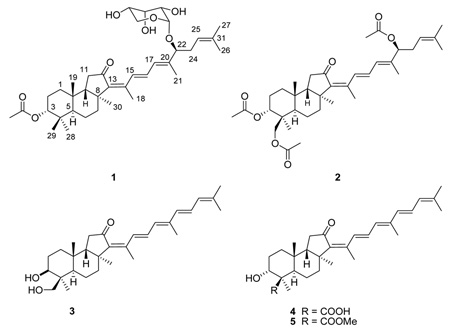
Abstract
Four isomalabaricane triterpenoids were isolated from an extract of the sponge Rhabdastrella globostellata that was active in an assay measuring stabilization of the binding of DNA with DNA polymerase β. The known compounds stelliferin riboside (1), and 3-epi-29-acetoxystelliferin E (2) were shown to induce 29% and 23% binding respectively at 28 µg/mL, while the new compound stellettin J (3), induced 5% binding at 28 µg/mL. The new compound stellettin K (4) had no activity in the binding assay. The compounds were characterized by spectroscopic methods. These compounds displayed varying levels of activity toward the A2780 ovarian cancer cell line, revealing structure-based effects on both the level of cytotoxicity and DNA–polymerase β binding. This is the first report of natural products with the ability to promote stabilization of the DNA–polymerase β covalent binary complex.
DNA-damaging agents such as bleomycins and ionizing radiation are used clinically for the treatment of a variety of tumor types. Although this has been successful, these agents are often accompanied by undesirable toxic side effects. Improving the efficacy of clinically used DNA-damaging agents is desirable, so that patients can benefit from the therapeutic treatment while limiting side effects. Combination therapy using multiple antitumor agents concurrently is standard procedure in anticancer chemotherapy. One possible combination treatment would be to combine the administration of a DNA-damaging agent with an inhibitor of DNA repair.
An interesting target for the inhibition of DNA repair is the DNA repair enzyme DNA polymerase β. Polymerase β is a key enzyme in base excision repair, which is required if a purine or pyrimidine base is damaged or excised within a strand of DNA. It has been shown that naturally occurring inhibitors of polymerase β can potentiate the activity of bleomycins in mammalian cell culture.1
In previous work we have reported the isolation of naturally occurring inhibitors of the lyase2 and polymerase3 activities of polymerase β. We have recently implemented a gel mobility shift assay to quantify the binding of a synthetic DNA oligonucleotide substrate with polymerase β. Just as camptothecin is known to bind to the topoisomerase I–DNA covalent binary complex, and thereby alters the equilibrium between free and DNA-bound enzyme, so promoting polymerase β–DNA binding should disrupt and effectively inhibit the DNA repair activity of polymerase β. Herein we report the isolation, characterization, and biological evaluation of the natural products 1–4 that are believed to stabilize the covalent binary complex formed between DNA and polymerase β, which is an obligatory intermediate in the lyase reaction.
The genus Rhabdastrella is part of the family Ancorinidae, and consists of marine sponge species that are distributed around the south Pacific region. Several chemical studies of Rhabdastrella species have been performed.4–8 Compounds from the genus Rhabdastrella have been shown to have a variety of biological activities with good potency. In addition, chemical studies have been carried out on some sponges that were not initially classified as Rhabdastrella sp. but were subsequently identified as such, as for example a collection originally identified as Jaspis stellifera.4 (Sentence deleted here) The compounds isolated most commonly from Rhabdastrella globostellata (Carter, 1883) are isomalabaricane triterpenoids.4,6–7 These compounds are prone to photoisomerization that can adversely affect bioactivity,5 and so work on them must be performed under conditions of subdued lighting.
Results and Discussion
Isolation and Characterization of Compounds 1 – 4
The crude bioactive MeOH/CH2Cl2 extract of R. globostellata was fractionated initially by use of an aminopropyl SPE cartridge. This first step was selected because acidic compounds from the genus Rhabdastrella have been shown to be cytotoxic, and aminopropyl SPE cartridges are useful for the selective retention of carboxylic acids.9 This fractionation by aminopropyl SPE afforded four fractions, one of which (fraction A, 2:1 CHCl3-i-PrOH wash) was active in the binding assay. The putative carboxylic acid-containing fraction (fraction B, 2% HOAc-ethyl ether wash) was not active in the polymerase β binding assay, but, as expected, it was active in the A2780 cytotoxicity assay.
Fraction A was further fractionated by reversed-phase HPLC to afford nine fractions (fractions A-1 through A-9), six of which were active in the binding assay. Fraction A contained compounds with significant UV absorbance in the region 340–400 nm, based on comparisons of chromatograms from a photodiode array (PDA) and an evaporative light scattering detector (ELSD), consistent with the possible presence of isomalabaricane-type compounds. Stelliferin riboside (1) was isolated by reversed-phase HPLC from the major active fraction A-3, and 3-epi-29-acetoxystelliferin E (2) was isolated in very low yield by repeated cyano normal-phase and C18 reversed-phase HPLC of fraction A-4. Stellettin J (3) was isolated from fraction A-5 by the same procedure as was used for 2, and stellettin K (4) was isolated from fraction B as the major cytotoxic component by C18 reversed-phase HPLC.
Compound 1 was isolated as a pale yellow amorphous solid. Sentence deleted here. Following full spectroscopic analysis, 1 was identified as the isomalabaricane triterpenoid stelliferin riboside based on comparison with data reported previously by Tabudravu and Jaspars.10 The published structure for stelliferin riboside left the stereochemistry at C-22 undefined. The small amount of sample available coupled with the need to preserve material for the biological studies described below precluded direct determination of this stereochemistry, but we have assigned the stereochemistry as 22S based on analogy to the absolute stereochemistry of 2, as explained below. Compound 1 was thus assigned as 3-epi-stelliferin A 22-α-ribopyranoside.
Compound 2 was isolated as an amorphous pale yellow solid. Sentence deleted here. Following a full spectroscopic analysis, a comparison of the 1H NMR spectrum (recorded in C6D6) and HRFABMS data for 2 with literature data revealed that 2 is identical to an isomalabaricane triterpenoid prepared as a semisynthetic derivative of a natural product by Oku et al.5 The absolute stereochemistry for 2 has been determined previously, and the reported optical rotation for 2 agreed well with that determined in the present study ( −45°, c 0.08, MeOH; lit.: −50°, c 0.05, MeOH). Thus, the absolute stereochemistry for 2 was assigned as shown. From a biogenetic perspective, the stereochemistry of the ring systems and of C-22 for 1 and 2 are most probably the same. On this basis, compounds 1 and 2 were assigned the same absolute configuration at C-22, and 2 was assigned as 3-epi-29-acetoxystelliferin E.
Compound 3 was isolated as an amorphous bright yellow solid. Positive-ion HRFABMS analysis indicated that it possesses the molecular formula C30H44O3, and its UV spectrum, with λmax 396 nm (log ε 4.46) in MeOH, was consistent with the presence of a conjugated pentaenone. The 1H NMR spectrum of 3 had many similarities to the analogous spectra of 1 and 2. Six olefinic signals were detected (δH 5.94, 6.23, 6.28, 6.52, 6.98 and 8.05), along with two oxygenated methylene protons (δH 3.57 and 3.79), an oxygenated methine proton (δH 4.09), and seven methyl singlets (δH 1.02, 1.10, 1.36, 1.82, 1.83,1.96 and 2.02). 1H NMR and 13C NMR spectroscopic data for 3 are shown in Table 1. From the 13C NMR spectrum of 3, nine sp2 carbon signals were detected, of which one (δC 126.2) consisted of two overlapped signals. A carbonyl signal (δC 206.7), and two oxygenated carbon signals (δC 68.2 and 71.8) were also observed, along with eighteen other signals. Based on the UV absorbance and the NMR data, 3 also appeared to be an isomalabaricane triterpenoid like 1 and 2, but the data for 3 did not match those of any known compound.
Table 1.
NMR Data for Compounds 3–4 (CDCl3).
| position | 3 | 4 | ||||
|---|---|---|---|---|---|---|
| δC | δH | multiplicity | δC | δH | multiplicity | |
| 1 | 29.1 | 1.23 | m | 28.8 | 1.14 | m |
| 1.85 | m | 1.86 | m | |||
| 2 | 26.4 | 1.71 | m | 27.9 | 1.69 | m |
| 1.86 | m | 2.20 | bd, | |||
| 3 | 71.8 | 4.09 | dd, 2.4, 7.8 | 70.8 | 4.16 | br s |
| 4 | 43.3 | 47.9 | ||||
| 5 | 42.4 | 2.23 | m | 40.6 | 2.46 | br d, 11.2 |
| 6 | 19.4 | 1.42 | m | 20.3 | 1.87 | m |
| 1.66 | m | |||||
| 7 | 38.8 | 2.07 | m | 38.9 | 2.12 | m |
| 8 | 44.8 | 45.0 | ||||
| 9 | 50.3 | 1.75 | m | 49.7 | 1.84 | m |
| 10 | 34.9 | 36.2 | ||||
| 11 | 37.1 | 2.19 | m | 37.2 | 2.24 | m |
| 12 | 206.7 | 206.9 | ||||
| 13 | 145.8 | 145.9 | ||||
| 14 | 143.1 | 143.1 | ||||
| 15 | 132.3 | 8.05 | d, 15.3 | 132.4 | 8.05 | d, 15.2 |
| 16 | 131.4 | 6.98 | dd, 11.5, 15.3 | 131.4 | 6.98 | dd, 11.2, 15.2 |
| 17 | 131.7 | 6.28 | d, 11.5 | 131.8 | 6.29 | d, 11.6 |
| 18 | 16.2 | 2.02 | s | 16.2 | 2.04 | s |
| 19 | 25.3 | 1.02 | S | 19.9 | 0.91 | s |
| 20 | 139.3 | 139.3 | ||||
| 21 | 13.2 | 1.96 | S | 13.2 | 1.96 | s |
| 22 | 135.0 | 6.23 | d, 15.2 | 135.0 | 6.24 | d, 15.2 |
| 23 | 126.2 | 6.52 | dd, 11.1, 15.2 | 126.2 | 6.52 | dd, 11.2, 15.2 |
| 24 | 126.2 | 5.94 | d, 11.0 | 126.2 | 5.94 | d, 11.2 |
| 25 | 137.1 | 137.0 | ||||
| 26 | 18.8 | 1.82 | S | 18.8 | 1.82 | s |
| 27 | 26.5 | 1.83 | S | 26.5 | 1.83 | s |
| 28 | 19.9 | 1.10 | S | 23.8 | 1.33 | s |
| 29 | 68.2 | 3.57 | d, 10.4 | 183.3 | ||
| 3.79 | d, 10.4 | |||||
| 30 | 24.6 | 1.36 | S | 24.9 | 1.40 | s |
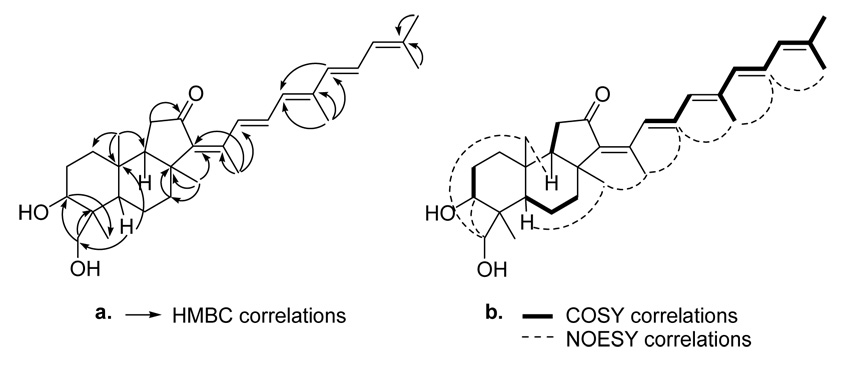
COSY NMR correlations (Figure 1) for the olefinic proton signals of 3 revealed two spin systems, one of which displayed allylic coupling to two methyl singlets. Also, the oxygenated methine proton at δH 4.09 was coupled to proton signals at δH 1.71 and 1.86, suggesting the presence of a CH2-CH(OH)-C moiety. Key HMBC correlations (Figure 1) depicted for 3 showed the proximity of the hydroxylated carbons (δC 68.2 and 71.8) in 3. Also, correlations from one olefinic proton signal (δH 6.23) to a carbon in a different spin system (δC 131.4) allowed the two allylic spin systems apparent from the COSY analysis to be connected. The structure of the remainder of the skeleton was assigned based on HMBC data and was found to be consistent with that of an isomalabaricane triterpenoid.
Figure 1.
Key 2D NMR correlations for 3. (a) HMBC correlations. (b) COSY and NOESY correlations.
NOESY correlations (Figure 1) confirmed the assignment of the double bond configurations as shown (15E, 17E, 22E). The downfield shift of the olefinic proton signal at δH 8.05 suggested that this proton was deshielded by the ketone at C-12, thus indicating the double bond configuration at C-13 as 13Z. NOESY correlations suggested that the methane protons at H-9, H-19, and the methylene protons H-29 were all on the same side of the tricyclic skeleton. NOESY data also implied that the methyl protons at H-30 were on the same face of the tricyclic skeleton as the methine proton at H-5. Finally, the signal for H-3 showed a strong correlation with the oxygenated methine proton signal for H-29b, which suggested that the hydroxyl group at C-3 was in an axial configuration. These correlations together suggested that 3 possesses an isomalabaricane skeleton.5 The absolute stereochemistry of the tricyclic ring system was assumed to be the same as that determined for 2, based on the fact that the two molecules likely arise from a common biosynthetic precursor. Compound 3 is thus (13Z,15E,17E,22E)-3α,29-dihydroxy-12-oxoisomalabarica-13,15,17,22,24-pentaene, to which we have assigned the name stellettin J in keeping with the convention of naming similar isomalabaricane triterpenoids.
Compound 4 was isolated as a bright yellow amorphous solid from fraction B of the aminopropyl SPE separation and was the major component of the crude extract. Although not active in the binding assay, fraction B had an IC50 of 0.9 µg/mL in the A2780 ovarian cancer cell line cytotoxicity assay. The 1H and 13C NMR data for 4 (Table 1) revealed a striking similarity to those of 3, indicating that 4 is likely a congener of 3. Compound 4 had λmax 401 nm (log ε 4.59) in MeOH, supporting the structural similarity of 3 and 4. Positive-ion HRFABMS analysis indicated that the molecular formula of 4 is C30H42O4. Since the aminopropyl fractionation procedure putatively isolates carboxylic acids in the 2% HOAc-ethyl ether wash (fraction B), and based on the molecular formula for 4, it appeared that 4 might be a carboxylic acid analog of 3. This was supported by the presence of a carboxyl carbonyl signal at δC 183.3 that was not observed for 3. A search of the literature revealed that methyl (13Z,15E,17E,22E)-3α-hydroxy-12-oxoisomalabarica-13,15,17,22,24-pentaen-29-oate, which is the C-29 methyl ester of 4, had been isolated by purification of a fully methylated crude extract from the sponge Jaspis stellifera, but that the free acid had not been characterized.11 A comparison of the NMR data for 4 with those of methyl (13Z,15E,17E,22E)-3β-hydroxy-12-oxoisomalabarica-13,15,17,22,24-pentaen-29-oate revealed great similarities, with the only major differences being attributable to the replacement of a COOH group (δc 183.3) by a COOCH3 group (δc 177.9). The structure shown for 4 was confirmed by analyses of HSQC, COSY, HMBC, and NOESY NMR spectra, and all the observed correlations were essentially identical to those observed for 3. Compound 4 was thus assigned the structure (13Z,15E,17E,22E)-3β-hydroxy-12-oxoisomalabarica-13,15,17,22,24-pentaen-29-oic acid.
Biological Characterization of Compounds 1 – 4
A radiolabeled DNA substrate for polymerase β containing a single apurinic acid lesion was incubated in the presence and absence of polymerase β. Polyacrylamide gel electrophoresis of each mixture afforded a single major band. When incubated in the presence of compound 1, a new band appeared on the native gel. In some experiments the same band appeared transiently in the absence of any stabilizer; it is believed to represent the covalent binary complex formed as an obligatory intermediate between the lyase center of polymerase β and the DNA lesion that is excised from damaged DNA.3 Similar experiments were also carried out on compounds 2 – 3, and the results are shown in Table 2. A photograph of the mobility shift assay gel plate for 1 is available as Supporting Information.
Table 2.
Bioassay Data for Compounds 1–4.
| compound | polymerase β-DNA binary complex (%) |
cytotoxicity toward A2780 cells (µM) |
|---|---|---|
| 1 | 29 | 7.3 |
| 2 | 23 | 27 |
| 3 | 8 | 1.2 |
| 4 | ND | 0.28 |
| Actinomycin D | ND | 0.0017 |
Compounds 1–4 were also evaluated for their cytotoxicities toward cultured A2780 cells (Table 2). All compounds isolated were shown to be cytotoxic toward cultured A2780 cells. It is interesting to note that the compounds promoting DNA–polymerase β stabilization (1 and 2) have side chains that are not fully unsaturated. Perhaps the flexibility afforded to the side chain by the absence of the C-22:C-23 double bond allows for more efficient interaction with the enzyme–DNA covalent binary complex. At the same time, the flexibility of the side chain was correlated with reduced cytotoxicity, arguing that stabilization of the enzyme–DNA binary complex was not a primary cause of cytotoxicity, at least in the absence of a DNA-damaging agent. Though little can be concluded concerning the mechanism of cytotoxicity of 1–4, it is interesting to note that previous studies have shown that stellettins have a cytotoxicity profile in the NCI 60-cell line panel that is most similar to that of the schweinfurthins, which are prenylated stilbene-type compounds with side chains similar to those of 3 and 4.12
Experimental Section
General Experimental Procedures
Optical rotations were measured on a Perkin-Elmer 241 polarimeter. UV spectra were recorded using a Shimadzu UV-1201 spectrophotometer. IR spectra were recorded for neat samples with a MIDAC M-series FTIR spectrophotometer. NMR spectra were obtained on either a Varian Inova 400 spectrometer operating at 399.9 MHz for 1H and 100.6 MHz for 13C, or a JEOL Eclipse+ 500 spectrometer operating at 500.2 MHz for 1H and 125.8 MHz for 13C. HRFABMS were recorded with a JEOL HX-110 spectrometer. The [α-32P] ddATP (3000 Ci/mmol) and terminal deoxynucleotidyltransferase for 3'-end labeling were purchased from Amersham Pharmacia Biotech. Uracil-DNA glycosylase (1000/mL) and AP endonuclease were from New England Biolabs. Distilled, deionized water from a Milli-Q system was used for all aqueous manipulations. Polyacrylamide gel loading solution contained 10 M urea, 1.5 mM EDTA, 0.05% (w/v) xylene cyanol and 0.05% (w/v) bromophenol blue. Gels were visualized and quantified using a Molecular Dynamics PhosphorImager with ImageQuant version 3.2 software.
Marine Sample Extraction
The sponge sample used in this work was collected for the National Cancer Institute by Pat Colin of the Coral Reef Research Foundation in Fiji on October 30, 1996, at 12 meters depth. The taxonomist was Michele Kelly (National Institute of Water and Atmospheric Research, Auckland, New Zealand) and a voucher is at the Smithsonian Department of Worms under the collector number 0CDN4278. A photograph of the sample is available as Supporting Information. The deep-frozen sample was pulverized at the National Cancer Institute in dry ice by use of a worm-fed grinder (hamburger mill), the powder produced was allowed to stand at −30 °C until the CO2 sublimed, and the mass was then extracted at 4 °C with deionized water (1 L) by stirring (30 rpm) for 30 min. The mixture was centrifuged at room temperature and the supernatant lyophilized to give the aqueous extract. The insoluble portion from the centrifugation was lyophilized and then statically extracted overnight at room temperature with 1 L of 1:1 MeOH-CH2Cl2. The organic phase was filtered, the pellet was washed with a 10% volume of fresh MeOH, and the combined organic phase was concentrated to dryness at < 35 °C by rotary evaporation and then finally dried under vacuum at room temperature to give the organic extract as a gum. An extract of this sponge was received from the National Cancer Institute as sample number C016375 (3.0 g).
Extraction and Isolation
Isolation procedures were performed under darkened laboratory conditions with aluminum foil-covered glassware, and fractions were stored dry at −20 °C in order to avoid light and temperature sensitivity problems. The crude extract (314 mg) was fractionated by use of an aminopropyl SPE cartridge (Supelco) to afford four fractions: a 2:1 CHCl3-i-PrOH wash (fraction A, 91 mg), a 2% acetic acid-ethyl ether wash (fraction B, 109 mg), a methanol wash (fraction C, 96 mg), and an 8% acetic acid-ethyl ether wash (fraction D, 6 mg). Fraction A was active in the polymerase β binding assay (18% binding), while fraction B was active in the A2780 cytotoxicity assay. Fractions C and D were inactive in the binding assay, and were not investigated further. Fraction A was further fractionated by use of preparative C18 reversed-phase HPLC (isocratic, 92% aq. MeOH) to afford nine fractions, six of which were active. The most active fraction (fraction A-3, 11.5 min., 8 mg) was purified further by C18 reversed-phase HPLC (89% aq. MeOH, to afford 1 (4 mg, 29% binding). From another active fraction (fraction A-4, 13.5 min, 2.5 mg), repeated cyano normal-phase (isocratic, 84:16 hexane-i-PrOH) and C18 reversed-phase (isocratic, 92% aq. MeOH) HPLC afforded 2 (0.4 mg, 23% binding; separation later scaled up to afford 0.9 mg). A third active fraction (fraction A-5) was unstable, so after scaling up and repeating the separation, fraction A-5 (15 min, 28 mg) was isolated. Compound 3 (4 mg, 5.3% binding) was isolated from fraction A-5 after repeated cyano normal-phase and C18 reversed-phase HPLC (same conditions as above). From fraction B of the aminopropyl separation, compound 4 was isolated by reversed-phase HPLC (isocratic, 92% aq. MeOH).
3-epi-29-Acetoxystelliferin E (2)
Light yellow powder; −45° (c 0.08, MeOH); UV (MeOH) (log ε 4.34) λmax 342; IR (neat film) νmax 2915, 2849, 1732, 1693, 1373, 1234, 1018, 798 cm−1; 1H NMR (CDCl3), 1.00 (3H, s, H-28), 1.02 (3H, s, H-19), 1.21 (1H, m, H-1), 1.41 (s, H-30), 1.52 (1H, m, H-6), 1.54 (1H, m, H-1), 1.61 (s, H-27), 1.68 (s, H-26), 1.70 (1H, m, H-6), 1.78 (1H, m, H-2), 1.82 (s, H-21), 1.85 (1H, m, H-9), 1.99 (1H, m, H-2), 2.01 (s, H-18), 2.10 (1H, m, H-7), 2.21 (1H, m, H-11), 2.30 (1H, m, H-23), 2.37 (1H, m, H-23), 2.39 (1H, m, H-5), 4.01 (1H, d, J = 11.2 Hz, H-29), 4.17 (1H, d, J = 11.6 Hz, H-29), 4.98 (1H, bs, H-24), 5.00 (1H, t, J = 6.8 Hz, H-3), 5.15 (1H, t, J = 6.6 Hz, H-22), 6.25 (1H, d, J = 10.8 Hz, H-17), 6.81 (1H, dd, J = 11.2, 15.6 Hz, H-16), 7.99 (1H, d, J = 15.2 Hz, H-15), 2.05 (3H, s, CH3CO-29), 2.06 (3H, s, CH3CO-22), 2.08 (3H, s, CH3CO-3); 13C NMR (CDCl3), 13.7 (C-21), 16.2 (C-18), 18.1 (C-27), 19.0 (C-6), 22.4 (C-28), 22.5 (C-19), 24.5 (C-2), 24.5 (C-30), 26.0 (C-26), 29.3 (C-1), 32.0 (C-23), 35.5 (C-10), 36.8 (C-11), 38.9 (C-7), 41.5 (C-4), 42.3 (C-5), 44.7 (C-8), 50.4 (C-9), 67.3 (C-29), 73.7 (C-3), 78.6 (C-22), 118.9 (C-24), 127.9 (C-16), 130.1 (C-17), 132.9 (C-15), 134.8 (C-25), 139.2 (C-20), 142.6 (C-14), 146.3 (C-13), 206.4 (C-12), 21.5 (CH3CO-3), 170.4 (CH3CO-3), 21.4 (CH3CO-22), 170.4 (CH3CO-22), 21.1 (CH3CO-29), 171.5 (CH3CO-29) (C-19 and C-28, and acetate ester carbons may be interchanged); HRFABMS (positive ion) m/z 596.3748 ([M]+ calcd for C36H52O7: 596.3714).
Stellettin J (3)
Bright yellow powder; −13° (c 0.3, CHCl3); UV (MeOH) (log ε 4.46) λmax 396; IR (neat film) νmax 3432, 2917, 2849, 1674, 1555, 1536, 1449, 1378, 1205, 1028, 970 cm−1; 1H NMR (CDCl3), see Table 1; 13C NMR (CDCl3), see Table 1; HRFABMS (positive ion) m/z 453.3368 ([M+H]+ calcd for C30H45O3: 453.3369).
Stellettin K (4)
Bright yellow powder; +56° (c 0.1, CHCl3); UV (MeOH) (log ε 4.59) λmax 401; IR (neat film) νmax 3453, 2926, 1688, 1559, 1536, 1449, 1209, 1163, 974 cm−1; 1H NMR (CDCl3), see Table 1; 13C NMR (CDCl3), see Table 1; HRFABMS (positive ion) m/z 467.3174 ([M+H]+ calcd for C30H43O4: 467.3161).
DNA Binding Mobility Shift Assay
The affinity of polymerase β for a radiolabeled 36-nulceotide DNA substrate containing an apurinic site at position 20 was studied using a gel mobility assay in the presence and absence of the polymerase β inhibitors. Rat DNA polymerase β (30 nM) was incubated with 200 nM radiolabeled DNA substrate, and the tested samples (30 – 500 µM, dissolved in DMSO) in buffer containing 10 mM K Hepes, pH 7.4, 50 mM KCl, 5 mM MgCl2 and 10 mg/mL BSA (10 µL total volume) at 37 °C for 2 h. Samples were loaded onto a 12% native polyacrylamide gel and visualized by autoradiography. Bound protein was quantified using ImageQuant software, after scanning the gel using a Molecular Dynamics Phosphorimager model 450.
The 36-nucleotide oligodeoxyribonucleotide containing a uridine at position 20 on one strand was labeled at its 3'-end with terminal deoxynucleotidyltransferase + [α-32P]ddATP. The product was then purified by 20% denaturing polyacrylamide gel electrophoresis. The band of interest was visualized by autoradiography and excised from the gel. After removal by the “crush and soak ” method, the oligodeoxyribonucleotide was annealed to its complementary strand by heating the solution at 70 °C for 3 min, followed by slow cooling to 25 °C.
The apurinic site was created in the DNA substrate in a reaction mixture (200 µL total volume) that contained 354 nM [α-32P]-labeled double-stranded oligodeoxynucleotide having a uridine at position 20 in 10 mM K Hepes, pH 7.4, 50 mM KCl, 5 mM MgCl2, 10 mg/mL bovine serum albumin, 3 units AP endonuclease, and 2.4 units uracil-DNA glycosylase. After incubation at 37 °C for 20 min, the [α-32P]-labeled double-stranded oligodeoxynucleotide containing an AP site at position 20 was ready for DNA binding mobility shift assay.
A2780 cytotoxicity assay
The A2780 ovarian cancer cell line cytotoxicity assay was performed by Mr. Andrew Norris at Virginia Polytechnic Institute and State University as previously reported.13
Supplementary Material
1H NMR spectra for 1 and 2, a photograph of R. globostellata, and a photograph of the binding assay gel plate for 1. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgement
This work was funded by a National Cooperative Drug Discovery Group grant awarded by the National Cancer Institute to the University of Virginia (U19 CA 50771, Dr. S. M. Hecht, Principal Investigator), and this support is gratefully acknowledged. We thank the Natural Products Branch, National Cancer Institute, for the supply of the marine extract. We thank Andrew Norris for cytotoxicity data, William R. Bebout, Sr. (VPISU) for mass spectrometric data, and Thomas E. Glass (VPISU) for assistance with NMR spectroscopy.
Footnotes
Dedicated to Dr. Norman R. Farnsworth of the University of Illinois at Chicago for his pioneering work on bioactive natural products.
References and Notes
- 1.Sun D-A, Deng J-Z, Starck SR, Hecht SM. J. Am. Chem. Soc. 1999;121:6120–6124. [Google Scholar]
- 2.Cao S, Gao Z, Thomas SJ, Hecht SM, Lazo JS, Kingston DGI. J. Nat. Prod. 2004;67:1716–1718. doi: 10.1021/np049849+. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SM. Pharm. Biol. 2003;41 Suppl.:68–77. [Google Scholar]
- 4.Tasdemir D, Mangalindan GC, Concepción GP, Verbitski SM, Rabindran S, Miranda M, Greenstein M, Hooper JNA, Harper MK, Ireland CM. J. Nat. Prod. 2002;65:210–214. doi: 10.1021/np0104020. [DOI] [PubMed] [Google Scholar]
- 5.Oku N, Matsunaga S, Wada S, Watabe S, Fusetani N. J. Nat. Prod. 2000;63:205–209. doi: 10.1021/np990333d. [DOI] [PubMed] [Google Scholar]
- 6.Rao Z, Deng S, Wu H, Jiang S. J. Nat. Prod. 1997;60:1163–1164. [Google Scholar]
- 7.Bourguet-Kondracki M-L, Longeon A, Debitus C, Guyot M. Tetrahedron Lett. 2000;41:3087–3090. [Google Scholar]
- 8.Lv F, Deng Z, Li J, Fu H, van Soest RWM, Proksch P, Lin W. J. Nat. Prod. 2004;67:2033–2036. doi: 10.1021/np040145+. [DOI] [PubMed] [Google Scholar]
- 9.Blevins DD, Burke MF, Good TJ, Harris PA, Van Horne KC, Simpson N, Yago LS, editors. Varian Sorbent Extraction Technology Handbook. Harbor City, CA: Varian Sample Preparation Products; 1993. pp. 82–84. [Google Scholar]
- 10.Tabudravu JN, Jaspars M. J. Nat. Prod. 2001;64:813–815. doi: 10.1021/np010019v. [DOI] [PubMed] [Google Scholar]
- 11.Ravi BN, Wells RJ. Aust. J. Chem. 1982;35:39–50. [Google Scholar]
- 12.Meragelman KM, McKee TC, Boyd MR. J. Nat. Prod. 2001;64:389–392. doi: 10.1021/np000478g. [DOI] [PubMed] [Google Scholar]
- 13.Louie KG, Behrens BC, Kinsella TJ, Hamilton TC, Grotzinger KR, McKoy WM, Winker MA, Ozols RF. Cancer Res. 1985;45:2110–2115. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1H NMR spectra for 1 and 2, a photograph of R. globostellata, and a photograph of the binding assay gel plate for 1. This material is available free of charge via the Internet at http://pubs.acs.org.