Summary
SCFFbx4 was recently identified as the E3 ligase for cyclin D1. We now describe cell cycle-dependent phosphorylation and dimerization of Fbx4 that is regulated by GSK3β and defective in human cancer. We present data demonstrating that a pathway involving Ras-Akt-GSK3β controls the temporal phosphorylation and dimerization of the SCFFbx4 E3 ligase. Inhibition of Fbx4 activity results in accumulation of nuclear cyclin D1 and oncogenic transformation. The importance of this regulatory pathway for normal cell growth is emphasized by the prevalence of mutations in Fbx4 in human cancer that impair dimerization. Collectively, this data reveals that inactivation of the cyclin D1 E3 ligase will likely contribute to cyclin D1 overexpression in a significant fraction of human cancer.
Keywords: Cyclin D1, Fbx4, esophageal cancer, GSK3β
Significance
Cyclin D1 overexpression occurs frequently in human cancer; however contributing mechanisms remain ambiguous as the since the frequency of gene amplification is lower than the incidence of overexpression. Herein we demonstrate that attenuation of cyclin D1 SCFFbx4 E3 ubiquitin ligase activity occurs frequently in human cancer and may represent a common mechanism of overexpression and deregulation of cyclin D1. Significantly, inactivation of the Fbx4 ligase is the result of mutations in N-terminal regulatory regions of Fbx4 that disrupt ligase dimerization thereby revealing the biological significance of SCF ligase oligomerization. In summary, our study establishes Fbx4 as a tumor suppressor in human cancer, function of which is abrogated by a unique category of mutations that target E3 ligase activity.
Introduction
Cell cycle progression requires the orderly accumulation of proteins that drive cell division with the coordinated inactivation or loss of proteins that inhibit this activity. Progression through G1 phase ensues upon activation of CDK4, CDK6 and CDK2, through association with their regulatory subunits, cyclin D (D1, D2, or D3 depending upon the tissue type) and cyclin E. Accumulation of cyclins is determined largely through coordinated cycles of synthesis and destruction (Sherr et al., 1992); (Winston and Pledger, 1993; Geng et al., 1996). Efforts to define the proteolytic machinery led to the identification of SCF (Skp1-Cul1-F-box protein) E3 ligases as the primary regulators of G1 cyclin proteolysis. The F-box component of SCF ligases determines substrate specificity and generally binds to substrates in a phosphorylation-dependent manner (Skowyra et al., 1997; Willems et al., 1999; Craig and Tyers, 1999). More specifically, cyclin D1 proteolysis is directed by the SCFFbx4/αB-crystallin ligase, whereas cyclin E ubiquitination is directed by SCFFbw7 ligase (Lin et al., 2006; Koepp et al., 2001; Strohmaier et al., 2001; Welcker et al., 2003). Accumulation of cyclin D1 is acutely sensitive to growth factor stimulation. Cyclin D1 initially accumulates in G1 phase through growth factor dependent transcription and translation (Sherr et al., 1992). At the G1/S boundary, cyclin D1 is phosphorylated by GSK3β at the residue Thr-286 followed by CRM1-dependent nuclear export to the cytoplasm where cyclin D1 is recognized by SCFFbx4/αB-crystallin ligase, ubiquitinated and targeted to the 26S proteasome (Diehl et al., 1997; Alt et al., 2000; Lin et al., 2006).
Cyclin D1 is overexpressed frequently in different human cancers such as those originating from the esophagus, head/neck and breast, and overexpression is considered an early, causative event in some of these cancers (Diehl, 2002). Overexpression was thought initially to result from chromosomal translocations or gene amplification (Dickson et al., 1995); (Zukerberg et al., 1995); (Nakagawa et al., 1995). Because the frequency of cyclin D1 overexpression exceeds the frequency of such large chromosomal alterations, it was proposed that alterations in turnover of cyclin D1 protein might also contribute to aberrant cyclin D1 levels (Russell et al., 1999; Diehl, 2002). Indeed, recent work revealed mutations in cyclin D1 that directly inhibit phosphorylation-dependent proteolysis (Moreno-Bueno et al., 2004; Benzeno S, 2006). However, such mutations are rare, and imply other mechanisms. Investigations that might reveal such mechanisms were hindered in part by our lack of knowledge regarding the E3 ligase that directs cyclin D1 ubiquitin-mediated proteolysis. With the recent identification of Fbx4-αB crystallin as the specificity factor that directs cyclin D1 ubiquitination (Lin et al., 2006), it is now possible to ascertain whether components of the cyclin D1 E3 ligase, Fbx4 or αB crystallin are subjected to genetic alterations in human cancers and may represent potential tumor suppressor proteins in this context.
Since SCF recognition generally depends upon substrate phosphorylation, it has been inferred that SCF ligases are constitutively active, with regulation occurring mainly at the level of substrate phosphorylation. However, recent work has revealed that many F-box proteins can function as homodimers, thereby providing a potential means of ligase regulation. Fbw7, an E3 ligase that ubiquitinates cyclin E, c-Myc and Notch, was demonstrated to form dimers but whether dimerization regulates ligase activity or substrate recognition is controversial (Zhang and Koepp, 2006); (Welcker and Clurman, 2007; Tang et al., 2007). β-Trcp, an E3 ligase for IκB, forms both homo- and heterodimers between two isoforms, β-Trcp1 and β-Trcp2, with only the homodimers forming productive ligases while heterodimerization is inhibitory (Suzuki et al., 2000). The mechanism by which the formation of F-box protein dimers promotes substrate ubiquitination is unclear, but possible scenarios include increased concentration of substrate in the area of high E2 activity as well as positioning of substrate in a manner that facilitates the ubiquitination of multiple lysine residues (Tang et al., 2007). Dimerization of F-box proteins is directed through the so-called D-domain. The D-domain is localized N-terminal to the F-box region and critical hydrophobic residues (leucine and isoleucine) are conserved spatially in distinct F-box proteins such as Fbw7, β-Trcp and Fbw2 (Zhang and Koepp, 2006). Although the phenomenon of ubiquitin ligase substrate adaptors self-association is established, the functional significance of this and whether the dimerization is constitutive or regulated remains to be elucidated.
This work provides evidence that Fbx4 contains an N-terminal dimerization domain (D-domain) and that dimerization is regulated by phosphorylation of a conserved serine (serine 11, mouse; serine 12, human) residue proximal to the D-domain. We demonstrate that dimerization deficient Fbx4 mutants fail to support efficient cyclin D1 degradation. Mutation of this serine residue to alanine attenuates dimerization of Fbx4 and significantly impairs cyclin D1 ubiquitination and proteolysis. Importantly, the data provided reveal that phosphorylation of serine 11 is temporally regulated during cell cycle progression with maximal phosphorylation and dimerization occurring during S phase. Strikingly, Fbx4 phosphorylation is GSK3β-dependent, thereby revealing that GSK3β controls not only substrate availability, through phosphorylation of cyclin D1, but also activation of the E3 ligase that promotes cyclin D1 ubiquitination. Finally, we demonstrate that inhibition of SCFFbx4/αB-crystallin ligase activity leads to accumulation of cyclin D1 in the nuclei and promotes cellular transformation.
RESULTS
Fbx4 contains an N-terminal dimerization domain
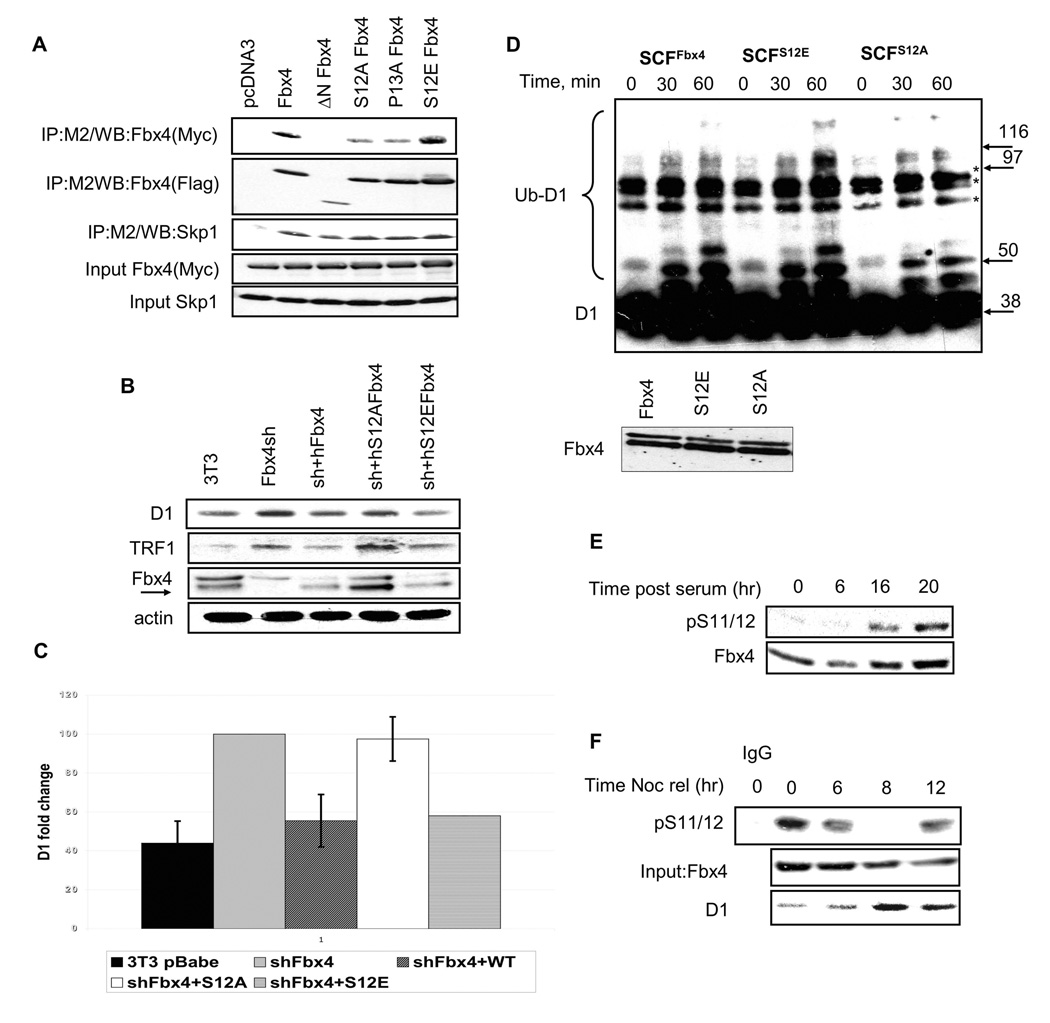
To test Fbx4 dimerization, a series of Fbx4 deletion alleles with an N-terminal Flag-tag (Fig. S1A) were co-expressed with Myc-tagged wild type Fbx4 in 293T cells and interactions were assessed by anti-Flag precipitation. Myc-Fbx4 was detected in precipitates containing wild type Flag-Fbx4 (Fig. 1A) revealing homo-typic dimerization. While neither the F-box nor the C-terminal regions were required, removal of the N-terminal 50 amino acids of Fbx4 abrogated dimerization (Fig. 1A; ΔNFbx4). Critically, ΔNFbx4 still bound Skp1 (Fig. 2A), suggesting that this deletion did not result in global folding defects. Mutation of leucine/isoleucine residues, within Fbx4 (Fig. S1B), that are spatially conserved with leucines that contribute to dimerization of Fbw7 (Zhang and Koepp, 2006), defined leucine 23 as essential for Fbx4 dimerization (Fig. 1B). As with dimerization defective Fbw7 (Zhang and Koepp, 2006; Welcker and Clurman, 2007), Fbx4 mutants retained their capacity to associate with cyclin D1 (Fig. 1C) and αB-crystallin (Fig. S1C) (Lin et al., 2006).
Figure 1. Dimerization of Fbx4 occurs through its N-terminus.
A and B. Lysates were prepared from 293T cells transfected with vectors encoding human wild type Myc-tagged Fbx4 and the indicated Flag-Fbx4 mutant constructs. Protein complexes were isolated by M2 affinity chromatography and detected by immunoblot with a Fbx4 antibody. C. 293T cells were co-transfected with Fbx4 constructs along with cyclin D1, CDK4 and αB-crystallin. 24hrs post-transfection cells were treated with MG132 for 6 hrs followed by cyclin D1 immunoprecipitation. Cyclin D1 was detected with the 13G11 monoclonal antibody.
Figure 2. Fbx4 serine 12 is required for Cyclin D1 ubiquitination and proteolysis.
A. 293T cells were transfected with plasmids encoding Flag-tagged Fbx4 mutants and wild type Myc-tagged Fbx4. Complexes were isolated by affinity chromatography using the M2 conjugated agarose and individual components detected by immunoblot with Fbx4 and Skp1 antibodies. B. NIH-3T3 cells, wherein Fbx4 had previously been knocked down by shRNA, were transfected with wt, S12A or S12E Fbx4 pcDNA3 followed by G418 selection of stably expressing clones. Asynchronous cells were harvested and subjected to Western blotting with cyclin D1, TRF1 and Fbx4 antibodies. C. Quantification of B. Error bars indicate +/−SD. D. Ligase complexes (purified protein shown in bottom panel) were purified from stable cell lines expressing wt, S12E and S12A Fbx4 (described in B). In vitro ubiquitination reactions were performed using GST-tagged purified cyclin D1. Ubiquitin-conjugated cyclin D1 was detected by immunoblot. Non-specific complexes are denoted by asterisk. E. NIH3T3 cells were serum starved for 48 hrs and released into 10% FBS containing media for the indicated intervals. Immunoblot analysis was performed using pS11/12-Fbx4 and a total Fbx4 antibody. F. Cells synchronized via a nocodazole block were harvested at the indicated intervals following nocodazole release. Fbx4 phosphorylation was detected by precipitation with the pS11/12 antibody and immunoblot with the total Fbx4 antibody. Total cyclin D1 and Fbx4 levels were assessed by direct western.
Serine 12 of Fbx4 contributes to Fbx4 dimerization
Serine 12 of Fbx4 is a predicted proline-directed phosphorylation motif (Beausoleil et al., 2004). Due to the proximity of Ser12 with the D-domain, we considered the possibility that phosphorylation of serine 12 could regulate dimerization of Fbx4. We therefore generated both non-phosphorylatable (S12A) and phosphomimetic (S12E) mutations in Fbx4 as well as a P13A mutant to address whether a proline directed kinase targeted this site; all mutants were then assessed for dimerization potential. Both Fbx4S12A and Fbx4P13A exhibited reduced dimerization potential when compared to wild type Fbx4 or Fbx4S12E (Fig. 2A). We next reconstituted murine NIH3T3 cell lines wherein endogenous Fbx4 had been stably knocked down (Lin et al., 2006), with either wild type (human) Fbx4 or the serine 12 mutants of Fbx4 followed by G418 selection to establish stable cell lines. We previously demonstrated that Fbx4 knockdown results in overexpression of endogenous cyclin D1 (Fig. 2B, 2C). Reconstitution of cells with either wild type Fbx4 or Fbx4S12E resulted in a reduction of cyclin D1 to levels comparable to parental NIH3T3, while Fbx4S12A had reduced activity. Significantly, levels of Trf1, the only other reported Fbx4 substrate (Lee et al., 2006), mimicked that of cyclin D1. To assess the contribution of serine-12 to Fbx4-dependent ubiquitination activity, we purified wild type or mutant SCF Fbx4 ligases from NIH3T3 stable cell lines by anti-Flag affinity purification (Fig. 2D; Fbx4 levels shown in the lower panel). Both, wild type and Fbx4S12E catalyzed in vitro ubiquitination of cyclin D1, while activity of Fbx4S12A was attenuated. Thus, disruption of serine 12 leads to decreased dimerization of Fbx4 and decreased ubiquitination of cyclin D1.
Phosphorylation of S12 is induced in S phase of cell cycle
Since S12 is important for ligase function, we considered a scenario wherein phosphorylation of serine 12 regulates Fbx4 dimerization and ultimately ligase activity. To test this notion, we first generated phospho-serine 11/12 (serine 11 in mouse, 12 human) reactive antibodies (Fig. 2SA–B). If phosphorylation of Ser-12 regulates Fbx4 dimerization and ubiquitination activity, we anticipated temporal regulation of Ser-12 phosphorylation. Because cyclin D1 ubiquitination is triggered upon G1/S transition following phosphorylation-dependent nuclear export, we predicted peak phosphorylation of Fbx4 at the G1/S boundary. To test this premise, NIH-3T3 cells were synchronized by serum starvation; following mitogen triggered cell cycle reentry, p-S12 of Fbx4 was assessed at 0, 6, 16 and 20h using our phospho-specific antibody. Phosphorylation of Ser-12 was low during cell cycle entry, but increased dramatically upon S-phase entry (16h) and increased further during S-phase (20h; Fig. 2E). Cells were also synchronized with nocodazole to assess Ser-12 phosphorylation in cells traversing the G2/M-G1 transition. Ser-12 phosphorylation was high during G2/M and early G1 phase (Fig. 2F); 0, 6h; points at which cyclin D1 levels remain low. By contrast, Ser-12 phosphorylation was reduced dramatically during mid/late G1 phase when cyclin D1 levels increase (8h) and was again high in early S-phase (12h; Fig. 2F).
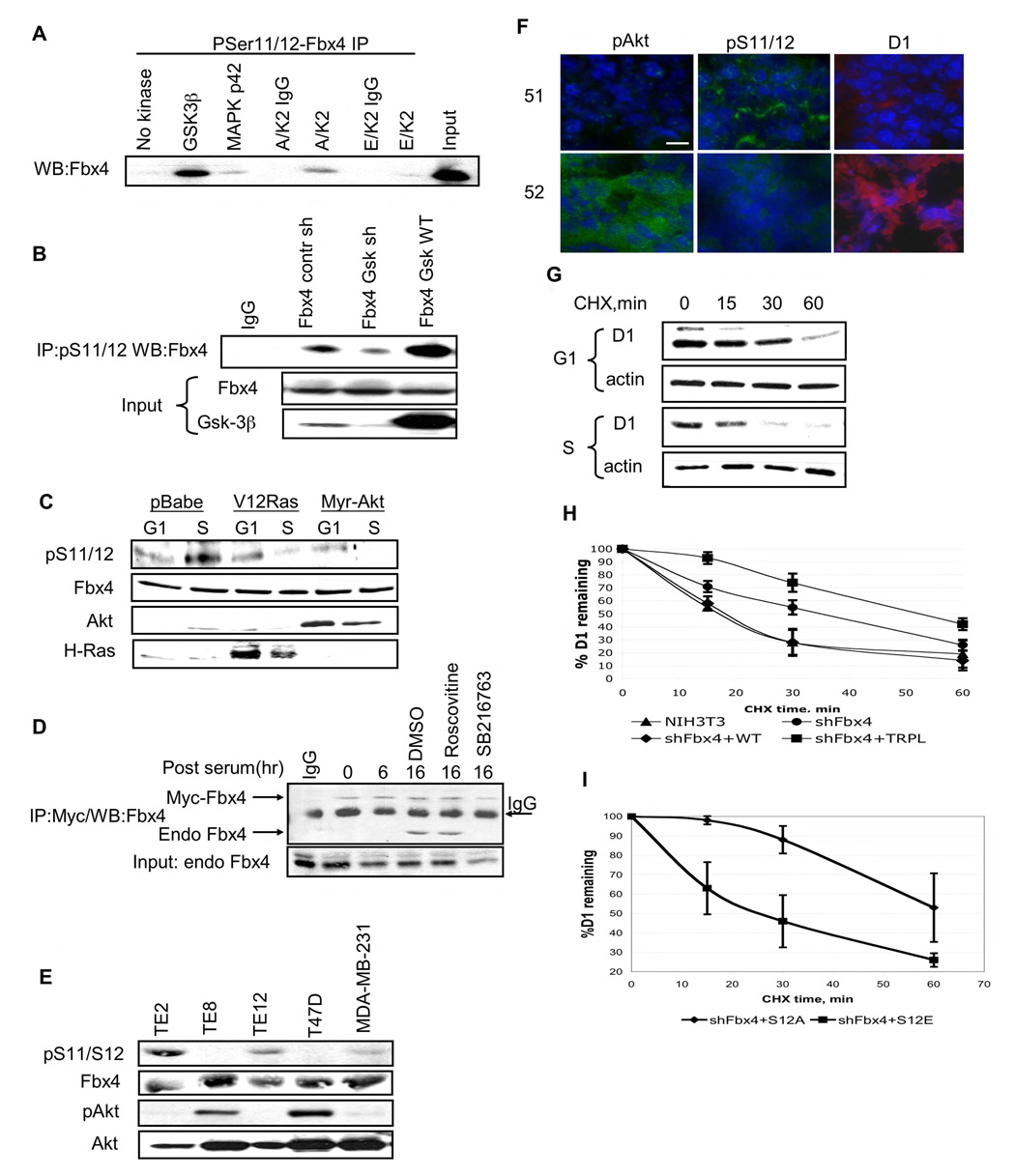
Given the proximity of Ser-12 and Pro-13, we reasoned that a proline-directed kinase likely regulates phosphorylation of this serine residue. To identify the kinase responsible for Ser-12 phosphorylation, we focused on proline-directed protein kinases known to be active or activated at the G1/S phase boundary. Strikingly, only GSK3β efficiently phosphorylated Ser-12 in vitro as determined by IP/Western (Fig. 3A). Phosphorylation of Fbx4 by GSK3β was also confirmed with recombinant proteins in an in vitro kinase assay (Fig. S3A). Additionally, while GSK3β efficiently phosphorylated S12, GSK3α did not exhibit detectable activity toward this residue in Fbx4 (Fig. S3B). This result is consistent with the capacity of GSK3β to function as a proline directed kinase, while GSK3α does not. We evaluated the role of CDK2 (roscovitine), GSK3β (LiCl, SB216763) in vivo. Both LiCl and SB216763 reduced pS12 while roscovitine did not (Fig. S4A).
Figure 3. GSK3β phosphorylates S11/12 of Fbx4 in S phase of cell cycle.
A. In vitro kinase assay was performed using purified GST-tagged Fbx4 and indicated kinases. Following in vitro phosphorylation, samples were precipitated with the pS11/12–Fbx4 antibody and subjected to immunoblot with a Fbx4 antibody. B. 293T cells were transfected with wild type Fbx4 along with control shGFP vector, a GSK3β shRNA vector, or a plasmid encoding wild type GSK3β Fbx4 was immunoprecipitated from cellular extracts using pS11/12-Fbx4 antibody 48hrs post–transfection and detected by immunoblot with the total Fbx4 antibody. C. NIH3T3 cells were transfected with an empty vector, or RasV12 or Myr Akt expressing constructs. Cells were synchronized by serum starvation and released into complete medium for 6hrs (G1) or 16hrs(S). Protein extracts were processed for immunoblot using pS11/12-Fbx4, total Fbx4, total Akt and H-Ras antibodies. D. NIH3T3 cells were transfected with vector encoding Myc-tagged Fbx4 and 24 hrs later were synchronized in G0. Cells were released into cell cycle for the indicated intervals (cells were treated with kinase inhibitors for 6 hrs before harvesting samples at 16 hrs time point). Myc-Fbx4 pull down was performed using Myc-agarose. Myc-Fbx4 associated endogenous Fbx4 was detected by immunoblot with total Fbx4 antibody. E. Total cell extracts from esophageal carcinoma cell lines (TE2, TE8 and TE12) and breast carcinoma cell lines (T47D and MDA-MB-231) were subjected to precipitaion with the pS11/12 antibody followed by immunoblot with the Fbx4 antibody. Direct Western blotting was performed using pAkt (Ser473) and total Akt antibodies. F. Frozen sections from MMTV-Neu breast tumors were analyzed by IHC using following antibodies: pS11/12-Fbx4, pAkt (Ser473) and cyclin D1 (bar, 8µM). G. NIH3T3 cells were arrested by serum starvation following by replating in complete media. Cycloheximide was added for the indicated intervals during G1 (6 hrs post-release) and S phases of cell cycle (16 hrs post-release). Cyclin D1 turnover was analyzed by Western analysis using cyclin D1 antibody. H. NIH3T3 shFbx4 stable cell line was transfected with either wild type or Fbx4 trpl. Cycloheximide chase was performed in asynchronous cells 48 hrs after transfection. Quantification of 3 independent experiments is provided, error bars represent +/− SD. I. Fbx4S12A or Fbx4S12E were individually re-introduced into Fbx4 knockdown cells and cyclin D1 turnover was assessed in S phase cells; quantification of 3 independent experiments is provided, error bars represent +/− SD.
To establish the relevance of GSK3β in vivo, we used several independent approaches. Initially, we determined whether overexpression of kinase-dead GSK3β in insect cells inhibited Ser-12 phosphorylation. Expression of kinase-dead GSK3β efficiently inhibited phosphorylation in a dose-dependent manner and when total Fbx4 was immunoprecipitated from cells, kinase-dead GSK3β stably associated with Fbx4 (Fig. S4B), demonstrating a direct interaction. Second, we utilized a previously validated shRNA vector(Jin et al., 2003) to knockdown GSK3β in human cells. Knockdown of GSK3β reduced pS12 relative to a non-specific shGFP RNA, while overexpression of wild type GSK3β increased pS12 (Fig. 3B). Finally, we determined whether the inhibition of GSK3β activity through expression of activated Ras (Ras-V12) or Akt (Myr-Akt) (Rodriguez-Viciana et al., 1994); (Cross et al., 1995) would inhibit phosphorylation of S12. Both Ras-V12 and Myr-Akt inhibited pS12 (Fig. 3C), consistent with a model wherein the Ras-Akt-GSK3β cascade regulates Ser-12 phosphorylation of Fbx4.
If phosphorylation of Ser-12 regulates Fbx4 dimerization, then dimerization of Fbx4 should parallel phosphorylation and increase during S-phase. NIH-3T3 cells were transfected with a plasmid encoding 6XMyc-Fbx4, synchronized by serum deprivation and released into the cell cycle. Dimerization of endogenous Fbx4 with Myc-Fbx4 was assessed by precipitation with anti-myc antibodies from lysates prepared from cells harvested in G0, G1 and S-phase of cell cycle followed by immunoblot with antiserum directed against total Fbx4. This approach allows the simultaneous detection of both endogenous and ectopic Fbx4 on the same gel and thereby allows assessment of stoichoimetry. Stoichiometric binding of endogenous Fbx4 to Myc-Fbx4 was detected during S-phase (16h) and was inhibited by incubation of cells with a small molecule GSK3β inhibitor demonstrating that dimerization is induced during S-phase and correlates with Ser-12 phosphorylation by GSK3β (Fig. 3D). To corroborate GSK-dependent regulation of Fbx4 dimerization a plasmid encoding RasV12 or the shRNA targeting GSK3β was introduced into 293T cells along with constructs encoding Myc-Fbx4 and Flag-Fbx4. Dimerization was analyzed by Flag pulldown and western blotting with antisera directed against Fbx4 allowing detection of both tagged forms on a single western blot. Inhibition of GSK3β activity through overexpression of RasV12 or GSK3β knockdown resulted in an inhibition of S12 phosphorylation and decreased Fbx4 dimerization (Fig. S4C). To determine whether Fbx4 S12 phosphorylation is regulated in an Akt-dependent manner in vivo, we utilized a panel of esophageal and breast cancer cells that have differential Akt activity levels. In cells with high Akt activity (Fig 3E, TE8 and T47D) there was no detectable S12 phosphorylation. This finding was also supported by the absence of immunostaining with pS11/12 antibody in MMTV-Neu mouse tumors, which exhibit high Akt activity (Fig. 3F, tumor 51 vs. 52). Consistent with increased Akt activity down regulating ligase phosphorylation (and thus activity), cyclin D1 levels were also increased in tumors with high Akt activity.
The data presented suggest that dimerization, while not essential for Fbx4-dependent ubiquitination, does potentiate this activity. If so, cyclin D1 should exhibit increased turnover during S-phase. Indeed the half-life of cyclin D1 was faster during S-phase (t1/2<15 min) than in G1 (t1/2>40 min) (Fig. 3G). Finally, we assessed the ability of phosphorylation deficient or phosphomimetic Fbx4 mutants to rescue cyclin D1 turnover in NIH3T3 cells wherein endogenous Fbx4 is knocked down. We transfected the stable shFbx4 NIH-3T3 line with human cDNAs encoding either wild type Fbx4 or Fbx4S12A/S12E (stable cell lines, presented in Fig 2B), synchronized cells by serum starvation and measured the half-life of cyclin D1 during S phase. The cyclin D1 half-life is extended to almost 40 minutes in Fbx4 knockdown cells (Fig. 3H). In cells wherein wild type Fbx4 was re-introduced cyclin D1 turnover was restore (t1/2~ 15 min) such that it is comparable with parental cells (Fig. 3H, S5). Strikingly, the Fbx4S12A mutant was not able to restore normal kinetics of cyclin D1 turnover while Fbx4S12E was proficient in restoring the rate cyclin D1 proteolysis (Fig. 3I). Consistent with dimerization being critical for Fbx4-dependent cyclin D1 turnover, the dimerization deficient Fbx4L trpl also failed to restore cyclin D1 turnover (Fig. 3I, S5). Therefore, both phosphorylation of S12 and dimerization of Fbx4 are required for sufficient cyclin D1 turnover in S phase cells.
Inhibition of cyclin D1 proteolysis leads to its nuclear accumulation and transformed phenotype
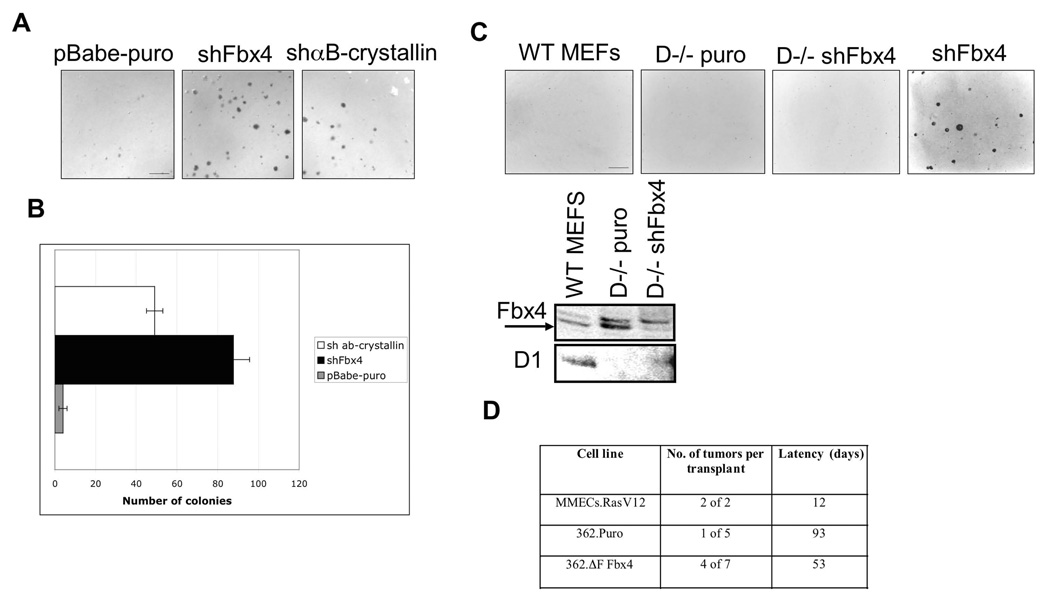
Because Fbx4 is a critical regulator of cyclin D1 accumulation and thus CDK4 activity, we considered it likely that Fbx4 would exhibit tumor suppressor activities. Indeed knock down of either Fbx4 or αB-crystallin in mouse fibroblasts facilitated growth of cells in soft agar, a characteristic of neoplastic transformation (Fig. 4A–4B). In contrast, knockdown of Fbx4 in cyclin D1 null fibroblasts failed to induce growth in soft agar (Fig. 4C) indicating that cyclin D1 is a critical target for SCFFbx4 . We also assessed the capacity of inhibition of Fbx4 to cooperate with cyclin D1 in tumor growth. For this experiment, mammary tumors were harvested from MMTV-D1 mice (Lin et al., 2007) and cultured in vitro. We have found that over the course of serial cultivation, transgene expression is reduced and the tumorigenic phenotype of the cells is lost; we reasoned that inhibition of cyclin D1 turnover might restore tumorgenicity. We therefore infected mammary epithelial cells with control empty vector or a vector encoding a dominant negative Fbx4 allele (ΔF-Fbx4), which extended the half-life of cyclin D1 (Fig. S6A). In this system, we were unable to achieve any significant knockdown of Fbx4 using shRNA, and therefore, we focused on ΔF-Fbx4 as a basis for abrogating Fbx4 function. Cells were injected in fat pads of NOD/SCID mice and tumor growth assessed. While only 1 of 4 MMTV-D1 formed tumors, 4 of 7 MMTV-D1/ΔF-Fbx4 formed established tumors (Fig. 4D).
Figure 4. Inhibition of SCFFbx4/αB-crystallin activity leads to neoplastic growth.
A. Soft agar colony formation assay was performed using pBabe-puro, shFbx4 and shαB-crystallin knockdown cell lines. Cells were plated in soft agar and grown for 21 days (bar, 250µM). B. Quantification of A, error bars represent +/− SD. C. Same as A using wt and cyclin D1−/− MEFs infected with control (puro) or shFbx4 retrovirus (bar, 250µM). Levels of Fbx4 and cyclin D1 proteins are provided in the bottom panel. D. 106 cells were transplanted into cleared mammary fat pads of 3 week-old NOD-SCID mice. Mice were monitored for palpable tumor formation bi-weekly.
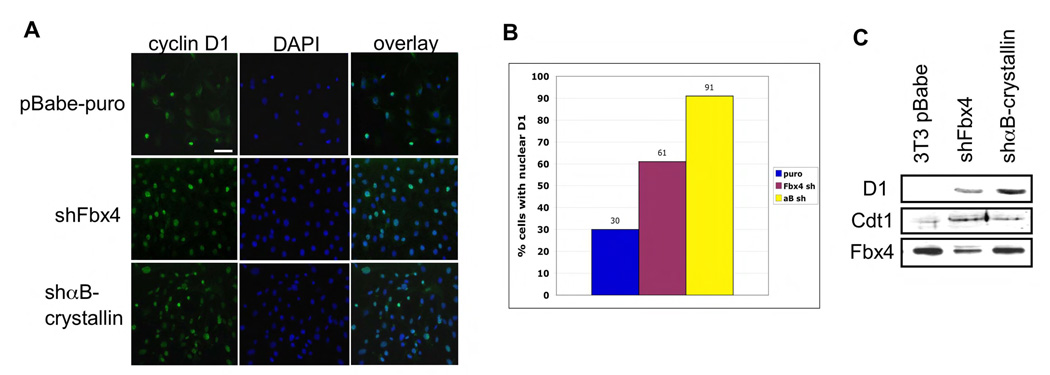
Past work has provided evidence that accumulation of cyclin D1-dependent kinase in the nucleus during S-phase disrupts temporal control of DNA replication through stabilization of Cdt1 that in turn induces DNA re-replication and genomic instability (Aggarwal et al., 2007). We therefore addressed the fate of cyclin D1 in cells that have decreased Fbx4 or αB-crystallin activity. Indeed cyclin D1 accumulated in the nucleus in cells wherein endogenous Fbx4 or αB-crystallin were reduced by shRNA (Fig. 5A–5B). The differences in nuclear accumulation in Fbx4 versus αB-crystallin knockdown likely reflect knockdown efficiency, as our degree of Fbx4 knockdown is routinely 60% while αB-crystallin is reduced to undetectable levels (Fig. 5C). As expected with the nuclear accumulation of cyclin D1, Cdt1 was overexpressed following knockdown of either Fbx4 or αB-crystallin (Fig. 5C). Thus, inhibition of cyclin D1 SCFFbx4 activity leads to nuclear accumulation of cyclin D1 and cell transformation.
Figure 5. Disruption of SCFFbx4/αB-crystallin activity leads to nuclear accumulation of cyclin D1.
A. Immunofluorescence was performed on the indicated cell lines using the 13G11 cyclin D1 monoclonal antibody and counterstained with DAPI (bar, 25 µM). B. Quantification of A. C. Protein extracts from stable NIH3T3 cell lines expressing shFbx4 or shαB-crystallin were analyzed by Western Blot using Cdt1 and cyclin D1 antibodies.
Fbx4 is mutated in human esophageal carcinoma
In a majority of cancers harboring cyclin D1 overexpression defects proteolytic machinery have been implicated as a molecular basis. We therefore assessed primary esophageal cancers, which are known to frequently overexpress cyclin D1(Nakagawa et al., 1995); (Ikeguchi et al., 2001) for mutations in Fbx4 and αB crystallin. Fbx4 DNA sequence was assessed in 116 primary esophageal carcinomas and in matched adjacent normal tissue. We identified and verified hemizygous, missense mutations in 14% of the primary tumors (Table 1). No mutations were identified in αB crystallin and cyclin D1 was wild type in tumors harboring inactivating Fbx4 mutations. A majority of the mutations in Fbx4 occur in the N-terminal regulatory domain and interfere with Fbx4 dimerization. Mutations that impair phosphorylation (S12L; P13S) and indirectly influence dimerization along with mutations that target critical residue within the dimerization motif (L23Q) were detected. In addition, a proline to threonine mutation was identified at residue 76 within the F-box (P76T); P76T impairs binding to Skp1 (Fig. S6B) resulting in a dominant negative allele. In addition to the described above mutations, numerous S8R mutations were identified. Mutations in this domain, like S12L, impair Fbx4-dependent regulation of cyclin D1 degradation (Fig. S6C).
Table 1. Analysis of Fbx4 mutations in esophageal tumors.
Full-length cDNA encoding Fbx4, cyclin D1 and αB-crystallin were generated by first-strand RT-PCR using oligo dT primer followed by PCR using specific primers. 116 totals samples were sequenced in both directions. Cyclin D1 protein expression was analyzed by immunohistochemistry. ESCC-esophageal squamous cell carcinoma, EAC-esophageal adenocarcinoma. N/A indicates that paraffin embedded tumor tissue was not available for IHC analysis of cyclin D1 accumulation.
| tumor # | Fbx4 | cyclin D1 | CryAB | cyclin D1 protein levels |
|---|---|---|---|---|
| 1 ESCC | L23Q | wt | wt | na |
| 4 EAC | P13S | wt | wt | na |
| 7 ESCC | S12L | wt | wt | +++ |
| 8 ESCC | L23Q | wt | wt | +++ |
| 9 ESCC | wt | wt | wt | + |
| 10 ESCC | wt | wt | wt | +++ |
| 11 ESCC | wt | wt | wt | ++ |
| 12 ESCC | wt | wt | wt | na |
| 14 ESCC | wt | wt | wt | na |
| 15 ESCC | S8R | wt | wt | +++ |
| 16 ESCC | wt | wt | wt | na |
| 17 ESCC | wt | wt | wt | na |
| 18 ESCC | wt | wt | wt | + |
| 19 ESCC | P76T | wt | wt | +++ |
| 21 EAC | wt | wt | wt | na |
| 22 EAC | L23Q | wt | wt | na |
| 23 EAC | P76T | wt | wt | na |
| 24 EAC | L23Q | wt | wt | na |
| 38 ESCC | S8R | na | na | na |
| 39 ESCC | G30N | na | na | na |
| 40 ESCC | exon 1 24 bp insert | na | na | na |
| 41 ESCC | S8R | na | na | na |
| 42 ESCC | S8R | na | na | na |
| 43 ESCC | S8R | na | na | na |
| 44 ESCC | codon 18 CA insertion | na | na | na |
Because these mutations should reduce total SCF Fbx4 activity, we expected that there should be a corresponding increase in the accumulation of downstream substrates. Immunohistochemical analysis revealed overexpression of cyclin D1 and TRF1 in Fbx4 mutant tumors consistent with a substrate-ligase relationship (Fig. 6A–B; Table 1). Additionally we assessed S12L and P13SFbx4 dimerization properties. Both mutants exhibited reduced dimerization potential, compared with wtFbx4 (Fig. S6D) and consequentially were unable to catalyze rapid cyclin D1 proteolysis (Fig. S6E). We also entertained the notion that a second ligase implicated in targeting cyclin D1 for ubiquitination in cultured cells, Fbw8, might also be inactivated in esophageal caners. However, our analysis of Fbw8 expression in normal esophageal epithelium and tumor tissue revealed that Fbw8 is not expressed in either normal esophageal epithelium or a associated tumors (data not shown). Given the lack of detectable expression of Fbw8 in this tissue, we conclude that an Fbw8 based E3 ligase is unlikely to contribute to cyclin D1 proteolysis.
Figure 6. Analysis of Fbx4 mutations in human esophageal tumors.
A. and B. Immunohistochemical analysis of cyclin D1 or TRF1 expression in esophageal tumors: S8R Fbx4, P76T Fbx4, S12L and wtFbx4, normal esophagus (bars, 50 (A) and 25 (B) µM). C. shFbx4 NIH3T3 lines were transfected with indicated Fbx4 expression vectors (immunoblot of Fbx4 levels is presented in the bottom panel) were assessed for growth in soft agar (21 days, bar, 250 µM). D. Quantification of C, error bars represent +/− SD.
Since all identified mutations in Fbx4 are hemizygous, we set out to determine whether mutant Fbx4 alleles exhibit dominant negative activity. NIH3T3 fibroblasts were transfected with wtFbx4 with or without Fbx4 mutants. Overexpression of wtFbx4 led to the reduction of total cyclin D1 protein levels (Fig. S6F, compare lanes 1–2), while co-expression of Fbx4-S8R, S12L or P13SFbx4 (transfected in 1:1 ratio relative to wtFbx4) attenuated Fbx4-dependent cyclin D1 loss (Fig. S6F, lanes 3–5).
We next assessed the ability of wtFbx4 or Fbx4 mutants, S8R, S12A and L23A, restore normal growth characteristics of Fbx4 knockdown fibroblasts. shFbx4 NIH3T3 cells were transfected with indicated constructs (Fig. 6C, lower panel) and their capacity to proliferate in soft agar was assessed. Re-expression of wtFbx4 significantly reduced the number of colonies, while ligase defective Fbx4 mutants did not exhibit significant activity (Fig. 6C–D). Altogether, we provide the evidence that cancer derived Fbx4 mutations that target GSK3β phosphorylation site in Fbx4 and affect its dimerization have reduced activity toward cyclin D1. The mechanism, by which these mutations contribute to the tumor formation in vivo is a subject of the great interest for future studies.
Discussion
To date, there is little evidence that SCF ligase activity is regulated. Rather, recognition of substrates by the F-box component of SCF ligases generally relies on substrate phosphorylation. Recent work, however, has revealed the presence of a dimerization domain in F-box proteins (Zhang and Koepp, 2006; Welcker and Clurman, 2007), thereby providing a platform for potential regulation through F-box protein (and thus, SCF) dimerization. The work described herein reveals the presence of a D-domain within Fbx4, an F-box only subtype; the conservation of this domain suggests that dimerization is a common feature of the SCF ligase. Strikingly, our data demonstrates that dimerization of Fbx4 is regulated through growth factor-dependent phosphorylation of Ser-12 (serine 11 in the mouse). Furthermore, Ser-12 phosphorylation and Fbx4 dimerization are required and contribute to an increased rate of cyclin D1 proteolysis during S-phase. Surprisingly, phosphorylation of Fbx4, like cyclin D1 (Diehl et al., 1998), is catalyzed by GSK3β.
As might be expected of a negative regulator of an oncogene, cyclin D1, our data provide evidence that Fbx4 has biological properties consistent with a tumor suppressor. First, inactivation or a reduction in Fbx4 levels results in the cellular acquisition of a transformed phenotype in vitro. Of equal importance, analysis of human esophageal cancer, where cyclin D1 overexpression is a common event, revealed mutations in Fbx4 that directly abrogate Fbx4 dimerization or inhibit binding to core SCF components (Skp1). All mutations identified interfere with ubiquitin ligase activity. In aggregate, the data presented reveal a unique mechanism of growth factor-dependent regulation of cyclin D1 and G1/S transition through phosphorylation-dependent dimerization of SCFFbx4. Our data also demonstrate direct abrogation of this pathway in human esophageal cancer and are suggestive of such potential in other cancers that are highlighted by cyclin D1 overexpression.
While SCF ligases are considered constitutively active, accumulating evidence suggests regulation is much more sophisticated than initially appreciated. Skp2 levels are determined in part through E2F-dependent transcription (Zhang and Wang, 2006). Likewise, βTrcp levels can be modulated by stress activated protein kinases (Spiegelman et al., 2001). Dimerization represents another potential regulatory apex. In fact Skp2, Fbw7, and βTrcp1/2 all dimerize (Zhang and Koepp, 2006; Suzuki et al., 2000; Welcker and Clurman, 2007; Chew et al., 2007). Our data support a role for temporally regulated Fbx4 dimerization in the regulation of substrate accumulation. Like Fbw7, Fbx4 contains a D-domain adjacent to the F-box. We have defined leucine/isoleucine residues (leu-23/28; iso-27) in Fbx4 that are essential for homo-dimerization. The spatial organization of these residues is conserved spatially with leu-246 and val-251, which contribute to the dimerization of Fbw7 (Tang et al., 2007). What distinguishes Fbx4 is the role of serine-12/proline-13, which acts as a phospho-acceptor residue for GSK3β. The generation of a Ser-12 phospho-specific antibody reveals GSK3β-dependent phosphorylation of this residue. Strikingly, mutation of this residue to a non-phosphorylatable residue attenuated homo-dimerization and significantly attenuated cyclin D1 ubiquitination and destruction. Taken together, these data suggest phosphorylation contributes to regulated self-association. The significance of this is highlighted by mutations of serine 12 and proline 13 in human cancer.
We noted that peak of Fbx4 phosphorylation/dimerization occurs at the G1/S boundary providing evidence for temporal regulation of Fbx4 dimerization. Significantly, at the G1/S boundary cyclin D1 undergoes nuclear export and levels are reduced through phosphorylation-dependent proteolysis (Diehl et al., 1997) and half-life measurements reveal that Fbx4 dimerization coincides with increased cyclin D1 turnover during S-phase. Thus, through this mechanism, growth factor availability and cell cycle progression, are coordinated via GSK3β, which in turn targets both substrate (cyclin D1) and E3 ligase (SCFFbx4).
Accumulating data suggest that oncogenic functions of cyclin D1 depend upon its nuclear accumulation during S-phase (Lin et al., 2007; Gladden AB, 2006). The nuclear deregulation of cyclin D1/CDK4 activity disrupts temporal regulation of DNA replication by directly impairing proteolysis of the replication licensing factor Cdt1 (Aggarwal et al., 2007). If nuclear overexpression of cyclin D1 is in fact the oncogenic event, why does loss of components of the D1 E3 ligase promote neoplastic growth? Critically, impaired Fbx4 or αB-crystallin, also promotes nuclear accumulation of cyclin D1 and a consequent stabilization of Cdt1, a direct consequence of nuclear cyclin D1/CDK4.
The significance of Fbx4 activity for cell homeostasis is emphasized by the relative frequency of mutations that impair its biochemical activity in human cancer. We identified 16 mutations in 116 esophageal tumors screened. Importantly, the fact that these mutations are hemizygous, implies that a reduction in Fbx4 dosage is sufficient to stabilize cyclin D1 and trigger neoplastic growth. Consistent with this interpretation, our knockdowns of Fbx4 are typically achieve an approximate 60% reduction in Fbx4 levels and this is sufficient to trigger neoplastic growth. Alternatively, our data reveals that co-expression of mutants with wild type Fbx4 can attenuate Fbx4 mediated cyclin D1 turnover. This result is expected in the case of the P76 mutation, which disrupts Skp1 binding resulting in a dominant negative allele similar to those generated through deletion of the F-box. Given that the other mutations disrupt oligomerization rather than Skp1-Cul-Rbx1 recruitment, the dominant negative activity may reflect titration of these ligase components into inactive complexes.
In all tumor specimen that harbor Fbx4 mutations, cyclin D1 protein levels were elevated and accumulated in the nuclei. This observation, together with the afore-mentioned transformation phenotype, strongly supports the supposition that abrogation of cyclin D1 E3 ligase activity is oncogenic. It also underscores the importance of Fbx4 dimerization for ligase activity, since the disruption of dimerization is observed in human esophageal cancer, in contrast to Fbw7 where all identified mutations directly impair substrate binding (O'Neil et al., 2007; Thompson et al., 2007). One can infer from the strong selection for such mutations that complete loss of Fbx4 may in fact be catastrophic for the cell.
Methods
Cell culture, plasmids and transfections
All mammalian cells were maintained in DMEM medium containing 10% of fetal bovine serum, glutamine and antibiotics. G0 synchronization was achieved by culture in medium containing 0.1% fetal bovine serum for 24 hours. Where indicated, 24 hours before transfection, cells were plated at optimal density and the following day transfected using Lipofectamine Plus reagent (Invitrogen, Carlsbad, CA). Stable cell lines expressing Fbx4 constructs were generated by G418 (Calbiochem, San Diego, CA) selection (1mg/ml) for 21 days and subsequently cultured in medium containing 500µg/ml G418. Insect Sf9 cells were maintained as described elsewhere. Human Fbx4 Myc-tagged vector was generated using PCR of human Fbx4 with primers designed for directional cloning into pCS2-MT plasmid (in frame with 6 Myc tags at the N-terminus of Fbx4).
Site directed mutagenesis
Flag-tagged Fbx4 mutants were constructed using QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using pcDNA3-Fbx4 plasmid as template. PCR reactions were performed following manufacturer instructions. Clones were sequenced in their entirety to confirm the presence of mutations.
Immunoprecipitation and western analysis
Cells were harvested in buffer containing following components: 50 mM HEPES (pH 8.0), 150mM NaCl, 2.5mM EGTA, 1mM EDTA, 0.1% Tween 20, protease, and phosphatase inhibitors (1mM PMSF, 20U/ml aprotinin, 5mg/ml leupeptin, 1mM DTT, 0.4mM NaF, 10mM β-glycerophosphate, and 100nM okadaic acid), protein concentration of samples was determined by BCA assay, and Fbx4 was precipitated using either M2 agarose (Sigma-Aldrich, St. Lois, MO) or a cyclin D1–mouse monoclonal antibody, D1-72-13G. Proteins were resolved by SDS-PAGE, transferred to nitrocellulose membrane and analyzed by immunoblot. Antibodies used in these studies were as follows: Fbx4 rabbit polyclonal antibody (Rockland, Gilbertsville, PA), αB crystallin (SPA 223) rabbit polyclonal antibody (Stressgen, Ann Arbor, MI), H-Ras (Santa Cruz Biotechnology, Inc, Santa Cruz, CA), pSer473 Akt and Akt (Cell Signaling, Danvers, MA), cyclin D1 mouse monoclonal D1-72-13G, cyclin D1 mouse anti-human (AB3, Calbiochem, San Diego, CA), β-actin (Sigma-Aldrich, St. Louis, MO), TRF1 rabbit polyclonal (Abcam Inc, Cambridge, MA), mouse anti Skp1 (BD Transduction Labs, Palo Alto, CA). Anti-phospho Ser11-Fbx4 (mouse), corresponding to Ser-12 (human), antibody was generated by immunizing rabbits with peptides containing p-Ser11 (murine based peptide; Yenzym antibodies, LLC, Burlingame, CA) followed by affinity purification.
Immunofluorescence
NIH3T3 derived cell lines were plated at optimal density on glass coverslips. 24 hours after splitting cells were permeabilized with Methanol:Acetone (1:1), washed with PBS and incubated in primary antibody (cyclin D1 17-13G11) for 2 hours. After washing and secondary anti-mouse FITC conjugated antibody application, slides were mounted using anti-fade DAPI reagent and analyzed by fluorescent microscopy using Nikon Eclipse E800 microscope.
Soft agar transformation assay
Anchorage-independent growth was determined by analyzing cellular growth in semisolid medium. Cells were seeded in Iscove's media containing 0.65% noble agar/10% FCS. Cells were grown for 21 days in 8% CO2.
In vitro kinase assay
Purified GST-tagged Fbx4 was used as a substrate for in vitro kinase reactions (30 minutes at 30°C). GSK3β and MAPK p38 were purchased from Cell Signaling, GSK3α from Abcam. Cdk2 complexes were purified from Sf9 cells infected with indicated baculoviruses.
In vitro ubiquitination
NIH3T3 derived stable cell lines expressing wild type Fbx4 and S12 mutants were used to purify Fbx4 SCF complexes. The SCF complexes were immunoprecipitated from the lysates using M2 agarose and incubated with purified GST-tagged cyclin D1 phosphorylated in vitro by recombinant GSK3β (5U), E1, E2 (UbcH5A), ATP, and ubiquitin for indicated times at 37°C. Proteins were resolved on 10% SDS-PAGE and visualized by Western blotting with anti cyclin D1 antibody D1-72-13G11.
Analysis of Fbx4 mutations in human tumors
Fbx4, cyclin D1 and αB-crystallin polymerase chain reaction products from normal and tumor-derived mRNA samples were generated using human Fbx4, cyclin D1 and αB-crystallin specific primers. All mutations were confirmed via bidirectional sequencing. All esophageal cancer tissues were obtained with informed consent and with IRB approval from the University of Pennsylvania, Kitano hospital and Peking University. Patient de-identifiers were used.
Supplementary Material
Acknowledgements
The authors wish to thank Margarita Romero for technical assistance; to C.J. Sherr for providing the RCC antiserum and the D1-72-13G11 hybridoma; Y. Naomoto for providing ESCC and EAC samples; and S. Fuchs and R. Assoian for critical reading of this manuscript. This work was supported by CA11360 (NIH) and a Leukemia & Lymphoma Scholar award (JAD); P01-CA098101 (NIH; AKR, HN, JAD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aggarwal P, Lessie MD, Lin DI, Pontano L, Gladden AB, Nuskey B, Goradia A, Wasik MA, Klein-Szanto AJ, Rustgi AK, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21:2908–2922. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt JR, Cleveland JL, Hannink M, Diehl JA. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14:3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzeno S, L F, Guo M, Barbash O, Zhang F, Herman JG, Klein PS, Rustgi A, Diehl JA. Identification of mutations that disrupt phosphorylation-dependent nuclear export of cyclin D1. Oncogene. 2006 doi: 10.1038/sj.onc.1209644. [DOI] [PubMed] [Google Scholar]
- Chew EH, Poobalasingam T, Hawkey CJ, Hagen T. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells--evidence for cullin dimerization. Cell Signal. 2007;19:1071–1080. doi: 10.1016/j.cellsig.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Craig KL, Tyers M. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol. 1999;72:299–328. doi: 10.1016/s0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Dickson C, Fantl V, Gillett C, Brookes S, Bartek J, Smith R, Fisher C, Barnes D, Peters G. Amplification of chromosome band 11q13 and a role for cyclin D1 in human breast cancer. Cancer Lett. 1995;90:43–50. doi: 10.1016/0304-3835(94)03676-a. [DOI] [PubMed] [Google Scholar]
- Diehl JA. Cycling to Cancer with Cyclin Dl. Cancer Biol Ther. 2002;1:226–231. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- Geng Y, Eaton EN, Picon M, Roberts JM, Lundberg AS, Gifford A, Sardet C, Weinberg RA. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- Gladden AB, W R, Aggarwal P, Wasik MA, Diehl JA. Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene. 2006;25:998–1007. doi: 10.1038/sj.onc.1209147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeguchi M, Sakatani T, Ueta T, Kaibara N. Cyclin D1 expression and retinoblastoma gene protein (pRB) expression in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2001;127:531–536. doi: 10.1007/s004320100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, Harper JW. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ. The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J Biol Chem. 2006;281:759–768. doi: 10.1074/jbc.M509855200. [DOI] [PubMed] [Google Scholar]
- Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ, Rustgi A, Fuchs SY, Diehl JA. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24:355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DI, Lessie MD, Gladden AB, Bassing CH, Wagner KU, Diehl JA. Disruption of cyclin D1 nuclear export and proteolysis accelerates mammary carcinogenesis. Oncogene. 2007;27:1231–1242. doi: 10.1038/sj.onc.1210738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Bueno G, Rodriguez-Perales S, Sanchez-Estevez C, Marcos R, Hardisson D, Cigudosa JC, Palacios J. Molecular alterations associated with cyclin D1 overexpression in endometrial cancer. Int J Cancer. 2004;110:194–200. doi: 10.1002/ijc.20130. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Zukerberg L, Togawa K, Meltzer SJ, Nishihara T, Rustgi AK. Human cyclin D1 oncogene and esophageal squamous cell carcinoma. Cancer. 1995;76:541–549. doi: 10.1002/1097-0142(19950815)76:4<541::aid-cncr2820760402>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- Russell A, Thompson MA, Hendley J, Trute L, Armes J, Germain D. Cyclin D1 and D3 associate with the SCF complex and are coordinately elevated in breast cancer. Oncogene. 1999;18:1983–1991. doi: 10.1038/sj.onc.1202511. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Matsushime H, Roussel MF. Regulation of CYL/cyclin D genes by colony-stimulating factor 1. Ciba Found Symp. 1992;170:209–219. doi: 10.1002/9780470514320.ch13. discussion 219–226. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Spiegelman VS, Stavropoulos P, Latres E, Pagano M, Ronai Z, Slaga TJ, Fuchs SY. Induction of beta-transducin repeat-containing protein by JNK signaling and its role in the activation of NF-kappaB. J Biol Chem. 2001;276:27152–27158. doi: 10.1074/jbc.M100031200. [DOI] [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Chiba T, Suzuki T, Fujita T, Ikenoue T, Omata M, Furuichi K, Shikama H, Tanaka K. Homodimer of two F-box proteins betaTrCP1 or betaTrCP2 binds to IkappaBalpha for signal-dependent ubiquitination. J Biol Chem. 2000;275:2877–2884. doi: 10.1074/jbc.275.4.2877. [DOI] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Singer J, Loeb KR, Grim J, Bloecher A, Gurien-West M, Clurman BE, Roberts JM. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell. 2003;12:381–392. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- Willems AR, Goh T, Taylor L, Chernushevich I, Shevchenko A, Tyers M. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos Trans R Soc Lond B Biol Sci. 1999;354:1533–1550. doi: 10.1098/rstb.1999.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Pledger WJ. Growth factor regulation of cyclin D1 mRNA expression through protein synthesis-dependent and -independent mechanisms. Mol Biol Cell. 1993;4:1133–1144. doi: 10.1091/mbc.4.11.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang C. F-box protein Skp2: a novel transcriptional target of E2F. Oncogene. 2006;25:2615–2627. doi: 10.1038/sj.onc.1209286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Koepp DM. Fbw7 isoform interaction contributes to cyclin E proteolysis. Mol Cancer Res. 2006;4:935–943. doi: 10.1158/1541-7786.MCR-06-0253. [DOI] [PubMed] [Google Scholar]
- Zukerberg LR, Yang WI, Gadd M, Thor AD, Koerner FC, Schmidt EV, Arnold A. Cyclin D1 (PRAD1) protein expression in breast cancer: approximately one-third of infiltrating mammary carcinomas show overexpression of the cyclin D1 oncogene. Mod Pathol. 1995;8:560–567. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.