Abstract
Interaction between the cytochrome caa3 respiratory chain complex and F1F0-ATP synthase from extremely alkaliphilic Bacillus pseudofirmus OF4 has been hypothesized to be required for robust ATP synthesis by this alkaliphile under conditions of very low protonmotive force. Here, such an interaction was probed by differential scanning calorimetry (DSC) and by saturation transfer electron paramagnetic resonance (STEPR). When the two purified complexes were embedded in phospholipids vesicles individually [(caa3) × PL, (F1F0) × PL)] or in combination [(caa3+F1F0) × PL] and subjected to DSC analysis, they underwent exothermic thermodenaturation with transition temperatures at 69, 57, and 46/75 °C, respectively. The enthalpy change, ΔH, (−8.8 Kcal/mmol) of protein-phospholipid vesicles containing both cytochrome caa3 and F1F0 was smaller than that (−12.4 Kcal/mmol) of a mixture of protein-phospholipid vesicles formed from each individual electron transfer complex [(caa3 × PL) + (F1F0 × PL)]. The rotational correlation time of spin-labeled caa3 (65 µs) in STEPR studies increased significantly when the complex was mixed with F1F0 prior to being embedded in phospholipids vesicles (270 µs). When the complexes were reconstituted separately then mixed together, or either mitochondrial cytochrome bc1 or F1F0 was substituted for the alkaliphile F1F0, the correlation time was unchanged (65–70 µs). Varying the ratio of the two alkaliphile complexes in both the DSC and STEPR experiments indicated that the optimal stoichiometry is 1:1. These results demonstrate a specific interaction between the cytochrome caa3 and F1F0-ATP synthase from B. pseudofirmus OF4 in a reconstituted system. They support the suggestion that physical association between these complexes may contribute to sequestered proton transfers during alkaliphile oxidative phosphorylation at high pH.
INTRODUCTION
During oxidative phosphorylation (OXPHOS)2 in mitochondria and most bacteria, energy from the exergonic oxidation-reduction reactions of the electron transport chain is conserved in an electrochemical gradient of protons across the membrane, alkaline and negative inside relative to outside. This Δp is used to energize ATP synthesis by the proton-coupled ATP synthase (1). Protons moving downhill through the integral membrane a- and c-subunits of the F0 sector of the synthase lead to rotation of the c-subunit rotor and the associated γ-subunit, causing conformational changes in the catalytic F1 sector that result in ATP synthesis (2,3). The nature and involvement of the Δp as the crucial intermediate form of energy in OXPHOS was a major contribution of Mitchell’s chemiosmotic hypothesis (1). The chemiosmotic formulation further posited that the proton path from the proton-extruding respiratory chain complexes to the ATP synthase was through the bulk liquid phase outside the mitochondrion or bacterial cell. By contrast, Williams and others proposed that greater efficiency of energy-coupling would be achieved by transfer or protons without full equilibration with the bulk phase (4–7). Williams considered the possibility of actual protein-protein interactions, perhaps facilitated by special adaptations of the proteins and participation of membrane phospholipids (6). Other proposals have involved proton trapping at the outside membrane surface that creates a delocalized surface Δp that is functionally larger than the bulk force and obviates the need for specific adaptations of participating respiratory chain elements or the ATP synthase (6,8,9). The expectation is that the drugs or genetic variations that effect OXPHOS will depend upon the specifics of the proton path, i.e. (i) involvement of membrane lipid and protein surfaces versus (ii) additional involvement of protein-protein interactions versus (iii) only a bulk energization pathway of protons. Therefore, further definition of the proton path is important.
OXPHOS by alkaliphilic Bacillus species that carry out proton-coupled ATP synthesis at pH values ≥ 10.5 has been an important experimental system for assessing models of the proton path (10–12). As studied in extremely alkaliphilic Bacillus pseudofirmus OF4, two properties of alkaliphile OXPHOS suggest that above pH 9.5, ATP synthesis requires the bulk transmembrane electrical potential, the ΔΨ, but also requires specific adaptations of the synthase that support proton transfer from the respiratory chain to the synthase without full equilibration with the bulk (13–15). First, OXPHOS is more robust at pH 10.5 than at pH 7.5 although the bulk Δp is about 3-times lower at the higher pH because of the large, chemiosmotically adverse ΔpH (~ 2.3 pH units, acid in relative to out) that is sustained by the Na+/H+ antiporter-dependent pH homeostasis mechanism (13). This capacity for OXPHOS at pH 10.5 depends upon alkaliphile-specific sequence features of the F0-ATP synthase. Mutation of two of these features to the Bacillus consensus sequence resulted in a fully functional enzyme that supported OXPHOS at pH 7.5 but not at pH 10.5 (15). Second, large artificially imposed diffusion potentials fail to energize ATP synthesis above pH 9.5, where non-fermentative growth and respiration-dependent OXPHOS are optimal (13). This property reflects a blockage of proton flux both to and from the bulk phase at pH ≥ 9.5 that protects against cytoplasmic alkalinization during sudden exposure to high pH. Two residues of the F0-ATP synthase a-subunit have been shown to participate in this pH-dependent proton “gating”, one of which is also required for ATP synthesis at high pH (15). These findings led us to propose that a kinetically sequestered proton path from the respiratory chain to the ATP synthase at high pH is supported by properties of the participating complexes as well as the membrane and may involve dynamic protein-protein interactions between the ATP synthase and one of the terminal oxidases of the respiratory chain, the proton-pumping caa3 oxidase. The expression of this oxidase is up-regulated by either high pH or low Δp resulting from protonophore treatment (16,17) and mutations that reduce cytochrome caa3 activity also prevent non-fermentative growth at high pH (18). Moreover, alignments show that alkaliphile cytochrome caa3 has alkaliphile-specific sequence features that might contribute to interactions with the ATP synthase (e.g. an alkaliphile-specific T79 of subunit I and an unusually acidic region in subunit II in the putative proton path through the oxidase). Before initiating mutagenesis study of such sequence features of cytochrome caa3, we sought evidence for the putative protein-protein interactions.
Using methods of differential scanning calorimetry (DSC) and saturation transfer electron paramagnetic resonance (STEPR), protein-protein interactions were detected between the bovine heart mitochondrial cytochrome c oxidase and F1F0-ATP synthase in the native membrane state (19), but the bovine system does not lend itself to analysis of mutations in which in vivo OXPHOS patterns are correlated with the capacity for interaction. In this study, we use these methods to study the interaction between cytochrome caa3 and ATP synthase from alkaliphilic B. pseudofirmus OF4 to probe protein-protein interactions of these alkaliphile complexes. The DSC study is based on the assumption that if two lipoprotein complexes exist separately in a phospholipid vesicle, no difference in thermotropic properties will be observed between protein-phospholipid vesicles formed from a mixture of two complexes and a mixture of protein-phospholipid vesicles formed individually from each complex. Differences in the thermodenaturation temperatures and enthalpy changes would suggest formation of a physical complex between cytochrome caa3 and ATP synthase. In the STEPR study, the formation of a physical complex between cytochrome caa3 and ATP synthase will be indicated by an increase of rotational correlation time of spin-labeled cytochrome caa3. Herein, we report experimental details and results of DSC and STEPR studies with cytochrome caa3 and ATP synthase embedded in phospholipid vesicles. The results of DSC and STEPR indicate that cytochrome caa3 does interact directly with the ATP synthase in a reconstituted membrane vesicle system.
MATERIAL AND METHODS
Materials
Spin label, 4-maleimide-2,2,6,6-tetramethyl-1-piperidinyl-N-oxyl (MSL), cytochrome c (horse heart, type III) and sodium cholate were purchased from Sigma. N-dodecyl-β-D-maltoside (DM) and N-octyl-β-D-glucoside were from Anatrace. Asolection was obtained from Associated Concentrates, Inc., and purified according to the procedure reported by Kagama et. al (20). Centriprep-30 and Centricon-30 were bought from Amicon. Other chemicals were of the highest purity commercially available.
Enzyme Preparations
The F1F0-ATP synthase from alkaliphilic B. pseudofirmus OF4 was purified from everted membrane vesicles isolated from pH 10.5-grown cells essentially as reported previously (21), except that the ammonium sulfate fraction used for sucrose gradient centrifugation was the pellet obtained between 37% and 60% saturation (P37–60). During purification, the hydrolytic activity of the preparations was assayed by the phosphate formation method of LeBel (22) in the presence of octylglucoside and Na2SO3 (21). The specific activity of preparations ranged from 30–40 µmoles Pi released min−1 mg protein−1.
The four-subunit cytochrome caa3 was purified from the resulting supernatant (S37–60) by the following modifications of the original protocol (17). During purification, the activity of the preparations was monitored by assays of TMPD oxidase in a 20 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0 buffer using 1 mM TMPD (23). Unless otherwise specified, all the purification steps were carried out at 4 C. The S37–60 was sequentially brought to 70% and 80% ammonium sulfate saturation and the P70–80 was removed by ultracentrifugation. The S70–80 was diluted with 4 volumes of buffer A (10% glycerol, 10 mM NaPi, pH 8.0, 1 mM EDTA, 0.05% dodecyl maltoside, 0.2 mM phenylmethylsulfonyl fluoride) and loaded on a hydroxyapatite column (Bio-Rad) equilibrated in the same buffer. After washing the column with two bed volumes of buffer A, the column was equilibrated at room temperature and washed sequentially with 0.2 (2 bed volumes), 0.4 (3 bed volumes) and 0.6 M NaPi, pH 8.0 in buffer A (3–4 bed volumes). The 0.6 M fraction, containing the bulk of the cytochrome caa3, was diluted with 6 volumes of buffer B (10% glycerol, 20 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.05% dodecyl maltoside, and 0.2 mM phenylmethylsulfonyl fluoride) and loaded on a Q sepharose anion exchange column equilibrated in the same buffer. This column was washed with 5 bed volumes of buffer B and then sequentially with 0.2 M (7 bed volumes), 0.3 M (10 bed volumes), 0.4 M (7 bed volumes), and 0.5 M NaCl (4 bed volumes) in buffer B. Cytochrome caa3 eluted principally in the 0.4 M fraction with the 0.5 M fraction containing about 30% of the eluted oxidase. The 0.4 M fraction was concentrated and used in the reconstitution experiments. The heme a + a3 content of the purified oxidase preparations was >9.0 nmoles mg protein−1, based on an extinction coefficient of 20.5 mM at A600–622 (from the mitochondrial value given in (24)).
Preparation of Maleimide Spin-Labeled (MSL) Cytochrome caa3
Alkaliphilic B. pseudofirmus OF4 cytochrome caa3, 20 mg/ml in 20 mM Tris-Cl, pH 8.0, containing 0.35 M NaCl, 1 mM EDTA, 0.1 mM PMSF, 0.05 % dodecyl maltoside, and 10 % glycerol was incubated with a 5 molar excess of MSL for 1 hour at room temperature. The stock solution of MSL (10 mM) was made in 10 mM Tris-HCl /sucrose buffer, pH 8.0, containing 20% methanol. After incubation, the unreacted MSL was removed by passage through a D-Salt™ Excellulose Desalting Column from Pierce, equilibrated with 10 mM Tris-HCl buffer, containing 0.05 % dodecyl maltoside. Fractions containing MSL-cytochrome caa3 were pooled and concentrated by Centriprep-30 and Centrico-30 to a protein concentration of approximately 30 mg/ml. MSL-cytochrome caa3 obtained by this method was verified to contain no free spin-label by the conventional EPR spectra.
Preparation of Cytochrome caa3 and F1F0 Complex-Phospholipid Vesicles
The protein-phospholipid vesicles were prepared by the cholate-dialysis method reported by Racker (25). Cytochrome caa3 complex, with or without MSL labeling, alone or in combination with F1F0, at a protein concentration of approximately 30 mg/ml, was mixed with an asolectin micellar solution (20 mg/ml in 50 mM phosphate buffer, pH 7.4) and a sodium cholate solution [20% (w/v) in water]. The final solution contained 7 mg/ml protein, 10 mg/ml sodium cholate, and 10.5 mg/ml asolectin. After incubation at 4°C for 60 min, the solution was dialyzed overnight against 500 volumes of 50 mM phosphate buffer, pH 7.4, with four changes of buffer to form vesicles. The protein-phospholipid vesicles formed were collected by centrifugation at 80,000 g for 1 h and the precipitates were re-suspended together in 50 mM phosphate buffer, pH 7.4, to an appropriate protein concentration depending on the assay to be used. The suspensions were used for the DSC and STEPR experiments. Although not anticipated, the vesicles prepared by the cholate-dialysis procedure described above are quite uniform in sizes, with diameters between 20 to 40 nm (28). The relatively high protein to phospholipid ratio used might contribute to this uniformity. The vesicles may not be completely sealed as judged by their low oxidation control index (data not shown). This, however, does not present any complication on the work of DSC and STEPR as the essential lipid environment of the protein complex is provided.
Differential Scanning Calorimetry
All calorimetric measurements were performed with a CSC 6100 NanoII DSC from Calorimetry Science Corp. The reference and sample solutions were carefully degassed under vacuum for 15 min prior to use. A 0.50-ml sample in 50 mM K+/Na+ phosphate buffer, pH 7.4, was placed in the sample capillary cell, and the same amount of buffer was placed in the reference capillary cell. All DSC scans reported in this study were run at a rate of 2°C/min. After the first scan, the samples were cooled to the original temperature and rescanned. Since after the first scan the protein was completely and irreversibly denatured, no thermotransition peaks were observed in the second scan and the second scan could be used as a baseline. All thermodynamic analyses were carried out according to the program known as CpCal from the Nano DSC program group.
EPR Measurements
All EPR measurements were made with a Bruker EMX EPR spectrometer, using an aqueous quartz flat cell. The temperature of the microwave cavity was controlled by circulation of cooled nitrogen gas from a modified variable temperature housing assembly equipped with an electric temperature sensor. Conventional EPR spectra were recorded with instrument settings as follows: field modulation frequency, 100 kHz; modulation amplitude, 8 G; microwave frequency, 9.757 GHz; microwave power, 10.78 mW; time constant, 1310.72 ms. Saturation transfer EPR spectra were recorded using the same instrument settings as those described by Thomas et al. (26) and Poore et al. (27). A field modulation of 8 G and microwave frequency of 9.757 GHz were employed with phase-sensitive detection at 100 Hz (second harmonic) 90° out of phase. Incident microwave power was 107.80 mW. The phase was adjusted to minimize the second harmonic signal. Approximate rotational correlation time (τ2) was obtained from the ratio of the two field lines (L”/L). The calibration curve of Thomas et al.(26). derived from isotropic tumbling of MSL-labeled hemoglobin was used in the calculation.
Other Analytical Methods
Protein concentration was determined by the biuret method, using bovine serum albumin as the standard (assuming 1 mg/ml has an A279 of 0.667). Absorption spectra were measured in a Shimadzu UV-2401 PC spectrophotometer.
RESULTS AND DISCUSSION
Thermotropic Properties of Cytochrome caa3 and F1F0-synthase Embedded in Phospholipid Vesicles
To unambiguously study the interaction between cytochrome caa3 and F1F0-synthase from alkaliphilic B. pseudofirmus OF4, DSC studies were carried out with both complexes embedded in phospholipid (asolectin) vesicles, because these enzymes in protein-phospholipid vesicles would have an environment similar to that in membrane. The isolated complexes, singly or in combination, were embedded in phospholipids vesicles by the cholate dialysis method (25). A constant phospholipid to protein ratio of 1.5 was used. The ratio between F1F0-synthase and cytochrome caa3 varied from 0 to 1.5. If the two lipoprotein complexes have no specific interaction, then no difference in DSC characteristics should be observed between phospholipids vesicles embedded with a mixture of two complexes and a mixture of phospholipids vesicles embedded with one or the other complex, i.e. differences in the thermodenaturation temperatures (Tm) and enthalpy changes (ΔH) would suggest formation of a physical complex between these two lipoproteins. The protocol for vesicle preparation included a fixed incubation period of 60 minutes for the single or mixed complexes in solution before the reconsitution into vesicles. It is therefore possible that interactions between complexes during this period contribute to any overall interactions that are observed. However, any such contribution is likely to be minor since earlier assays of interactions of these two alkaliphile complexes in solution were negative (D. Hicks, unpublished data).
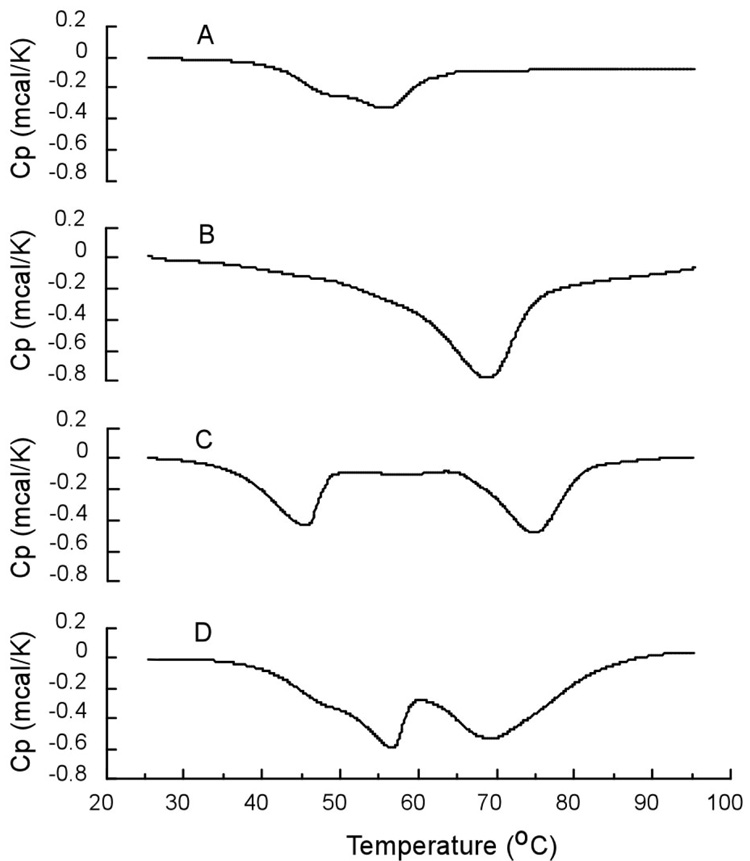
Figure 1 shows the differential scanning calorimetric curves of alkaliphilic B. pseudofirmus OF4 F1F0-synthase and cytochrome caa3 embedded in phospholipids singly or in combination. When F1F0-synthase was embedded into phospholipid vesicles and subjected to DSC analysis, an exothermic peak at 57.2 °C with a small shoulder at 45 °C and an enthalpy change of −7.3 Kcal/mmol of protein was observed (see Fig. 1A). Purified cytochrome caa3 also showed a single transition with ΔH = −17.8 Kcal/mmol of protein and Tm = 68.5 °C when it was embedded into phospholipids vesicles (Fig.1B). As expected, when the mixture of proteinphospholipid vesicles formed individually from F1F0-synthase and cytochrome caa3 was subjected to DSC analysis under identical conditions, two exothermic transient peaks at 56.8 and 68.9 °C with the ΔH of −12.4 Kcal/mmol of protein were observed (see Fig. 1D). The data from the mixed vesicles equaled the sum of the thermotropic properties of F1F0-synthase-phospholipid vesicles and cytochrome caa3-phospholipid vesicles. By contrast, the protein-phospholipid vesicles formed from a mixture of F1F0-synthase and cytochrome caa3 exhibited Tm1 = 45.5 °C and Tm2 = 74.6 °C with ΔH = −8.8 Kcal/mmol of protein (see Fig. 1C). These data for the embedded mixture are significantly different from those observed in a mixture of phospholipids vesicles embedded individually with F1F0-synthase or cytochrome caa3, suggesting that there is an interaction between these two complexes. In the vesicles of the supercomplex (prepared by embedding the mixture of complexes), the component F1F0-synthase and cytochrome caa3 complexes, respectively, undergo thermodenaturation at lower and higher temperatures than in the vesicles in which the complex is embedded alone. Since, when a mixture of F1F0-synthase and cytochrome caa3 is embedded in phospholipid vesicles, the F1F0-synthase undergoes thermodenaturation at 45 °C (Tm1) (Fig. 1C), the small shoulder around 45 °C in the DSC analysis of F1F0-synthase vesicles alone (Fig. 1A) suggests that a small portion of F1F0-synthase complex had characteristics similar to that present in the vesicles of supercomplex of F1F0-synthase and cytochrome caa3. This could result from trace contamination of F1F0-synthase complex preparation by an interacting subunit of cytochrome caa3 that was not detectable in redox spectra.
Figure 1.
DSC curves of alkaliphilic Bacillus pseudofirmus OF4 F1F0 and cytochrome caa3 embedded in phospholipids singly or in combination. The molar ratio of F1F0 and caa3 is one and the weight ratio of phospholipids to protein is 1.5 in all cases. These vesicles are prepared by the cholate dialysis method. Curve A shows the exothermic thermodenaturation of 0.5 mg F1F0 embedded in phospholipid vesicles. Curve B is the DSC thermogram of 0.105 mg/ml caa3 embedded in phospholipid vesicles. Curve C is the DSC profile of phospholipid vesicles embedded with a mixture of 0.5 mg F1F0 and 0.105 mg caa3. Curve D is a mixture of phospholipid vesicles, embedded individually with either 0.5 mg F1F0 or 0.105 mg caa3. A total of twenty-one assays were conducted on two independent preparations. The standard deviations for the transition temperatures were in the range of 0.1–0.3 degrees and for enthalpy changes were 0.5–0.8 Kcal/mmol.
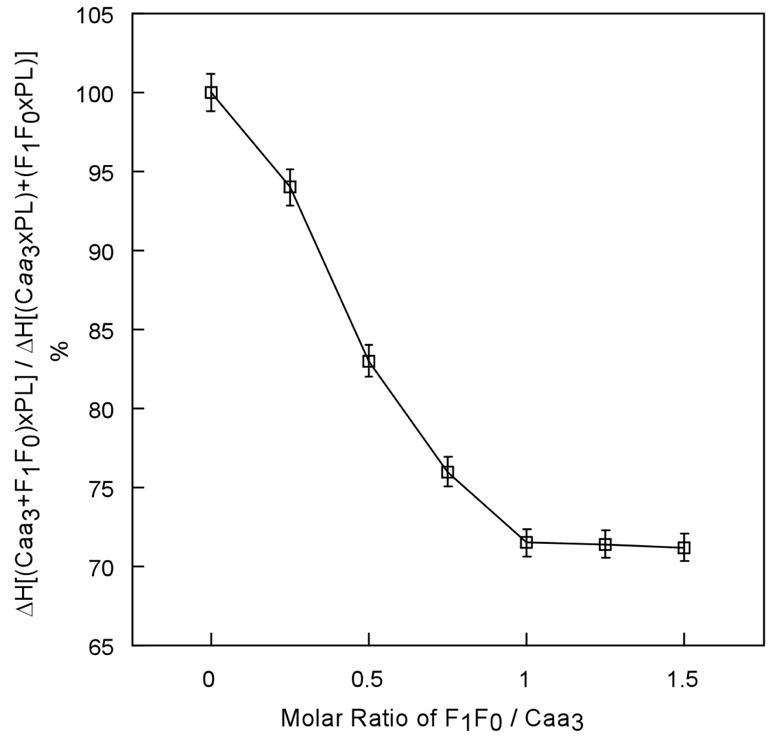
Figure 2 compares the thermodenaturation enthalpy changes of phospholipid vesicles formed with mixtures of cytochrome caa3 and F1F0-synthase at various molar ratios and of mixtures of phospholipid vesicles of individual complexes. The value of the difference in ΔH increases as F1F0-synthase is increased. The maximum difference is obtained when approximately one mol of F1F0-synthase per mol cytochrome caa3 is used. This suggests that the interaction between cytochrome caa3 and F1F0-synthase is specific.
Figure 2.
Comparison of enthalpy changes of thermodenaturation of phospholipids vesicles formed with mixtures of cytochrome caa3 and F1F0-synthase from alkaliphilic Bacillus pseudofirmus OF4 at various molar ratios and of mixtures of phospholipids vesicles of individual complexes. The molecular masses used in calculation of molar ratios were 517,000 and 105, 500 daltons for F1F0-synthase and cytochrome caa3, respectively. The ratio of phospholipids to protein was 1.5 by weight in all cases. The data presented in error bars are averages of three assays conducted on two independent preparations.
As discussed earlier (19,28–30), the energy for the exothermic transition of a protein complex embedded in phospholipid vesicles comes from the collapse, upon thermodenaturation, of a strained interaction between unsaturated fatty acyl groups of phospholipids and a protein surface on the protein complex. The surface may be exposed during isolation by removal of another protein with which the protein complex interacts in the native membrane. Little exothermic transition was observed in mitochondrial or submitochondrial preparations because there is no such exposed area in the native complex or supercomplex (19). When two interacting complexes are mixed together before being embedded in phospholipids vesicles, the exposed area on the protein surface is greatly diminished through the protein-protein interaction. Therefore, less strained interaction occurs upon vesicle formation, and less enthalpy change of exothermic denaturation is observed. It has been suggested that heat release that accompanies thermodenaturation of the mitochondrial membrane under aerobic conditions may be attributable to autooxidation of iron-sulfur proteins (31). This suggestion is not germane to the current study because there are no iron-sulfur proteins in either cytochrome caa3 or F1F0-ATP synthase complex.
STEPR Studies of Spin-labeled Cytochrome caa3 Embedded in Phospholipid Vesicles in the Absence and Presence of F1F0-synthase
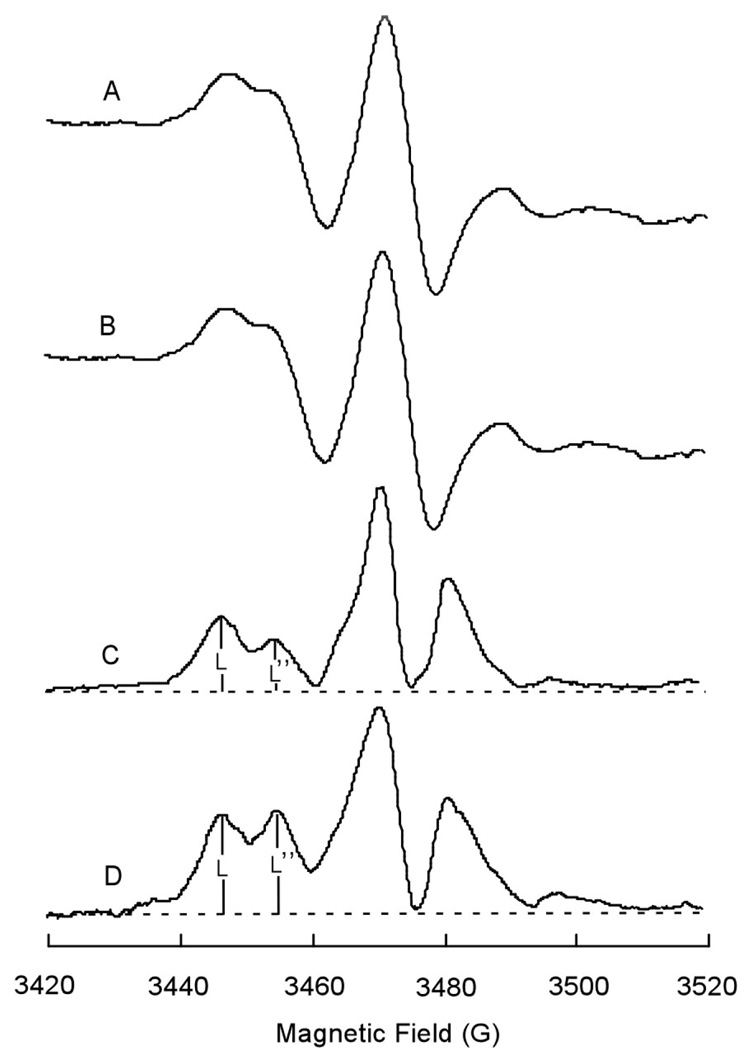
To confirm the existence of a specific interaction between alkaliphile cytochrome caa3 and F1F0-synthase, cytochrome caa3 was labeled with 4-maleimide-2,2,6,6-tetramethyl-1-piperidinyl-N-oxyl (MSL) as described under Materials and Methods. The MSL-cytochrome caa3, which is enzymatically active, was embedded in phospholipids vesicles alone or together with F1F0-synthase. The electron paramagnetic resonance (EPR) measurements of these electron transfer complex-phospholipid vesicles showed typical spin-immobilized spectra (see spectra A and B of Figure 3). The spectra were identical regardless of whether the protein-phospholipid vesicles contained only cytochrome caa3 or cytochrome caa3 and F1F0-synthase complexes (Fig. 3A & B). This suggested that the difference in mobility of the spin-label on cytochrome caa3, in the absence and presence of F1F0-synthase, is too small to be measured by conventional EPR. Therefore, the protein rotational diffusion of the spin-labeled complex was measured by saturation transfer electron paramagnetic resonance (STEPR). From the change of the ratio of two low-field signals (L’’/L) (see spectra C and D of Figure 3), rotational correlation times (τ2) can be calculated according to reported methods (26, 27). Table I shows the effect of the addition of F1F0-synthase on the rotational correlation time of spin-labeled cytochrome caa3. When mixed with F1F0-synthase from alkaliphilic B. pseudofirmus OF4 before being embedded in phospholipids vesicles, a significant increase in τ2 was observed compared to that of spin-labeled cytochrome caa3 embedded in phospholipids vesicles alone. By contrast, the τ2 of spin-labeled cytochrome caa3 was not affected when cytochrome bc1 complex or ATP-synthase from bovine heart mitochondria was substituted for the alkaliphile ATP synthase prior to the formation of vesicles; and the mixture of spin-labeled cytochrome caa3 complex and F1F0-synthase phospholipids vesicles showed the same τ2 as that of cytochrome caa3 phospholipid vesicles alone (see Table I).
Figure 3.
EPR spectra of spin-labeled alkaliphilic Bacillus pseudofirmus OF4 cytochrome caa3 in the presence and absence of OF4 F1F0-synthase complex. Spectra A and B are conventional EPR spectra of spin-labeled OF4 cytochrome caa3 embedded in phospholipid vesicles in the absence or presence of OF4 F1F0-synthase complex. Spectra C and D are the saturation transfer EPR spectra of the same samples. The protein concentrations were 6 and 36 mg/ml for caa3 and (caa3 + F1F0) vesicles, respectively. The patterns shown are typical of a total of twenty-one assays were conducted on two independent preparations.
Table I.
Effect of additions on the rotational correlation time (τ2) of spin-labeled cytochrome caa31
| Preparations | L’’/L | τ2 (µs) |
|---|---|---|
| (MSL-caa3 × PL) | 0.70 ± 0.01 | 65 ± 2 |
| [(MSL-caa3 + bF1F0) × PL] | 1.06 ± 0.01 | 270 ± 1 |
| [(MSL-caa3 × PL) + (bF1F0 × PL] | 0.71 ± 0.01 | 70 ± 2 |
| [(MSL-caa3 + mF1F0) × PL] | 0.71 ± 0.01 | 70 ± 2 |
| [(MSL-caa3 + mbc1) × PL] | 0.70 ± 0.01 | 65 ± 2 |
The molar ratio of cytochrome caa3 to other proteins used was 1:1.
The data presented are averages of three assays conducted on two independent preparations.
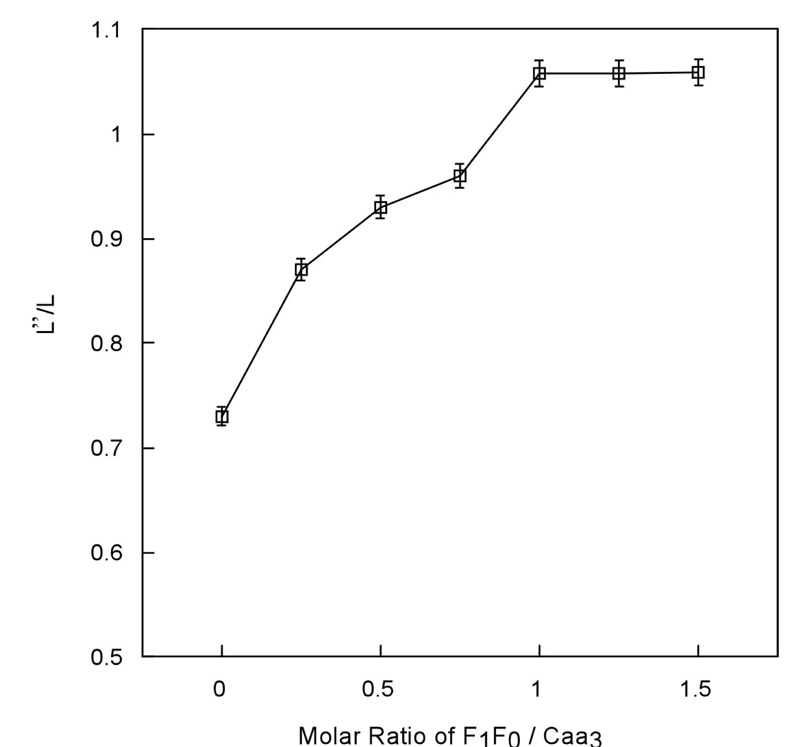
A similar effect of succinate-Q reductase on τ2 of spin-labeled ubiquinol-cytochrome c reductase (29) and of F1F0-synthase on τ2 of spin-labeled cytochrome c oxidase from bovine mitochondria (19) has been reported from this group. It is conceivable that at least part of the observed effect resulted from a change in the fluidity of the membrane by inclusion of protein complexes other than the spin-labeled complex. To ensure that the observed τ2 increase upon mixing alkaliphile F1F0-synthase with spin-labeled cytochrome caa3 is indeed due to the specific interaction between these two complexes, and not due to the change of protein concentration or self-aggregation upon addition of F1F0-synthase, a titration of spin-labeled cytochrome caa3 with F1F0-synthase was carried out. If a specific interaction between these two complexes exists, it is expected that a maximum τ2 will be obtained. As shown in Figure 4, the break point in τ2 was obtained when the ratio of alkaliphile F1F0-synthase to cytochrome caa3 approached one, which is in consistent with the number obtained from the DSC data. We note that the rotational correlation time obtained from STEPR is only an approximate value; it is based on the calibration curve derived from the isotropic motion of spin-label. The values obtained, however, agree with those obtained by other methods, such as flash photolysis (32). Moreover, although our main interest in this study was the relative τ2 of spin-labeled cytochrome caa3 in the absence and presence of the F1F0-synthase from alkaliphilic B. pseudofirmus OF4, the τ2 values obtained were in agreement with the DSC data (i.e. the titration curve of the τ2 values shows the same value of one for the saturation as that obtained from the DSC data).
Figure 4.
Effect of F1F0-synthase on STEPR of spin-labeled cytochrome caa3. Increasing amounts of F1F0-synthase were added to a constant amount of spin-labeled cytochrome caa3. The solutions were incubated for 60 min at 4 °C before being embedded in phospholipid vesicles. 1.5 mg of phospholipid/mg of protein was used in all cases. L’’/L was calculated from the saturation transfer EPR spectra of each sample. Instrument settings are given under Materials and Methods. The data shown in error bars are averages of three assays conducted on two independent preparations.
From the results of DSC and STEPR experiments, we conclude that cytochrome caa3 and F1F0-synthase from alkaliphilic B. pseudofirmus OF4 may form a supermacromolecule complex in the membrane. Although these interactions could be dynamic, this model differs significantly from the free diffusible model of electron transfer complexes derived from results of membrane fusing (33) and fluorescence recovery after photobleaching (FRAP) measurements (34). Rather, the current results are in line with a growing body of work that now suggests that some mitochondrial electron-transfer complexes specifically interact to form supermolecular structures called supercomplexes in organisms ranging from the yeast Saccharomyces cerevisiae (35,36), beef (19,29,35,37), and plants (38–42). Similar supermolecular structures have also been described for the respiratory chains of bacteria (43–47). The roles that have been attributed to respiratory supercomplexes are substrate-channeling, catalytic enhancement, sequestration of reactive intermediates (35), stabilization of protein complexes (48), increasing the capacity of the inner mitochondrial membrane for protein insertion (36), and generating mitochondrial cristae morphology (49). Furthermore, the dynamic formation of such supercomplexes is speculated to serve some regulatory function in the energy generation in mitochondria from plants (39) and beef (19). It should be mentioned that supercomplex forms from two or more lipoprotein complexes isolated from detergent solublized membrane depends on the kind of detergent used and the method used in the detection. In the blue gel system, non-ionic detergents are normally used and non native environment is provided, the interaction detected may be detergent dependent. The interaction among electron transfer complexes detected by the described DSC and STEPR methods is less influenced by detergents used in the isolation of complexes as they are removed during the formation of vesicles. In the vesicles the protein complexes are in a membrane environment similar to the native system, which may facilitate detection of interactions that are dynamic. The information obtained is therefore more reliable. Since a stoichiometrical interaction between F1F0 synthase and cytochrome oxidase of beef heart mitochondria is also observed, the supercomplex formation between these two complexes may not be a specific design only for alkaliphiles but also for other organisms in general.
In the alkaliphile context, the specific interaction of the cytochrome oxidase and the ATP synthase observed here in a reconstituted system is relevant to models of the proton path during OXPHOS that posit direct protein-protein interactions. The alkaliphile system can now be used to dissect the elements involved in the interactions using mutants whose effects upon OXPHOS in vivo can also be determined. This will make it possible to test whether one or more of the alkaliphile-specific features of the ATP synthase and cytochrome caa3 facilitate the interactions observed here and/or apparent proton sequestration in bioenergetic experiments. A candidate in the alkaliphile ATP synthase would be the unusually polar loop on the external side of the a-subunit, near the putative proton uptake pathway, that was shown to be required for optimal OXPHOS at high pH but not critically involved in the proton-gating function (15). A candidate of interest in the cytochrome caa3 is the unusually acidic patch that was noted when the operon encoding the complex was first cloned and sequenced (17). This acidic domain is predicted to be near the region of subunit II just above where the pumped protons are modeled to emerge (50,51). Also of interest will be the roles of specific membrane lipids, especially cardiolipin (8), and additional respiratory chain components in supercomplex formation or proton transfer. Goto et al. (52) suggest that cytochrome c may support rapid proton translocation to a cytochrome oxidase-ATP synthase complex in alkaliphiles.
Footnotes
This work was supported in part by grants GM30721 (to CAY) and GM028454 (to TAK) from the National Institutes of Health.
The abbreviations used are: OXPHOS, oxidative phosphorylation; Δp, electrochemical proton gradient across the membrane; ΔΨ, transmembrane electrical potential (negative in), Caa3, cytochrome caa3; DSC, differential scanning calorimetry; EPR, electron paramagnetic resonance; FRAP, fluorescence recovery after photobleaching; MSL, maleimide spin label (4-maleimide-2, 2, 6, 6-tetramethyl-1- piperidinyl-N-oxyl); STEPR, saturation transfer electron paramagnetic resonance.
REFERENCES
- 1.Mitchell P. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 2.Boyer PD. Annu. Rev. Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 3.Stock D, Gibbons C, Arechaga I, Leslie AG, Walker JE. Curr. Opin. Struct. Biol. 2000;10:672–679. doi: 10.1016/s0959-440x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 4.Rottenberg H. Modern Cell Biol. 1985;4:47–83. [Google Scholar]
- 5.Williams RJP. J. Theor. Biol. 1961;1:1–17. doi: 10.1016/0022-5193(61)90023-6. [DOI] [PubMed] [Google Scholar]
- 6.Williams RJ. Biochim. Biophys. Acta. 1978;505:1–44. doi: 10.1016/0304-4173(78)90007-1. [DOI] [PubMed] [Google Scholar]
- 7.Slater EC. Eur. J. Biochem. 1987;166:489–504. doi: 10.1111/j.1432-1033.1987.tb13542.x. [DOI] [PubMed] [Google Scholar]
- 8.Haines TH, Dencher NA. FEBS Lett. 2002;528:35–39. doi: 10.1016/s0014-5793(02)03292-1. [DOI] [PubMed] [Google Scholar]
- 9.Mulkidjanian AY, Cherepanov DA, Heberle J, Junge W. Biochemistry (Mosc) 2005;70:251–256. doi: 10.1007/s10541-005-0108-1. [DOI] [PubMed] [Google Scholar]
- 10.Dimroth P, Cook GM. Adv. Microb. Physiol. 2004;49:175–218. doi: 10.1016/S0065-2911(04)49004-3. [DOI] [PubMed] [Google Scholar]
- 11.Krulwich TA, Ito M, Gilmour R, Hicks DB, Guffanti AA. Adv. Microb. Physiol. 1998;40:401–438. doi: 10.1016/s0065-2911(08)60136-8. [DOI] [PubMed] [Google Scholar]
- 12.Krulwich TA, Hicks DB, Swartz TH, Ito M. In: Physiology and Biochemistry of Extremophiles. Gerday C, Glansdorff N, editors. Washington, DC: ASM Press; 2006. in press. [Google Scholar]
- 13.Krulwich TA. Mol. Microbiol. 1995;15:403–410. doi: 10.1111/j.1365-2958.1995.tb02253.x. [DOI] [PubMed] [Google Scholar]
- 14.Guffanti AA, Krulwich TA. J. Biol. Chem. 1994;269:21576–21582. [PubMed] [Google Scholar]
- 15.Wang Z, Hicks DB, Guffanti AA, Baldwin K, Krulwich TA. J. Biol. Chem. 2004;279:26546–26554. doi: 10.1074/jbc.M401206200. [DOI] [PubMed] [Google Scholar]
- 16.Quirk PG, Guffanti AA, Plass RJ, Clejan S, Krulwich TA. Biochim. Biophys. Acta. 1991;1058:131–140. doi: 10.1016/s0005-2728(05)80229-4. [DOI] [PubMed] [Google Scholar]
- 17.Quirk PG, Hicks DB, Krulwich TA. J. Biol. Chem. 1993;268:678–685. [PubMed] [Google Scholar]
- 18.Krulwich TA, Ito M, Gilmour R, Sturr MG, Guffanti AA, Hicks DB. Biochim. Biophys. Acta. 1996;1275:21–26. doi: 10.1016/0005-2728(96)00044-8. [DOI] [PubMed] [Google Scholar]
- 19.Qiu ZH, Yu L, Yu CA. Biochemistry. 1992;31:3297–3302. doi: 10.1021/bi00127a036. [DOI] [PubMed] [Google Scholar]
- 20.Kagawa Y. Tanpakushitsu Kakusan Koso. 1971;16:775–786. [PubMed] [Google Scholar]
- 21.Hicks DB, Krulwich TA. J. Biol. Chem. 1990;265:20547–20554. [PubMed] [Google Scholar]
- 22.LeBel D, Poirier GG, Beaudoin AR. Anal. Biochem. 1978;85:86–89. doi: 10.1016/0003-2697(78)90277-4. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto J, Matsumoto A, Oobuchi K, Sone N. FEMS Microbiol. Lett. 1996;143:151–158. doi: 10.1111/j.1574-6968.1996.tb08474.x. [DOI] [PubMed] [Google Scholar]
- 24.van Gelder BF. Biochim. Biophys. Acta. 1966;118:36–46. doi: 10.1016/s0926-6593(66)80142-x. [DOI] [PubMed] [Google Scholar]
- 25.Racker E. J. Membr. Biol. 1972;10:221–235. doi: 10.1007/BF01867856. [DOI] [PubMed] [Google Scholar]
- 26.Thomas DD, Dalton LR, Hyde JS. J. Chem. Phys. 1976;65:3006–3024. [Google Scholar]
- 27.Poore VM, Fitzsimons JT, Ragan CI. Biochim. Biophys. Acta. 1982;693:113–124. doi: 10.1016/0005-2736(82)90477-1. [DOI] [PubMed] [Google Scholar]
- 28.Gwak SH, Yu L, Yu CA. Biochim. Biophys. Acta. 1985;809:187–198. doi: 10.1016/0005-2728(85)90062-3. [DOI] [PubMed] [Google Scholar]
- 29.Gwak SH, Yu L, Yu CA. Biochemistry. 1986;25:7675–7682. doi: 10.1021/bi00371a059. [DOI] [PubMed] [Google Scholar]
- 30.Yu CA, Gwak SH, Yu L. Biochim Biophys Acta. 1985;815:656–664. doi: 10.1016/0005-2736(85)90258-5. [DOI] [PubMed] [Google Scholar]
- 31.Tsong TY, Knox BE. Biophys. J. 1984:296a. [Google Scholar]
- 32.Cherry RJ. Biochim. Biophys. Acta. 1979;559:289–327. doi: 10.1016/0304-4157(79)90009-1. [DOI] [PubMed] [Google Scholar]
- 33.Schneider H, Lemasters JJ, Hochli M, Hackenbrock CR. J. Biol. Chem. 1980;255:3748–3756. [PubMed] [Google Scholar]
- 34.Gupte S, Wu ES, Hoechli L, Hoechli M, Jacobson K, Sowers AE, Hackenbrock CR. Proc. Natl. Acad. Sci. USA. 1984;81:2606–2610. doi: 10.1073/pnas.81.9.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schagger H, Pfeiffer K. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schagger H. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schagger H, Pfeiffer K. J. Biol. Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt B, McCracken J, Ferguson-Miller S. Proc. Natl. Acad. Sci. USA. 2003;100:15539–15542. doi: 10.1073/pnas.2633243100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eubel H, Jansch L, Braun HP. Plant Physiol. 2003;133:274–286. doi: 10.1104/pp.103.024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eubel H, Heinemeyer J, Braun HP. Plant Physiol. 2004;134:1450–1459. doi: 10.1104/pp.103.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP. Proc. Natl. Acad. Sci. USA. 2005;102:3225–3229. doi: 10.1073/pnas.0408870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krause F, Reifschneider NH, Vocke D, Seelert H, Rexroth S, Dencher NA. J. Biol. Chem. 2004;279:48369–48375. doi: 10.1074/jbc.M406085200. [DOI] [PubMed] [Google Scholar]
- 43.Berry EA, Trumpower BL. J. Biol. Chem. 1985;260:2458–2467. [PubMed] [Google Scholar]
- 44.Iwasaki T, Matsuura K, Oshima T. J. Biol. Chem. 1995;270:30881–30892. doi: 10.1074/jbc.270.52.30881. [DOI] [PubMed] [Google Scholar]
- 45.Niebisch A, Bott M. J. Biol. Chem. 2003;278:4339–4346. doi: 10.1074/jbc.M210499200. [DOI] [PubMed] [Google Scholar]
- 46.Sone N, Sekimachi M, Kutoh E. J. Biol. Chem. 1987;262:15386–15391. [PubMed] [Google Scholar]
- 47.Stroh A, Anderka O, Pfeiffer K, Yagi T, Finel M, Ludwig B, Schagger H. J. Biol. Chem. 2004;279:5000–5007. doi: 10.1074/jbc.M309505200. [DOI] [PubMed] [Google Scholar]
- 48.Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, Moraes CT, Enriquez JA. Mol. Cell. 2004;13:805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hosler JP, Ferguson-Miller S, Mills DA. Annu. Rev. Biochem. 2006;75:165–187. doi: 10.1146/annurev.biochem.75.062003.101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Megehee JA, Hosler JP, Lundrigan MD. Microbiology. 2006;152:823–829. doi: 10.1099/mic.0.28723-0. [DOI] [PubMed] [Google Scholar]
- 52.Goto T, Matsuno T, Hishinuma-Narisawa M, Yamazaki K, Matsuyama H, Inoue N, Yumoto I. J. Biosci. Bioeng. 2005;100:365–379. doi: 10.1263/jbb.100.365. [DOI] [PubMed] [Google Scholar]