Abstract
Phylogenetic analysis of the reverse transcriptase (RT) and protease of 117 published complete human immunodeficiency virus (HIV) type 1 genome sequences demonstrated that these genes cluster into distinct subtypes. There was a slightly higher proportion of informative sites in the RT (40.4%) than in the protease (34.8%; P = .03). Although most variation between subtypes was due to synonymous nucleotide substitutions, several subtype-specific amino acid patterns were observed. In the protease, the subtype-specific variants included 7 positions associated with drug resistance. Variants at positions 10, 20, 36, and 82 were more common in non-B isolates, whereas variants at positions 63, 77, and 93 were more common in subtype B isolates. In the RT, the subtype-specific mutations did not include positions associated with anti-retroviral drug resistance. RT and protease sequences from 2246 HIV-infected persons in northern California were also examined: 99.4% of the sequences clustered with subtype B, whereas 0.6% clustered with subtype A, C, or D.
During its spread among humans, the main (M) group of human immunodeficiency virus type 1 (HIV-1) has developed an extraordinary degree of genetic diversity [1-4]. To genetically classify global group M HIV-1 isolates, researchers initially developed a subtyping system based on env and gag sequences. However, it was soon realized that certain isolates belonged to different subtypes, depending on the region of the genome analyzed. In 2000, a more comprehensive system based on complete genome analysis categorized viruses into 9 pure subtypes (A, B, C, D, F, G, H, J, and K), 6 circulating recombinant forms (CRFs; CRF01_AE, CRF02_AG, CRF03_AB, CRF04_cpx, CRF05_DF, and CRF06_cpx), and incidental viral variants [1, 2].
HIV-1 protease and reverse-transcriptase (RT) sequencing is used increasingly to help clinicians select antiretroviral drugs for their patients [5, 6], and these 2 genes are now the most frequently sequenced HIV-1 genes. The available anti-HIV drugs were developed by drug screening, and susceptibility testing by use of subtype B isolates, and characterization of drug resistance mutations has been done predominantly for subtype B isolates. However, subtype B accounts for only a small proportion of HIV-1 isolates worldwide, and non-B isolates have been identified with increased frequency, particularly in Europe.
As HIV-1 protease and RT sequence data accumulate, it is essential to be able to distinguish intersubtype changes from changes resulting from selective drug pressure [7-9]. In addition, most available quantitative HIV-1 RNA assays have not been optimized for non-B isolates [10]. The finding that a patient’s virus belongs to a non-B subtype may have implications for interpreting the results of quantitative virus load assays.
We examined sequence data of previously published global HIV-1 isolates to determine whether the protease and RT genes cluster into subtypes and, if so, to determine whether these subtypes correlate with those of other genes in the same isolate. We also examined the correlation between different subtypes and specific amino acid patterns with respect to the possible implications for anti-HIV drug resistance. Finally, we examined whether subtype consensus sequences could be used to simplify HIV-1 protease and RT subtyping. For this analysis, we examined previously published HIV-1 sequences and sequences from a clinic-based population of HIV-1-infected persons in northern California.
Materials and Methods
Previously published sequences of complete HIV-1 genomes
A list of complete HIV-1 genomes published before 1 July 2000 was compiled by searching GenBank and the Los Alamos National Laboratory HIV Sequence Database [11]. The list was then short-ened to include just 1 sequence per person. If multiple clones of an isolate were sequenced, we selected 1 clone. If multiple isolates were available from a person, one of the isolates was selected. The Appendix following the text includes the GenBank accession numbers of the completely sequenced genomes analyzed in this study.
Phylogenetic analyses
Phylogenetic trees were constructed from the complete protease sequence, the complete RT sequence, the 5′ polymerase-coding region of the RT (codons 1-300), and the combination of the protease and RT by use of Phylogenetic Analysis Using Parsimony (PAUP*) [12] and fastDNAml [13] programs. Neighbor-joining, maximum parsimony, and maximum likelihood trees were created with a variety of nucleotide substitution models, including one with no corrections, one that takes into account transition/transversion bias [14], and one that takes into account transition/transversion bias, variable base frequency, and site variation in substitution rate (HKY85 + Γ) [15, 16].
An outgroup sequence was created by taking the consensus sequence of an alignment of representative nonrecombinant reference sequences (M:A-92UG037, M:A-U455, M:B-RF, M:B-HXB2R, M:C-92BR025, M:C-ETH2220, M:D-94UG114, M:D-NDK, M:F1-VI850, M:F2-MP255C, M:G-SE6165, M:G-92NG083, M:H-90CF056, M:H-VI991, M:J-SE91733, and M:J-SE92809) [1]. This consensus sequence has been considered to be a parsimonious candidate for the group M ancestral sequence and has been used in an attempt to avoid bias resulting from oversampling some clades relative to others [3]. This consensus sequence, however, may not be completely unbiased—this would be true only if the sampling of sequences and the phylogenetic sampling of surviving lineages were balanced. Nonetheless, it is not necessary for our analysis to reconstruct the ancestral sequence; moreover, we obtained similar phylogenetic trees by using a group O isolate sequence as an outgroup. Synonymous and nonsynonymous nucleotide distances between and within subtypes were calculated with the Synonymous Nonsynonymous Analysis Program (SNAP) available on line (http://hiv-web.lanl.gov/) from the Los Alamos HIV Sequence Database [17, 18].
Subtype-specific amino acid sequence patterns
To investigate the amino acid sequences of non-B viruses, a list of published RT and protease sequences from untreated persons was compiled by searching GenBank. The consensus of each non-B sequence was compared with the subtype B consensus sequence [19], and the patterns of polymorphisms in each non-B sequence were compared with the patterns of polymorphisms in the subtype B sequences.
Protease and RT isolates from northern California
The Stanford University Hospital (SUH) Diagnostic Virology Laboratory has performed HIV-1 protease and RT sequencing at the request of physicians in northern California since June 1997. Between June 1997 and June 2000, isolates from 2246 patients were sequenced as described elsewhere [20]. RNA was extracted from 0.2 mL of plasma by using the guanidine-thiocyanate lysis reagent in the AMPLICOR HIV Monitor test kit (Roche Diagnostic Systems). Reverse strand cDNA was generated from viral RNA, and first-round polymerase chain reaction (PCR) was done with Superscript One-Step RT-PCR (Life Technologies). Direct PCR (population-based) cycle sequencing was performed by using AmpliTaq DNA fluorescent sequencing polymerase and dRhodamine terminators (ABI).
The protease and RT of each of the northern California sequences were classified by use of neighbor-joining trees [1]. The trees were created by using each of the 2246 patient sequences together with the protease and RT sequences of the previously published completely sequenced genomes. Sequence distances were computed by using both uncorrected nucleotide sequence comparisons and an adjusted nucleotide distance, which ignored positions associated with drug resistance, including positions 10, 24, 30, 32, 46-48, 50, 53, 54, 71, 73, 82, 84, 88, and 90 in the protease and positions 41, 62, 65, 67, 70, 74, 75, 77, 98, 100, 101, 103, 106, 108, 115, 116, 151, 179, 181, 184, 188, 190, 210, 215, 219, 225, and 236 in the RT [21, 22]. Sequences from the northern California patients were submitted to GenBank. The accession numbers for the subtype B sequences were AY030410-AY032587, AF085086-AF085138, and AF088078-AF088130; the accession numbers for the non-B sequences were AF331676-AF331703).
Results
Protease and RT subtypes of completely sequenced HIV-1 genomes
We analyzed complete HIV-1 genome sequences from 117 persons. On the basis of previously published analyses, 77 of these sequences belonged to 1 subtype throughout their entire genome, and 40 sequences were recombinants. Of the 77 sequences of 1 subtype, 10 were subtype A, 29 were subtype B, 15 were subtype C, 6 were subtype D, 6 were subtype F (4 subsubtype F1 and 2 subsubtype F2), 4 were subtype G, 3 were subtype H, 2 were subtype J, and 2 were subtype K isolates. The 40 recombinant isolates included 29 that contained regions belonging to 2 subtypes and 11 with regions belonging to ≥3 subtypes.
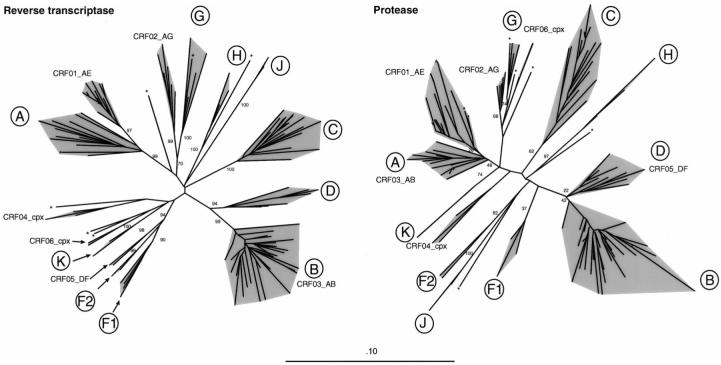
To determine whether the protease and RT gene sequences clustered into groups similar to those of the complete genomes, we created phylogenetic trees by using the complete protease gene and first 300 codons (the 5′ polymerase coding region) of the RT gene. Figure 1 shows neighbor-joining trees constructed by using the HKY85+ Γ nucleotide substitution model; nearly identical trees were obtained by use of maximum parsimony and maximum likelihood methods. The complete genome subtypes of the sequences are indicated, and bootstrap values are shown for the ancestral nodes of each subtype. Highly mosaic variants and incidental viral variants that did not fit into the revised classification were not included in the bootstrap analysis.
Figure 1.
Neighbor-joining trees constructed from reverse-transcriptase and protease sequences from 117 completely sequenced human immunodeficiency virus type 1 genomes. Trees were constructed with the HKY85 nucleotide-substitution model, with a Γ distribution to allow for site variation in substitution rate. Bootstrap values are shown at ancestral nodes of each subtype and circulating recombinant form. Branches with asterisks represent isolates that were complex recombinants and incidental variants not included in the bootstrap analysis. The scale bar represents a 10% nucleotide difference.
On the basis of the clustering pattern in the neighbor-joining trees, we assigned protease and RT subtypes to 103 of the 117 published sequences (table 1). For the nonrecombinant viruses, the subtypes assigned to the protease and RT were, in all cases, the same as the subtype of the complete genome. For the 2-subtype recombinants, the subtype assigned to the protease and RT were, in all cases, 1 of the 2 subtypes present in the complete genome. For 14 of 26 2-subtype recombinants (11 CRF01_AE, 2 AC, and 1 AD), both the protease and RT had the same subtype as gag. For 6 of 26 2-subtype recombinants (6 CRF02_AG), the protease and RT were mosaic but closest to G in the protease and in the first 300 codons of RT. For 4 of 26 2-subtype recombinants, the protease had the same subtype as gag and the RT had the same subtype as env. In 1 recombinant, the RT had the same subtype as gag and the protease had the same subtype as env. In another recombinant, both the protease and RT had the same subtype as env.
Table 1.
Distribution of isolates based on neighbor-joining phylogenetic analysis
| Genome subtype | No. | Protease |
RT |
Description of recombination in protease or RT | ||
|---|---|---|---|---|---|---|
| Subtype | No. | Subtype | No. | |||
| A | 10 | A | 10 | A | 10 | — |
| B | 29 | B | 29 | B | 29 | — |
| C | 15 | C | 15 | C | 15 | — |
| D | 6 | D | 6 | D | 6 | — |
| F | 6 | F | 6 | F | 6 | — |
| G | 4 | G | 4 | G | 4 | — |
| H | 3 | H | 3 | H | 3 | — |
| J | 2 | J | 2 | J | 2 | — |
| K | 2 | K | 2 | K | 2 | — |
| CRF01_AE | 11 | A | 11 | A | 11 | No evidence of recombination in protease or RT. |
| CRF02_AG | 6 | G | 6 | G | 6 | For all 6 isolates, protease is closest to subtype G. First 300 codons of RT are closest to subtype G but contain small mosaic region closer to subtype A; rest of RT is closest to subtype A [23]. |
| CRF03_AB | 2 | A | 2 | B | 2 | For both isolates, protease and about first 50 codons of RT are closest to subtype A; rest of RT is closest to subtype B [24]. |
| CRF05_DF | 2 | D | 2 | F | 2 | For both isolates, protease is closest to subtype D; RT is closest to subtype F [25]. |
| BF | 1 | B | 1 | B | 1 | No evidence of recombination in protease or RT. |
| AC | 3 | A C A |
1 1 1 |
A C C |
1 1 1 |
One isolate is closest to subtype A in both protease and RT (AC_SE9488; GenBank accession no. AF071474), 1 to subtype C in both protease and RT (AC_21301; GenBank AF067156), and 1 isolate (AC_RW009; GenBank U88823) is closest to subtype A in protease and in first 100 codons of RT but is closest to subtype C in rest of RT [26]. |
| AD | 1 | D | 1 | D | 1 | No evidence of recombination in protease or RT. |
NOTE. Subtype of complete genome of isolates is based on previously published analyses, compared with subtype indicated by clustering within protease and reverse-transcriptase (RT) neighbor-joining trees, respectively. Rows with recombinant isolates, on the basis of complete genome analysis, also contain short comment. CRF04_cpx and CRF06_cpx were omitted because of complexity.
The branching patterns of the protease and RT trees were similar, but the bootstrap values were lower in the protease tree. In both trees, there was a starlike branching structure, with the highest bootstrap values at the ancestral nodes for each subtype and at the node for the B-D ancestor. In the RT tree, the bootstrap values at the nodes of each of the recognized subtypes were as follows: A (99%), B (99%), C (100%), D (94%), F1 (90%), F2 (99%), G (70%), H (100%), J (100%), and K (100%). In the protease, bootstrap values at the ancestral nodes were, in all but 1 case, lower than those in the RT tree: A (48%), B (42%), C (62%), D (22%), G (85%), H (97%), J (100%), and K (74%). In the protease, sequences of F1 and F2 isolates did not form a clade; their most recent common ancestor was shared with isolates belonging to subtypes B, D, and J. The higher bootstrap values in the RT tree were probably due to the greater number of phylogenetically informative sites in the RT (364) than in the protease (103). There was also a higher proportion of informative sites in the RT (364 [40.4%] of 900) than in the protease (103 [34.7%] of 297; P = .03).
Each CRF formed clades; however, only CRF01_AE and CRF02_AG sequences formed lineages that were independent of other subtypes in both trees. In contrast, CRF03_AB protease sequences were within the subtype A clade, and CRF03_AB RT sequences were within the subtype B clade. CRF04_cpx RT sequences clustered with an incidental variant. CRF05_DF protease sequences were within the subtype D clade. CRF06_cpx protease and RT sequences clustered with incidental variants.
To determine the extent to which differences between and within subtypes were due to amino acid substitutions or to silent nucleotide substitutions, we measured the synonymous and nonsynonymous nucleotide distances between the sequenced isolates. About 21% of the RT nucleotides and 23% of the protease nucleotides had the potential for synonymous substitutions. In the RT, the mean intersubtype ratio of synonymous and nonsynonymous substitutions (DS:DN) was 7.1, and the mean intrasubtype DS:DN was 11.0. In the protease, the mean intersubtype DS:DN was 5.1; the mean intrasubtype DS:DN was 6.8.
Amino acid patterns in nonsubtype B isolates
Although most variation between subtypes was caused by synonymous nucleotide substitutions, there also were significant differences in the amino acid patterns between isolates belonging to different subtypes. To further characterize this variation, we searched GenBank for additional published sequences of non-B protease and RT group M isolates from untreated persons. We found 236 protease and 139 RT isolates that were not included in the original set of 117 completely sequenced isolates, for a total of 353 protease and 256 RT sequences.
The 353 protease sequences included 93 subtype A, 48 subtype C, 35 subtype D, 31 subtype F1/F2, 11 subtype G, 3 subtype H, 6 subtype J, 2 subtype K, 29 CRF01_AE, 86 CRF02_AG, and 9 sequences that belonged to CRF03-CRF06 or were incidental variants. Among these sequences, there were 14 positions at which 1 consensus sequence differed from the consensus B sequence (positions 12-15, 19, 20, 35, 36, 41, 57, 69, 82, 89, and 93) [19]. Each of these 14 nonconsensus positions, however, was at least somewhat polymorphic among subtype B isolates (ranging from 1% at positions 82 and 89 to 21% at position 35).
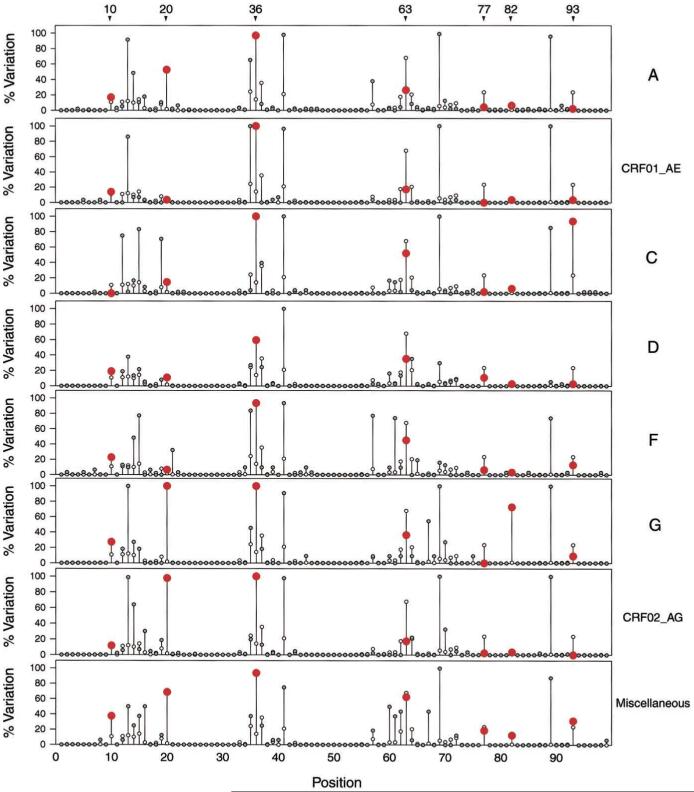
Figure 2 illustrates the protease amino acid variation in those subtypes for which the most sequences were available. In this figure, the amino acid variation within each subtype is compared with the variation within 991 subtype B isolates from untreated persons. Among positions associated with drug resistance, variants at positions 10, 20, 36, and 82 were more common in non-B isolates; whereas variants at positions 63, 77, and 93 were more common in subtype B isolates. Variants at other positions associated with drug resistance were uncommon. There were 3 variants at position 46 (2 subtype A and 1 subtype F) and 1 at position 30 (subtype C).
Figure 2.
Comparison of non-subtype B protease sequences with subtype B consensus sequence. White circles, percentage of subtype B sequences (n = 991) that differ from consensus B sequence; gray circles, percentage of non-B sequences that differ from subtype B consensus sequence; non-B percentage is red at positions associated with drug resistance that have significant differences between subtype B and non-B sequences. Non-B subtypes include subtype A (n = 95), subtype CRF01_AE (n = 29), subtype C (n = 48), subtype D (n = 37), subtype F (including F1 and F2 [n = 31]), subtype G (n = 11), subtype CRF02_AG (n = 86), and all other subtypes (H, J, K, CR04_cpx, and CRF06_cpx [n = 16]). Variants at positions 10, 20, 36, and 82 were more common in non-B isolates; variants at positions 63, 77, and 93 were more common in subtype B isolates.
Variation from subtype B at positions 20 (47% vs. 2%, P < .001) and 36 (94% vs. 14%, P < .001) was more common in each non-B subtype than in subtype B isolates. A variant in the substrate cleft, V82I, was found in 8 (73%) of 11 subtype G sequences, compared with 8 (1%) of 991 subtype B sequences (P < .001). Variants at position 10 occurred in 14% of non-B isolates and in 10% of subtype B isolates (P = .04) but not in the consensus sequence of any of the subtypes.
Variants at position 63 (L has long been considered to be the consensus, even though P is now reported more frequently) were more common in subtype B (68%) than in non-B isolates (33%; P < .001). V77I occurred in 24% of subtype B isolates and in 5% of non-B isolates (P < .001). I93L was more common in subtype C than in subtype B isolates (94% vs. 24%, P < .001) but was more common in subtype B than in the remaining non-B isolates.
The 256 RT sequences included 27 subtype A, 38 subtype C, 21 subtype D, 33 subtype F1/F2, 16 subtype G, 3 subtype H, 6 subtype J, 2 subtype K, 29 CRF01_AE, 72 CRF02_AG, and 9 RT that belonged to CRF03-CRF06 or were incidental variants. Among these sequences, at 26 positions ≥1 non-B consensus sequence differed from the consensus B sequence (positions 11, 35, 36, 39, 40, 48, 49, 60, 122, 135, 162, 173, 174, 177, 178, 200, 207, 211, 245, 248, 272, 277, 286, and 291-293). Each of the nonconsensus positions was at least somewhat polymorphic in subtype B isolates (from 1% at positions 36, 40, and 48 to >40% at positions 122 and 211). None of the 26 nonconsensus positions has been associated with RT inhibitor drug resistance.
HIV-1 protease and RT subtypes of isolates from northern California
Between July 1997 and June 2000, the SUH Diagnostic Virology Laboratory sequenced HIV-1 protease and RT genes from 2246 patients. For 801 patients, treatment histories were known. Of these 801, 556 had received RT inhibitors and 403 had received protease inhibitors. To assess how well these sequences clustered with sequences of known subtype, neighbor-joining trees were constructed by using the SUH sequences and the 103 previously published noncomplex sequences. In the RT, sequences from 2233 (99.42%) of 2246 persons clustered with subtype B, 9 (0.41%) with subtype C, 3 with subtype A (0.13%), and 1 with subtype D (0.04%). The protease sequences of isolates from subjects with non-B RT sequences were of the same subtype as the RT sequence.
We also determined the drug resistance-adjusted nucleotide distance of the 103 published sequences and the 2246 northern California sequences by comparing them with a set of reference isolates and disregarding positions associated with drug resistance [1]. In all cases, the subtype of the most similar reference sequence was the same as the subtype with which the sequence clustered in the neighbor-joining tree. For both the protease and RT, the mean adjusted nucleotide distance between the northern California sequences and the reference sequence of the same subtype was between 3.9% and 6.6%—with the highest intrasubtype distances occurring within subtype A. The mean adjusted nucleotide distance between the northern California sequences and the reference sequences of different subtypes (subtypes A-D) was 6.5%-10.9%; the lowest intersubtype distances occurred between subtypes B and D.
Discussion
HIV-1 RT and protease are sequenced frequently in clinical settings to detect drug resistance and to help physicians select antiretroviral treatment regimens. Because most HIV-1 isolates in the United States and Europe belong to subtype B, it has been standard procedure to describe a new protease or RT sequence by reporting the amino acid differences from the consensus B sequence. Thus, it is important that sequencing laboratories and consulting physicians be aware of the distinction between intersubtype sequence variation and sequence variation resulting from selective drug pressure.
Although it might be expected that the functional constraints of the protease and RT enzymes would prevent their genes from displaying much intersubtype variation, we and others have shown that the RT and protease genes of global isolates are sufficiently different from one another to allow subtype grouping [7-9]. We also found that the subtypes based on protease and RT sequences are usually but not always the same as the subtype of the gag genes of these isolates—probably because ancestral recombination events were less likely between the 5′ part of pol and gag, compared with the 5′ part of pol and the more distant env gene.
The high intersubtype DS:DN ratios indicate that intersubtype variation is predominantly due to synonymous nucleotide substitution. Nonsynonymous nucleotide changes encoding polymorphic amino acid residues contribute, to a lesser extent, to intersubtype variation. The larger number of phylogenetically informative sites in the RT (364), compared with the protease (103), explains the higher bootstrap values in the RT trees.
Intersubtype nucleotide distances were sufficiently high to permit subtype determination by comparing a sequence with a set of reference sequences and by assigning the subtype of the most similar reference sequence. This approach correctly categorized isolates from each of the pure subtypes and from the 2 most common CRFs (CRF01_AE and CRF02_AG). The remaining CRFs were less common and did not form lineages that were independent of other subtypes. Bootscanning programs are likely to be more useful in identifying the CRFs that have known breakpoints within protease or RT [23-28].
There were amino acid differences between subtype B and non-B isolates in both the protease and RT. However, protease sequence variation was more likely to occur at positions associated with drug resistance. V82I was particularly common in subtype G isolates. Position 82 is in the substrate cleft, and several mutations at this position, including V82A/T/F/S, have been associated with in vitro and in vivo resistance to several protease inhibitors [5]. However, several studies suggest that V82I confers minimal or no resistance to the available protease inhibitors [29-31]. Variants at the accessory drug-resistance positions 10, 20, and 36 also occurred more commonly in non-B than in subtype B isolates. However, variants at the accessory resistance positions 63, 77, and 93 occurred more commonly in subtype B than in non-B isolates.
It has been hypothesized that persons with multiple accessory mutations may be at a greater risk of virologic failure during protease inhibitor therapy than are patients who lack such mutations [7, 8]. However, results of clinical studies to date have been inconclusive [32-35]. Although most studies found that non-B isolates are as susceptible as subtype B isolates to currently available protease inhibitors [36-39], an insufficient number of non-B isolates have been studied.
HIV-1 subtyping of protease and RT sequences from 2246 persons in northern California indicated that 99.4% of sequences clustered with subtype B, whereas 0.6% clustered with subtypes A, C, or D. Even though a large proportion of these patients were receiving antiretroviral therapy, all the protease and RT sequences were ≤9% distant from the closest reference sequence. The finding that only ∼1% of isolates belong to non-B subtypes is consistent with previous US studies [40, 41], but differs from recent European studies in which the prevalence of non-B isolates is often >10% [9, 42-45]. Like several previously published studies, this study used isolates from patients undergoing HIV resistance testing and thus was biased toward persons with access to antiretroviral therapy and susceptibility testing. Although there are no data indicating that persons with non-B subtypes are less likely to have access to antiretroviral therapy, continued monitoring is necessary to determine the role of non-B viruses in treated and in untreated populations in the United States.
Acknowledgments
We thank Christina Trevino, Chris Imazumi, and Marilyn Marr, for doing the gene sequencing at Stanford.
Financial support: National Institutes of Health (AI-46148 to M.J.G. and R.W.S.).
Appendix
Table A1.
GenBank accession numbers of completely sequenced genomes analyzed
Footnotes
Human experimentation guidelines of the US Department of Health and Human Services and of the Stanford University institutional review board were followed in the conduct of the research.
References
- 1.Robertson DL, Anderson JL, Bradac JA, et al. HIV-1 nomenclature proposal: a reference guide to HIV-1 classification. In: Kuiken CL, Foley B, Hahn BH, et al., editors. Human retroviruses and AIDS: a compilation and analysis of nucleic and amino acid sequences. Los Alamos National Laboratory; Los Alamos, NM: 2000. pp. 492–505. [Google Scholar]
- 2.Robertson DL, Anderson JP, Bradac JA, et al. HIV-1 nomenclature proposal [letter] Science. 2000;288:55–6. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 3.Korber B, Muldoon M, Theiler J, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–96. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 4.Peeters M, Sharp PM. Genetic diversity of HIV-1: the moving target. AIDS. 2000;14:S129–40. [PubMed] [Google Scholar]
- 5.Hirsch MS, Brun-Vezinet F, D’Aquila RT, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. JAMA. 2000;283:2417–26. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- 6.EuroGuidelines Group for HIV Resistance Clinical and laboratory guidelines for the use of HIV-1 drug resistance testing as part of treatment management: recommendations for the European setting. AIDS. 2001;15:309–20. doi: 10.1097/00002030-200102160-00003. [DOI] [PubMed] [Google Scholar]
- 7.Vergne L, Peeters M, Mpoudi-Ngole E, et al. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: evidence of many minor drug resistance mutations in treatment-naive patients. J Clin Microbiol. 2000;38:3919–25. doi: 10.1128/jcm.38.11.3919-3925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pieniazek D, Rayfield M, Hu DJ, et al. HIV Variant Working Group Protease sequences from HIV-1 group M subtypes A-H reveal distinct amino acid mutation patterns associated with protease resistance in protease inhibitor-naive individuals worldwide. AIDS. 2000;14:1489–95. doi: 10.1097/00002030-200007280-00004. [DOI] [PubMed] [Google Scholar]
- 9.Yahi N, Fantini J, Tourres C, Tivoli N, Koch N, Tamalet C. Use of drug resistance sequence data for the systematic detection of non-B human immunodeficiency virus type 1 (HIV-1) subtypes: how to create a sentinel site for monitoring the genetic diversity of HIV-1 at a country scale. J Infect Dis. 2001;183:1311–7. doi: 10.1086/319859. [DOI] [PubMed] [Google Scholar]
- 10.Parekh B, Phillips S, Granade TC, Baggs J, Hu DJ, Respess R. Impact of HIV type 1 subtype variation on viral RNA quantitation. AIDS Res Hum Retroviruses. 1999;15:133–42. doi: 10.1089/088922299311556. [DOI] [PubMed] [Google Scholar]
- 11.Kuiken CL, Foley B, Hahn BH, et al. Human retroviruses and AIDS: a compilation and analysis of nucleic and amino acid sequences. Los Alamos National Laboratory; Los Alamos, NM: 1999. [Google Scholar]
- 12.Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates; Sunderland, MA: 1998. [Google Scholar]
- 13.Olsen GJ, Matsuda H, Hagstrom R, Overbeek R. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–8. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 14.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–74. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Kumar S. Approximate methods for estimating the pattern of nucleotide substitution and the variation of substitution rates among sites. Mol Biol Evol. 1996;13:650–9. doi: 10.1093/oxfordjournals.molbev.a025625. [DOI] [PubMed] [Google Scholar]
- 17.Ganeshan S, Dickover RE, Korber BT, Bryson YJ, Wolinsky SM. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–77. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–26. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 19.Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–60. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 20.Shafer RW, Warford A, Winters MA, Gonzales MJ. Reproducibility of human immunodeficiency virus type 1 (HIV-1) protease and reverse transcriptase sequencing of plasma samples from heavily treated HIV-1-infected individuals. J Virol Methods. 2000;86:143–53. doi: 10.1016/s0166-0934(00)00144-0. [DOI] [PubMed] [Google Scholar]
- 21.Hammond J, Calef C, Larder B, Schinazi RF, Mellors J. Mutations in retroviral genes associated with drug resistance. In: Kuiken CL, Foley B, Hahn BH, et al., editors. Human retroviruses and AIDS: a compilation and analysis of nucleic and amino acid sequences. Los Alamos National Laboratory; Los Alamos, NM: 1999. [Google Scholar]
- 22.Shafer RW, Stevenson D, Chan B. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 1999;27:348–52. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carr JK, Salminen MO, Albert J, et al. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- 24.Lukashov VV, Huismans R, Rakhmanova AG, et al. Circulation of subtype A and gagA/envB recombinant HIV type 1 strains among injecting drug users in St. Petersburg, Russia: correlates with geographical origin of infections. AIDS Res Hum Retroviruses. 1999;15:1577–83. doi: 10.1089/088922299309874. [DOI] [PubMed] [Google Scholar]
- 25.Laukkanen T, Carr JK, Janssens W, et al. Virtually full-length subtype F and F/D recombinant HIV-1 from Africa and South America. Virology. 2000;269:95–104. doi: 10.1006/viro.2000.0214. [DOI] [PubMed] [Google Scholar]
- 26.Gao F, Robertson DL, Carruthers CD, et al. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–98. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salminen MO, Carr JK, Burke DS, McCutchan FE. Identification of break-points in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–5. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 28.Van der Auwera G, Heyndricks L, Larder B, et al. The use of protease and reverse transcriptase sequences for the identification of intersubtype recombinant HIV strains [abstract MoPeA 2067]; Program and abstracts of the 13th International AIDS Conference (Durban, South Africa); Stockholm: International AIDS Society. 2000. [Google Scholar]
- 29.King RW, Winslow DL, Garber S, et al. Identification of a clinical isolate of HIV-1 with an isoleucine at position 82 of the protease which retains susceptibility to protease inhibitors. Antiviral Res. 1995;28:13–24. doi: 10.1016/0166-3542(95)00033-i. [DOI] [PubMed] [Google Scholar]
- 30.Descamps D, Apetrei C, Collin G, Damond F, Simon F, Brun-Vezinet F. Naturally occurring decreased susceptibility of HIV-1 subtype G to protease inhibitors [letter] AIDS. 1998;12:1109–11. [PubMed] [Google Scholar]
- 31.Brown AJ, Precious HM, Whitcomb J, et al. Reduced susceptibility of HIV-1 to protease inhibitors from patients with primary HIV infection by three distinct routes [abstract 424]; Program and abstracts of the 8th Conference on Retroviruses and Opportunistic Infections (Chicago); Alexandria, VA: Foundation for Retrovirology and Human Health. 2001.p. 169. [Google Scholar]
- 32.Bossi P, Mouroux M, Yvon A, et al. Polymorphism of the human immunodeficiency virus type 1 (HIV-1) protease gene and response of HIV-1-infected patients to a protease inhibitor. J Clin Microbiol. 1999;37:2910–2. doi: 10.1128/jcm.37.9.2910-2912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perno CF, Balotta C, Cozzi Lepri A, et al. Selected secondary mutations in the protease region can be independent predictors of virologic failure in antiretroviral-naive patients treated with protease inhibitor-containing HAART regimens [abstract 4] Antivir Ther. 2000;5:129. [Google Scholar]
- 34.Harrigan PR, Alexander C, Dong W, Jahnke N, O’Shaughnessy MV, Montaner JS. Prevalence of resistance-associated mutations in patients starting antivirals: virologic response after approximately one year of therapy [abstract 124] Antivir Ther. 1999;4(Suppl 1):88. [Google Scholar]
- 35.Servais J, Lambert C, Fontaine E, et al. Variant human immunodeficiency virus type 1 proteases and response to combination therapy including a protease inhibitor. Antimicrob Agents Chemother. 2001;45:893–900. doi: 10.1128/AAC.45.3.893-900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shafer RW, Eisen JA, Merigan TC, Katzenstein DA. Sequence and drug susceptibility of subtype C reverse transcriptase from human immunodeficiency virus type 1 seroconverters in Zimbabwe. J Virol. 1997;71:5441–8. doi: 10.1128/jvi.71.7.5441-5448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shafer RW, Chuang TK, Hsu P, White CB, Katzenstein DA. Sequence and drug susceptibility of subtype C protease from human immunodeficiency virus type 1 seroconverters in Zimbabwe. AIDS Res Hum Retroviruses. 1999;15:65–9. doi: 10.1089/088922299311727. [DOI] [PubMed] [Google Scholar]
- 38.Palmer S, Alaeus A, Albert J, Cox S. Drug susceptibility of subtypes A, B, C, D, and E human immunodeficiency virus type 1 primary isolates. AIDS Res Hum Retroviruses. 1998;14:157–62. doi: 10.1089/aid.1998.14.157. [DOI] [PubMed] [Google Scholar]
- 39.Tanuri A, Vicente AC, Otsuki K, et al. Genetic variation and susceptibilities to protease inhibitors among subtype B and F isolates in Brazil. Antimicrob Agents Chemother. 1999;43:253–8. doi: 10.1128/aac.43.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wegner SA, Larder B, Hertogs K, Carr JK, McCutchan FE. Use of a drug resistance sequence database to determine the prevalence of non-subtype B HIV-1 infections in US and Euopean clinical cohorts. Antivir Ther. 2000;5(Suppl 3):136. [Google Scholar]
- 41.Irwin KL, Pau CP, Lupo D, et al. Centers for Disease Control and Prevention-Bronx Lebanon HIV Serosurvey Team Presence of human immunodeficiency virus (HIV) type 1 subtype A infection in a New York community with high HIV prevalence: a sentinel site for monitoring HIV genetic diversity in North America. J Infect Dis. 1997;176:1629–33. doi: 10.1086/517343. [DOI] [PubMed] [Google Scholar]
- 42.Van der Auwera G, Hertogs K, Larder B, Janssens W, Van der Groen G. Subtype analysis of 1831 European HIV-1 protease and reverse transcriptase pol gene sequences collected from clinical studies. Antivir Ther. 2000;5(Suppl 3):132. [Google Scholar]
- 43.Alaeus A, Leitner T, Lidman K, Albert J. Most HIV-1 genetic subtypes have entered Sweden. AIDS. 1997;11:199–202. doi: 10.1097/00002030-199702000-00010. [DOI] [PubMed] [Google Scholar]
- 44.Barin F, Courouce AM, Pillonel J, Buzelay L, Retrovirus Study Group of the French Society of Blood Transfusion Increasing diversity of HIV-1M serotypes in French blood donors over a 10-year period (1985-1995) AIDS. 1997;11:1503–8. doi: 10.1097/00002030-199712000-00015. [DOI] [PubMed] [Google Scholar]
- 45.Dietrich U, Ruppach H, Gehring S, et al. Large proportion of non-B HIV-1 subtypes and presence of zidovudine resistance mutations among German seroconvertors [letter] AIDS. 1997;11:1532–3. [PubMed] [Google Scholar]