Abstract
Cytochrome P450s (P450s) are important enzymes involved in the metabolism of xenobiotics, particularly clinically used drugs, and are also responsible for metabolic activation of chemical carcinogens and toxins. Many xenobiotics can activate nuclear receptors that in turn induce the expression of genes encoding xenobiotic metabolizing enzymes and drug transporters. Marked species differences in the expression and regulation of cytochromes P450 and xenobiotic nuclear receptors exist. Thus obtaining reliable rodent models to accurately reflect human drug and carcinogen metabolism is severely limited. Humanized transgenic mice were developed in an effort to create more reliable in vivo systems to study and predict human responses to xenobiotics. Human P450s or human xenobiotic-activated nuclear receptors were introduced directly or replaced the corresponding mouse gene, thus creating “humanized” transgenic mice. Mice expressing human CYP1A1/CYP1A2, CYP2E1, CYP2D6, CYP3A4, CY3A7, PXR, PPARα were generated and characterized. These humanized mouse models offers a broad utility in the evaluation and prediction of toxicological risk that may aid in the development of safer drugs.
Introduction
Foreign chemicals that enter the body are subject to metabolism by a number of xenobiotic metabolizing enzymes, a process which functions primarily to facilitate their elimination. Cytochromes P450 (P450) are among the most important enzymes responsible for the oxidative metabolism of a diverse range of xenobiotics, including therapeutic drugs, carcinogens and toxicants, as well as endogenous compounds such as steroid hormones and bile acids (Guengerich, 2003). In particular, the CYP1A, CYP2C, CYP2D, CYP2E1 and CYP3A subfamilies play central roles in the metabolism and disposition of drugs (Guengerich, 2003). The majority of P450 enzymes are expressed in the liver, while some are specifically expressed in extrahepatic tissues. P450-mediated drug metabolism exhibits considerable inter-individual variation, due in part to genetic polymorphisms and factors such as age, sex, body weight and disease state. Consequently, altered pharmacokinetics, pharmacodynamics and clearance of drugs are a result. Transcriptional regulation of P450s by xenobiotics acting through receptors such as the arylhydrocarbon receptor (AHR) and nuclear receptors such as pregnane-X-receptor (PXR), constitutive androstane receptor (CAR) and peroxisome proliferator-activated receptor α (PPARα) can also account for this variability. The nuclear receptor proteins all contain a core DNA binding domain and a ligand-binding domain. Ligands can activate the receptor by binding to the protein; the receptor then undergoes conformational changes allowing release of corepressors and binding of coactivators enabling recruitment of RNA polymerase II and other accessory proteins allowing transcriptional activation.
Striking species differences in the response to xenobiotics exist, notably between rodents and humans. In particular, expression and catalytic activity differences between human CYP2D6 and rodent Cyp2d, and between human CYP3A4 and rodent Cyp3a have been documented (Bogaards et al., 2000). These differences in P450 expression and nuclear receptor ligand binding affinities between human and rodent xenobiotic receptors may explain these differences between rodents and humans. Therefore typical rodent models may not be of value in the prediction of human responses to xenobiotics.
Species differences in drug metabolism
The underlying reasons for interspecies differences in drug metabolism were originally proposed to be a species’ inability to carry out a metabolic reaction (such as N-hydroxylation of aliphatic amines in rats), restriction of the occurrence of a reaction to a particular species or competing reactions by which a compound may be metabolized (Caldwell, 1981). A classic example of species-specific difference in metabolism by competing reactions is the biotransformation of 2-acetyl-amino-fluorene, a potent carcinogen in a number of species. The carcinogenicity of 2-acetyl-amino-fluorene depends on the ratio of N-hydroxylation (bioactivation pathway) and aromatic hydroxylation (detoxification pathway). The degree of N-hydroxylation in rat, rabbit and dog is considerable, and therefore this chemical is considered a carcinogen in those species. In guinea-pigs no N-hydroxylated metabolites are formed and so 2-acetyl-amino-fluorene is not carcinogenic in this species (Razzouk et al., 1982).
It is now recognized that rodents metabolize xenobiotics differently from humans due in part to species differences in the expression and catalytic activities of P450s (Caldwell, 1981; Bogaards et al., 2000). Functional orthologs of almost all human genes exist within the mouse genome. In particular 36 orthologous pairs of P450 genes have been identified in the genomes of mice and humans which have similar or identical functions in both species (Nelson et al., 2004). 102 putative functional P450 genes have been identified in the mouse genome compared to 57 in humans (Nelson et al., 2004). It can be assumed that differences in P450 isoforms between species are a major cause of species differences in drug metabolism. The anti-hypertensive drug debrisoquine for example, is hydroxylated to 4-hydroxydebrisoquine in both humans and Sprague-Dawley rats primarily by CYP2D6 and CYP2D respectively (Corchero et al., 2001a) However, in mice no significant formation of 4-hydroxy debrisoquine was detected (Masubuchi et al., 1997) suggesting that the murine Cyp2d genes do not have the same enzymatic activity as in humans (Bogaards et al., 2000).
Total P450 induction by the anti-tuberculcosis drug rifampicin in humans was demonstrated as early as 1973 by Hebert Remmer, and in 1975 it was reported that this was a species specific induction, as it did not occur in the rat (reviewed in Bolt, 2004). The human CYP3A4 was shown to be the major drug metabolizing enzyme induced by rifampicin (Li et al., 1995) and further studies revealed distinct species differences in CYP3A induction. Pregnelolone-16α-carbontirile (PCN) and dexamethasone were powerful inducers of CYP3A in rodents but not in humans, whereas the opposite is true of rifampicin which is an effective inducer of CYP3A4 in humans but not in rodents, thus confirming the results of Herbert Remmer’s group in the 1970s (reviewed in (Bolt, 2004). The xenobiotic nuclear receptor PXR was identified as the major determinant of CYP3A gene regulation by xenobiotics (Kliewer et al., 1998). Species differences in ligand specificities of PXR were demonstrated: for example, human PXR but not the mouse ortholog is activated by rifampicin, clotrimazole and diethylstilbestrol. Conversely, dexamethasone, PCN and 17α-hydroxypregnenolone are strong activators of mouse but not human PXR (Kliewer et al., 1998; Lehmann et al., 1998). The mouse versus human PXR activation profiles generally correspond with the species-specific CYP3A induction profiles.
Peroxisome proliferators cause liver tumors when chronically administered to rats and mice (Reddy and Krishnakantha, 1975) through a mechanism involving the nuclear receptor PPARα as revealed by PPARα-null mice (Peters et al., 1997). However, epidemiological studies on patients receiving fibrate drugs, PPARα agonists, suggest that humans are resistant to the carcinogenic effects of these peroxisome proliferators (Gonzalez et al., 1998). There exists a striking species differences in the levels of PPARα expression (Palmer et al., 1998) and ligand affinities between human and mouse forms (Sher et al., 1993) which may possibly explain this differential response to peroxisome proliferators.
These examples of species differences in drug metabolism described demonstrate that use of regular rodent models may not be effective in understanding and predicting the human response to drugs and more complex animal models resembling the human pathology need to be established.
Generation of humanized mouse models
The safety assessments of drugs or xenobiotics to which humans are exposed are largely based upon extrapolation of animal experiments to the human situation. Although the validity of animal testing to predict drug efficacy and safety in humans has been questioned, it is commonly believed that pharmacokinetic data can be reasonably extrapolated to humans. Allometric scaling approaches have been applied, where anatomical, physiological and biochemical variables in mammals can be scaled across species as a power function of body weight (Martignoni et al., 2006). This methodology has been used to predict plasma concentration-time profiles and the main pharmacokinetic parameters (Martignoni et al., 2006). In vitro studies using human liver microsomes, human hepatocytes, liver slices and recombinant enzymes are also important methods used in assessing human drug metabolism. However, they alone cannot predict how the variable processes of absorption, distribution, metabolism and excretion will modulate the expression of pharmacologic activity in vivo. The only way this modulation can be estimated is by studying the new drug in intact animals. Rodent models are widely accepted as useful experimental tools for the evaluation of chemical carcinogenicity and/or toxicity, metabolism and pharmacology of xenobiotics, such as therapeutic and agrochemical agents. Since rodents have a short lifespan, this allows for the growth of large numbers of animals in a short period of time and thus the feasibility of many studies. Mice in particular have many practical advantages over other rodents, short gestation, large litter sizes, fast breeding and lower animal husbandry and maintenance costs.
In an approach to overcome the species differences in P450 and xenobiotic receptor expression and regulation inherent between rodents and humans, transgenic humanized mouse models were developed. Several transgenic strategies can be employed to generate mice expressing the particular human protein. The P450 or xenobiotic receptor cDNA can be placed behind a promoter in an expression vector and introduced into mice by the standard pronuclei injection methodology. Tissue specific promoters are used to direct expression of the protein, for example, the mouse albumin promoter (transthyretin) or the liver-enriched activator protein promoter (LAP or CEBP/β), will deliver liver hepatocyte specific expression. Alternatively, the entire human gene, containing all important regulatory elements, exons and introns can be introduced into mice by using genomic clones derived from λ phage, bacterial artificial chromosome (BAC) or P1 phage artificial chromosomes (PAC). With the gene under control of their intrinsic regulatory systems, it is assumed that the human tissue specific regulation and induction patterns will be maintained in the mouse, since most transcription factors are conserved in mammals. However, since the endogenous murine genes are present that are orthologous or homologous to the introduced human transgene, they may exhibit overlapping functions and confound the effect of the human transgene. Therefore, these human transgenes would ideally be introduced onto the corresponding null mouse background by breeding with the gene knockout mouse or by incorporating the human transgene directly into the site of the endogenous mouse gene and thereby causing its instantaneous disruption (knock-in strategy). The human cDNA is often used as the transgene in the knock-in approach since the production of a recombination vector that contains the complete human gene and sufficient flanking murine sequences to promote recombination with the native mouse gene is technically difficult. A different approach to generating humanized models has been described that uses human hepatocytes transplanted into livers of chimeric mice thus generating humanized livers (Tateno et al., 2004).
Transgenic technology can therefore be used to generate mice that contain the human target protein or improved models of human disease that are better at mimicking the specific human pathology. Mice humanized for xenobiotic receptors or drug metabolism pathways may be used to investigate and predict potential overt toxicological or metabolic problems.
P450-humanized mouse models
CYP1A1- and CYP1A2-humanized mice
CYP1A1 and CYP1A2 are inducible upon exposure to xenobiotics such as 2,3,7,8-tetrachlorodibenzo-p-dioxin, 3-methylcholanthrene (3-MC), polycyclic aromatic hydrocarbons and polychlorinated biphenyls (Nebert et al., 2004). Many of these xenobiotics act as ligands for AHR, which activates a battery of genes encoding enzymes including P450s that are primarily involved in the metabolic activation of procarcinogens such as arylamines, polycyclic aromatic hydrocarbons and heterocyclic amines (Nebert et al., 2004). Heterocyclic amines are produced during the cooking of meats and fish, with the most abundant being, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Metabolic activation of PhIP is principally mediated by CYP1A2, followed by esterification with N-acetyltransferase or sulfotransferase to produce the unstable esters that can rearrange to electrophiles that bind covalently to DNA to form adducts. Formation of DNA adducts is considered a necessary initial step in chemical carcinogenesis. PhIP-DNA adducts have been identified in the colon, heart and lung of rats treated with the carcinogen (Lin et al., 1992) and in the liver, colon and mammary gland of mice (Snyderwine, 2002). PhIP-DNA adducts have also been detected in colonic tissue of humans (Friesen et al., 1994). A higher CYP1A2 activity in combination with higher N-acetyltransferase activity has been associated with an elevated risk for colon cancer in individuals eating well cooked meats, a rich source of heterocyclic amines (Le Marchand et al., 2001).
Species differences in the oxidative metabolism of PhIP have been observed between humans and rodents (Turteltaub et al., 1999). Experimental animals are able to both activate and detoxify these amines, whereas, humans convert them predominantly to their reactive genotoxic metabolites. In rodents, metabolism of PhIP is predominantly oxidation in the ring system followed by conjugation. However, in humans, N2-hydroxylation to the proximate mutagen N2-hydroxy-PhIP is the major metabolic pathway for PhIP followed by glucuronidation (Turteltaub et al., 1999). So clearly, large differences exist in the metabolism of PhIP between humans and rodents and thus appropriate extrapolation of cancer risk from experimental animals to humans is of concern, particularly in establishing safe thresholds for human exposure to PhIP. By generating humanized mouse lines where both human CYP1A1 and CYP1A2 are incorporated into the mouse genome in the absence of the mouse Cyp1a1 and/or mouse Cyp1a2 genes, suitable models can be provided for human health risk assessment studies of PhIP, other dietary or environmental toxicants and also drugs that are substrates for CYP1A1 or CYP1A2.
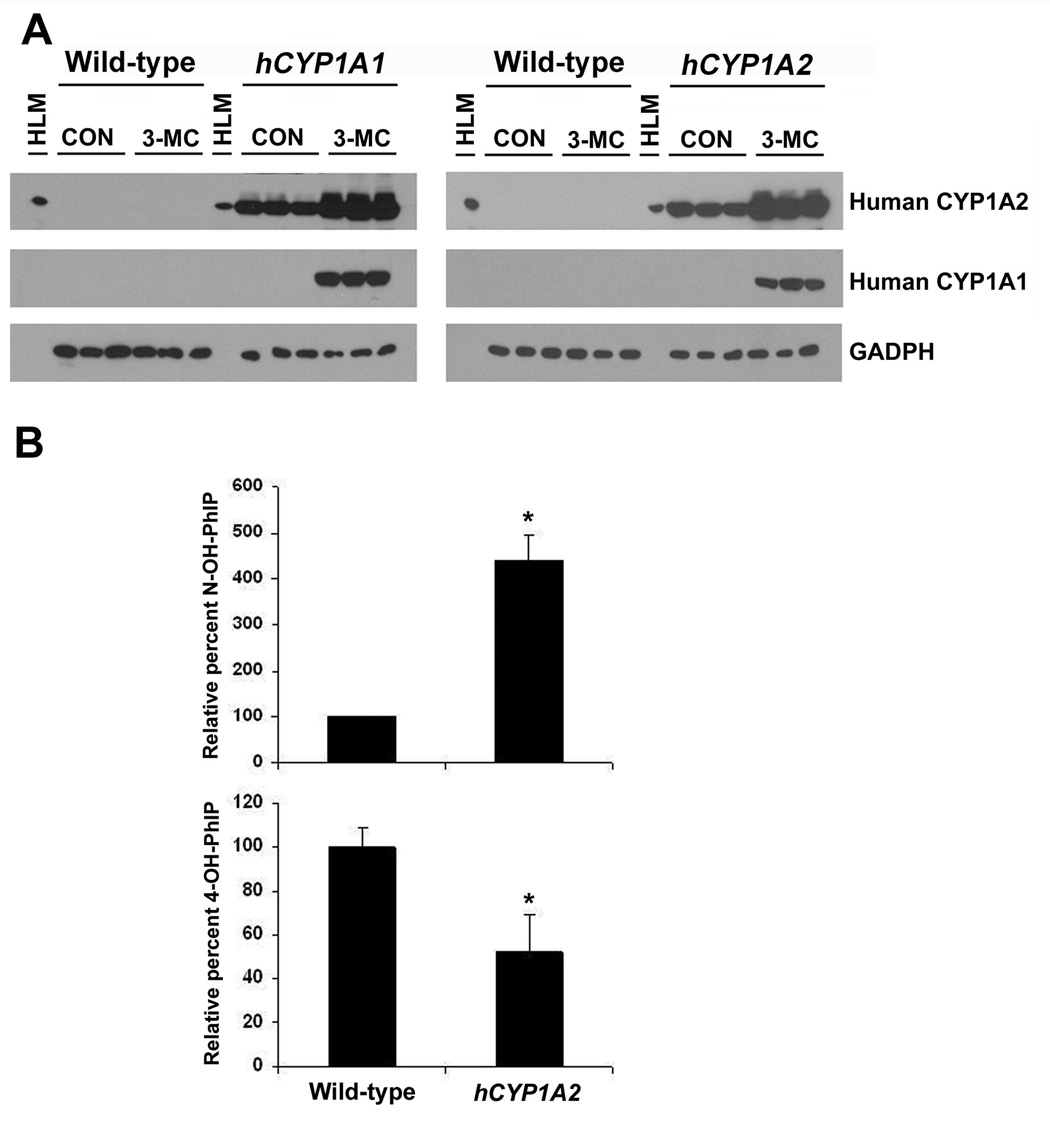
A BAC clone containing both the human CYP1A1 and CYP1A2 genes (Corchero et al., 2001b) was used to generate a transgenic mouse line that was bred into either a Cyp1a1-null or a Cyp1a2-null background, creating CYP1A1-humanized (hCYP1A1) and CYP1A2-humanized (hCYP1A2) mice respectively (Cheung et al., 2005a; Jiang et al., 2005). Both CYP1A1 and CYP1A2 proteins were demonstrated to be functional in the hCYP1A1 and hCYP1A2 mice and 3-MC treatment resulted in their induction (Figure 1A) (Cheung et al., 2005a). In hCYP1A2 mice, CYP1A2 protein expression was predominantly expressed in the liver with considerably lower levels in extrahepatic tissues such as lung, kidney colon and heart (Cheung et al., 2005a). In particular, hCYP1A2 mice were shown to accurately express CYP1A2 protein reflective of their expression in humans. When compared to wild-type mice, preferential N2-hydroxylation of PhIP was demonstrated in these hCYP1A2 mice (Figure 1B), a pathway for PhIP metabolism that in vitro studies revealed as predominant with the human ortholog. This was demonstrated both in vivo by measuring the relative total of N2-hydroxy-PhIP-glucuronides in the urine and in vitro using liver microsomes (Cheung et al., 2005a). Therefore, the hCYP1A2 mouse provides a more appropriate model than wild-type mice with which to perform human risk evaluations and estimation of safe exposure levels to this dietary mutagen. Recently, a transgenic mouse line was developed that also contained both human CYP1A1 and CYP1A2 genes but was deficient in both the mouse Cyp1a1 and Cyp1a2 genes (Dragin et al., 2007). This model would also be of value in human risk assessment studies involving drugs that are largely detoxified or activated by CYP1A1 or CYP1A2.
Figure 1. Characterization of hCYP1A1 and hCYP1A2 mice.
(A) Assessing the induction of hepatic human CYP1A1 and CYP1A2 proteins following 3-methylcholanthrene treatment in hCYP1A1 and hCYP1A2 mice using western blot analysis of liver microsomes. Transgenic mice containing both CYP1A1 and CYP1A2 genes were bred into a mouse Cyp1a1-null background and were designated hCYP1A1 mice. Transgenic mice containing both CYP1A1 and CYP1A2 genes in a mouse Cyp1a2-null background were designated hCYP1A2 mice. Monoclonal antibodies specific to human CYP1A1, human CYP1A2, rat CYP1A2 and GAPDH were used. HLM, human liver microsomes; 3-MC, 3-methylcholanthrene; CO, corn oil.
(B) Differential metabolism of PhIP in vivo in hCYP1A2 mice. N2-hydroxylation and 4’-hydroxylation of PhIP was examined in the urine of wild-type and hCYP1A2 mice which had been treated with an oral dose of PhIP. The relative amount of N-OH PhIP include the N2-OH-PhIP-N2-glucuronide and N2-OH-PhIP-N3-glucuronide. The relative amount of 4-OH PhIP was present by total free 4-OH PhIP and de-conjugated 4-OH-PhIP. The relative percentage of metabolite detected was calculated based on the mean value of the wild-type mice (defined as 100%). The pooled urine samples from three mice were analyzed in triplicate (values are mean ± SE). *P<0.05 compared to wild-type.
CYP2D6-humanized mice
Human CYP2D6 is involved in the metabolism of up to 20% of clinical drugs belonging to several therapeutic classes that include many centrally acting antidepressants and codeine (Zanger et al., 2004). In humans, the CYP2D subfamily has a single active member, CYP2D6 that is highly polymorphic, whereas, rats and mice have at least five Cyp2d genes, none of which encodes a protein with the same enzymatic activity as human CYP2D6 (Bogaards et al., 2000). There are more than 90 known allelic variants of CYP2D6 to date (www.imm.ki.se/CYPalleles/cyp2d6.htm). From 5 to 10% of Caucasians are poor metabolizers (Zanger et al., 2004) and this can lead to individual variation in response to many drugs due to impaired metabolism by CYP2D6. A lack of robust animal models to study the CYP2D6 polymorphism led to development of a CYP2D6-humanized mouse line that could be used to model CYP2D6 poor and extensive metabolizer phenotypes (Mahgoub et al., 1977).
CYP2D6-humanized mice were generated using a λ phage genomic clone containing the whole human CYP2D6 gene and its regulatory sequences (Corchero et al., 2001a). While the endogenous murine Cyp2d genes were present in these transgenic mice, not of these gene products had the same catalytic activity and substrate specificity as CYP2D6. Functional CYP2D6 was expressed in the transgenic mouse liver resulting in enhanced metabolism and disposition of the anti-hypertensive β-adrenoceptor blocking drug, debrisoquine. Urinary metabolic ratios (debrisoquine/4-hydroxydebrisoquine) following debrisquine treatment have been used to estimate the extent of debrisoquine metabolism in humans (Mahgoub et al., 1977). Indeed, treatment of CYP2D6-humanized mice (both homozygous and hemizygous) with debrisoquine revealed that this metabolic ratio was approximately 1, a value similar to that found in human extensive metabolizers, whereas in wild-type mice, the metabolic ratio was approximately 10, which is typical of human poor metabolizers. The CYP2D6-humanized mouse model is therefore similar to human extensive metabolizers and wild-type mice are similar to human poor metabolizers of debrisoquine (Corchero et al., 2001a). This mouse line was used to determine the mechanism of autoimmune response to CYP2D6 (Holdener et al., 2008). The CYP2D6-humanized mouse is also of value for screening for dietary and endogenous substrates revealing a role for CYP2D6 as a 5-methoxyindolethylamine O-demethylase (Yu et al., 2003). The CYP2D6-humanized model provides a useful in vivo tool to further investigate the physiological and pathological significance of CYP2D6 endogenous substrates and the association with its genetic polymorphism.
CYP2E1-humanized mice
CYP2E1 metabolizes and activates a wide spectrum of toxicologically important chemicals (acetone, ethanol, benzene, halothane, carbon tetrachloride), clinically used drugs (acetaminophen (APAP), disulfiram, chlorzoxazone), and carcinogens such as the low molecular weight nitrosoamines (Kessova and Cederbaum, 2003). It is expressed abundantly in human liver, with some lower level extrahepatic expression in the kidney, lung and brain (Lieber, 1997). CYP2E1 was suggested to play a definitive role in alcohol-induced liver damage associated with oxidative stress (Caro and Cederbaum, 2004).
Most studies on the pharmacological and biochemical actions of CYP2E1 are derived from studies with rodent, rabbits and cultured hepatocytes and thus extrapolation of these results to humans can be difficult. Creating a humanized transgenic mouse model by introducing the human CYP2E1 gene into Cyp2e1-null mice can circumvent this disadvantage. A transgenic mouse line was described that expresses the human CYP2E1 cDNA under the control of the mouse albumin enhancer promoter (Morgan et al., 2002), however, this transgenic model contains endogenous mouse CYP2E1 and is therefore not a true humanized model.
Transgenic mice were generated using a BAC clone containing the entire human CYP2E1 gene and these were bred into a mouse Cyp2e1-null background, thus creating CYP2E1 humanized mice (Cheung et al., 2005b). Functional differences between human and mouse CYP2E1 can therefore be directly compared using this model. Human CYP2E1 activity was demonstrated in these transgenic mice by increased chlorzoxazone 6-hydroxylation and increased p-nitrocatechol formation, the activities of which were inhibited using a CYP2E1 monoclonal antibody. There was no significant difference in chlorzoxazone 6-hydroxylase activity between CYP2E1-humanized and wild-type mice, but a higher O-hydroxylation of p-nitrophenol was observed in CYP2E1-humanized mice compared to wild-type. Inducible human CYP2E1 was shown in the CYP2E1-humanized mice using the known CYP2E1 inducer acetone. The response to the widely used analgesic APAP was explored in the CYP2E1-humanized mice. Therapeutic doses of APAP are considered safe, however, an APAP overdose can result in severe hepatotoxicity. Using Cyp2e1-null mice, a critical role of CYP2E1 in APAP toxicity was demonstrated since they were found to be resistant to APAP-induced liver toxicity (Lee et al., 1996). CYP2E1 metabolizes APAP to a highly reactive metabolite, N-acetyl-p-benzoquinone imine (NAPQI) that can covalently bind to cellular nucleophiles such as DNA, RNA and proteins. NAPQI is typically inactivated by glutathione S-transferases, however, in an overdose of APAP, hepatic glutathione becomes depleted and formation of the reactive intermediate metabolites outstrips their detoxification. Interestingly, a 200 to 400 mg/kg dose of APAP administered to CYP2E1-humanized mice revealed no centrilobular hepatic necrosis lesions, whereas in wild-type mice, mild-to moderate degrees of centrilobular hepatic necrosis lesions were observed with this same dose (Table 1) (Cheung et al., 2005b). These results were correlated with measurements of serum alanine aminotransferase levels, elevated levels of which serve as a marker of liver damage. These data establish that in this transgenic model, human CYP2E1 is functional and these mice exhibit altered sensitivity to the hepatotoxicity of APAP. The CYP2E1-humanized mice will be an excellent in vivo tool to delineate the role of human CYP2E1 in ethanol-induced oxidative stress and alcoholic liver damage and also for predicting drug metabolism, disposition and drug-drug interactions (DDIs) of chemicals that are substrates for human CYP2E1. This transgenic model allows direct assessment of the human CYP2E1 function in a whole animal model system and thus may be of importance in the evaluations of toxicological and pharmacological risk assessment studies for human CYP2E1 substrates.
Table 1. Degree of centrilobular hepatic necrosis in mice treated with saline or APAP.
The livers were examined histologically and classed according to the degree of centrilobular hepatocyte necrosis, marked as none, mild, moderate or severe. Mice found dead after 24hr of APAP treatment were noted. The numbers indicate how many mice were classed with which degree of hepatocyte necrosis per total number of mice examined in each group (total numbers of mice in each group n=5 to 10).
| GENOTYPE | SALINE | 200 mg/kg APAP | 400 mg/kg APAP |
|---|---|---|---|
| Wild | None 5/5 | None 3/5 | Moderate 5/7 |
| Mild 1/5 | Found dead 2/7 | ||
| Moderate 1/5 | |||
| Cyp2e1-null | None 5/5 | None 6/6 | Normal 5/5 |
| CYP2E1-humanized | None 8/8 | None 9/9 | Mild 3/10 |
| Moderate 2/10 | |||
| Severe 4/10 | |||
| Found dead 1/10 |
Human CYP3A4 transgenic mice
There are four CYP3A isoforms expressed in humans, CYP3A4, CYP3A5, CYP3A7 and CYP3A43 (Nelson et al., 2004). Interestingly, mice have eight complete Cyp3a genes and 3 pseudogenes (Nelson et al., 2004), illustrating the complexity of P450 gene families between species. CYP3A4 is the predominant P450 expressed in human liver; it is also the major expressed in non-hepatic tissues, most notably the intestine. CYP3A4 is central to the metabolism of a wide array of drugs (e.g. antiretrovirals, anticancer), chemicals, and dietary constituents. CYP3A gene expression can be regulated by xenobiotic receptors such as the ligand activated nuclear receptors, PXR or CAR (Willson and Kliewer, 2002). Since CYP3A4 is estimated to be responsible for the full or partial metabolism of over fifty percent of clinically used drugs, it has a significant impact on drug metabolism and pharmacokinetics (Guengerich, 1999). This is of particular concern in the development of new drugs, as many are used in combination therapies and the co-administration with other drugs can lead to DDIs. Drugs such as the HIV protease inhibitor ritonavir and food constituents such as grapefruit juice have been shown to profoundly inhibit CYP3A and affect the pharmacokinetic and pharmacodynamics of CYP3A substrate drugs (Dresser et al., 2000). During preclinical development, drug candidates can be screened using recombinant P450s to determine if the compound will be metabolized by CYP3A4. In vivo models to study CYP3A4 would aid the assessment of drugs that are CYP3A4 substrates.
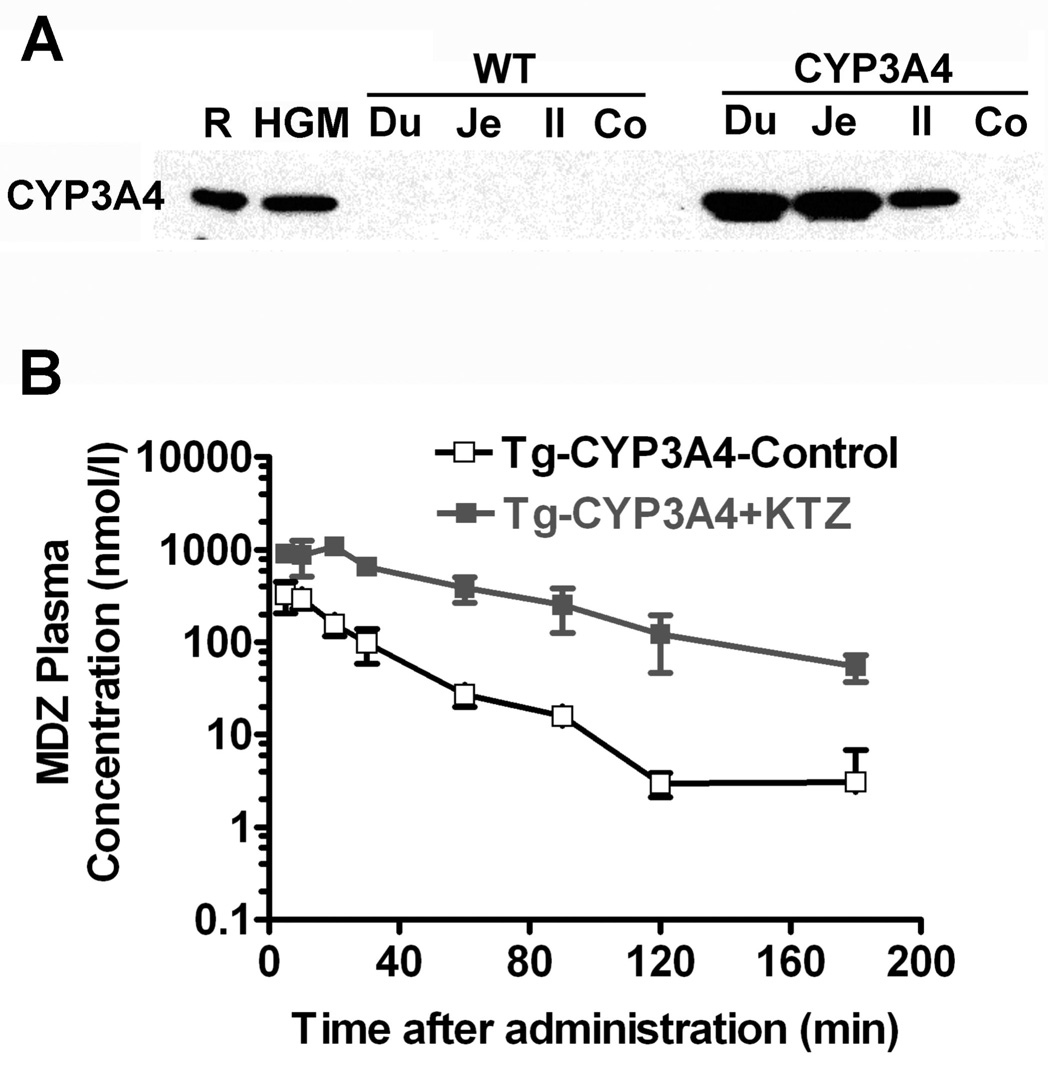
To generate mice that express human CYP3A4, a BAC clone containing the complete CYP3A4 gene including the upstream PXR-binding sites was used as a transgene (Granvil et al., 2003). This mouse line (designated hCYP3A4) expressed high levels of CYP3A4 in the small intestine, comparable to that found in human gut microsomes (Figure 2A). Expression of CYP3A4 in the small intestine influenced the metabolism and pharmacokinetics of the orally administered drug midazolam, which is commonly used as a standard probe for CYP3A4 activity in humans. A higher rate of midazolam metabolism and elimination was observed in the hCYP3A4 mice as compared to non-CYP3A4 expressing wild-type mice. No differences were observed between hCYP3A4 and wild-type mice when the drug was administered intravenously (Granvil et al., 2003). The potent CYP3A4 inhibitor, ketoconazole markedly lowered the rate of midazolam metabolism and elimination when co-administered orally with midazolam when compared to midazolam alone (Figure 2B). This demonstrates a possible DDI between midazolam and ketoconozole.
Figure 2. CYP3A4 expression and catalytic activity in hCYP3A4 mice.
(A) Western blot analysis of the expression of human CYP3A4 in human gut microsomes (HGM), duodenum (Du), jejunum (Je), Ileum (Il) and colon (Co) of wild-type and hCYP3A4 mice. Recombinant human CYP3A4 (R).
(B) Pharmacokinetics of midazolam (MDZ) in hCYP3A4 mice orally administered MDZ in the absence and presence of the CYP3A4 inhibitor ketaconozole.
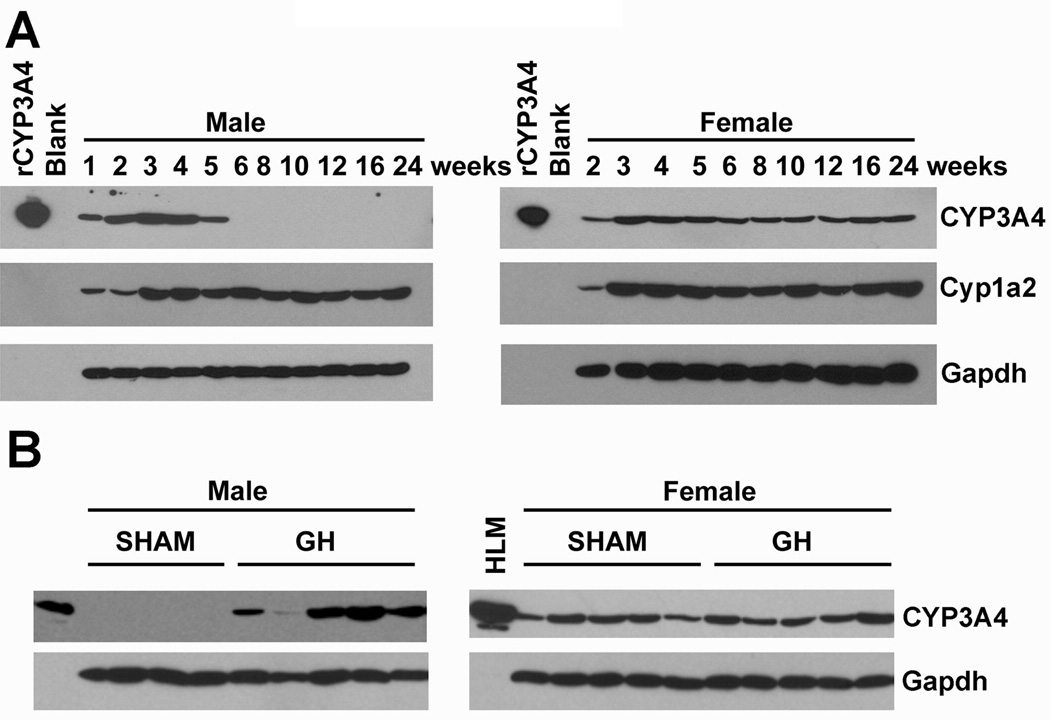
Surprisingly little CYP3A4 expression was observed in the liver, the major site of CYP3A4 expression in humans (Granvil et al., 2003). However these studies were carried out on mature male mice. Later studies using mice generated from a BAC clone containing both CYP3A4 and CYP3A7 genes (Cheung et al., 2006), revealed constitutive CYP3A4 expression in the livers of immature male and adult female mice, indicating that the transgene is developmentally regulated in a gender-specific manner (Yu et al., 2005; Cheung et al., 2006). Indeed, human studies have indicated higher CYP3A4 expression in females (Wolbold et al., 2003). In both males and females, CYP3A4 expression is present as early as 1 week after birth (Figure 3A). In 6-week-old male mice, CYP3A4 expression became undetectable, but remained in females (Figure 3A). In contrast, the developmental expression pattern for murine Cyp1a2 showed no differences between sexes from ages 3 to 24 weeks (Figure 3A). Gender-specific regulation of P450s has been extensively studied in rodents and is suggested to be due to differences in growth hormone secretory patterns between males and females (Waxman and O'Connor, 2006). Female mice secrete high constant levels of growth hormone from the pituitary gland. In male mice, lower levels of growth hormone are secreted in a pulsatile manner (Waxman and O'Connor, 2006). Continuous infusion of growth hormone was shown to convert the male growth hormone secretory pattern to that of the female pattern in hCYP3A4 mice (Cheung et al., 2006). This was demonstrated by implanting growth hormone releasing pellets into the mice, thus resulting in the 6-week-old male hCYP3A4 mice to now express CYP3A4 (Figure 3B). Growth hormone-dependent changes in hepatic P450 expression may significantly alter the pharmacokinetics and pharmacodynamics of drugs metabolized by CYP3A4 leading to differing drug efficacies and possibly adverse effects from interactions between growth hormone and CYP3A4-metabolized drugs.
Figure 3. Gender-dependent regulation of the constitutive expression of CYP3A4 by growth hormone.
(A) CYP3A4 developmental expression in livers of male and female hCYP3A4 mice. Liver tissues were collected from transgenic male and female mice of different ages (4–5 livers in each group). Pooled samples were used for Western blot analysis of CYP3A4. Antibodies against CYP1A2 and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) were used as controls.
(B) Stimulation of CYP3A4 protein expression in male livers by continuous growth hormone treatment. Mice (6–7 weeks old) were implanted with a growth hormone pellet that provided a continuous infusion for 7 days. Sham-treated mice were used as controls. Western blot analysis of the liver microsomes was carried out.
Interestingly, one line of hCYP3A4 mice exhibited low estradiol levels in the serum of pregnant females, resulting in underdeveloped mammary glands, low milk protein production and low-pup survival rate due to impaired lactation phenotype (Yu et al., 2005). The low estrogen may be a result of enhanced metabolism of estradiol and its precursor testosterone, to the inactive 2- and 4-hydroxylated estradiol metabolites in the gut by CYP3A4 (Down et al., 2007). The possibility that this increased metabolism occurs in the gut during the course of enterohepatic circulation of estradiol is intriguing since untreated hCYP3A4 mice express high levels of the enzyme in the intestine (Granvil et al., 2003). Enterohepatic recycling of estrogen has been demonstrated in humans (Sher and Rahman, 2000). These hCYP3A4 mice will facilitate in the in vivo analysis of orally administered drugs that are CYP3A4 substrates and be potentially of great value in the prediction of drug interactions.
The hCYP3A4 mice described above have a mouse Cyp3a background. In order for a fully humanized mouse line to be produced, the murine Cyp3a locus needs to be disrupted. An ideal humanized model would be to have the transgenic mice containing the entire human CYP3A4 gene, which expresses CYP3A4 both in the liver and intestine and is under the control of its native promoter (Cheung et al., 2006), bred into a Cyp3a-knockout background. Recently, Cyp3a-knockout mice lacking all functional murine Cyp3a genes were generated and shown to display greatly impaired metabolism of the anticancer drug docetaxel (van Herwaarden et al., 2007). These Cyp3a-knockout mice were bred with transgenic mice expressing human CYP3A4 cDNA under the control of a villin or ApoE promoter (directing expression of CYP3A4 to the intestine or liver respectively) to generate humanized models (van Herwaarden et al., 2005; van Herwaarden et al., 2007). In particular, these humanized models were used to investigate the relative importance of intestinal versus hepatic Cyp3a in first-pass metabolism of docetaxel. Expression of CYP3A4 in the intestine of Cyp3a-knockout mice dramatically increased absorption of docetaxel into the bloodstream, while hepatic expression increased systemic docetaxel clearance (van Herwaarden et al., 2007). The combination of these transgenic mice can provide excellent models for assessment of toxicity, drug efficacy, drug-drug and drug-food interaction risks.
Xenobiotic receptor-humanized mice
PXR-humanized mouse model
Pregnane X receptor (PXR) is a ligand-activated nuclear receptor that transcriptionally activates an array of genes in the liver and intestine that are involved in various aspects of detoxification and elimination of xenobiotics from the body and therefore has a significant impact on drug metabolism. Such genes include those that encode the P450s CYP3A, CYP2B6, and CYP2C, and the conjugation enzymes glutathione S-transferase, UDP-glucuronosyltransferase, sulfotransferase and carboxylesterase and the drug transporters organic anion transporting polypeptide 2 (OATP2), multidrug resistance-associated protein 2 (MRP2) and multidrug resistance protein 1 (MDR1) (Kliewer, 2003). PXR is activated by a large number of xenobiotics and clinically used drugs such as antibiotics, HIV protease inhibitors, anti-cancers and anti-hypertensives. Since multi-therapy regimens are commonly used in patients with cancer, HIV, cardiovascular disease and diabetes, the most common clinical implication of PXR activation is the occurrence of DDIs. The CYP3A subfamily of enzymes, predominantly CYP3A4, is responsible for the metabolism of over 50% of clinically marketed drugs. Since many of the drugs that are ligands for PXR are also substrates for CYP3A4, DDIs can arise, resulting in decreased drug efficacy and increased drug toxicity. Species differences in PXR ligand specificity occur between humans and mice (Jones et al., 2000). Drugs such as rifampicin, clotrimazole and troglitazone strongly activate human PXR, but are weak activators of rodent PXR. In contrast, the synthetic steroid PCN and the drug dexamethasone preferentially activate rodent PXR but not human PXR (Schuetz et al., 2000). Cell based reporter gene assays represent a simple and rapid in vitro strategy for screening human PXR ligands and CYP3A inducers (Raucy et al., 2002), however, limitations exist in extrapolation of in vitro data to the clinical in vivo situation. Therefore, a more realistic tool to study in vivo effects of human PXR would be the development of a humanized PXR mouse line (Xie et al., 2000).
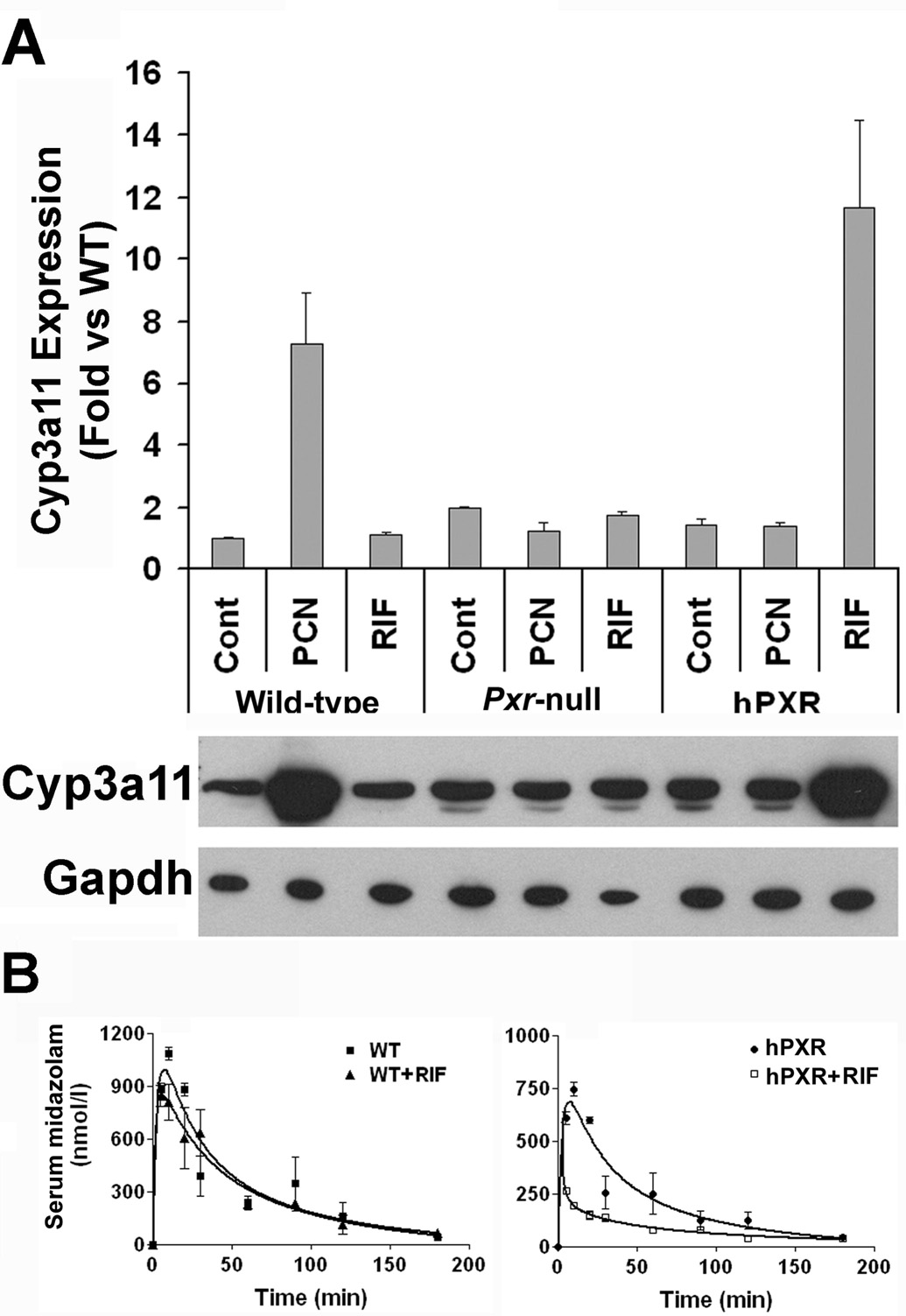
Previous studies have developed humanized PXR models by creating a transgenic mouse expressing the human PXR cDNA under the direct control of a tissue specific promoter (albumin or FABP) and breeding this transgene into a Pxr-null mouse background (Xie et al., 2000). More recently, a PXR humanized mouse line (designated as hPXR mice) was generated using a similar strategy, except the complete human PXR gene is under the control of its native promoter (Ma et al., 2007). A transgenic mouse was generated using a BAC clone containing the complete human PXR gene, including the 5’- and 3’-flanking sequences and then this mouse was bred with Pxr-null mice to create hPXR mice. In this model, PXR was expressed selectively in the liver and intestine, the same tissue pattern as CYP3A. Activation of PXR by rifampicin and PCN was investigated by monitoring mouse CYP3A11 expression. Rifampicin treatment induced CYP3A11 mRNA in the livers of hPXR mice, but not wild-type or Pxr-null mice (Figure 4A) (Ma et al., 2007). PCN treatment resulted in a marked induction of CYP3A11 mRNA in wild-type mice, but not in hPXR or Pxr-null mice (Figure 4A). This demonstrates a clear difference between wild-type and hPXR mice in response to PXR ligands, suggesting the use of this BAC transgenic hPXR mouse model in investigating human PXR function in vivo. The potential usefulness of this hPXR model in studying induction of CYP3As and thus identifying associated DDIs in vivo was examined using midazolam and rifampicin as a paradigm. Midazolam is a short acting hypnotic-sedative drug metabolized primarily by CYP3A (Gorski et al., 1994). Loss of midazolam pharmacodynamic effects were reported in tuberculosis patients undergoing rifampicin treatment, with the activation of human PXR and induction of CYP3A suggested as important factors in causing this DDI (Backman et al., 1996). In wild-type mice, rifampicin had no significant effects on the pharmacokinetics of midazolam (Figure 4B & Table 2). In contrast, in hPXR mice pretreated with rifampicin, the maximal midazolam serum concentration (Cmax) was decreased by 64% and the area under the curve (AUC0–180min) was decreased by 60% (Figure 4B & Table 2). Since this interaction between rifampicin and midazolam could not have been predicted from studies using wild-type mice, as murine PXR is not activated by rifampicin, this hPXR model represents a strong in vivo tool to investigate potential clinical pharmacokinetic DDIs between PXR ligand drugs and CYP3A substrates.
Figure 4. Analysis of PXR function in hPXR mice.
(A) Hepatic CYP3A regulation and expression in hPXR mice. Mice were treated with PXR ligands, rifampicin (RIF) or pregnenolone 16α-carbonitrile (PCN) with corn oil used as a control. Induction of CYP3A11 mRNA was measured by real-time quantitative PCR analysis and the induction of mouse CYP3A protein by western blot analysis.
(B) Effect of RIF on midazolam (MDZ) pharmacokinetics in wild-type and hPXR mice. Mice were pretreated with RIF, 10 mg/kg/day for 3 days followed by an oral dose of midazolam, 5 mg/kg. The timecourse of serum MDZ was analyzed by LC-MS/MS in wild-type (WT) mice and in hPXR mice with or without RIF treatment.
Table 2. Pharmacokinetics of midazolam (MDZ) in wild-type and PXR-humanized mice pretreated with or without rifampicin (RIF), at 10 mg/kg/day for 3 days.
Serum MDZ was detected by LC-MS/MS. Pharmacokinetic parameters were estimated from the serum concentration-time (three mice at each time point) data by a noncompartmental approach using WinNonlin, Data is expressed as means ± S.D.
| Genotype | MDZ | Control | RIF | RIF (% Control) |
|---|---|---|---|---|
| Wild-type | Cmax (nmol/l) | 1090 ± 50 | 908 ± 3 | 83.6 |
| AUC0–180 (nmol · min/l) | 55,300 ± 5200 | 50,200 ± 2900 | 90.7 | |
| hPXR | Cmax (nmol/l) | 748 ± 45 | 266 ± 7* | 35.6 |
| AUC0–180 (nmol · min/l) | 38,000 ± 3000 | 14,800 ± 493** | 40.0 |
P<0.0001 compared to control
P = 0.0002 compared to control
PPARalpha-humanized mouse model
Peroxisome proliferators are a structurally diverse group of chemicals including naturally occurring steroids and lipids, herbicides, pesticides, industrial plasticizers, lipid and cholesterol lowering fibrate drugs (e.g. fenofibrate, gemfibrozil). They are so named since they induce the size and number of peroxisomes (peroxisome proliferation) in the liver of rats and mice to which they are administered (Reddy and Krishnakantha, 1975). This is coincident with an increase in fatty acid catabolism as a result of elevated expression of genes encoding proteins involved in fatty acid β-oxidation and lipid transport (Reddy, 2001). These effects are mediated by the nuclear receptor peroxisome proliferator-activated receptor (PPAR) α as confirmed by using Ppara-null mice (Lee et al., 1995). Peroxisome proliferators have been shown to act as non-genotoxic carcinogens. Long-term administration of these chemicals to rats and mice results in the formation of hepatocellular carcinomas, however, humans appear to be resistant to the induction of peroxisome proliferation and the development of liver tumors by such chemicals (Gonzalez et al., 1998). The mechanism of action of PPARα in causing hepatocarcinogenesis and the species difference in response to peroxisome proliferators is largely unknown, but it may be related to differences in expression, activity or ligand affinity between rats/mice and human PPARα. Indeed, it has been demonstrated that human livers express one-tenth the level of PPARα mRNA and functional DNA binding capacity than do mouse livers (Palmer et al., 1998). Fibrate drugs are widely prescribed clinically and new generation drugs with higher PPARα affinities (agonist activity, EC50, of more than 100-fold greater than presently used fibrates) are under development by the pharmaceutical industry (Brown et al., 2001). Thus, it is of importance to investigate the mechanism of species differences in response to peroxisome proliferators and the mechanism of action of PPARα ligands in causing carcinogenesis. Therefore, PPARα-humanized mice were developed as an approach to assess human risk to peroxisome proliferators.
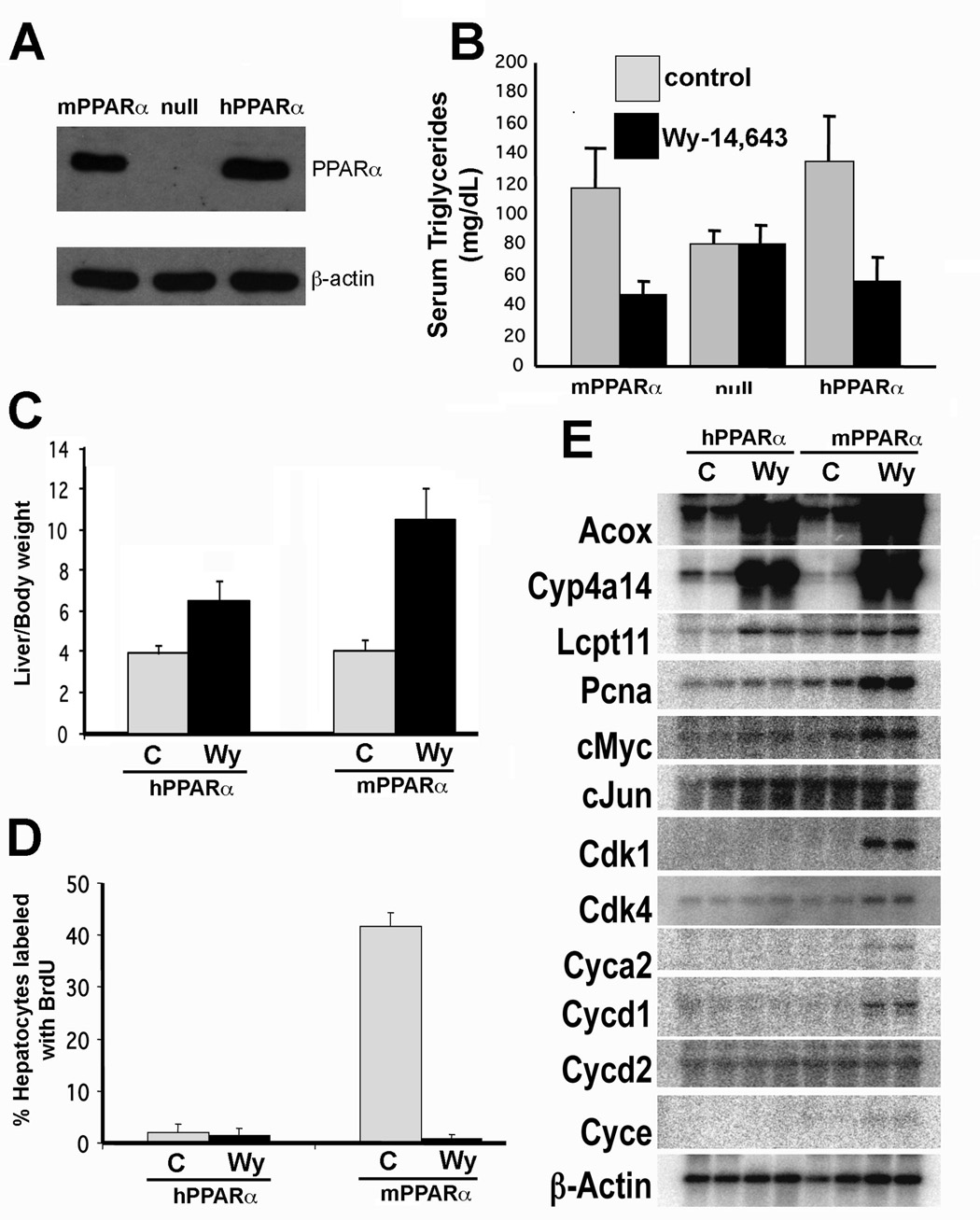
A PPARα-humanized mouse line was created that expressed human PPARα in the liver in a PPARα null background and was designated hPPARα TetOff (Cheung et al., 2004). In this mouse line, human PPARα cDNA was placed under the control of the tetracycline responsive regulatory system (Tet-off) with the liver-specific LAP (C/EBPβ) promoter. In the absence of doxycycline (a tetracycline derivative) human PPARα was constitutively expressed, the protein levels of which were comparable to that expressed in wild-type mice (Figure 5A). In the presence of doxycycline, no human PPARα protein is expressed (Cheung et al., 2004). Administration of the peroxisome proliferator Wy-14,643, revealed an induction of genes encoding peroxisomal acyl-CoA oxidase (ACOX), bifunctional enzyme (BIEN), and thiolase (THIOL), microsomal (CYP4A), mitochondrial short-, medium-, long-, very long-chain, acyl-CoA dehydrogenases (MCAD, LCAD, VLCAD) fatty acid metabolizing enzymes, as well as fatty acid transport proteins fatty acid translocase (CD36), liver fatty acid binding protein (L-FABP) demonstrating that human PPARα is functionally active in these mice (Cheung et al., 2004). Wy-14,643 treated hPPARαTetOff mice also demonstrated lower fasting serum total triglycerides similar to wild-type mice expressing native mouse PPARα (Figure 5B). Marked hepatomegaly was observed following Wy-14,643 treatment due to enhanced cell proliferation and cell hypertrophy resulting from an increase in the size and number of peroxisomes (Figure 5C); the extent of this increase was less in the hPPARαTetOff mice compared to wild-type. Surprisingly, only the wild-type mice but not the hPPARαTetOff mice exhibited hepatocellular proliferation following Wy-14,643 treatment, as revealed by the increased incorporation of 5-bromo-2’-deoxyuridine into hepatocyte nuclei (Figure 5D) and the elevation of cell cycle control genes (Figure 5E). hPPARαTetOff mice were found to be resistant to Wy-14,643-induced hepatocarcinogenesis following 44 weeks of Wy-14,643 feeding in contrast to the wild-type mouse group (Table 3) (Morimura et al., 2006). These findings suggest that the species specific difference in response to peroxisome proliferators such as fibrates are likely due to differences in profiles of genes activated by mPPARα compared to hPPARα. This species specific regulation of gene expression is probably responsible for the differential susceptibility to the development of hepatocarcinomas observed after fibrate treatment.
Figure 5. Differential response to Wy-14,643 treatment in hPPARαTetOff compared to wild-type mice.
(A) Human PPARα protein expression in the livers of hPPARαTetOff mice as revealed by western blot analysis using antibodies to PPARα and actin (loading control).
(B) Serum total triglyceride levels in wild-type, PPARα-null and hPPARαTetOff mice after treatment with the PPARα ligand Wy-14,643. Mice were fed a 0.1% (w/w) Wy-14,643 containing diet for 2 weeks, fasted overnight and serum triglycerides measured.
(C) Lack of Wy-14,643 induced hepatocyte proliferation in hPPARαTetOff mice. Mice were fed a Wy-14,643 diet for 8 weeks and 1 week before the termination of the experiment, mice were subcutaneously implanted with an osmotic pump releasing bromodeoxyuridine (BrdUrd). BrdUrd incorporation into the nuclei of hepatocytes, as a measure of replicative DNA synthesis was determined by immunohistochemistry using an anti-BrdUrd antibody. Labelling index of BrdUrd incorporation was calculated as a percentage of hepatocytes with BrdUrd incorporated.
(D) Lack of induction of cell cycle control genes in Wy-14,643 treated hPPARαTetOff mice. Northern analysis of genes involved in fatty acid oxidation (peroxisomal acyl-CoA oxidase, ACOX; cytochrome CYP4A family, CYP4A; liver carnitine palmitoyltransferase, LCPT) and cell cycle control (proliferating cellular nuclear antigen, PCNA; cyclin-dependent kinase, CDK).
Table 3 . Incidences of liver tumours in hPPARαTetOff and wild-type (mPPARα) mice treated with Wy-14,643.
Mice were fed 0.1% (w/w) Wy-14,643 for up to 44 weeks.
| Genotype | Treatment | Mice number | Tumors (%) |
|---|---|---|---|
| hPPARα | Control | 10 | 0 (0) |
| hPPARα | Wy-14,643 | 20a | 1 (5) |
| mPPARα | Control | 9 | 0 (0) |
| mPPARα | Wy-14,643 | 7a | 5 (71)* |
Mice that became moribund before termination of the experiment were euthanized and the gross pathology and histological examination of the liver was carried out to score tumour incidences.
P<0.01 versus values of hPPARα given Wy-14,643.
Recently a second PPARα-humanized mouse line was generated using a PAC clone containing the entire human PPARα gene and was designated hPPARαPAC mice (Yang et al., 2008). hPPARαPAC mice treated with the fibrate drug fenofibrate demonstrated peroxisome proliferation, lowering of fasted serum total triglycerides and induction of PPARα target genes encoding enzymes involved in fatty acid metabolism. In contrast to liver-specific expression of the hPPARαTetOff mice, the hPPARαPAC mice revealed PPARα target gene induction in extrahepatic tissues (kidney and heart) as well as in the liver. The hPPARαPAC mice provide an ideal in vivo platform for use in drug development and a model for human risk assessment to peroxisome proliferators, and also to elucidate the mechanisms of species differences mediated by PPARα.
Other humanized transgenic mice
Multiple reports of other transgenic mice expressing human P450s and xenobiotic receptors exist and are beyond the scope of this review. These transgenic mice were generated using similar techniques as discussed in this article. Examples and appropriate references are described in a previous review (Gonzalez and Yu, 2006) and include transgenic models of human CYP1B1, CYP1A2, CYP4B1, CYP3A7, CYP7A1, CYP27, CYP2C18/19 (Lofgren et al., 2008), CYP19, AHR and CAR. Transgenic mice containing the regulatory promoter for the human CYP1A2, CYP3A4 and CYP27B1 (Hendrix et al., 2004) have been generated to investigate the regulation of the P450 gene in mice (Gonzalez and Yu, 2006). However, of particular interest is the recent development of chimeric mice with humanized liver and their use for predictive drug metabolism and pharmacokinetic studies (Katoh et al., 2008). Instead of selectively introducing one or two transgenes as described in the humanized models above, this approach results in the almost complete repopulation of the mouse liver with human hepatocytes and so introduces all human drug metabolizing enzymes that are contained within the donor hepatocytes. An urokinase-type plasminogen activator+/+/severe combined immunodeficient transgenic mouse line was established that exhibits a low degree of rejection of transplanted cells and this model was used to develop a nearly complete humanized liver by transplantation of the liver with 80–90% human hepatocytes (Tateno et al., 2004). In the liver of chimeric mice, human phase I enzymes (including P450s), phase II enzymes and drug transporters were shown to be expressed and have similar drug metabolizing capacity as the donor hepatocytes, as well as P450 induction potencies of rifampicin, rifabutin and 3-mehylcholanthrene (Katoh et al., 2008). Human specific metabolites were detected in the serum, suggesting the use of these chimeric mice as human drug metabolism and pharmacokinetic models (Katoh et al., 2008). Using these chimeric mice to study drug metabolism is advantageous as the combined effects of not just human P450s, but human conjugation enzymes and human drug transporters can be collectively studied. Since genetic polymorphisms from the donor can be retained in chimeric mice, this also allows these human genetic polymorphisms to be studied in vivo. Even though generation of these chimeric mice requires a relatively small (5 × 105 cells) input of healthy human hepatocytes, acquisition of suitable donor hepatocytes remains a limiting factor in establishing this model. The donor population is largely heterogeneous with respect to age, gender, disease status, drugs/medication taken or exposure to xenobiotics and therefore variability amongst hepatocyte preparations can result. These factors should be carefully considered when analyzing studies using such donor hepatocytes to establish chimeric mouse humanized livers. Another limiting factor is unlike the transgenic humanized models, which once established are maintained through breeding, chimeric mice must be generated de novo as needed (Muruganandan and Sinal, 2008). As this approach results in only humanization of the liver, other extra-hepatic organs still contain the endogenous mouse enzymes and so the contribution of a humanized intestine, for example, cannot be examined. This is not the case in the transgenic humanized models, where the human transgene can be expressed in the liver and intestine. Therefore, pharmacokinetics of oral drug administration in humans cannot be effectively modeled using these chimeric mice as only the contribution of the liver to human drug metabolism can be recapitulated. However, the generation of chimeric mice with humanized livers does represent an important advance in the human drug metabolism research and with further validation this model should prove to be an immensely valuable tool for predicting human drug metabolism and in drug development.
Conclusions
The studies reviewed herein show that P450 and xenobiotic receptor humanized mice can be generated using cDNAs or complete human genes introduced into mice with BAC or PAC clones and breeding them into the respective knockout background. These transgenic mouse models were characterized and shown to accurately express the corresponding enzymes and exhibit catalytic activities at levels comparable to or higher to that found in human tissues.
Perspectives
The development of these humanized mouse models provides an approach to overcome species differences in drug metabolism, which are in part associated with differences in the genes encoding P450 drug metabolizing enzymes or xenobiotic receptors. These mice have proven to be very stable, with some lines having been maintained for several years without loss of the transgene. Such humanized mouse models have shown to be valuable tools for studying the function and regulation of P450s and xenobiotic receptors in the whole animal system. They offer broad applications in the evaluation and prediction of human drug and carcinogen metabolism, pharmacokinetics, pharmacodynamics, drug toxicities and DDIs. These novel mouse lines also offer the opportunity to investigate endogenous substrates for P450s, screen for nuclear receptor ligands, identify target genes and to define physiological and pharmacological pathways mediated by xenobiotic nuclear receptors. Combinations of these mouse lines, for example CYP3A4/CYP2D6 (Felmlee et al., 2008) or CYP3A4/PXR double humanized mice could ultimately provide even better models for preclinical drug development.
Abbreviations
- CYP450
cytochromes P450
- PXR
pregnane-X-receptor
- PPARα
peroxisome proliferator-activated receptor α
- CAR
constitutive androstane receptor
- PCN
Pregnelolone-16α-carbontirile
- BAC
bacterial artificial chromosome
- PAC
P1 phage artificial chromosome
- AHR
arylhydrocarbon receptor
- 3-MC
3-methylcholanthrene
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- APAP
acetaminophen
- NAPQI
N-acetyl-p-benzoquinone imine
- DDIs
drug-drug interactions
- Wy-14,643
[[4-chloro-6-[(2,3-dimethylphenyl)amino]-2-pyrimidinyl]thio]-acetic acid
References
- Backman JT, Olkkola KT, Neuvonen PJ. Rifampin drastically reduces plasma concentrations and effects of oral midazolam. Clin Pharmacol Ther. 1996;59:7–13. doi: 10.1016/S0009-9236(96)90018-1. [DOI] [PubMed] [Google Scholar]
- Bogaards JJ, Bertrand M, Jackson P, Oudshoorn MJ, Weaver RJ, van Bladeren PJ, Walther B. Determining the best animal model for human cytochrome P450 activities: a comparison of mouse, rat, rabbit, dog, micropig, monkey and man. Xenobiotica. 2000;30:1131–1152. doi: 10.1080/00498250010021684. [DOI] [PubMed] [Google Scholar]
- Bolt HM. Rifampicin, a keystone inducer of drug metabolism: from Herbert Remmer's pioneering ideas to modern concepts. Drug Metab Rev. 2004;36:497–509. doi: 10.1081/dmr-200033432. [DOI] [PubMed] [Google Scholar]
- Brown PJ, Stuart LW, Hurley KP, Lewis MC, Winegar DA, Wilson JG, Wilkison WO, Ittoop OR, Willson TM. Identification of a subtype selective human PPARalpha agonist through parallel-array synthesis. Bioorg Med Chem Lett. 2001;11:1225–1227. doi: 10.1016/s0960-894x(01)00188-3. [DOI] [PubMed] [Google Scholar]
- Caldwell J. The current status of attempts to predict species differences in drug metabolism. Drug Metab Rev. 1981;12:221–237. doi: 10.3109/03602538108994030. [DOI] [PubMed] [Google Scholar]
- Caro AA, Cederbaum AI. Oxidative stress, toxicology, and pharmacology of CYP2E1. Annu Rev Pharmacol Toxicol. 2004;44:27–42. doi: 10.1146/annurev.pharmtox.44.101802.121704. [DOI] [PubMed] [Google Scholar]
- Cheung C, Akiyama TE, Ward JM, Nicol CJ, Feigenbaum L, Vinson C, Gonzalez FJ. Diminished hepatocellular proliferation in mice humanized for the nuclear receptor peroxisome proliferator-activated receptor alpha. Cancer Res. 2004;64:3849–3854. doi: 10.1158/0008-5472.CAN-04-0322. [DOI] [PubMed] [Google Scholar]
- Cheung C, Ma X, Krausz KW, Kimura S, Feigenbaum L, Dalton TP, Nebert DW, Idle JR, Gonzalez FJ. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in mice humanized for CYP1A1 and CYP1A2. Chem Res Toxicol. 2005a;18:1471–1478. doi: 10.1021/tx050136g. [DOI] [PubMed] [Google Scholar]
- Cheung C, Yu AM, Chen CS, Krausz KW, Byrd LG, Feigenbaum L, Edwards RJ, Waxman DJ, Gonzalez FJ. Growth hormone determines sexual dimorphism of hepatic cytochrome P450 3A4 expression in transgenic mice. J Pharmacol Exp Ther. 2006;316:1328–1334. doi: 10.1124/jpet.105.094367. [DOI] [PubMed] [Google Scholar]
- Cheung C, Yu AM, Ward JM, Krausz KW, Akiyama TE, Feigenbaum L, Gonzalez FJ. The cyp2e1-humanized transgenic mouse: role of cyp2e1 in acetaminophen hepatotoxicity. Drug Metab Dispos. 2005b;33:449–457. doi: 10.1124/dmd.104.002402. [DOI] [PubMed] [Google Scholar]
- Corchero J, Granvil CP, Akiyama TE, Hayhurst GP, Pimprale S, Feigenbaum L, Idle JR, Gonzalez FJ. The CYP2D6 humanized mouse: effect of the human CYP2D6 transgene and HNF4alpha on the disposition of debrisoquine in the mouse. Mol Pharmacol. 2001a;60:1260–1267. doi: 10.1124/mol.60.6.1260. [DOI] [PubMed] [Google Scholar]
- Corchero J, Pimprale S, Kimura S, Gonzalez FJ. Organization of the CYP1A cluster on human chromosome 15: implications for gene regulation. Pharmacogenetics. 2001b;11:1–6. doi: 10.1097/00008571-200102000-00001. [DOI] [PubMed] [Google Scholar]
- Down MJ, Arkle S, Mills JJ. Regulation and induction of CYP3A11, CYP3A13 and CYP3A25 in C57BL/6J mouse liver. Arch Biochem Biophys. 2007;457:105–110. doi: 10.1016/j.abb.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Dragin N, Uno S, Wang B, Dalton TP, Nebert DW. Generation of 'humanized' hCYP1A1_1A2_Cyp1a1/1a2(−/−) mouse line. Biochem Biophys Res Commun. 2007;359:635–642. doi: 10.1016/j.bbrc.2007.05.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser GK, Spence JD, Bailey DG. Pharmacokinetic-pharmacodynamic consequences and clinical relevance of cytochrome P450 3A4 inhibition. Clin Pharmacokinet. 2000;38:41–57. doi: 10.2165/00003088-200038010-00003. [DOI] [PubMed] [Google Scholar]
- Felmlee MA, Lon HK, Gonzalez FJ, Yu AM. Cytochrome P450 expression and regulation in CYP3A4/CYP2D6 double transgenic humanized mice. Drug Metab Dispos. 2008;36:435–441. doi: 10.1124/dmd.107.018838. [DOI] [PubMed] [Google Scholar]
- Friesen MD, Kaderlik K, Lin D, Garren L, Bartsch H, Lang NP, Kadlubar FF. Analysis of DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5- b]pyridine in rat and human tissues by alkaline hydrolysis and gas chromatography/electron capture mass spectrometry: validation by comparison with 32P-postlabeling. Chem Res Toxicol. 1994;7:733–739. doi: 10.1021/tx00042a004. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Peters JM, Cattley RC. Mechanism of action of the nongenotoxic peroxisome proliferators: role of the peroxisome proliferator-activator receptor alpha. J Natl Cancer Inst. 1998;90:1702–1709. doi: 10.1093/jnci/90.22.1702. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Yu AM. Cytochrome P450 and xenobiotic receptor humanized mice. Annu Rev Pharmacol Toxicol. 2006;46:41–64. doi: 10.1146/annurev.pharmtox.45.120403.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JC, Hall SD, Jones DR, VandenBranden M, Wrighton SA. Regioselective biotransformation of midazolam by members of the human cytochrome P450 3A (CYP3A) subfamily. Biochem Pharmacol. 1994;47:1643–1653. doi: 10.1016/0006-2952(94)90543-6. [DOI] [PubMed] [Google Scholar]
- Granvil CP, Yu AM, Elizondo G, Akiyama TE, Cheung C, Feigenbaum L, Krausz KW, Gonzalez FJ. Expression of the human CYP3A4 gene in the small intestine of transgenic mice: in vitro metabolism and pharmacokinetics of midazolam. Drug Metab Dispos. 2003;31:548–558. doi: 10.1124/dmd.31.5.548. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Cytochromes p450, drugs, and diseases. Mol Intervent. 2003;3:194–204. doi: 10.1124/mi.3.4.194. [DOI] [PubMed] [Google Scholar]
- Hendrix I, Anderson P, May B, Morris H. Regulation of gene expression by the CYP27B1 promoter-study of a transgenic mouse model. J Steroid Biochem Mol Biol. 2004;89–90:139–142. doi: 10.1016/j.jsbmb.2004.03.093. [DOI] [PubMed] [Google Scholar]
- Holdener M, Hintermann E, Bayer M, Rodrigo E, Rhode A, Johnson EF, Gonzalez FJ, Manns M, von Herrath M, Christen U. Breaking tolerance to the human autoantigen cytochrome P450 2D6 by virus infection. J Exp Med. 2008;205:1409–1422. doi: 10.1084/jem.20071859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Dalton TP, Jin L, Wang B, Tsuneoka Y, Shertzer HG, Deka R, Nebert DW. Toward the evaluation of function in genetic variability: characterizing human SNP frequencies and establishing BAC-transgenic mice carrying the human CYP1A1_CYP1A2 locus. Hum Mutat. 2005;25:196–206. doi: 10.1002/humu.20134. [DOI] [PubMed] [Google Scholar]
- Jones SA, Moore LB, Shenk JL, Wisely GB, Hamilton GA, McKee DD, Tomkinson NC, LeCluyse EL, Lambert MH, Willson TM, Kliewer SA, Moore JT. The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- Katoh M, Tateno C, Yoshizato K, Yokoi T. Chimeric mice with humanized liver. Toxicology. 2008;246:9–17. doi: 10.1016/j.tox.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Kessova I, Cederbaum AI. CYP2E1: biochemistry, toxicology, regulation and function in ethanol-induced liver injury. Curr Mol Med. 2003;3:509–518. doi: 10.2174/1566524033479609. [DOI] [PubMed] [Google Scholar]
- Kliewer SA. The nuclear pregnane X receptor regulates xenobiotic detoxification. J Nutr. 2003;133:2444S–2447S. doi: 10.1093/jn/133.7.2444S. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, Perlmann T, Lehmann JM. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Le Marchand L, Hankin JH, Wilkens LR, Pierce LM, Franke A, Kolonel LN, Seifried A, Custer LJ, Chang W, Lum-Jones A, Donlon T. Combined effects of well-done red meat, smoking, and rapid N-acetyltransferase 2 and CYP1A2 phenotypes in increasing colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:1259–1266. [PubMed] [Google Scholar]
- Lee SS, Buters JT, Pineau T, Fernandez-Salguero P, Gonzalez FJ. Role of CYP2E1 in the hepatotoxicity of acetaminophen. J Biol Chem. 1996;271:12063–12067. doi: 10.1074/jbc.271.20.12063. [DOI] [PubMed] [Google Scholar]
- Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AP, Kaminski DL, Rasmussen A. Substrates of human hepatic cytochrome P450 3A4. Toxicology. 1995;104:1–8. doi: 10.1016/0300-483x(95)03155-9. [DOI] [PubMed] [Google Scholar]
- Lieber CS. Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev. 1997;77:517–544. doi: 10.1152/physrev.1997.77.2.517. [DOI] [PubMed] [Google Scholar]
- Lin D, Kaderlik KR, Turesky RJ, Miller DW, Lay JO, Jr., Kadlubar FF. Identification of N-(Deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine as the major adduct formed by the food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, with DNA. Chem Res Toxicol. 1992;5:691–697. doi: 10.1021/tx00029a016. [DOI] [PubMed] [Google Scholar]
- Lofgren S, Baldwin RM, Hiratsuka M, Lindqvist A, Carlberg A, Sim SC, Schulke M, Snait M, Edenro A, Fransson-Steen R, Terelius Y, Ingelman-Sundberg M. Generation of mice transgenic for human CYP2C18 and CYP2C19. Characterization of the sexually dimorphic gene-and enzyme expression. Drug Metab Dispos. 2008;36:955–962. doi: 10.1124/dmd.107.019349. [DOI] [PubMed] [Google Scholar]
- Ma X, Shah Y, Cheung C, Guo GL, Feigenbaum L, Krausz KW, Idle JR, Gonzalez FJ. The PREgnane X receptor gene-humanized mouse: a model for investigating drug-drug interactions mediated by cytochromes P450 3A. Drug Metab Dispos. 2007;35:194–200. doi: 10.1124/dmd.106.012831. [DOI] [PubMed] [Google Scholar]
- Mahgoub A, Idle JR, Dring LG, Lancaster R, Smith RL. Polymorphic hydroxylation of Debrisoquine in man. Lancet. 1977;2:584–586. doi: 10.1016/s0140-6736(77)91430-1. [DOI] [PubMed] [Google Scholar]
- Martignoni M, Groothuis GM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin Drug Metab Toxicol. 2006;2:875–894. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- Masubuchi Y, Iwasa T, Hosokawa S, Suzuki T, Horie T, Imaoka S, Funae Y, Narimatsu S. Selective deficiency of debrisoquine 4-hydroxylase activity in mouse liver microsomes. J Pharmacol Exp Ther. 1997;282:1435–1441. [PubMed] [Google Scholar]
- Morgan K, French SW, Morgan TR. Production of a cytochrome P450 2E1 transgenic mouse and initial evaluation of alcoholic liver damage. Hepatology. 2002;36:122–134. doi: 10.1053/jhep.2002.33720. [DOI] [PubMed] [Google Scholar]
- Morimura K, Cheung C, Ward JM, Reddy JK, Gonzalez FJ. Differential susceptibility of mice humanized for peroxisome proliferator-activated receptor alpha to Wy-14,643-induced liver tumorigenesis. Carcinogenesis. 2006;27:1074–1080. doi: 10.1093/carcin/bgi329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruganandan S, Sinal C. Mice as Clinically Relevant Models for the Study of Cytochrome P450-dependent Metabolism. Clin Pharmacol Ther. 2008;83:818–828. doi: 10.1038/clpt.2008.50. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SMG, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Palmer CN, Hsu MH, Griffin KJ, Raucy JL, Johnson EF. Peroxisome proliferator activated receptor-alpha expression in human liver. Mol Pharmacol. 1998;53:14–22. [PubMed] [Google Scholar]
- Peters JM, Cattley RC, Gonzalez FJ. Role of PPAR alpha in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis. 1997;18:2029–2033. doi: 10.1093/carcin/18.11.2029. [DOI] [PubMed] [Google Scholar]
- Raucy J, Warfe L, Yueh MF, Allen SW. A cell-based reporter gene assay for determining induction of CYP3A4 in a high-volume system. J Pharmacol Exp Ther. 2002;303:412–423. doi: 10.1124/jpet.102.038653. [DOI] [PubMed] [Google Scholar]
- Razzouk C, Batardy-Gregoire M, Roberfroid M. Metabolism of N-hydroxy-2-acetylaminofluorene and N-hydroxy-2-aminofluorene by guinea pig liver microsomes. Cancer Res. 1982;42:4712–4718. [PubMed] [Google Scholar]
- Reddy JK. Nonalcoholic steatosis and steatohepatitis. III. Peroxisomal beta-oxidation, PPAR alpha, and steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G1333–G1339. doi: 10.1152/ajpgi.2001.281.6.G1333. [DOI] [PubMed] [Google Scholar]
- Reddy JK, Krishnakantha TP. Hepatic peroxisome proliferation: induction by two novel compounds structurally unrelated to clofibrate. Science. 1975;190:787–789. doi: 10.1126/science.1198095. [DOI] [PubMed] [Google Scholar]
- Schuetz EG, Schmid W, Schutz G, Brimer C, Yasuda K, Kamataki T, Bornheim L, Myles K, Cole TJ. The glucocorticoid receptor is essential for induction of cytochrome P-4502B by steroids but not for drug or steroid induction of CYP3A or P-450 reductase in mouse liver. Drug Metab Dispos. 2000;28:268–278. [PubMed] [Google Scholar]
- Sher A, Rahman MA. Enterohepatic recycling of estrogen and its relevance with female fertility. Arch Pharm Res. 2000;23:513–517. doi: 10.1007/BF02976582. [DOI] [PubMed] [Google Scholar]
- Sher T, Yi HF, McBride OW, Gonzalez FJ. cDNA cloning, chromosomal mapping, and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry. 1993;32:5598–5604. doi: 10.1021/bi00072a015. [DOI] [PubMed] [Google Scholar]
- Snyderwine EG. Mammary gland carcinogenesis by food-derived heterocyclic amines: metabolism and additional factors influencing carcinogenesis by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Environ Mol Mutagen. 2002;39:165–170. doi: 10.1002/em.10053. [DOI] [PubMed] [Google Scholar]
- Tateno C, Yoshizane Y, Saito N, Kataoka M, Utoh R, Yamasaki C, Tachibana A, Soeno Y, Asahina K, Hino H, Asahara T, Yokoi T, Furukawa T, Yoshizato K. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol. 2004;165:901–912. doi: 10.1016/S0002-9440(10)63352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turteltaub KW, Dingley KH, Curtis KD, Malfatti MA, Turesky RJ, Garner RC, Felton JS, Lang NP. Macromolecular adduct formation and metabolism of heterocyclic amines in humans and rodents at low doses. Cancer Lett. 1999;143:149–155. doi: 10.1016/s0304-3835(99)00116-0. [DOI] [PubMed] [Google Scholar]
- van Herwaarden AE, Smit JW, Sparidans RW, Wagenaar E, van der Kruijssen CM, Schellens JH, Beijnen JH, Schinkel AH. Midazolam and cyclosporin a metabolism in transgenic mice with liver-specific expression of human CYP3A4. Drug Metab Dispos. 2005;33:892–895. doi: 10.1124/dmd.105.004721. [DOI] [PubMed] [Google Scholar]
- van Herwaarden AE, Wagenaar E, van der Kruijssen CM, van Waterschoot RA, Smit JW, Song JY, van der Valk MA, van Tellingen O, van der Hoorn JW, Rosing H, Beijnen JH, Schinkel AH. Knockout of cytochrome P450 3A yields new mouse models for understanding xenobiotic metabolism. J Clin Invest. 2007;117:3583–3592. doi: 10.1172/JCI33435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman DJ, O'Connor C. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol. 2006;20:2613–2629. doi: 10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- Willson TM, Kliewer SA. PXR, CAR and drug metabolism. Nat Rev Drug Discov. 2002;1:259–266. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- Wolbold R, Klein K, Burk O, Nussler AK, Neuhaus P, Eichelbaum M, Schwab M, Zanger UM. Sex is a major determinant of CYP3A4 expression in human liver. Hepatology. 2003;38:978–988. doi: 10.1053/jhep.2003.50393. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-Tetri BA, Brunt EM, Guzelian PS, Evans RM. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- Yang Q, Nagano T, Shah Y, Cheung C, Ito S, Gonzalez FJ. The PPAR alpha-humanized mouse: a model to investigate species differences in liver toxicity mediated by PPAR alpha. Toxicol Sci. 2008;101:132–139. doi: 10.1093/toxsci/kfm206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, Fukamachi K, Krausz KW, Cheung C, Gonzalez FJ. Potential role for human cytochrome P450 3A4 in estradiol homeostasis. Endocrinology. 2005;146:2911–2919. doi: 10.1210/en.2004-1248. [DOI] [PubMed] [Google Scholar]
- Yu AM, Idle JR, Herraiz T, Kupfer A, Gonzalez FJ. Screening for endogenous substrates reveals that CYP2D6 is a 5-methoxyindolethylamine O-demethylase. Pharmacogenetics. 2003;13:307–319. doi: 10.1097/01.fpc.0000054094.48725.b7. [DOI] [PubMed] [Google Scholar]
- Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:23–37. doi: 10.1007/s00210-003-0832-2. [DOI] [PubMed] [Google Scholar]