Abstract
We have shown previously that Dipeptidyl Peptidase 2 (DPP2) activity is essential for the survival of quiescent, but not activated, lymphocytes. The specific requirement of DPP2 activity for non-dividing cells is indicative of cell cycle specific regulation of this gene product. In the present study, we tested this hypothesis by looking at contact and serum dependence of Dpp2 transcription. We found that transfected promoter-reporter activity, as well as endogenous Dpp2 transcripts, were enhanced in NIH-3T3 cells upon contact-inhibition or serum starvation. Since Lung Kruppel-like factor (KLF2), a transcription factor, and TOB1, a transcriptional co-activator, have been shown to be important in maintaining T-lymphocyte quiescence and are both downregulated upon cellular activation, we also looked at the contributions of these factors to Dpp2 transcription. Using a Dpp2 promoter-reporter system, we demonstrate that KLF2 and TOB1 activate the mouse Dpp2 promoter. Finally, we show that in human PBMC, there is a decrease in levels of endogenous DPP2 transcripts upon T cell receptor activation when compared to resting cells. These results demonstrate that Dpp2 transcription is serum and contact-dependent and link two quiescence-specific transcriptional elements to the quiescencespecific requirement of DPP2 enzymatic activity.
Keywords: T Cells, Transcription Factors, Cell Differentiation
Introduction
Cellular quiescence in lymphocytes is represented by smaller cell size, lack of DNA synthesis and reduced metabolic activity (Freitas and Rocha, 2000; Rathmell et al., 2003; Yusuf and Fruman, 2003). However, despite the perceived inaction during this state, quiescence is an actively maintained process, regulated by ongoing signaling and de novo protein synthesis. Cell-cell and cell-autonomous (Smith and Cancro, 2003; Torcia et al., 1996) activation of surface receptors have been proposed as mediators of lymphocyte quiescence. CD8 single positive T cells require MHC class I contact for long term survival (Tanchot et al., 1997). Naïve B cell survival has been linked to “tickling” by the B cell antigen receptor (Lam et al., 1997; Meffre and Nussenzweig, 2002). These signaling events culminate in a regulated, quiescence-specific transcriptional program mediated, in part, by factors such as Lung Kruppel-like factor (KLF2) (Kuo et al., 1997) and TOB1 (Tzachanis et al., 2001). However, specific targets of these transcription factors that are important in maintaining cellular quiescence have not been well characterized.
We previously reported that lymphocyte quiescence is dependent on the enzymatic activity of Dipeptidyl peptidase 2 (DPP2), a serine protease with an amino terminal dipeptidase activity (Underwood et al., 1999). Inhibition of DPP2 in resting, but not activated, T cells results in apoptosis (Chiravuri et al., 1999). It was thus our hypothesis that DPP2 promoter activity is controlled by quiescence-specific factors, such as KLF2 and TOB1. The transcription factor KLF2 is necessary for T cell quiescence (Kuo and Leiden, 1999). KLF2 is a zinc-finger transcription factor that is required for lung development (Wani et al., 1999), as well as the development of single positive T cells (Kuo et al., 1997). Over-expression of KLF2 in normally cycling Jurkat cells causes these cells to resemble quiescent cells (Buckley et al., 2001). KLF2 is expressed in naïve and memory lymphocytes, and KLF2 mRNA is transcriptionally downregulated upon cellular activation (Buckley et al., 2001; Schober et al., 1999). It exerts its quiescence-promoting effect partially through suppression of the protooncogene C-MYC. Leiden and his co-workers (Buckley et al., 2001) have shown that ectopic expression of a chimeric suppressor of MYC function, MAD-MYC (Berns et al., 1997), resembles the effect of KLF2 over-expression.
The transactivator of ErbB2, TOB1, is a member of the BTG family of anti-proliferative proteins (Matsuda et al., 2001; Tirone, 2001). It has also been implicated in maintaining lymphocyte quiescence (Tzachanis et al., 2001). Exogenous expression of TOB1 and other BTG family-member proteins in fibroblasts has growth-suppressive effects (Matsuda et al., 2001). Like KLF2, TOB1 is expressed primarily in naïve and memory T cells. TOB1 inhibits T cell proliferation and downregulates IL-2 transcription through SMAD22 and SMAD4, and loss of TOB1 reduces the threshold for T cell activation (Tzachanis et al., 2001).
We report here analysis of the mouse Dpp2 promoter activity. We show that Dpp2 promoter activity is enhanced during growth-inhibiting conditions and repressed upon proliferation. We also show that Dpp2 transcription is enhanced by the quiescence-specific transcription factor KLF2 and the transcriptional co-factor TOB1. Furthermore, human DPP2 transcripts are significantly reduced upon activation of PBMC when compared to resting cells. Thus, DPP2 is an integral part of the machinery maintaining lymphocyte quiescence that is regulated by quiescence-specific transcription.
Materials and Methods
Cell Culture and stimulation
NIH3T3 fibroblasts (ATCC, Manassas, VA) were maintained in a 37° C incubator with 5% CO2. Cells were cultured in Dulbecco’s Modified Eagle Medium, DMEM (Gibco, Grand Island, NY), supplemented with 10% Fetal Bovine Serum (Atlanta Biologicals, Norcross, GA) and 20 mM HEPES, Sodium Pyruvate, Penicillin-Streptomycin and 2-mercaptoethanol (all from Gibco). Whole blood from healthy donors was acquired by venipuncture in accordance with the Institutional Review Board at Tufts University School of Medicine. PBMCs were acquired by Ficoll-Hypaque (GE Healthcare) separation. Cells were cultured in RPMI 1640 (Gibco) supplemented with 10% FBS, Hepes, and Sodium Pyruvate. PBMCs were stimulated on plates coated with Protein-A bound anti-CD3 antibody alone, or in combination with anti-CD28 antibody, or an isotype matched control IgG for 96 h.
Plasmid Constructs and Transfections
A 2 kb upstream region of the mouse Dpp2 gene was amplified by PCR from BALB/c genomic DNA, using the primers 5'CCGCTCGAGCTGGAGTGCCTGAAGACAGCTAC3’ and 5’GCTCTAGAGCTTGATTCTGAGCCGGGCGCT3’ (Tufts University Core Facility). This fragment was cloned into the XhoI and BglII sites of the luciferase expression vector, pGL-3 Basic (Promega, Madison, WI). pCDNA-LKLF-HA (Buckley et al., 2001), and pCDNA-TOB1 (Tzachanis et al., 2001) vectors were gifts of J. Leiden, and V. Boussiotis, respectively. The MADMYC expression vector was a gift of R. Bernards (Berns et al., 1997). Empty pGL3-Basic vector was used as a control, and a Renilla expression vector, pRL-TK (Promega), was used for normalization. Cells were trypsinized and plated in 12-well plates (Costar, Corning, NY) at 50% density the day before transfection. Cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA), and the lysates were assayed for luciferase activity 48 h post transfection. For contact inhibition or serum-starvation experiments, cells were split 4 h after transfection and replated at 100%, 50%, 25% or 10% densities. 4 h post-splitting, the cells were washed and serum starved with 1% or 0.1% serum.
SDS-PAGE and western blotting
Lysates were run on 4–20% gradient polyacrylamide Criterion XT gels (Bio-Rad, Hercules, CA) and transferred onto PVDF membranes (Millipore, Billerica, MA). Membranes were blocked in 0.4% I-Block (Tropix, Foster City, CA) in TBS and 0.1% Tween-20 (TBS-Tween) for 2 h at room temperature and probed with primary antibodies against Tob (4B1, Sigma, St. Louis, MO), HA epitope tag (HA.11, Covance), Myc (9E10, Santa Cruz Biotechnology, Santa Cruz, CA) in 0.2% I-Block in TBS-Tween for overnight at 4° C. Blots were washed three times for 5 min each with 0.2% I-Block in TBS-Tween and then incubated in the same buffer containing horseraddish peroxidase conjugated secondary antibody (Amersham, Piscataway, NJ), for 1 h at room temperature. Blots were developed with ECL chemiluminescent substrate (Amersham) and exposed on film (Kodak, Rochester, NY).
Luciferase Reporter Assays and real-time RT-PCR
Cells in 12-well plates were washed once with PBS and lysed in 200 µl of Passive Lysis Buffer (Promega). 5 µl of the lysate was used to measure firefly and Renilla luciferase activities with Dual-Luciferase Reporter Assay System (Promega) and a Turner Designs (Sunnyvale, CA) luminometer. All reported luciferase values are normalized to the Renilla luciferase control activity. Real-time RT-PCR analyses were performed on total RNA isolated from cells using mouse Dpp2 (primer pair: GGAGGCCCTGCTTGTCTTT and CACCGAACGGAAGCGATTT C; TaqMan MGB probe: 6-FAM-CTGAGCACCGGTACTATG-NFQMGB) and human DPP2 TaqMan MGB probe (#MGB4316034) and RT-PCR reagents (#4304971) (Applied Biosystems) and run and analyzed on ABI 7200 sequence detection system. Probe for 18S RNA (#4308329, Applied Biosystems) was used to normalize individual samples. The calculation is based on the relative differences ddC(t) method. Briefly, one cycle difference in real-time PCR represents a two-fold difference; the relative difference of two samples can be calculated mathematically by taking 2 to the power of the differences of the Ct (ddC(t)) between these two samples. The relative differences of DPP2, as well as the housekeeping gene r18S RNA are first calculated independently. The relative difference of the DPP2 signal is then divided by the relative difference of the r18S RNA signal to give the normalized difference for each sample.
Results
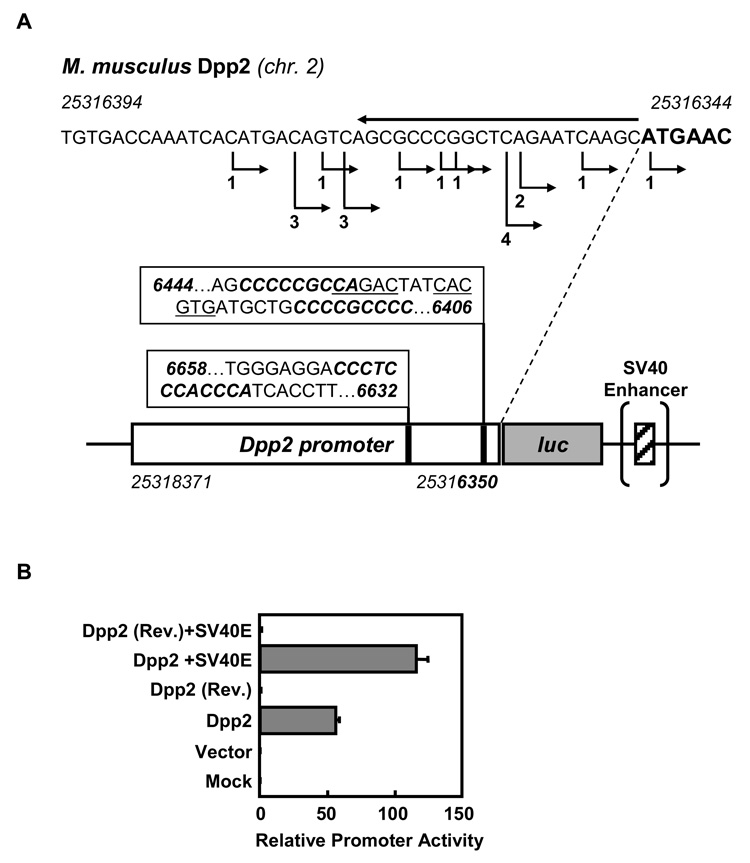
Since DPP2 inhibition leads to lymphocyte apoptosis in quiescent cells, but not in activated cells (Chiravuri et al., 1999), we hypothesized that Dpp2 promoter activity is cell cycle dependent. To test this working model, we cloned a 2,021 bp segment directly upstream of the translation start site of mouse Dpp2 into a luciferase reporter construct (Figure 1A). Data from DBTSS (http://dbtss.hgc.jp) show 18 out of 19 full-length cDNAs to have their transcriptional start-sites (TSS) just before the first codon (Figure 1A). GC-rich regions containing a core binding site for KLF proteins, CACCC (pos. 25316639–25316650) (Turner and Crossley, 1999), as well as a consensus SMAD binding element, CAGACA (pos. 23516430–25316434) (Jonk et al., 1998), are both present within this DNA segment, slightly upstream of the TSS. Also present within this sequence is a MYC/MAX binding site (25316421–25316426) (see Figure 1A). When transfected into 293T cells, our reporter construct exhibits directional promoter activity that is enhanced approximately two-fold by the addition of an SV40 enhancer (Figure 1B).
Figure 1. 2 kb upstream region of murine Dpp2 contains promoter activity.
A) 2 kb region directly upstream of the translation initiation site (ATG codon) was cloned into a firefly luciferase expression vector, pGL3, by PCR (3’ primer is overlined on top of the sequence). Transcription start sites in the DBTSS (http://dbtss.hgc.jp) database are indicated with right-angled arrows below the sequence, and by numbers of full-length cDNAs representing a particular TSS. A schematic representation of the cloned region is shown with predicted binding sites for SMAD transcription factors (CAGACA, underlined), for the transcription factor KLF2 (core binding site CACCC, italicized sequences) and binding site for MYC/MAX dimers (CACGTG, underlined). Nucleotide position numbers represent the numbers used by DBTSS. B) The promoter construct, with or without the SV40 enhancer, and in forward and reverse orientations, were transfected into 293T cells. Mean relative promoter activities (normalized by Renilla activity) 48 h post transfection are shown from duplicate transfections +/− S.D.
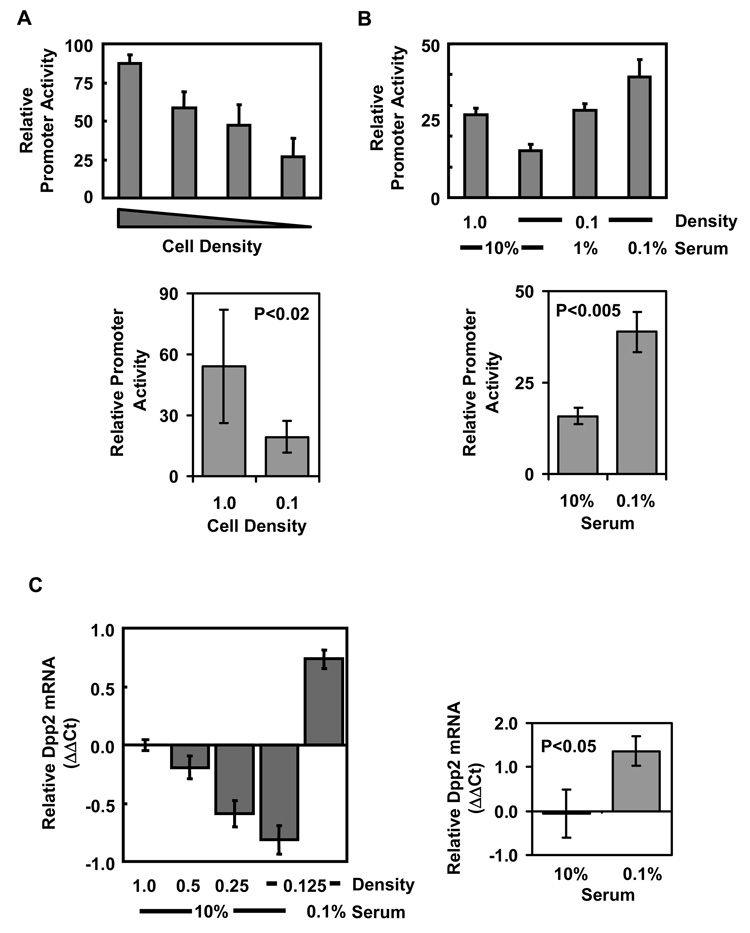
For further analysis of the promoter, we chose NIH-3T3 fibroblasts, because they can be driven into growth arrest through contact inhibition or serum starvation (Meisler, 1973). The Dpp2 promoter construct was transfected into NIH-3T3 fibroblasts, and the cells were either grown in increasing cell densities to induce contact inhibition, or they were serum-starved. As can be seen in Figure 2A, contact inhibition resulted in a significant increase in mouse Dpp2 promoter activity compared to that observed in sparsely grown cultures. We also found that the basal promoter activity in sparse cultures could be significantly enhanced by serum withdrawal (Figure 2B). To verify the transfected promoter-reporter results, we analyzed endogenous Dpp2 transcripts from NIH-3T3 cells, grown in different serum-concentrations and densities, by real-time RT-PCR and found that the endogenous Dpp2 RNA levels correlated with the promoter activity from the reporter construct (Figure 2C). We conclude that the Dpp2 promoter is activated by serum withdrawal and by contact-inhibition, resulting in an increase in Dpp2 transcript levels.
Figure 2. Dpp2 promoter activity is cell density and serum dependent.
NIH3T3 cells were transfected with the Dpp2 promoter luciferase vector. A) Transfected cells were split 4 h post transfection into wells in confluent (1.0) to sub-confluent (0.5, 0.25, 0.1) densities. Promoter activity was analyzed 48 h later and is reported as normalized luciferase activity (luciferase/renilla ratios). Dpp2 promoter activity is the highest in contact-inhibited conditions. B) Transfected cells were split 4 h post transfection into different cell density and serum conditions as shown, and promoter activity was analyzed 48 h post transfection. Dpp2 promoter activity is the highest in serum-starved conditions. Results are mean ±SD of duplicate transfections, representative of three independent experiments (top). Statistical differences between the groups were determined by Student’s two-tailed t-test from three experiments (bottom). C) Untransfected cells were split as in A and B, and RNA extracted from the cells 24 h later. Dpp2 RNA was measured by real-time RT-PCR, and shown as ΔΔCt, normalized against 18S RNA, and then against Dpp2 levels from confluent (1.0) samples grown in 10% serum. Results are mean +/− SD from triplicate samples from a representative experiment (left). Statistical differences between the groups were determined by Student’s two-tailed t-test from three experiments (right).
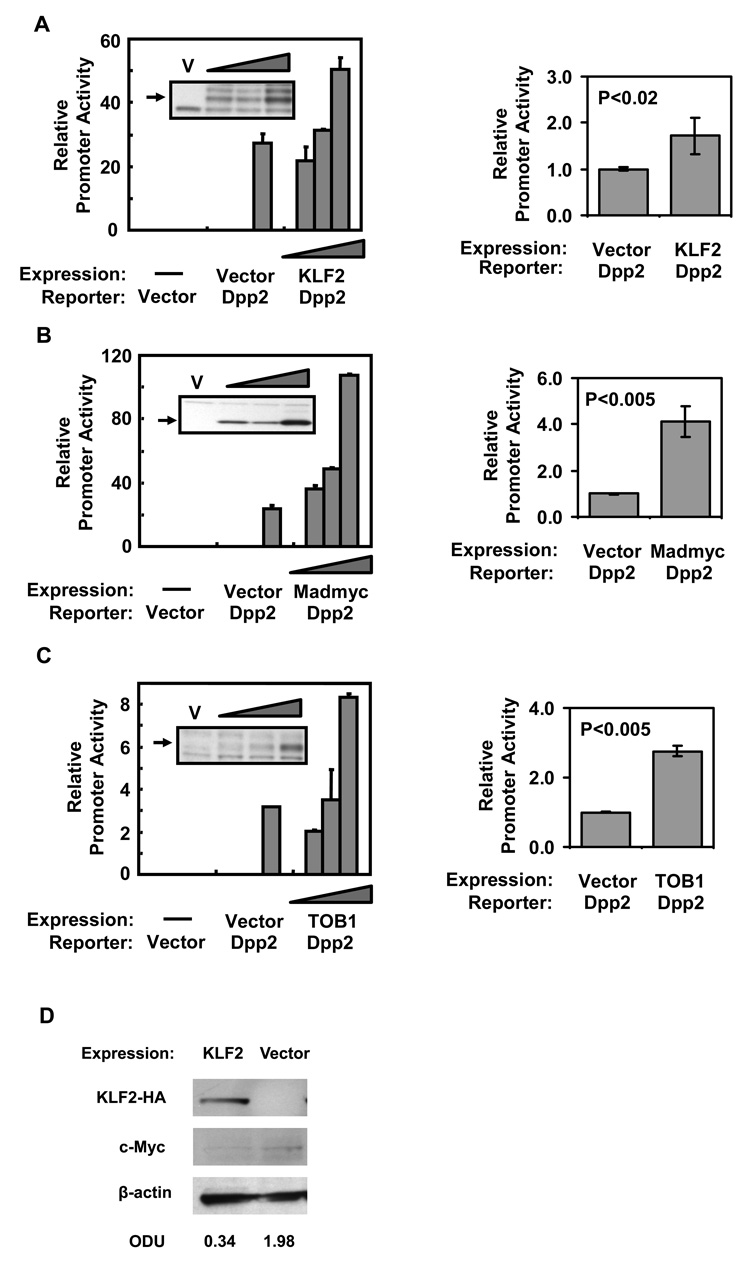
Since DPP2 enzymatic activity is required for the survival of quiescent lymphocytes (Chiravuri et al., 1999), we hypothesized that this protease is transcriptionally regulated by factors that maintain lymphocyte quiescence. To test this, NIH-3T3 fibroblasts were co-transfected with the Dpp2 promoter construct and a KLF2 expression construct. KLF2 significantly augmented Dpp2 promoter activity in a dose-dependent manner (Figure 3A). Since the quiescence promoting effect of KLF2 is mediated in part by the proto-oncogene MYC (Buckley et al., 2001), we theorized that MYC suppresses Dpp2 promoter activity. To test this, we co-transfected NIH-3T3 fibroblasts with the Dpp2 promoter construct and an expression construct for MADMYC, a chimeric protein that antagonizes MYC activity (Berns et al., 1997). We observed that expression of MADMYC increased Dpp2 promoter activity significantly and in a dose-dependent manner (Figure 3B). Since TOB1 is involved in lymphocyte quiescence through SMAD4 activity (Tzachanis et al., 2001), and the Dpp2 promoter region contains a consensus binding site for SMAD proteins (see Figure 1), we then wanted to test whether TOB1 had an effect on Dpp2 transcription. In co-transfection experiments with the Dpp2 promoter, TOB1 significantly increased the promoter activity in a dose-dependent manner (Figure 3C). As a further control, we measured whether ectopic expression of KLF2 would influence endogenous c-Myc levels. As can be seen in Fig. 3D, NIH3T3 cells transiently transfected with KLF2 had decreased levels of c-Myc, as normalized to β-actin.
Figure 3. KLF2 and TOB1 increase Dpp2 promoter activity.
NIH3T3 cells were transfected with a fixed amount (0.9 µg) of Dpp2 promoter luciferase construct and a construct expressing A) KLF2 (HA-tagged) or B) MADMYC, and C) TOB1 at varying amounts (0.25, 0.5 and 1.0 µg), normalized with empty expression vector (pCI-Neo) or 1.0 µg of empty vector alone. Lysates were analyzed for luciferase activity 48 h post transfection. An aliquot of the lysate was run on SDS-PAGE and western-blotted for HA, MYC, or TOB1 expression. KLF2 and MADMYC both increase the Dpp2 promoter activity. D) Transfection of KLF2 decreases endogenous c-Myc expression in NIH3T3 fibroblasts. c-Myc protein levels were normalized to β-actin. Data shown are mean ±SD of duplicate transfections from a single experiment, representative of three (left). Statistical differences between the groups were determined by Student’s two-tailed t-test from three experiments (right).
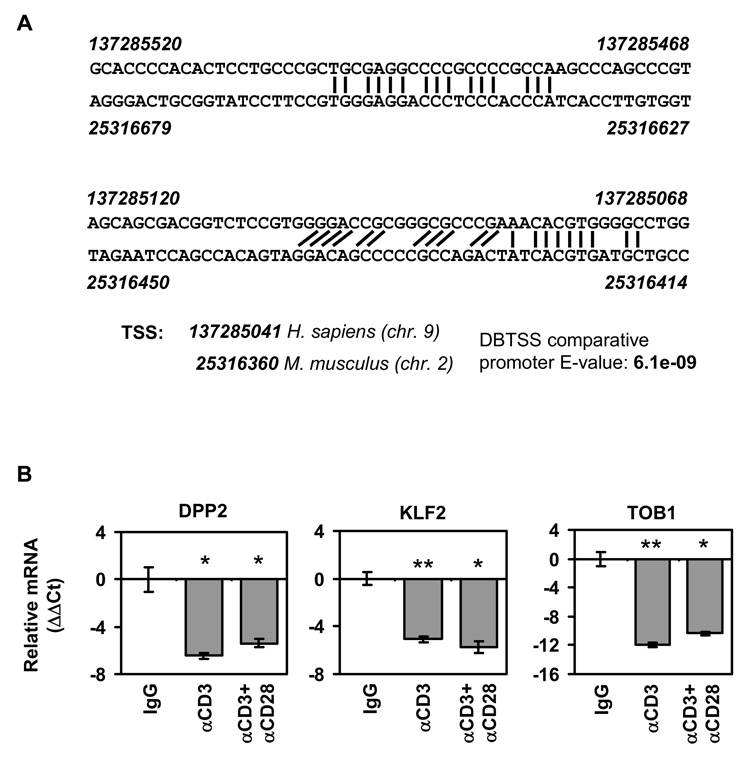
Finally, we wanted to test whether endogenous DPP2 mRNA in lymphocytes was regulated upon activation. Since mouse and human promoter regions share a significant homology (Figure 4A) and from our data that human proteins like KLF2 and TOB1 can regulate the mouse Dpp2 promoter (see Figure 3), it was likely that the quiescence factors we tested on the murine promoter-reporter constructs would also be relevant for human lymphocytes. We measured the relative levels of DPP2, KLF2 and TOB1 mRNA by real-time RT-PCR after activating PBMC with anti-CD3 alone or anti-CD3 plus anti-CD28 mAbs, which are T cell specific activators. As a control, isotype-matched IgG was used. In cells activated by plate-bound antibodies for 96 h, we found that DPP2, KLF2 and TOB1 mRNA were reduced significantly compared to control stimulated cells (Figure 4B).
Figure 4. Lymphocyte activation results in reduced levels of DPP2 mRNA.
A) GC-rich regions in proximal promoter region of mouse and human DPP2 are well-conserved. Numbers represent nucleotide positions in DBTSS (http://dbtss.hgc.jp). Transcription start sites, and expectation score (E-value) for the alignment of the two promoters are from DBTSS. B) Lymphocytes were stimulated for 96 h with anti-CD3 mAb alone or in combination with anti-CD28 mAb, or with isotype-matched control antibody (IgG). RNA was collected and real-time RT-PCR, performed using human DPP2, KLF1 or TOB1 TaqMan probes. Results are shown as mean ΔΔCt +/− SD from triplicate samples, normalized first against 18S RNA, then against the IgG control sample. *, p<0.05; Student’s two-tailed t test, **, p<0.005; Student’s two-tailed t test.
Discussion
The data presented here show that Dpp2 is transcriptionaly activated by two quiescence-specific elements, KLF2 and TOB1 (Kuo et al., 1997; Tzachanis et al., 2001). It has been shown previously that KLF2 expression is restricted to naïve and memory T cells, and, likewise, TOB1 is specifically expressed in naïve and anergic T cells and is down regulated upon activation. Thus, the two transcriptional elements were excellent candidates for regulating Dpp2 expression.
Limited transcriptional targets for KLF2 have been described previously; namely, KLF2 activates the VAV promoter (Denkinger et al., 2001) and represses the PPARG promoter (Banerjee et al., 2003). However, despite KLF2’s importance in maintaining lymphocyte quiescence, no physiologically relevant target for KLF2 in lymphocytes has been defined so far. Our data point to Dpp2 as the first instance of a target gene regulated by KLF2 that is functionally relevant for lymphocyte quiescence. KLF2 also negatively controls the transcription of MYC (Buckley et al., 2001), another transcription factor with diverse functions, including regulation of proliferation, differentiation and apoptosis (Dang, 1999; Levens, 2003). Thus, like KLF2, MADMYC, a chimeric suppressor of MYC function, induces quiescence in Jurkat cells (Buckley et al., 2001). Consistent with these results, we now demonstrate that KLF2 and MADMYC both activate the Dpp2 promoter. However, it is not possible to determine from our results whether the activation seen with KLF2 is direct or through suppression of MYC, mediated by KLF2.
The transcriptional co-factor TOB1, in conjunction with SMAD transcription factors, is involved in the down regulation of IL-2 transcription (Tzachanis et al., 2001). As the Dpp2 promoter contains a SMAD binding site, it is quite likely that SMADs are also involved in Dpp2 transcriptional activation. In addition, TOB1 upregulates CDKN1B (p27) expression upon TCR stimulation and can activate the p27 promoter in vitro (Tzachanis et al., 2001). We have shown that TOB1 expression increases the Dpp2 promoter activity (Figure 3C).
While we have shown Dpp2 transcriptional activation with T cell specific transcription factors, it still remains to be seen whether other KLF or BTG family members control Dpp2 transcription in other tissue or cell types. For instance, Gut-enriched Kruppel like factor (GKLF) has been implicated in B cell quiescence (Glynne et al., 2000; Yusuf and Fruman, 2003), and GKLF can inhibit cell cycle progression in RKO cells (Chen et al., 2001). In addition to T cell activation, TOB1 inhibits osteoblast proliferation, albeit using different SMAD transcription factors (Yoshida et al., 2000). Since DPP2 transcripts are detectable at significant levels in most tissue types (Underwood et al., 1999), it is conceivable that DPP2 is a part of a general quiescence program, although it might be controlled by diverse tissue-specific KLF and/or BTG family members. KLF2 transcription is regulated by cytokine stimulation (Schober et al., 1999), by the TNF-receptor associated factor, TRAF2 (Lin et al., 2003), and its ability to transactivate is inhibited by the E3-ubiquitin ligase WWP1 (Conkright et al., 2001). These findings suggest that KLF2 might itself be regulated by upstream signaling events. It remains to be seen whether DPP2 activity results in a feed-forward loop, regulating KLF2 and TOB1 transcription. It is also worth noting that the apoptosis caused by DPP2 inhibition is protected by blocking the ubiquitin-proteasome pathway (Chiravuri et al., 1999). Inhibition of DPP2 might, thus, act to deregulate quiescence-specific factors, like KLF2, through ubiquitination, leading to a loss of KLF2 function (Conkright et al., 2001). This is consistent with KLF2’s role as a quiescence-promoting factor, and with its role in the survival of naïve and memory T cells (Kuo et al., 1997; Schober et al., 1999).
Cellular quiescence is central to our understanding of immunological questions in naïve and memory cell homeostasis. The majority of lymphocytes in vivo are in a resting state, and although there is growing evidence that lymphocyte quiescence is an active process, very few mediators of this quiescence have been identified so far. We have provided a correlation between the transcriptional activators of lymphocyte quiescence, KLF2 and TOB1, and DPP2 expression, a requisite protease involved in quiescent T cell survival.
Acknowledgments
We thank Dr. Albert Tai for help with real-time PCR, and Dr. Mark Ryan for his help in discussion and in reading the manuscript. We also thank the members of the Ananda Roy laboratory at Tufts for help with luciferase assays, and Drs. Rene Bernards, Vassiliki Boussiotis, and Jeff Leiden for generous gifts of plasmids. The authors declare no competing financial interests.
Footnotes
This work was supported by NIH AI43469 to B.T.H. Additional support was provided by the GRASP center at Tufts University (GCRC grant M01-RR00054).
Abbreviations used in this paper: DPP2, Dipeptidyl Peptidase 2; GKLF, Gut-enriched Kruppel like factor; KLF2, Lung Kruppel-like factor; TSS, transcriptional start-site
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banerjee SS, Feinberg MW, Watanabe M, Gray S, Haspel RL, Denkinger DJ, Kawahara R, Hauner H, Jain MK. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem. 2003;278:2581–2584. doi: 10.1074/jbc.M210859200. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Bernards R. Repression of c-Myc responsive genes in cycling cells causes G1 arrest through reduction of cyclin E/CDK2 kinase activity. Oncogene. 1997;15:1347–1356. doi: 10.1038/sj.onc.1201280. [DOI] [PubMed] [Google Scholar]
- Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc--dependent pathway. Nat Immunol. 2001;2:698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiravuri M, Schmitz T, Yardley K, Underwood R, Dayal Y, Huber BT. A novel apoptotic pathway in quiescent lymphocytes identified by inhibition of a post-proline cleaving aminodipeptidase: a candidate target protease, quiescent cell proline dipeptidase. J Immunol. 1999;163:3092–3099. [PubMed] [Google Scholar]
- Conkright MD, Wani MA, Lingrel JB. Lung Kruppel-like factor contains an autoinhibitory domain that regulates its transcriptional activation by binding WWP1, an E3 ubiquitin ligase. J Biol Chem. 2001;276:29299–29306. doi: 10.1074/jbc.M103670200. [DOI] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkinger DJ, Cushman-Vokoun AM, Kawahara RS. Regulation of the vav proto-oncogene by LKLF. Gene. 2001;281:133–142. doi: 10.1016/s0378-1119(01)00792-2. [DOI] [PubMed] [Google Scholar]
- Freitas AA, Rocha B. Population biology of lymphocytes: the flight for survival. Annu Rev Immunol. 2000;18:83–111. doi: 10.1146/annurev.immunol.18.1.83. [DOI] [PubMed] [Google Scholar]
- Glynne R, Ghandour G, Rayner J, Mack DH, Goodnow CC. B-lymphocyte quiescence, tolerance and activation as viewed by global gene expression profiling on microarrays. Immunol Rev. 2000;176:216–246. doi: 10.1034/j.1600-065x.2000.00614.x. [DOI] [PubMed] [Google Scholar]
- Jonk LJ, Itoh S, Heldin CH, ten Dijke P, Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Leiden JM. Transcriptional regulation of T lymphocyte development and function. Annu Rev Immunol. 1999;17:149–187. doi: 10.1146/annurev.immunol.17.1.149. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- Levens DL. Reconstructing MYC. Genes Dev. 2003;17:1071–1077. doi: 10.1101/gad.1095203. [DOI] [PubMed] [Google Scholar]
- Lin Y, Ryan J, Lewis J, Wani MA, Lingrel JB, Liu ZG. TRAF2 exerts its antiapoptotic effect by regulating the expression of Kruppel-like factor LKLF. Mol Cell Biol. 2003;23:5849–5856. doi: 10.1128/MCB.23.16.5849-5856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Rouault J, Magaud J, Berthet C. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 2001;497:67–72. doi: 10.1016/s0014-5793(01)02436-x. [DOI] [PubMed] [Google Scholar]
- Meffre E, Nussenzweig MC. Deletion of immunoglobulin beta in developing B cells leads to cell death. Proc Natl Acad Sci U S A. 2002;99:11334–11339. doi: 10.1073/pnas.172369999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler AI. Studies on contact inhibition of growth in the mouse fibroblast, 3T3. II. Effects of amino acid deprivation and serum on growth rate. J Cell Sci. 1973;12:861–873. doi: 10.1242/jcs.12.3.861. [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Elstrom RL, Cinalli RM, Thompson CB. Activated Akt promotes increased resting T cell size, CD28-independent T cell growth, and development of autoimmunity and lymphoma. Eur J Immunol. 2003;33:2223–2232. doi: 10.1002/eji.200324048. [DOI] [PubMed] [Google Scholar]
- Schober SL, Kuo CT, Schluns KS, Lefrancois L, Leiden JM, Jameson SC. Expression of the transcription factor lung Kruppel-like factor is regulated by cytokines and correlates with survival of memory T cells in vitro and in vivo. J Immunol. 1999;163:3662–3667. [PubMed] [Google Scholar]
- Smith SH, Cancro MP. Cutting edge: B cell receptor signals regulate BLyS receptor levels in mature B cells and their immediate progenitors. J Immunol. 2003;170:5820–5823. doi: 10.4049/jimmunol.170.12.5820. [DOI] [PubMed] [Google Scholar]
- Tanchot C, Lemonnier FA, Perarnau B, Freitas AA, Rocha B. Differential requirements for survival and proliferation of CD8 naive or memory T cells. Science. 1997;276:2057–2062. doi: 10.1126/science.276.5321.2057. [DOI] [PubMed] [Google Scholar]
- Tirone F. The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J Cell Physiol. 2001;187:155–165. doi: 10.1002/jcp.1062. [DOI] [PubMed] [Google Scholar]
- Torcia M, Bracci-Laudiero L, Lucibello M, Nencioni L, Labardi D, Rubartelli A, Cozzolino F, Aloe L, Garaci E. Nerve growth factor is an autocrine survival factor for memory B lymphocytes. Cell. 1996;85:345–356. doi: 10.1016/s0092-8674(00)81113-7. [DOI] [PubMed] [Google Scholar]
- Turner J, Crossley M. Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci. 1999;24:236–240. doi: 10.1016/s0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- Tzachanis D, Freeman GJ, Hirano N, van Puijenbroek AA, Delfs MW, Berezovskaya A, Nadler LM, Boussiotis VA. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat Immunol. 2001;2:1174–1182. doi: 10.1038/ni730. [DOI] [PubMed] [Google Scholar]
- Underwood R, Chiravuri M, Lee H, Schmitz T, Kabcenell AK, Yardley K, Huber BT. Sequence, purification, and cloning of an intracellular serine protease, quiescent cell proline dipeptidase. J Biol Chem. 1999;274:34053–34058. doi: 10.1074/jbc.274.48.34053. [DOI] [PubMed] [Google Scholar]
- Wani MA, Wert SE, Lingrel JB. Lung Kruppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J Biol Chem. 1999;274:21180–21185. doi: 10.1074/jbc.274.30.21180. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Tanaka S, Umemori H, Minowa O, Usui M, Ikematsu N, Hosoda E, Imamura T, Kuno J, Yamashita T, Miyazono K, Noda M, Noda T, Yamamoto T. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell. 2000;103:1085–1097. doi: 10.1016/s0092-8674(00)00211-7. [DOI] [PubMed] [Google Scholar]
- Yusuf I, Fruman DA. Regulation of quiescence in lymphocytes. Trends Immunol. 2003;24:380–386. doi: 10.1016/s1471-4906(03)00141-8. [DOI] [PubMed] [Google Scholar]