Abstract
MicroRNAs are small non-coding RNAs that regulate the expression of protein-coding genes. To evaluate the involvement of microRNAs in prostate cancer, we determined genome-wide expression of microRNAs and mRNAs in 60 primary prostate tumors and 16 non-tumor prostate tissues. The mRNA analysis revealed that key components of microRNA processing and several microRNA host genes, e.g., MCM7 and C9orf5, were significantly up-regulated in prostate tumors. Consistent with these findings, tumors expressed the miR-106b-25 cluster, which maps to intron 13 of MCM7, and miR-32, which maps to intron 14 of C9orf5, at significantly higher levels than non-tumor prostate. The expression levels of other microRNAs, including a number of miR-106b-25 cluster homologues, were also altered in prostate tumors. Additional differences in microRNA abundance were found between organ-confined tumors and those with extraprostatic disease extension. Lastly, we found evidence that some microRNAs are androgen-regulated and that tumor microRNAs influence transcript abundance of protein-coding target genes in the cancerous prostate. In cell culture, E2F1 and p21/WAF1 were identified as targets of miR-106b, Bim of miR-32, and exportin-6 and protein tyrosine kinase 9 of miR-1. In summary, microRNA expression becomes altered with the development and progression of prostate cancer. Some of these microRNAs regulate the expression of cancer-related genes in prostate cancer cells.
Keywords: Prostate cancer, microRNA, gene expression profile
INTRODUCTION
Recently, a new class of small RNAs has been described, termed microRNAs, which was found to regulate mRNA function by modulating both mRNA stability and the translation of mRNA into protein (1,2). MicroRNA genes are expressed as large precursor RNAs, called pri-mRNAs, which may encode multiple microRNAs in a polycistronic arrangement (3). These precursors are converted into a mature microRNA of 19 to 25 nucleotides by the nuclear RNase III enzyme, Drosha, and the cytosolic RNase III enzyme, Dicer. These two enzymes and their cofactors, e.g., DGCR8/Pasha, TRBP, and EIF2C2/argonaute-2, are key components of microRNA processing activity. Changes to their expression levels can alter cell function and induce cellular transformation (4).
A crucial role of microRNAs in cancer has been demonstrated (5). Their expression is commonly altered in solid human tumors (6). MicroRNA expression profiles also classify tumors by developmental lineage and differentiation state (6,7). Multiple microRNAs have been shown to have oncogenic properties, or act like tumor suppressor genes (5,8). These microRNAs have been termed oncomiRs. An alteration in their expression is causatively linked to cancer development.
We investigated the microRNA profiles of 60 prostate tumors and 16 non-tumor tissues to evaluate the relationship between microRNA expression and prostate cancer. We also studied the global expression of mRNAs. That approach was used to find alterations in the expression of genes that regulate prostatic microRNA processing and to identify candidate mRNAs that are post-transcriptionally repressed by microRNAs in the prostate. Lastly, we studied the influence of an androgen on microRNA transcript abundance and used precursor and antisense microRNAs and luciferase reporter constructs to show that microRNAs regulate the expression of cancer-related genes in human prostate cancer cells.
SUBJECTS AND METHODS
Clinical samples
Sixty fresh-frozen prostate tumors (macrodissected) and patient’s clinicopathological information were received from the NCI Cooperative Prostate Cancer Tissue Resource (CPCTR) and the University of Maryland. Written informed consent was obtained from all donors. The tumors had not received any therapy prior to prostatectomy. Surrounding non-tumor prostate tissue was collected from 16 patients with prostate cancer.
RNA isolation, expression analysis by microarray, and quantitative Real-time PCR
See detailed information in Supplementary Notes.
Regulation of protein expression by microRNAs
LNCaP and PC3 human prostate cancer cells (ATCC, Manassas, VA) were grown to 50% confluency and transfected with either microRNA precursor or antisense microRNA inhibitor (Ambion, Austin, TX) at 100 nM final concentration using Lipofectamine 2000 reagent (Invitrogen). After 48 hours, cells were harvested by scraping and protein was extracted with RIPA buffer (Pierce Biotechnology, Rockford, IL). The following primary antibodies were for used to visualize protein expression by Western blot analysis: polyclonal rabbit anti-exportin-6 antibody, 1:200 (ProteinTech Group, Chicago, IL: 11408-1-AP); monoclonal mouse anti-PTK9 antibody, 1:500 (Abnova Corp., Taipei, Taiwan: clone 1E2); monoclonal mouse anti-E2F1 antibody, 1:200 (Santa Cruz Biotechnology, Santa Cruz, CA: sc-251); monoclonal mouse anti-p21/WAF1 antibody, 1:200 (Santa Cruz Biotechnology: sc-6246); and polyclonal rabbit anti-BIM antibody, 1:1000 (Cell Signaling/Santa Cruz Biotechnology: #2819). A quantification of protein expression was obtained with the AIDA Biopackage, 2D–Densitometry (raytest Isotopenmessgeraete GmbH, Straubenhardt, Germany).
Luciferase assays of reporter constructs containing the 3’UTR of E2F1, BCL2L11 and CDKN1A
The E2F1, BCL2L11, and CDKN1A 3’UTRs containing the predicted miR-106b and miR-32 target sequence, respectively, were amplified from genomic DNA (293T cells) and cloned into the pGL3 firefly luciferase control vector (Promega, Madison, WI) at the XbaI restriction site immediately downstream of the luciferase reporter gene. To generate E2F1 and BCL2L11 3’UTRs with a mutant target sequences, a deletion of the first 3 nucleotides was inserted into the miR-106b and miR-32 seed region complementary sites using the QuikChange-site-directed mutagenesis kit (Stratagene, La Jolla, CA). Translational inhibition of the luciferase reporter gene by either miR-106b or miR-32 was assayed in LNCaP prostate cancer cells. Briefly, 1.2×105 LNCaP cells per well were seeded in 24 well plates. The next day, cells were transfected with 500 ng of reporter plasmid, 2 ng Renilla reporter, and either microRNA negative control or precursor microRNA at a 100 nM final concentration using the lipofectamine 2000 reagent according the manufacturer’s instructions (Invitrogen). Transfections were performed in triplicates. After 24 hours, cells were lysed according to a Promega standard protocol, and the relative luciferase activity was determined using a DYNEX Technologies MLX luminometer. Reporter activity was normalized to the protein concentration in the cell extracts.
Apoptosis assay
We used the Caspase-GloR apoptosis assay, as described by the manufacturer (Promega). More details see Supplementary Notes.
Regulation of microRNA expression by androgen
Experiments are described in the Supplementary Notes.
RESULTS
Up-regulation of Dicer in prostate tumors
Prostate tumors were collected from African-American and European-American patients with localized disease (Table 1). After isolation of total RNA from these tumors and from 16 non-tumor tissues, the expression of about 13,000 protein-coding genes and 329 unique human microRNAs was determined with microarrays.
Table 1.
Clinical characteristics of the study population
| All Cases (n = 60) |
African-American (n = 30) |
European-American (n = 30) |
P value2 t-test |
|
|---|---|---|---|---|
| Age at prostatectomy median (range) n = 60 | 60 (47 – 73) | 61 (48 – 72) | 60 (47 – 73) | 0.91 |
| PSA at diagnosis median (range) n = 441 | 6.1 (1.3 – 47.7) | 6.0 (1.3 – 47.7) | 6.1 (4.0 – 20.0) | 0.67 |
| Largest individual nodule (grams) median (range) n = 511 | 1.6 (0.2 – 2.9) | 1.5 (0.8 – 2.9) | 1.6 (0.2 – 2.8) | 0.78 |
| N (%) | N (%) | N (%) | Fisher’s exact test | |
| Source of tissue | ||||
| NCI CPCTR | 52 (87) | 27 (90) | 25 (83) | |
| University of Maryland | 8 (13) | 3 (10) | 5 (17) | 0.71 |
| Gleason sum score | ||||
| < 7 (5–6) | 15 (25) | 8 (27) | 7 (23) | |
| ≥ 7 (7–9) | 45 (75) | 22 (73) | 23 (77) | 1.0 |
| Extraprostatic extension1 | ||||
| No | 35 (67) | 19 (70) | 16 (64) | |
| Yes | 17 (33) | 8 (30) | 9 (36) | 0.77 |
| Surgical margin status1 | ||||
| Negative | 30 (59) | 17 (63) | 13 (54) | |
| Positive | 21 (41) | 10 (37) | 11 (46) | 0.58 |
| Seminal vesicle invasion1 | ||||
| No | 43 (83) | 20 (74) | 23 (92) | |
| Yes | 9 (17) | 7 (26) | 2 (8) | 0.14 |
Cases with unknown status are not included.
P value for difference between African-Americans and European-Americans
Initially, the gene expression profiles of these samples were searched for cancer-related alterations in the expression of those mRNAs that have been shown to regulate the processing of microRNAs, e.g. mRNAs that encode Drosha or Dicer, among others. Our analysis revealed that Dicer is significantly higher expressed in prostate tumors (1.6-fold; FDR < 1%) when compared with non-tumor tissue. DGCR8, which encodes an essential cofactor for Drosha, was also up-regulated in tumors (1.2-fold; FDR < 1%) than Dicer. The increased expression of Dicer and DGCR8 in tumors was confirmed by qRT-PCR, which revealed a larger fold difference than indicated by the microarray (Supplementary Figure 1). Further analysis showed that Dicer and EIF2C2, both components of the RISC complex, were more highly expressed in tumors with a high Gleason sum score (score 7–9) than in tumors with a low Gleason sum score (score 5–6). However, these expression differences were rather modest (Dicer: 1.2-fold; EIF2C2: 1.3-fold). Because a frequent co-expression of host genes and intronic microRNAs has been found in human cells (9), we also investigated the expression of microRNA host genes in prostate tumors. Among those, the expression of five was found to be altered in prostate cancer (all FDR < 1%). Of those, C9orf5 (2.1-fold up-regulation), which is the host for miR-32, and MCM7 (1.7-fold up-regulation), which is the host for the miR-106b-25 cluster (miR-106b/miR-25/miR-93), were most highly over-expressed in tumors. NFYC (host of miR-30c-1), SMC4L1 (host of miR-15b and miR-16-2), and PTPRN2 (host of miR-153-2) showed a more moderate 30% to 40% increased expression in tumors when compared to non-tumor tissue.
MicroRNA gene signature of prostate cancer
We first searched for the microRNAs that showed differential expression between tumor and non-tumor tissue. As shown in Table 2, the expression of multiple microRNAs was altered in prostate tumors. Among the microRNAs with lower transcript levels in tumors than non-tumor tissues, miR-520h, miR-494, and miR-490 were most highly decreased. Two other notable microRNAs in this list were miR-1(-2) and miR-133a(-1). These two microRNAs are encoded by the same pri-mRNA. miR-32 was the most significantly up-regulated tumor microRNA, followed by miR-182, miR-31, and miR-26a. The list of more highly expressed tumor microRNAs also contained all members of the miR-106b-25 cluster (miR-106b/miR-93/miR-25) and two members of the miR-99b cluster, miR-99b and miR-125a. The up-regulation of both miR-32 and the miR-106b-25 cluster is consistent with the increased expression of their respective host genes, C9orf5 and MCM7, in prostate tumors. Statistical analysis of the microarray data confirmed that tissue transcript levels of C9orf5 and miR-32 are statistically significantly correlated (P = 0.0003). The Pearson coefficient indicated that this correlation was moderately strong across all samples (0.39; 95% confidence interval (CI): 0.18 to 0.57; n = 76). Similar data were obtained for the correlation between MCM7 and miR-106b-25 cluster transcript levels (miR-106b: 0.37 (Pearson coefficient), P = 0.001; miR-93: 0.35, P = 0.002; miR-25: 0.23, P = 0.04).
Table 2.
MicroRNAs differentially expressed between tumor and non-tumor tissue*
| Up-regulated in Tumors | Down-regulated in Tumors | ||||
|---|---|---|---|---|---|
| Gene Name | Fold Change | Chromosomal Location | Gene Name | Fold Change | Chromosomal Location |
| miR-32 | 2.1 | 9q31.3 | miR-520h | 0.3 | 19q13.42 |
| miR-182 | 1.9 | 7q32.2 | miR-494 | 0.4 | 14q32.31 |
| miR-31 | 1.8 | 9p21.3 | miR-490 | 0.4 | 7q33 |
| miR-26a-1/2 | 1.8 | 3p22.3/12q14.1 | miR-133a-1 | 0.5 | 18q11.2 |
| miR-200c | 1.7 | 12p13.31 | miR-1-2 | 0.6 | 18q11.2 |
| miR-375 | 1.6 | 2q35 | miR-218-2 | 0.6 | 5q34 |
| miR-196a-1/2 | 1.6 | 17q21.32/12q13.13 | miR-220 | 0.6 | Xq25 |
| miR-370 | 1.6 | 14q32.31 | miR-128a | 0.6 | 2q21.3 |
| miR-425 | 1.6 | 3p21.31 | miR-221 | 0.7 | Xp11.3 |
| miR-194-1/2 | 1.5 | 1q41/11q13.1 | miR-499 | 0.7 | 20q11.22 |
| miR-181a-1/2 | 1.5 | 1q31.3/9q33.3 | miR-329 | 0.7 | 14q32.31 |
| miR-34b | 1.5 | 11q23.1 | miR-340 | 0.7 | 5q35.3 |
| let-7i | 1.5 | 12q14.1 | miR-345 | 0.7 | 14q32.2 |
| miR-188 | 1.4 | Xp11.22 | miR-410 | 0.7 | 14q32.31 |
| miR-25 | 1.4 | 7q21.11 | miR-126 | 0.7 | 9q34.3 |
| miR-106b | 1.4 | 7q21.11 | miR-205 | 0.8 | 1q32.2 |
| miR-449 | 1.4 | 5q11.2 | miR-7-1/2 | 0.8 | 9q21.33/15q26.1 |
| miR-99b | 1.4 | 19q13.41 | miR-145 | 0.8 | 5q32 |
| miR-93 | 1.3 | 7q21.11 | miR-34a | 0.8 | 1p36.22 |
| miR-92-1/2 | 1.3 | 13q31.3/Xq26.2 | miR-487 | 0.8 | 14q32.31 |
| miR-125a | 1.3 | 19q13.41 | let-7b | 0.8 | 22q13.31 |
FDR < 5% and P value (t-test) < 0.01 for all microRNAs; Fold change: reference is non-tumor tissue.
We corroborated the microarray data by qRT-PCR analysis of selected microRNAs in a random subset of the tumor and nontumor tissues. Consistent with the microarrays, we found that mature miR-32 (average: 3.2-fold) and miR-106b (average: 3.0-fold) were higher expressed in tumors than nontumor tissues (Supplementary Figure 2). We also found that mature miR-1 was down-regulated (average: 0.44-fold) and miR-106a, a miR-106b homologue, was over-expressed (average: 3.7-fold) in the tumors when compared with nontumor tissues (Supplementary Figure 2).
Lastly, we performed a paired analysis of the microarray data for those 10 tumors in our study whose surrounding nontumor tissue was available. The paired analysis corroborated our previous findings. At a FDR <10%, miR-26a, miR-30c-1, miR-32, miR-146b, miR-181a, miR-182, miR-196a, miR-200c, miR-375, and all microRNAs of the miR-106b-25 cluster were found to be up-regulated in tumors (1.5-fold to 2.5-fold). The most significantly down-regulated tumor microRNAs were miR-494 (0.4-fold) and miR-126 (0.6-fold). However, the miR-1-133a cluster was not found to be significantly differently expressed in this tumor subset.
Association of microRNAs with extraprostatic extension
We next analyzed our dataset for differences in microRNA expression associated with extraprostatic extension of the tumors. At a FDR < 20%, we found 15 microRNAs with a difference in expression between tumors that showed an extraprostatic extension of the disease (n = 17) and those that did not (n = 35) (Table 3). miR-101 was the most consistently over-expressed microRNA in localized prostate tumors that spread out of the prostate gland (FDR < 1%). Extraprostatic extension shared a portion of its microRNA signature with the tumor signature. Two microRNAs, miR-99b and miR-196a, are common to both signatures. Two other microRNAs of the extraprostatic extension signature, miR-200a and miR-200b, have an extensive homology with miR-200c in the tumor signature. We could not identify a robust microRNA signature associated with Gleason score in our dataset.
Table 3.
MicroRNAs associated with extraprostatic disease
| Gene Name | Fold Change* | FDR | Chromosomal Location |
|---|---|---|---|
| miR-101-1/2 | 1.6 | < 1% | 1p31.3/9p24.1 |
| miR-200a | 1.6 | 10% to 15% | 1p36.33 |
| miR-200b | 1.6 | 10% to 15% | 1p36.33 |
| miR-196a-1/2 | 1.3 | 10% to 15% | 17q21.32/12q13.13 |
| miR-30c-1/2 | 1.3 | 10% to 15% | 1p34.2/6q13 |
| miR-484 | 1.3 | 10% to 15% | 16p13.11 |
| miR-99b | 1.3 | 10% to 15% | 19q13.41 |
| miR-186 | 1.3 | 10% to 15% | 1p31.1 |
| miR-195 | 1.3 | 10% to 15% | 17p13.1 |
| let-7f-2 | 1.3 | 10% to 15% | Xp11.22 |
| miR-34c | 1.2 | 10% to 15% | 11q23.1 |
| miR-371 | 0.7 | 15% to 20% | 19q13.42 |
| miR-373 | 0.7 | 10% to 15% | 19q13.42 |
| miR-410 | 0.7 | 15% to 20% | 14q32.31 |
| miR-491 | 0.7 | 15% to 20% | 9p21.3 |
Comparing tumors with and without (reference) extraprostatic extension. P value (t-test) < 0.01 for all microRNAs
Because the tumors in our study were collected from African-American and European-American patients that were well matched on clinicopathological parameters (Table 1), we compared the tumor microRNA signatures between African-Americans (n = 30) and European-Americans (n = 30). Few microRNAs were differently expressed (P < 0.01). At a FDR < 20%, miR-129, miR-196b, and miR-342 were found to be less abundant (20% to 30% lower) in tumors of African-Americans than in tumors of European-Americans. From this analysis, it does not appear that tumor microRNAs are very differently expressed by race/ethnicity.
Relationship between transcript abundance of microRNAs and their target mRNAs in prostate tissue
MicroRNAs regulate the expression of protein-coding genes by target-specific translational inhibition. However, it has recently been shown that some microRNAs, e.g., miR-1, can down-regulate the transcript levels of a large number of target genes in mammalian cells (1,2). Because miR-1 was among the down-regulated microRNAs in prostate tumors, we performed a correlation analysis between miR-1 expression levels and the expression levels of predicted miR-1 target genes in these tumors. This test was performed to identify candidate miR-1 target genes that become over-expressed in prostate tumors because of diminished miR-1 expression. The analysis yielded putative target mRNAs that were found to be up-regulated in prostate tumors (FDR < 1%) and inversely correlated with miR-1 expression (Supplementary Table 1). Among those, transcripts for WDR6, XPO6, and SMARCA4 showed the most significant inverse correlation with tumor miR-1 expression (each P < 1× 10−10). The relationship between XPO6 and miR-1 transcript levels in prostate tumors is shown in Supplementary Figure 3. We also found that XPO6 protein levels in the tumors are inversely correlated with miR-1 (−0.29, Spearman correlation coefficient; n = 8). However, not all predicted targets of miR-1 showed an inverse relationship with miR-1 transcript levels in the tumors. For example, TWF1 (also termed PTK9) was positively correlated with miR-1, suggesting that binding of microRNAs to its target sequence may sometimes lead to mRNA sequestration and cellular accumulation of the inhibited mRNA (10).
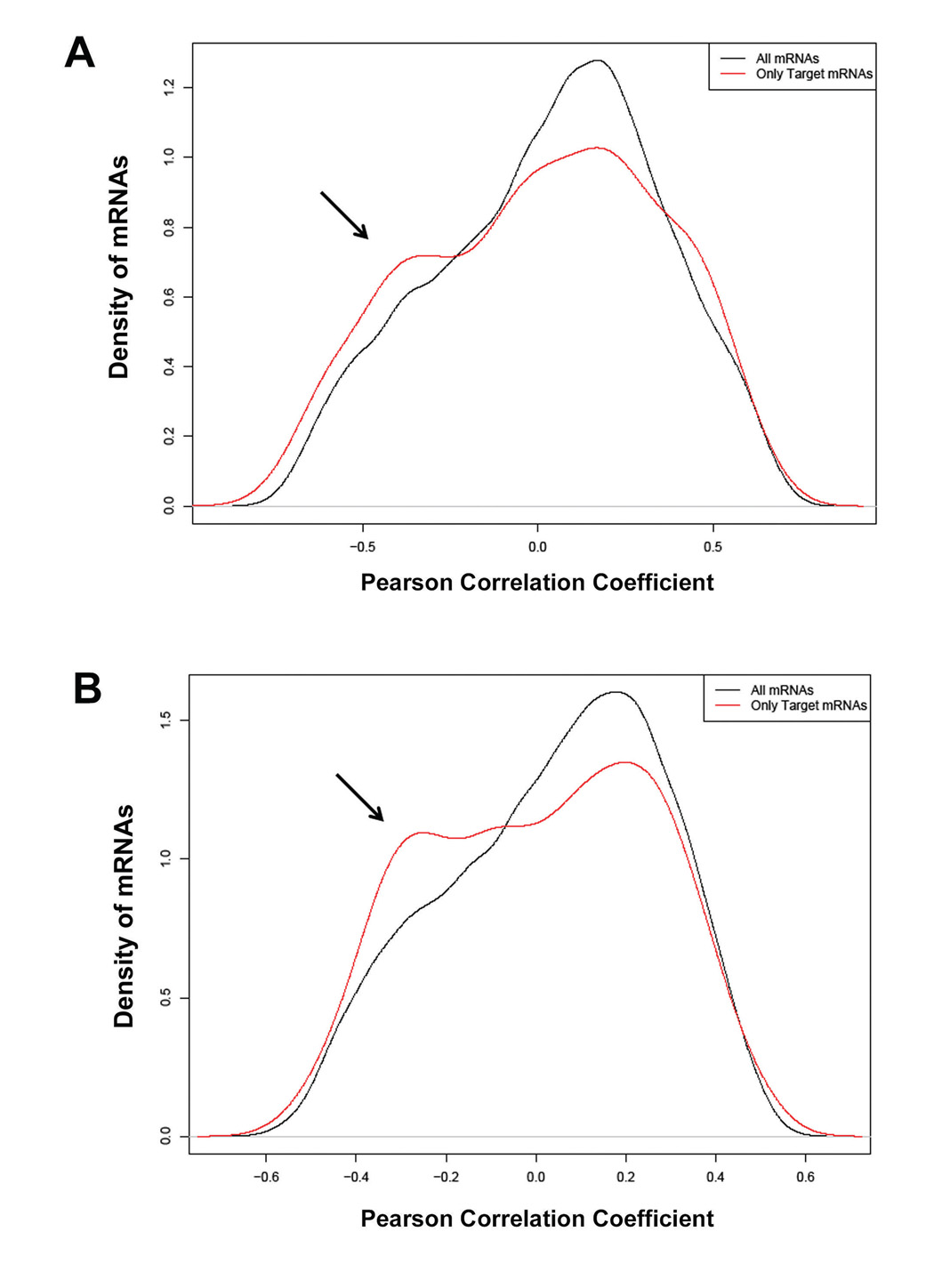
Our analyses were extended to other microRNAs that were either up- or down-regulated in prostate tumors. Here, we initially determined the global distribution of the Pearson correlation coefficients between the microRNA of interest and either all mRNAs that are probed by the HG-U133A 2.0 array or only those mRNAs that are predicted targets of the microRNA. For two microRNAs, miR-106b and miR-181a, the distribution of the correlation coefficients was notably different between all mRNA and those mRNA that are the predicted targets of miR-106b and miR-181a (Figure 1). The distribution curves for predicted target mRNAs of miR-106b and miR-181a showed a distinct shoulder that extended toward negative Pearson correlation coefficients. This pattern is a departure from a normal distribution and indicates that tissue transcript levels of a subset of mRNAs, which have a predicted microRNA target sequence in the 3’UTR, are reduced by miR-106b and miR-181a. A list of target genes that were significantly down-regulated in tumors (FDR < 1%) and whose transcript level inversely correlated with miR-181a expression is shown in Supplementary Table 2. A comparison of these target genes with a list of genes that correlated with miR-181a transcript levels in leukemia samples (11) showed that several, e.g., SLC9A6, RIN2, KLHL2, and GHITM, were negatively correlated with miR-181a in both lists.
Figure 1.
Analysis of the relationship between transcript levels of microRNAs and their respective target mRNAs in prostate tissue. Shown is the global distribution of the Pearson correlation coefficients between mRNAs and miR-106b (A) or miR-181a (B). The black-lined curves show the distribution of the correlation coefficients for all mRNAs. The red-lined curves show the correlation coefficient distribution for only those mRNAs that are a predicted target of either miR-106b or miR-181a. The red-lined curves have an additional shoulder (arrow) indicating an enrichment of target mRNAs, whose transcript levels are negatively correlated with the transcript levels of the microRNA.
Inhibition of protein expression by candidate oncomiRs in prostate cancer cells
Our results from the tumor studies suggest that miR-1, miR-32, and the mir-106b-25 cluster are oncomiRs in prostate cancer. miR-32 and miR-25 share a high degree of homology, and their predicted target genes are the same (www.targetscan.org). Moreover, the mir-106b-25 cluster is highly homologous to a known oncomiR, the miR-17-92 cluster (12), and the predicted targets of miR-17-5p and miR-106b are identical. A target of the miR-17-92 cluster is E2F1 (12).
We transfected two human prostate cancer cell lines, LNCaP and PC-3, with precursor and antisense microRNAs to examine whether miR-1, miR-32, and miR-106b regulate the protein expression of cancer-related genes in these cells. The endogenous expression of these microRNAs in the cell lines was miR-106b > miR-32 > miR-1 by qRT-PCR. miR-1 expression was at the detection limit. For miR-1, we tested whether the relationship between transcript abundance of microRNAs and their target mRNAs in tumor tissue is useful to identify microRNA targets and examined whether the protein expression of exportin-6 (XPO6) and protein tyrosine kinase 9 (TWF1) is regulated by miR-1. Transfection of the prostate cancer cells with miR-1 confirmed that it represses both exportin-6 and protein tyrosine kinase 9 on the protein level in both prostate cancer cell lines (Figure 2A). Neither miR-32 nor mir-106b altered the expression of these proteins (data not shown).
Figure 2.
Inhibition of protein expression by miR-1 (A) and miR-106b (B). LNCaP and PC-3 human prostate cancer cells were transfected with either microRNA precursor (miR-1 and miR-106b) or antisense microRNA (antisense miR-1 and antisense miR-106b), or their respective vector controls, scrambled precursor microRNA (Scrambled-P) and scrambled antisense microRNA (Scrambled-A). Protein extracts were prepared 48 hours after transfection and protein expression was examined by Western blot analysis. Loading: 50 µg protein per lane.
We next investigated the regulation of E2F1 and p21/WAF1 protein levels by miR-106b. Both proteins are encoded by mRNAs that have a predicted target sequence of miR-106b in their 3’UTRs. While E2F1 did not correlate with miR-106b on the transcript level in prostate tumors, a significant inverse correlation existed between the expression of CDKN1A (encodes p21/WAF1) and miR-106b in these tumors (−0.34; 95% CI: −0.09 to −0.55; P = 0.003). As shown in Figure 2B, transfected precursor miR-106b decreased p21/WAF1 protein levels and antisense miR-106b increased p21/WAF1 protein levels in the two cell lines. We obtained the same results for E2F1 after transfection of the prostate cancer cells with precursor and antisense miR-106b (Figure 3A). Next, we studied the effect of miR-32 on Bim protein expression. Bim is encoded by BCL2L11 and is a predicted target of miR-32. BCL2L11 and miR-32 transcript levels did not correlate in the tissue samples suggesting that miR-32 may regulate this target mostly by translation inhibition. Transfection of prostate cancer cells with precursor miR-32 decreased Bim protein levels while antisense miR-32 increased Bim protein levels (Figure 3C). The protein expression of E2F1 and p21/WAF1 was not influenced by miR-32, nor was the protein expression of Bim influenced by miR-106b in these cell lines (data not shown).
Figure 3.
miR-106b and miR-32 inhibit expression of E2F1 and Bim, respectively, by a 3’UTR-mediated mechanism. (A,C) LNCaP and PC-3 human prostate cancer cells were transfected with either microRNA precursor (miR-106b or miR-32) or antisense microRNA (antisense miR-106b or antisense miR-32), or their respective vector controls, scrambled precursor microRNA (Scrambled-P) and scrambled antisense microRNA (Scrambled-A). Protein extracts were prepared 48 hours after transfection and protein expression was examined by Western blot analysis. To obtain relative intensity values, E2F1 and Bim expression were normalized to β-actin. (B) pGL3 luciferase reporter constructs containing either the wild-type or mutant 3’UTR target sequence of miR-106b in the E2F1 gene were co-transfected into LNCaP cells with either precursor microRNA negative control or miR-106b precursor (each n = 3; mean ± standard deviation). For comparison, cells were also transfected with the pGL3 control vector that did not contain the 3’UTR. After 24 hours, luciferase activity was determined in the cell extracts. In the presence of the wild-type E2F1 3’UTR, transfection with precursor miR-106b lead to a significant inhibition of the luciferase reporter when compared with the vector control (P = 0.045, two-sided t-test). This inhibition was not observed if the reporter construct contained a mutant 3’UTR target sequence of miR-106b. (D) pGL3 luciferase reporter constructs containing either the wild-type or mutant 3’UTR target sequence of miR-32 in the BCL2L11 (Bim) gene were co-transfected into LNCaP cells with either precursor microRNA negative control or miR-32 precursor (each n = 3). In the presence of the wild-type BCL2L11 3’UTR, transfection with miR-32 lead to a significant inhibition of the luciferase reporter when compared with the vector control (P = 0.003, two-sided t-test). This inhibition was attenuated if the reporter construct contained a mutant 3’UTR target sequence of miR-32.
E2F1 and Bim are direct targets of miR-32 and miR-106b
To further corroborate our findings and to provide evidence that these proteins are direct targets of miR-32 and miR-106b, LNCaP cells were co-transfected with precursor microRNA and pGL3 luciferase reporter constructs containing either wild-type or mutant 3’UTR of two genes, E2F1 and BCL2L11, respectively. Mutant 3’UTRs contained a deletion of the first 3 nucleotides in the miR-106b and miR-32 seed region complementary sites. The 3’UTRs were placed at a position that would lead to a translational inhibition of the luciferase reporter when the microRNA binds to the target sequence. As shown in Figures 3B and 3D, co-transfection of either miR-106b with the reporter construct containing the wild-type 3’UTRs of E2F1 or miR-32 with the reporter construct containing the wild-type 3’UTRs of BCL2L11 resulted in a significant inhibition of the luciferase reporters when compared with the precursor microRNA negative control. There was no inhibition of the reporter by the microRNAs in the absence of the 3’UTR. The presence of a mutant 3’UTR either abolished or attenuated the effect of the microRNAs. The results are consistent with a direct effect of the microRNAs on protein translation by binding to their 3’UTR target sequence. Such a mechanism has also been established for the regulation of p21/WAF1 by miR-106b in human colon and gastric cancer cells (13,14). Accordingly, we observed that miR-106b inhibits a luciferase reporter by a CDKN1A 3’UTR-mediated mechanism in LNCaP cells (Supplementary Figure 4).
Inhibition of caspase activation by the miR-106b-25 cluster in 22Rv1 human prostate cancer cells
Our previous data indicated that miR-32, miR-106b, and their homologues (e.g, miR-25) may act as oncogenes because they target the pro-apoptotic function of Bim and E2F1. To evaluate the effect of the miR-106b-25 cluster on apoptosis induced by doxorubicin and etoposide, we infected 22Rv1 cells, a nonmetastatic human prostate cancer cell line, with a lentiviral expression construct encoding the miR-106b-25 cluster. Using the Caspase-GloR apoptosis assay, we observed a significant inhibition of caspase-3/7 activation by this cluster in anticancer drug-treated cells (Supplementary Figure 5). The data are consistent with an anti-apoptotic function of the miR-106b-25 cluster in prostate cancer cells.
Identification of androgen-regulated microRNAs
Androgens play a key role in physiology and tumor biology of the prostate. We examined the regulation of microRNAs by androgens in DU145 and LNCaP cells. Treatment of the androgen-insensitive DU145 cells with R1881 did not yield any significant changes in microRNA expression. In contrast, expression of several microRNAs was significantly changed (FDR < 5%) in androgen-sensitive LNCaP cells following the R1881-treatment (Supplementary Table 3). One microRNA, miR-338, was significantly up-regulated. The other microRNAs were down-regulated, including miR-126-5p, miR-146b, miR-219-5p, and all members of the miR181b-1, miR-181c, and miR-221 clusters. An analysis of the baseline microRNA expression in cultured DU145 and LNCaP cells showed that all members of the three microRNA clusters had a significantly higher expression in the androgen-insensitive DU145 cells than in the androgen-responsive LNCaP cells (FDR < 5%). Using a motif search in the Genomatix transcription factor binding site database, we found that the aforementioned microRNAs have putative androgen receptor binding sites in their flanking regions (Supplementary Table 4). We further corroborated the microarray results in experiments with LNCaP cells that were treated with either 1 nM or 10 nM R1881 for 12, 24 and 48 hours. qRT-PCR analysis of mature miR-338 and miR-221 showed that their expression level is androgen-regulated (Supplementary Figure 6).
DISCUSSION
The present study revealed a distinct microRNA expression signature in prostate tumors and alterations in the expression of genes that regulate tumor microRNA processing. Furthermore, we found evidence that the deregulation of microRNAs influences transcript abundance and protein expression of target mRNAs in the prostate. The results are consistent with a pathogenic role of altered microRNA expression in human prostate carcinogenesis.
This is the first study to use large-scale gene expression profiling of both microRNAs and protein-encoding RNAs to identify alterations in microRNA function that occur in human prostate tumors. We found an increased expression of Dicer and DGCR8 in prostate tumors, and of Dicer and EIF2C2, which encodes argonuate-2, in tumors with a high Gleason score. The observation that Dicer and other genes involved in microRNA processing are up-regulated in prostate cancer is consistent with a recent report (15) and may indicate that prostate tumors are more efficient than normal prostate tissue in processing microRNA precursors into mature microRNA. Nonetheless, Dicer has functions independent of microRNA processing, and future research is needed to address the relationship between Dicer expression and microRNA processing in prostate tumors.
MicroRNA expression profiles classify human cancers (6,7). Distinct signatures for several epithelial cancers, including breast, lung, pancreatic and gastric cancer, have been reported (14,16–18). Other studies explored microRNA expression in prostate cancer (19,20). Consistent with these studies, we observed that miR-145 and miR-221 are significantly down-regulated in prostate tumors. However, the previous studies were rather small and examined only few tumors when compared with our study. We identified a tumor gene signature that contained up- and down-regulated microRNAs. The most highly up-regulated microRNA was miR-32, followed by miR-182, miR-31, miR-26a, miR-200c, and miR-196a. The list of over-expressed tumor microRNAs also contained the miR-106b-25 cluster, which is consistent with the observed gain in copy number for mir-25, mir-93, and mir-106b in several human malignancies (21). The most significantly down-regulated microRNAs included miR-520h, miR-494, miR-490, and the miR-1-133a cluster.
Altered expression of microRNAs in human cancer has been observed in numerous studies. Up-regulation of microRNAs in tumors is common (5,6,8), and it is consistent with the known oncogenic activity of many microRNAs (22–25). Mechanisms of up-regulation include transcriptional activation and the increase in gene copy numbers. A decrease in the abundance of mature microRNA may result from altered processing, as shown recently (26), which would lead to an indiscriminate lower expression of mature microRNAs. We did not observe that in the present study. Alternatively, microRNA expression could be lost because of mutations or genomic alterations (21), or epigenetic silencing of microRNA loci (27,28). Epigenetic silencing is an important mechanism in prostate cancer (29), and future studies will have to address whether this mechanism impedes microRNA expression in prostate tumors.
Little is known about the function of most of the deregulated tumor microRNAs that we identified. miR-32 is a homologue of miR-25, miR-92, miR-363, and miR-367. Several of them were also up-regulated in the prostate tumors suggesting a particular significance of this microRNA family in prostate cancer development. miR-32 is increased in colon and pancreatic cancer (6), and is a mediator of the antiviral defense of human cells (30). It is this function of miR-32 that could be the causal link between its altered expression and prostate cancer development because several of the known prostate cancer susceptibility genes are also involved in host defense (31). As shown for other microRNAs, miR-32 should regulate protein expression of target genes. We made the novel observation that miR-32 inhibits the expression of Bim, a pro-apoptotic member of the BCL-2 family. This result is consistent with the observation that the miR-32 homologue, miR-25, suppresses Bim in gastric cancer cells (14). Bim has key roles in the apoptosis of epithelial tumors and mediates antitumor effects of chemotherapy (32). Thus, down-regulation of Bim by miR-32 may contribute to the resistance of tumor cells to apoptotic stimuli in the tumor environment.
Other notable microRNAs with a known function include miR-1, miR-133a, and mir-196a. The miR-1-133a cluster has been shown to regulate cell differentiation (33). miR-1 is a homologue of miR-206, which is a suppressor of metastasis in breast cancer (34). Our discovery that miR-1 is down-regulated in prostate tumors is consistent with the tumor suppressor function of its homologue. We observed that expression of miR-1 inhibits the expression of exportin-6 and protein tyrosine kinase 9 (also termed A6/twinfilin) in prostate cancer cells. Not much is known about the function of these two genes, but recent data suggest that both regulate cellular actin dynamics (35,36). miR-196a was identified as a repressor of HOXB8 (37), and elevated expression of miR-196a predicts poor survival in pancreatic cancer (18). This microRNA was common to both the tumor signature and the extraprostatic extension signature in our study, indicating that up-regulation of miR-196a in prostate cancer could be a factor in disease progression.
The analysis of the genomic location of microRNAs can provide clues about their putative function and the mechanisms that cause altered microRNA expression in tumors (38). Recent studies have shown that microRNAs are frequently located within introns of protein-coding genes and are co-expressed with these host genes (39). We investigated host gene expression in prostate tumors and found that several of them were increased in prostate tumors. C9orf5 and MCM7 were the two most highly up-regulated host genes, and their expression correlated with the expression of the intronic microRNAs, miR-32 and the miR-106b-25 cluster, respectively. The data suggest a common mechanism that leads to the up-regulation of host gene and co-transcripted microRNA in prostate tumors.
While the role of C9orf5 in cancer is unknown, MCM7 amplifications have previously been associated with prostate cancer. The MCM7 locus was found to be amplified in 88% of cases with cancer relapse (40). MCM7 over-expression is not restricted to prostate cancer and has been observed in other malignancies. Further studies will have to address if the miR-106b-25 cluster is over-expressed in these and other cancers, and whether the oncogenic effect of MCM7 locus amplification in human cancer is due to either MCM7 or miR-106b-25 cluster expression, or both. We examined whether miR-106b targets E2F1 and CDKN1A in prostate cancer cells and found that protein expression of these genes is inhibited by miR-106b. The miR-106b-25 cluster has extensive homologue with two other microRNA clusters that are candidate human oncogenes, the miR-17-92 cluster and the miR-106a-363 cluster (22,24). E2F1 is also a target of miR-17-5p and miR-20a in the miR-17-92 cluster (12), and it has both oncogene and tumor suppressor functions (41). Like Bim, translated E2F1 can be pro-apoptotic and cooperates with the tumor suppressor p53 to mediate apoptosis (42). Its overexpression induces apoptosis in LNCaP cells (43), which indicates that inhibition of E2F1 translation by miR-106b may protect prostate cancer cells from apoptosis in the tumor environment. p21/WAF1 is another mediator of p53-induced tumor suppression (44). The growth inhibitory effect of p21/WAF1 in prostate cancer has been shown (45), and it mediates cell cycle arrest in prostate carcinoma cells in response to anticancer agents (46,47). We tested whether the miR-106b-25 cluster has anti-apoptotic activity and found that it inhibits caspase activation by doxorubicin and etoposide in 22Rv1 cells. These data are consistent with an oncogenic function of the miR-106b-25 cluster in prostate cancer, in part, because of its ability to suppress E2F1 and p21/WAF1 protein expression.
Neither miR-1, miR-32, nor the miR-106b-25 cluster was regulated by androgen stimulation of LNCaP cells. However, we identified several other microRNAs that were up- or down-regulated by androgen treatment. Those included miR-338 and miR-126, and the miR-181b-1, miR-181c, miR-221 clusters, among others. A motif search showed that these microRNAs have putative androgen receptor binding sites in their flanking regions. miR-338 was the only significantly up-regulated microRNA. There are no reports on the function of this microRNA but it is located in a region with frequent copy number gains in three epithelial cancers (21). miR-181 family members influence hematopoietic lineage differentiation (48), and their expression is altered in leukemia and several solid tumors (6,11). The miR-221 cluster has been found to regulate the p27Kip1 tumor suppressor and may have oncogenic properties in prostate cancer (49). However, this cluster also inhibits the oncogene c-Kit and angiogenesis (50).
The identification of protein-coding genes that are regulated by a specific microRNA has been proven difficult despite the development of computational approaches to predict microRNA targets. The ability to find target mRNAs is further complicated by the fact that target selectivity of microRNAs may depend on the cellular microenvironment. We used an exploratory approach and conducted a correlation analysis between microRNA expression and mRNA expression in prostate tissue. This approach can be successful if the microRNA of interest affects transcript abundance of target mRNAs, but it will fail if the target genes are regulated only by translational inhibition. We found that the expression of miR-1 is inversely correlated with a number of computationally predicted target genes in prostate tumors, e.g., XPO6. However, we also found that tumor miR-1 expression correlated positively with the transcript level of predicted targets, e.g., TWF1. Subsequent validation of these observations in cell culture confirmed that XPO6 and TWF1 are both regulated by miR-1 in prostate cancer cells. The data provide new evidence that binding of microRNAs to 3’UTR sequences can lead to both degradation and accumulation of the targeted mRNA in mammalian cells, and that both an inverse and a positive correlation between a microRNA and a mRNA in a human tissue can be predictive of a microRNA target gene. Thus, correlation analysis of microRNA and mRNA expression in human tissue may prove useful in identifying mRNAs that are regulated by microRNAs.
In conclusion, our study identified alterations in microRNA expression that occur in human prostate tumors and correlate with expression variations of protein-coding genes in these tissues. Experiments in cell culture showed that tumor microRNAs regulate the expression of cancer-related genes in human prostate cancer cells. These results indicate a pathogenic role of microRNAs in prostate cancer biology.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by National Institutes of Health grants CA081534 and CA128609 (C. Croce). The authors thank CPCTR for providing tissue specimens and supporting data. We would also like to thank John Cottrel, Audrey Salabes, Donna Perlmutter, Raymond Jones, and other personnel at the University of Maryland and the Baltimore Veterans Administration for their contributions.
Abbreviations
- FDR
false discovery rate.
REFERENCES
- 1.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat.Rev.Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Wang F, Yang GH, et al. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys.Res Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 4.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat.Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat.Rev.Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc.Natl.Acad.Sci U.S.A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat.Rev.Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat.Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 11.Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 12.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 13.Ivanovska I, Ball AS, Diaz RL, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol.Cell Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrocca F, Visone R, Onelli MR, et al. E2F1-Regulated MicroRNAs Impair TGFbeta-Dependent Cell-Cycle Arrest and Apoptosis in Gastric Cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Chiosea S, Jelezcova E, Chandran U, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 17.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 18.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 19.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA Expression Profiling in Prostate Cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 20.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Huang J, Yang N, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc.Natl.Acad.Sci U.S.A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 24.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 25.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 26.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lujambio A, Ropero S, Ballestar E, et al. Genetic Unmasking of an Epigenetically Silenced microRNA in Human Cancer Cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 28.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 29.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. N.Engl.J Med. 2003;349:366–381. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 30.Lecellier CH, Dunoyer P, Arar K, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 31.De Marzo AM, Platz EA, Sutcliffe S, et al. Inflammation in prostate carcinogenesis. Nat.Rev.Cancer. 2007;7:256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan TT, Degenhardt K, Nelson DA, et al. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell. 2005;7:227–238. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr.Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 34.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vartiainen M, Ojala PJ, Auvinen P, Peranen J, Lappalainen P. Mouse A6/twinfilin is an actin monomer-binding protein that localizes to the regions of rapid actin dynamics. Mol Cell Biol. 2000;20:1772–1783. doi: 10.1128/mcb.20.5.1772-1783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helfer E, Nevalainen EM, Naumanen P, et al. Mammalian twinfilin sequesters ADP-G-actin and caps filament barbed ends: implications in motility. EMBO J. 2006;25:1184–1195. doi: 10.1038/sj.emboj.7601019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 38.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc.Natl.Acad.Sci.U.S.A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren B, Yu G, Tseng GC, et al. MCM7 amplification and overexpression are associated with prostate cancer progression. Oncogene. 2006;25:1090–1098. doi: 10.1038/sj.onc.1209134. [DOI] [PubMed] [Google Scholar]
- 41.Johnson DG, Degregori J. Putting the Oncogenic and Tumor Suppressive Activities of E2F into Context. Curr.Mol Med. 2006;6:731–738. doi: 10.2174/1566524010606070731. [DOI] [PubMed] [Google Scholar]
- 42.Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc.Natl.Acad.Sci U.S.A. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Libertini SJ, Tepper CG, Guadalupe M, Lu Y, Asmuth DM, Mudryj M. E2F1 expression in LNCaP prostate cancer cells deregulates androgen dependent growth, suppresses differentiation, and enhances apoptosis. Prostate. 2006;66:70–81. doi: 10.1002/pros.20314. [DOI] [PubMed] [Google Scholar]
- 44.El-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 45.Gotoh A, Shirakawa T, Wada Y, et al. The growth inhibitory effect of p21 adenovirus on androgen-dependent and -independent human prostate cancer cells. BJU.Int. 2003;92:314–318. doi: 10.1046/j.1464-410x.2003.04318.x. [DOI] [PubMed] [Google Scholar]
- 46.Roy S, Kaur M, Agarwal C, Tecklenburg M, Sclafani RA, Agarwal R, et al. p21 and p27 induction by silibinin is essential for its cell cycle arrest effect in prostate carcinoma cells. Mol Cancer Ther. 2007;6:2696–2707. doi: 10.1158/1535-7163.MCT-07-0104. [DOI] [PubMed] [Google Scholar]
- 47.Hour TC, Chen J, Huang CY, Guan JY, Lu SH, Pu YS. Curcumin enhances cytotoxicity of chemotherapeutic agents in prostate cancer cells by inducing p21(WAF1/CIP1) and C/EBPbeta expressions and suppressing NF-kappaB activation. Prostate. 2002;51:211–218. doi: 10.1002/pros.10089. [DOI] [PubMed] [Google Scholar]
- 48.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 49.Galardi S, Mercatelli N, Giorda E, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 50.Poliseno L, Tuccoli A, Mariani L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.