 (p < 0.00001, paired T-test). Based on these data, we conclude that R972Q is hypersensitive to certain coding flank sequences.
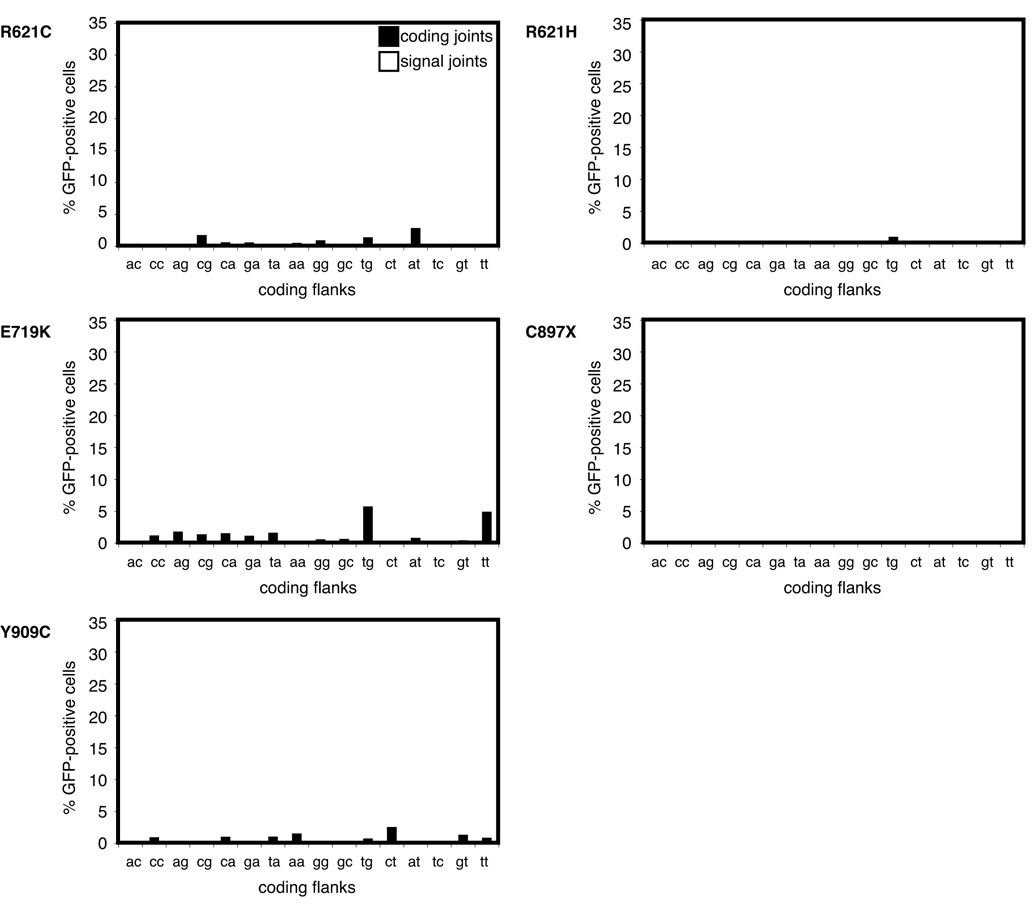
We asked whether other OS RAG1 mutations near known active site residues and/or the coding flank binding region (including R621C, R621H, E719K, C897stop, and Y909C) (8–10) might confer coding flank hypersensitivity. All of these mutants displayed extremely low activity across all 16 coding flank sequences, with no coding flank hypersensitivity (Figure 4). Note that the scales of the graphs in Figure 4 are the same as those in the left panel of Figures 3B–D. This behavior is in sharp contrast to R972Q, which displays much more robust recombination (average, 6%), but with hypersensitivity to certain flank sequences (Figure 3D, left panel).
Figure 4. Omenn Syndrome mutants display weak, hypomorphic activity with all coding flank sequences.
Plasmids containing RAG1 R621C, R621H, E719K, C897X, or Y909C and wild-type RAG2 plus each of sixteen coding joint substrates or sixteen signal joint substrates were transfected into CHO cells and analyzed by FACS. Percentage of GFP-positive cells is shown. Error bars represent standard error of the mean of ≥3 experiments.
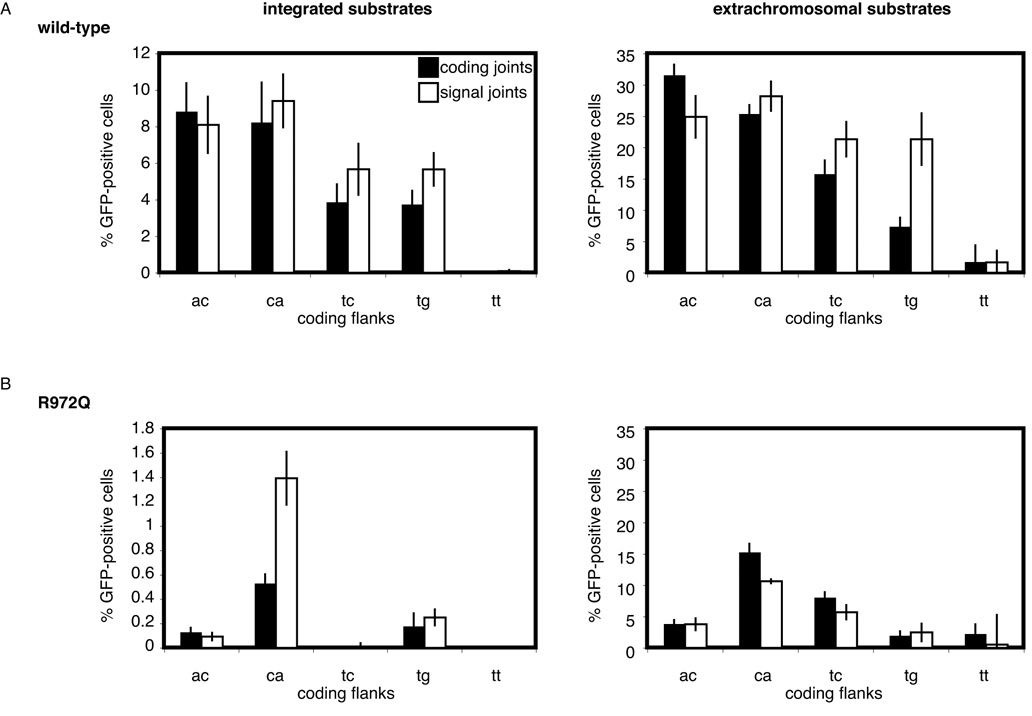
The coding flank hypersensitivity of R972Q was recapitulated using chromosomally integrated substrates, as shown in Figure 5. These data illustrate that the sequence sensitivity of R972Q is not limited to plasmid substrates but also holds in the context of chromatin, suggesting that R972Q is hypersensitive to coding flanks in vivo. Together, these data support two distinct mechanisms for the pathogenesis of OS derived from RAG mutants: uniformly hypomorphic activity and selective coding flank hypersensitivity.
Figure 5. Coding flank preferences of wild-type and R972Q RAG1 are recapitulated on integrated substrates.
(A and B) The percentage of GFP-positive cells is shown for cells containing integrated coding flank substrates transfected with plasmids containing RAG1 (wild-type or R972Q, respectively) and wild-type RAG2. The graph on the right depicts the extrachromosomal substrate data from Figure 3 for the same coding flank sequences. Error bars represent standard error of the mean of ≥3 experiments.
Discussion
Our data provide two new insights into the functions of the recombinase and the pathogenesis of OS. First, in addition to the canonical coding flank sensitive region (606–611), we have found that a mutation in the C-terminal coding flank binding region of RAG1 (amino acids 889–974) (35), R972 (near the catalytic E962), also affects coding flank preferences at the hairpin formation step. Of note, the R972 residue is part of a previously suggested hairpin-forming motif in RAG1, YKE962FR972K (31). Interestingly, a RAG2 mutant, K38A/R39A, appears to be sensitive to coding flank sequences at the nicking step in vitro, though, unlike D32 and R972Q, it is severely defective for coding and signal joint formation (20, 32) (SYW unpublished observations). Together, these findings indicate that multiple regions of the RAG recombinase contact and selectively cleave coding flank DNA during V(D)J recombination.
Second, our results separate RAG mutants found in OS patients into two distinct classes: hypomorphic mutants that show severely diminished recombination across all 16 dinucleotide coding flanks and at least one coding flank-sensitive mutant that is defective only at some coding flank sequences. These findings lead us to propose two pathogenetic mechanisms for the TCR repertoire restriction characteristic of OS. In one scenario, hypomorphic RAG mutants restrict the repertoire secondary to their crippled activity at all coding segments. The remaining recombination activity still allows a few, random V(D)J recombination events to occur; certain of these are amplified by exposure to antigens. In an alternative scenario, supported by the data reported here, repertoire restriction by R972Q (and possibly other, as yet unknown coding flank-sensitive mutants) is primary, a direct result of recombination only at those few coding segments that have preferred coding flank sequences.
Our analysis of murine TCRβ coding flank sequences reveals a number of coding flanks predicted to be poorly utilized by R972Q, including GG and GC at both Dβ 23RSSs. This should result in a severe restriction of the TCRβ repertoire as well as a substantial block to T cell development because of inefficient D-J joining. We also identified many IgH coding flanks that should be poor substrates for R972Q (both in mice and in humans), but given the presence of at least some preferred coding flanks at V, D, and J segments, we speculate that R972Q might skew the repertoire without causing such a severe block in B cell differentiation. This is in agreement with the clinical data from the OS patient bearing the corresponding R975Q mutation, which shows an almost normal peripheral B cell count—much higher than that usually observed in OS (9). This clinical observation is also consistent with our finding that R972Q is substantially more active (on most coding flanks) than other OS RAG mutants.
Toshio Hirano’s group showed that Vβ segment usage in CD4+ and CD8+ cells is skewed in the R972Q mouse compared to wild-type (14). This analysis was performed on T cells that have already passed through TCR selection and therefore not representative of all V(D)J recombination events. However, it is interesting to note that the “best” and “worst” Vβ coding flanks for R972Q, TG at Vβ8.1/2 and GC at Vβ12, were among the more highly and less represented segments, respectively, compared to wild-type, providing some confirmation that the activity we describe for R972Q recapitulates its activity in vivo. In addition, the variability of Vβ segment usage among mice bearing a RAG2 mutation, R229Q, was much higher than that observed in the R972Q mouse (12, 14). This supports our hypothesis that R972Q causes primary repertoire restriction through coding sequence selectivity rather than a secondary restriction due to rare, random recombination events.
Our data raise the possibility that other RAG mutations that confer coding flank hypersensitivity might have unexpected effects on the antigen receptor repertoire. Indeed, some mutants might skew the repertoire or cause selective repertoire defects, without manifesting overt signs of immunodeficiency. RAG mutants might thus be found in other inherited immunoregulatory disorders.
Acknowledgements
The authors thank J. Lafaille and members of the Roth laboratory, particularly G. Celli and L. Deriano, for critical reading and thoughtful comments.
Footnotes
This study was supported by National Institutes of Health Grant AI36420 (to D.B.R.) and Irene Diamond Foundation (to D.B.R.).
References
- 1.Omenn GS. Familial Reticuloendotheliosis with Eosinophilia. N Engl J Med. 1965;273:427–432. doi: 10.1056/NEJM196508192730806. [DOI] [PubMed] [Google Scholar]
- 2.Honig M, Schwarz K. Omenn syndrome: a lack of tolerance on the background of deficient lymphocyte development and maturation. Curr Opin Rheumatol. 2006;18:383–388. doi: 10.1097/01.bor.0000231907.50290.6f. [DOI] [PubMed] [Google Scholar]
- 3.de Saint-Basile G, Le Deist F, de Villartay JP, Cerf-Bensussan N, Journet O, Brousse N, Griscelli C, Fischer A. Restricted heterogeneity of T lymphocytes in combined immunodeficiency with hypereosinophilia (Omenn's syndrome) J Clin Invest. 1991;87:1352–1359. doi: 10.1172/JCI115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Signorini S, Imberti L, Pirovano S, Villa A, Facchetti F, Ungari M, Bozzi F, Albertini A, Ugazio AG, Vezzoni P, Notarangelo LD. Intrathymic restriction and peripheral expansion of the T-cell repertoire in Omenn syndrome. Blood. 1999;94:3468–3478. [PubMed] [Google Scholar]
- 5.Brooks EG, Filipovich AH, Padgett JW, Mamlock R, Goldblum RM. T-cell receptor analysis in Omenn's syndrome: evidence for defects in gene rearrangement and assembly. Blood. 1999;93:242–250. [PubMed] [Google Scholar]
- 6.Rieux-Laucat F, Bahadoran P, Brousse N, Selz F, Fischer A, Le Deist F, De Villartay JP. Highly restricted human T cell repertoire in peripheral blood and tissue-infiltrating lymphocytes in Omenn's syndrome. J Clin Invest. 1998;102:312–321. doi: 10.1172/JCI332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aleman K, Noordzij JG, de Groot R, van Dongen JJ, Hartwig NG. Reviewing Omenn syndrome. Eur J Pediatr. 2001;160:718–725. doi: 10.1007/s004310100816. [DOI] [PubMed] [Google Scholar]
- 8.Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, Gatta LB, Ochs HD, Schwarz K, Notarangelo LD, Vezzoni P, Spanopoulou E. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- 9.Villa A, Sobacchi C, Notarangelo LD, Bozzi F, Abinun M, Abrahamsen TG, Arkwright PD, Baniyash M, Brooks EG, Conley ME, Cortes P, Duse M, Fasth A, Filipovich AM, Infante AJ, Jones A, Mazzolari E, Muller SM, Pasic S, Rechavi G, Sacco MG, Santagata S, Schroeder ML, Seger R, Strina D, Ugazio A, Valiaho J, Vihinen M, Vogler LB, Ochs H, Vezzoni P, Friedrich W, Schwarz K. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 10.Corneo B, Moshous D, Gungor T, Wulffraat N, Philippet P, Le Deist FL, Fischer A, de Villartay JP. Identical mutations in RAG1 or RAG2 genes leading to defective V(D)J recombinase activity can cause either T-B-severe combined immune deficiency or Omenn syndrome. Blood. 2001;97:2772–2776. doi: 10.1182/blood.v97.9.2772. [DOI] [PubMed] [Google Scholar]
- 11.Sobacchi C, Marrella V, Rucci F, Vezzoni P, Villa A. RAG-dependent primary immunodeficiencies. Hum Mutat. 2006;27:1174–1184. doi: 10.1002/humu.20408. [DOI] [PubMed] [Google Scholar]
- 12.Marrella V, Poliani PL, Casati A, Rucci F, Frascoli L, Gougeon ML, Lemercier B, Bosticardo M, Ravanini M, Battaglia M, Roncarolo MG, Cavazzana-Calvo M, Facchetti F, Notarangelo LD, Vezzoni P, Grassi F, Villa A. A hypomorphic R229Q Rag2 mouse mutant recapitulates human Omenn syndrome. J Clin Invest. 2007;117:1260–1269. doi: 10.1172/JCI30928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakaguchi S, Ono M, Setoguchi R, Yagi H, Hori S, Fehervari Z, Shimizu J, Takahashi T, Nomura T. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 14.Khiong K, Murakami M, Kitabayashi C, Ueda N, Sawa S, Sakamoto A, Kotzin BL, Rozzo SJ, Ishihara K, Verella-Garcia M, Kappler J, Marrack P, Hirano T. Homeostatically proliferating CD4 T cells are involved in the pathogenesis of an Omenn syndrome murine model. J Clin Invest. 2007;117:1270–1281. doi: 10.1172/JCI30513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baccala R, Theofilopoulos AN. The new paradigm of T-cell homeostatic proliferation-induced autoimmunity. Trends Immunol. 2005;26:5–8. doi: 10.1016/j.it.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Krupica T, Jr, Fry TJ, Mackall CL. Autoimmunity during lymphopenia: a two-hit model. Clin Immunol. 2006;120:121–128. doi: 10.1016/j.clim.2006.04.569. [DOI] [PubMed] [Google Scholar]
- 17.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 18.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 19.Huye LE, Purugganan MM, Jiang MM, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol Cell Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 21.Gerstein RM, Lieber MR. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev. 1993;7:1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- 22.Boubnov NV, Wills ZP, Weaver DT. Coding sequence composition flanking either signal element alters V(D)J recombination efficiency. Nucleic Acids Res. 1995;23:1060–1067. doi: 10.1093/nar/23.6.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ezekiel UR, Sun T, Bozek G, Storb U. The composition of coding joints formed in V(D)J recombination is strongly affected by the nucleotide sequence of the coding ends and their relationship to the recombination signal sequences. Mol Cell Biol. 1997;17:4191–4197. doi: 10.1128/mcb.17.7.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu K, Lieber MR. Mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol.Cell.Biol. 1999;19:8094–8112. doi: 10.1128/mcb.19.12.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu K, Taghva A, Lieber MR. The cleavage efficiency of the human immunoglobulin heavy chain VH elements by the RAG complex: implications for the immune repertoire. J Biol Chem. 2002;277:5040–5046. doi: 10.1074/jbc.M109772200. [DOI] [PubMed] [Google Scholar]
- 26.Sadofsky MJ, Hesse JE, van Gent DC, Gellert M. RAG-1 mutations that affect the target specificity of V(D)J recombination: a possible direct role of RAG-1 in site recognition. Genes&Development. 1995;9:2193–2199. doi: 10.1101/gad.9.17.2193. [DOI] [PubMed] [Google Scholar]
- 27.Kale SB, Landree MA, Roth DB. Conditional RAG-1 mutants block the hairpin formation step of V(D)J recombination. Mol Cell Biol. 2001;21:459–466. doi: 10.1128/MCB.21.2.459-466.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roman CAJ, Baltimore D. Genetic evidence that the RAG1 protein directly participates in V(D)J recombination through substrate recognition. Proc.Natl.Acad.Sci.U.S.A. 1996;93:2333–2338. doi: 10.1073/pnas.93.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khoruts A, Fraser JM. A causal link between lymphopenia and autoimmunity. Immunol Lett. 2005;98:23–31. doi: 10.1016/j.imlet.2004.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carneiro-Sampaio M, Coutinho A. Tolerance and autoimmunity: lessons at the bedside of primary immunodeficiencies. Adv Immunol. 2007;95:51–82. doi: 10.1016/S0065-2776(07)95002-6. [DOI] [PubMed] [Google Scholar]
- 31.Lu CP, Sandoval H, Brandt VL, Rice PA, Roth DB. Amino acid residues in Rag1 crucial for DNA hairpin formation. Nat Struct Mol Biol. 2006;13:1010–1015. doi: 10.1038/nsmb1154. [DOI] [PubMed] [Google Scholar]
- 32.Lu CP, Posey JE, Roth DB. Understanding how the V(D)J recombinase catalyzes transesterification: distinctions between DNA cleavage and transposition. Nucleic Acids Res. 2008;36:2864–2873. doi: 10.1093/nar/gkn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, Shah S, Brandt VL, Meek K, Roth DB. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 34.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG-1 and RAG-2 identifies three active site amino acids in RAG-1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mo X, Bailin T, Sadofsky MJ. A C-terminal region of RAG1 contacts the coding DNA during V(D)J recombination. Mol Cell Biol. 2001;21:2038–2047. doi: 10.1128/MCB.21.6.2038-2047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]