SUMMARY
The effect of single amino acid substitutions associated with the Italian (E22K), Arctic (E22G), Dutch (E22Q) and Iowa (D23N) familial forms of Alzheimer’s disease (AD) and cerebral amyloid angiopathy on the structure of the 21-30 fragment of the Alzheimer amyloid β-protein (Aβ) is investigated using replica exchange molecular dynamics simulations. The 21-30 segment has been shown in our earlier work to adopt a bend structure in solution that may serve as the folding nucleation site for Aβ. Our simulations reveal that the 24-28 bend motif is retained in all E22 mutants, suggesting that mutations involving residue E22 may not affect the structure of the folding nucleation site of Aβ. Enhanced aggregation in Aβ with familial AD substitutions may result from the depletion of the E22-K28 salt bridge that destabilizes the bend structure. Alternately, the E22 mutations may affect longer-range interactions outside the 21-30 segment that can impact the aggregation of Aβ. Substituting at residue D23, on the other hand, leads to the formation of a turn rather than a bend motif, implying that in contrast to E22 mutants, the D23N mutant may affect monomer Aβ folding and subsequent aggregation. Our simulations suggest that the mechanisms by which E22 and D23 mutations affect the folding and aggregation of Aβ are fundamentally different.
Keywords: Alzheimer’s disease, amyloid β-protein, molecular dynamics simulations, replica exchange, familial Alzheimer’s disease, peptide folding and aggregation
INTRODUCTION
The self-assembly of the amyloid β-protein (Aβ) into aggregates, ranging from small oligomers to amyloid fibrils, is linked causally with Alzheimer’s disease (AD).1 Aβ is present in the brain in two main alloforms, Aβ(1-40) and Aβ(1-42). While most instances of AD are sporadic, a number of familial forms of AD and the related amyloidosis cerebral amyloid angiopathy have been identified which lead to early onset and increased severity of the disease. Interestingly, several familial forms of AD are characterized by single amino-acid mutations at residues E22 or D23 of Aβ. These include the Italian (E22K), Arctic (E22G), Dutch (E22Q), and Iowa (D23N) familial mutants. Aβ(1-40) and Aβ(1-42) variants containing these mutations have been found to be more neurotoxic and to aggregate more readily than the wild-type (WT) peptide in in vitro experiments.2
The first step in aggregation involves the rearrangement of the monomeric peptide into a conformation facilitating self-assembly. Experimental studies as well as fully atomic simulations on full length Aβ indicate that this peptide is mostly unstructured in solution, but bears some regions of structural order.3-5 To investigate further the regions that have intrinsic structure in this peptide, we performed limited proteolysis/mass spectrometry experiments on full-length monomeric Aβ. Limited proteolysis has been a useful technique for revealing domain structure (resistant regions) in folded proteins6-8 These experiments revealed a protease-resistant segment comprising residues 21-30 (sequence A21E22D23VGSNKGA30). In fact, NMR and molecular dynamics simulations of the Aβ(21-30) fragment showed that this peptide adopts a stable bend structure spanning residues 24-28 (see Fig. 1, top panel). A bend/loop structure in the central part of Aβ(21-30) was reported in many previous experimental and theoretical papers.9-13 The same 24-28 bend structure was also seen in simulations of monomeric Aβ fragments 12-28,14 15-28,15 10-3516-19 and of the full length Aβ(1-40) and Aβ(1-42) peptides.5 Although the precise population of the bend was seen to depend on the length of the Aβ fragment,5,10,15,16 its very existence implies that it may serve as the folding nucleation site for the full length Aβ peptide.9 It is striking that the Italian, Arctic, Dutch and Iowa familial mutants all involve mutations directly preceding the 24-28 region. This raised the possibility that mutations in this region could affect folding nucleation and hence the ability of Aβ to aggregate. To further explore this idea, we recently performed limited proteolysis and NMR experiments on familial mutants of the Aβ(21-30) fragment.20 Analysis of the kinetics of proteolysis revealed differences in the free energies of folding of the mutant Aβ peptides relative to the wild type peptide that reached 2.6 kT (a >10-fold decrease in stability). Decreased bend stability was accompanied by loss of resonances in the NMR spectra corresponding to increased K28 mobility and loss of E22-C-terminal interactions. Importantly, decreased stability correlated highly with increased oligomerization propensity.
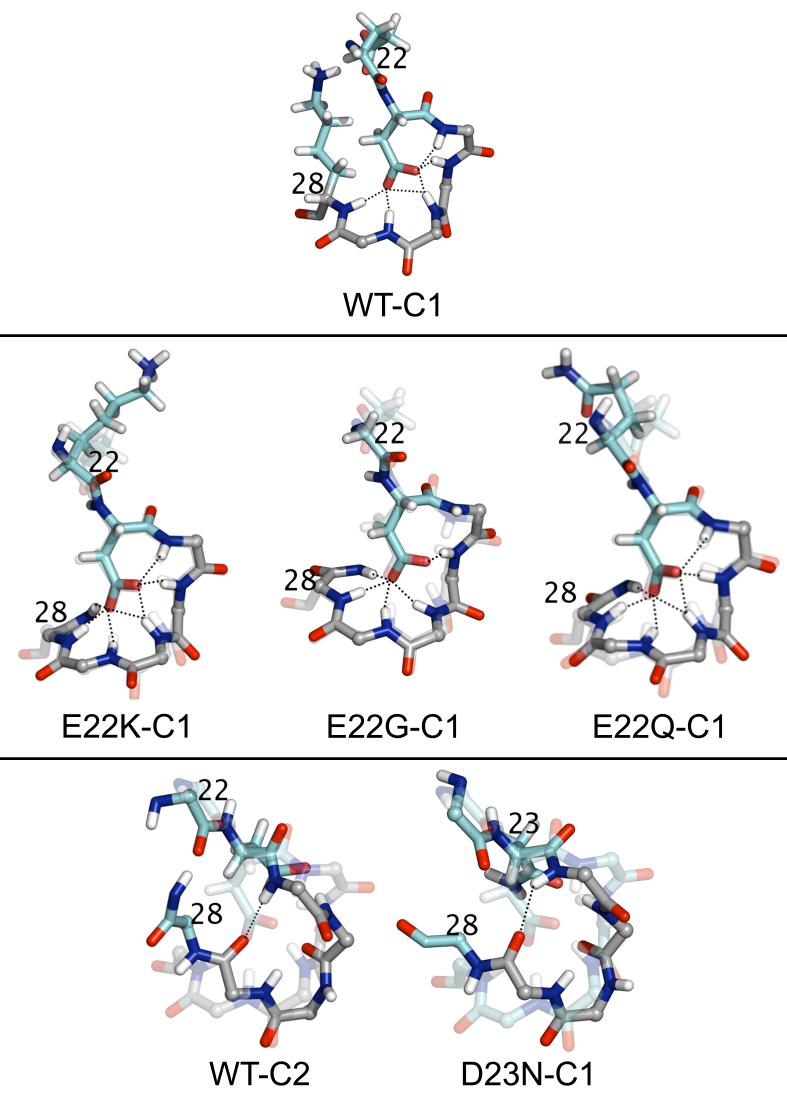
Fig. 1.
Shown in the figure are the central C1 structures of Aβ(21-30) WT and E22X mutants, WT C2 structure, and C1 structures of the D23N mutant. The WT C1 structure is shown transparently in the background of each mutant. Sidechains of the mutated residue and of residue 23 are shown. For E22X mutants, the bend region is highlighted in gray and for WT-C2 and D23N mutant the hydrogen-bonded turn region is highlighted in gray. Dotted lines denote key hydrogen bonds.
In the present work, we use replica exchange molecular dynamics (an enhanced sampling technique21-24) to provide the first structural characterization of the effect of the Italian, Arctic, Dutch, and Iowa mutations on the bend. Our simulations reveal how structural changes occurring as a result of familial mutations affect the folding nucleation of the protein. We anticipate that changes in the monomeric structure may affect the early stages of oligomerization. Further structural changes to this bend likely occur with increasing oligomer size25 and in the context of the final fibrillar structure.26-29
RESULTS AND DISCUSSION
The E22-K28 segment in the wild type Aβ(21-30) peptide adopts a bend motif. In an earlier publication,10 we used replica exchange molecular dynamics simulations to characterize the structural ensemble populated by the wild type (WT) Aβ(21-30) peptide. The salient features of that study are summarized below. Details of the simulation methodology used for the wild type and mutant peptides are given in the Methods and Model section.
The wild type Aβ(21-30) peptide samples a number of possible conformations at 300K. Clustering (as described in the Methods section) reveals that the peptide spends a majority of its time (44%) in a preferred conformation, which we denote as cluster C1. Other clusters are populated to a lesser extent (16% in the case of the second most populated cluster C2 and 5% in the case of the third most populated cluster C3). The population of each cluster is shown in Table 1.
Table 1.
Percentage of structures which belong to the 3 most populated cluster, C1, C2 and C3, for each peptide
| Peptide | C1 Population (%) | C2 population (%) | C3 population (%) |
|---|---|---|---|
| WT | 44 | 16 | 5 |
| E22K | 20 | 12 | 9 |
| E22G | 38 | 10 | 9 |
| E22Q | 33 | 8 | 8 |
| D23N | 24 | 16 | 6 |
A representative conformation belonging to the two most populated clusters from the simulations (C1 and C2) is shown in Figure 1 for the wild type peptide. In the WT C1 structure, the predominant structural motif involves a bend between residues 24-28. A bend is defined as a region with high geometric curvature such that the bond angle formed by the three Cα atoms of residues i-2,i,i+2 is at least 70°. 30 The C1 bend is stabilized by a network of hydrogen bonds between the D23 Oδ atoms and the backbone amide hydrogen atoms of residues 24-29. Hydrogen bonds are considered present when the donor and acceptor are within 3.5 Å and the donor-acceptor-hydrogen angle is <60°. A strong interaction between side chains of D23 and S26 is observed (see below). A salt bridge between a pair of aspartate/glutamate and lysine residues is considered formed when the distance between their respective Cδ/Cγ and Nζ atoms is <4.5 Å. An E22-K28 salt bridge is present within the WT (50% probability in the C1 cluster), with the D23-K28 salt bridge populated to a much smaller extent (<1% probability in the C1 cluster) (see Table 2). The C2 WT conformations show a turn rather than a bend structure, stabilized by a V24(O)-N27(NH) backbone hydrogen bond. A turn is defined as a region in which there exists a hydrogen bond between CO of residue i and NH of residue i+n.30 Our model of the folded Aβ(21-30) fragment satisfies all inter-proton constraints available from our previous NMR study.9 It also readily explains the origin of the anomalously high hydrogen exchange protection factors observed for residues G25-K28 by Maggio and collaborators in fragments Aβ(12-28) and Aβ(10-35).31,32 Our simulations indicate that the amide hydrogens of these residues engage in hydrogen bonds with the side chain of D23 and thus get shielded from water. Since the C1 cluster is significantly more populated than the other clusters (for the WT, and as can be seen in the next sections, for the mutants (see Table 1)), we will focus the major part of our analysis on the C1 cluster.
Table 2.
Comparison between most populated clusters of WT and mutant structures
| Peptide | RMSD from WT (Å)a | Probability of salt bridge formation (%)b | |
|---|---|---|---|
| E22-K28 | D23-K28 | ||
| WT | - | 50 | < 1 |
| E22K | 0.57 | - | < 1 |
| E22G | 0.45 | - | < 1 |
| E22Q | 0.47 | - | < 1 |
| D23N | 2.08 | 5 | - |
| D23G | 1.19 | 50 | - |
Measured over the Cα of residues 22-28 and averaged over all structures belonging to the most populated cluster (C1).
A salt bridge between a pair of aspartate/glutamate and lysine residues is considered formed when the distance between their respective Cδ/Cγ and Nζ atoms is below 4.5 Å.
Mutations at position E22 do not affect the structure of Aβ(21-30) and possibly leave the Aβ folding nucleus unaltered. A similar clustering analysis as for the WT was performed for the structures obtained from the replica exchange molecular dynamics simulations of the E22 mutants. The populations of each cluster are given in Table 1. Our simulations reveal that when residue E22 is mutated (“E22X,” where X is K, G, or Q) there is very little change in the structure of the peptide relative to the wild type. A representative conformation belonging to the most populated cluster for each mutant is shown in Figure 1. Table 2 displays the probability of salt bridge formation between residues E22-K28 and D23-K28 in the mutants and in the WT sequence, as well as the RMSD of each mutant from the WT C1 structure. The RMSD from the WT C1 central structure is <0.6 Å for each of these mutants. Each mutant has a high percentage of structures in the most populated cluster (C1) containing hydrogen bonds between residue D23 Oδ atoms and backbone amide hydrogen atoms, which serve to stabilize the bend motif present in the residue 24-28 region. Elimination of the E22-K28 salt bridge by substitution of E22 by a non-acidic residue (E22K, E22G, E22Q) preserves the bend of the backbone in the V24-K28 region (Fig. 1) indicating that it is primarily the hydrogen bond network involving the D23 side chain and the V24-K28 backbone that stabilizes the bend and not the E22-K28 salt bridge. Nevertheless, the fact that the population of cluster C1 is significantly larger for the WT than for the E22X mutants indicates that the additional E22-K28 salt bridge in the WT helps lock the V24-K28 bend in place, or equivalently, the absence of salt bridges in the E22X mutants destabilizes the bend, as seen in our earlier experimental work20 and in simulations of E22Q.12,33 The intensity of the E22-K28 contact in the most populated cluster (C1) of the WT sequence drops from 67% to <55% for the corresponding Q22-K28 contact of the E22Q mutant (see Fig. 2(a)). This is accompanied by a drop of 11% in the overall population of the C1 cluster upon mutation. The other E22X mutants yield similar data. Our results suggest that the effect of E22X mutations on Aβ self-assembly is not linked to an altered folding nucleation of Aβ. Indeed, the most populated clusters of the E22X mutants are almost identical in backbone structure to that of the WT peptide. We hypothesize that the E22X mutations have long-range effects, i.e., effects on other parts of the full-length peptide or intermolecular interactions, and affect peptide assembly in this manner. This idea is supported by our recent simulations on the Aβ(15-28) peptide,15 in which we observe that the E22Q mutation does not affect the 24-28 folding nucleus of the peptide, but rather the ability of the residues preceding residue 22 to adopt a β-strand conformation, which facilitates deposition of the peptide onto pre-existing fibrils.
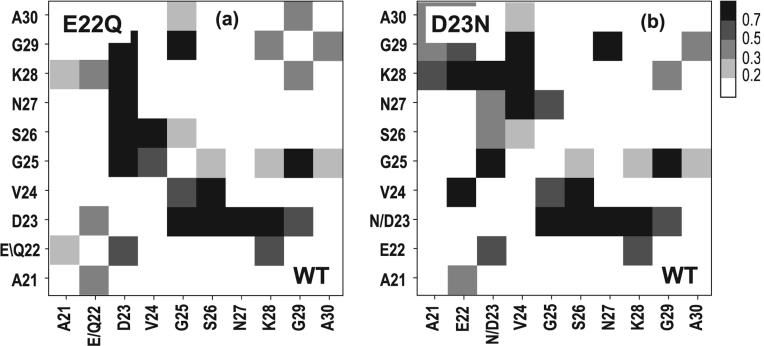
Fig. 2.
Probability map of side-chain contact formation obtained in the present work for the main cluster C1 of Aβ21-30WT (shown on the bottom right quadrant) and (a) E22Q (Dutch) mutant (shown on the upper left quadrant) and (b) D23N (Iowa) mutant (shown on the upper left quadrant). A contact between two side chains is considered formed when any atom of one side chain is within 4.5 Å of any atom of another side chain. Hα atoms were considered in the analysis of glycine residues. The axes correspond to the residues in the Aβ21-30 peptide. The darker the square on the plot, the higher the probability of side-chain contact formation (see color bar). The main effect of the E22Q mutation is to reduce the intensity of the 22-28 contact from 67% in the WT to 55% in the mutant. Consequently, the total amount of structuring in the middle part of the peptide is also reduced, with the population of the folded C1 conformation dropping from 44% in WT sequence to 33% in the mutant. Other E22X mutants studied in this work display similar behavior. In contrast, the D23N mutant displays a different pattern of inter-residue contacts.
Mutation at position D23 alters the structure of the Aβ(21-30) peptide and possibly affect Aβ folding nucleation
Mutating the aspartate residue at position 23 dramatically changes the structures present in the most populated cluster. A representative structure belonging to the most populated cluster for the D23N mutant is shown in Figure 1. The D23N mutant is found to have a turn between residues 24 and 27 stabilized by a backbone hydrogen bond between V24(NH) and N27(O) (Fig. 1). This hydrogen bond is found in 82% of the structures belonging to the most populated cluster (C1) of D23N. The RMSD from the WT C1 central structure is 2.08 Å, a significant increase over the values reported for the E22X substitutions studied (Table 2). Two possible hydrogen bonds can be formed involving the side chain of residue D23 and the peptide backbone of residues 24-29, Oδ-NH and NHδ-O. However, these hydrogen bonds are formed in very few of the D23N C1 structures. Furthermore, hydrogen bonding between the side chain of residue E22 and the peptide backbone was analyzed, however none is observed in the D23N mutant or the WT. The C1 structure of the D23N mutant is significantly different from the typical WT structure. In fact, not even the less populated clusters of D23N adopt a configuration similar to C1 of the WT peptide. The second most populated cluster of the WT and the third most populated cluster of the E22K and E22G mutants, on the other hand, contain a hydrogen-bonded turn structure similar to C1 of D23N. Our results suggest that the Iowa D23N mutation may affect aggregation by altering the folding nucleation of Aβ. The difference in folding between Aβ(21-30)WT and the D23N mutant is highlighted in the contact map shown in Fig. 2(b). In contrast to the E22X mutants (such as the E22Q mutant shown in Fig. 2(a)), the Iowa mutant displays a pattern of contact formation completely different from that of WT. Rather than being tightly folded around the D23 central part of the peptide, the D23N mutant exhibits a strong preference for forming contacts between K28 and the 3-residue segment A21-E22-N23. This segment directly precedes residue V24 in sequence space, a residue that forms a strong contact with residue N27. As mentioned earlier, this pair of residues share a hydrogen bond between their backbone atoms that helps stabilize the β-turn structure, the dominant structural motif in the conformational ensemble of the mutant peptide.
The E22 salt bridge in the wild type bend may enhance bend stability
Although no difference was seen between the backbone structures of the WT and E22X mutants, salt bridge formation in the E22X mutants may affect conformational dynamics in two ways. First, the presence of the salt bridge provides additional stabilization of the configuration already stabilized by side chain-backbone hydrogen bonds, effectively “locking” the bend in place. In contrast to the synthetic addition of a D23-K28 lactam bridge to Aβ(1-40), which irreversibly locks-in a monomer structure that promotes fibril formation,34 the E22-K28 salt bridge could provide additional stabilization to a structure that is resistant to aggregation. The observed bend structure in the WT must presumably rearrange itself to produce a conformer able to form fibrils.27-29 This could lead to a slower aggregation rate for the WT. Additionally, whereas in the WT C1 structures, E22 is involved in a salt bridge with K28, residue 22 of the Italian, Arctic, and Dutch mutant is more readily available for interaction with residues other than K28, as well as intermolecular interactions which may promote aggregation.
MODEL AND METHODS
All-atom explicit models of protein and solvent were used to model the conformations of Aβ(21-30) in water. The bulk of all simulations were performed using the OPLS/AA35 force-field for the peptide and TIP3P36 model for the solvent. All simulations were performed using the GROMACS software package,37,38 following the replica-exchange protocol described in our earlier work on the WT Aβ(21-30) peptide.10 To mimic the environment of the fragment within the context of the full length Aβ peptide and eliminate conformational artifacts due to interacting charged termini, we capped the N- and C-termini in all peptides considered by acetylation and carboxamidation, respectively. Charged systems were neutralized by adding a sodium ion (WT) or a chlorine ion (E22K). 40 replicas of the original system were exponentially spaced between 300 K and 600 K. Acceptance ratios for exchange between replicas at neighboring temperatures ranged from 15% to 30%. Covalent bonds in water molecules were kept constant using the SETTLE algorithm39 and those involving hydrogen atoms in the peptides were constrained according to the LINCS algorithm.40 The protocol of Nose-Hoover41 with the 0.05 ps time constant was used to maintain constant temperature in all our simulations. The total equilibrium sampling time in our simulations ranged between 30 and 40 ns. A total of 60,000-80,000 structures were generated per replica per simulated peptide. Structures recorded at T=300 K were used in the structural analysis. When dealing with large pools of conformations, it is important to determine what these conformations have in common. To test the possibility that common structural elements may not span the entire length of the peptide, we implemented the following structuring strategy. We assume that a structural motif of length M exists in all conformations of a peptide of length N>M. In this case, clustering over shorter segments NP<M within the motif should be successful and reveal a highly populated conformation, the one that belongs to the motif, while clustering over segments that include residues from outside of the motif should produce broad distributions of conformations with no dominating structure. Varying the length of the test segment NP allows one to locate structured parts in a diverse pool of conformations as well as determine their size (how many residues they encompass). We used this clustering strategy in our previous work16 to analyze structural preferences of Aβ(10-35) fragment. In the present work, we applied this strategy to analyze Aβ(21-30)WT and all its mutants. We start with a small size NP=5 and find all consecutive segments of this size contained in Aβ(21-30). We cluster conformations of these segments according to the Gromos algorithm42 and using RMSD among Cα as a measure of structural similarity. Clustering reveals regions with high and low extent of structuring. Highly structured regions are further tested with increasing values of the segment lengths. We continue increasing NP until the longest structural motifs are found. As an illustration of how our technique works, we find that populations P of 5-residue segments for the WT sequence are: P=21% for AEDVG, P=45% for EDVGS, P=34% for DVGSN, P=20% for VGSNK, P=12% for GSNKG and P=4% for SNKGA. Clearly, the best candidate for the structured motif in Aβ(21-30) is in the central 7-residue segment EDVGSNK while both C- and N-terminal segments are disordered. Clustering over the residues of this segment reveals that it does occupy a single predominant conformation (shown in Fig.1) with a population frequency of 44%. The highest-populated cluster C1 for all Aβ(21-30) mutants reported in this work were determined in a similar manner.
Convergence test
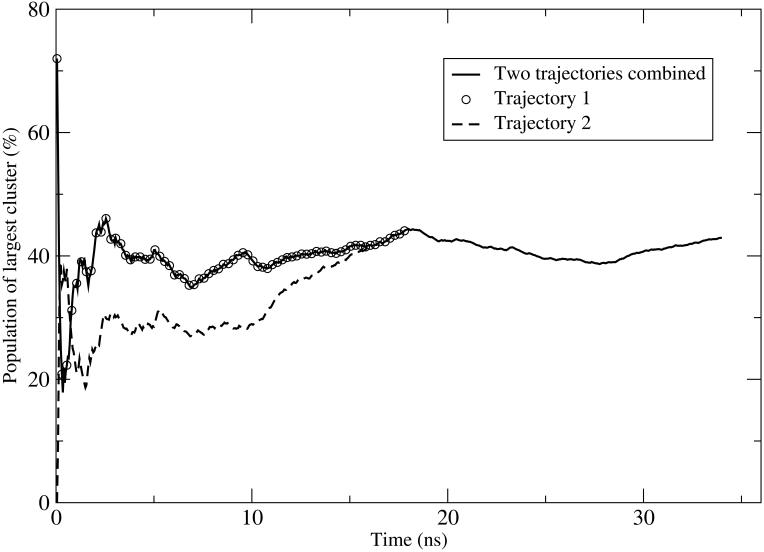
The major goal of this study was to determine occupation numbers of the most structured part in Aβ(21-30) peptide and to compare these numbers among Aβ(21-30) mutants. The accuracy of these numbers thus is of central importance. To improve conformational sampling in our simulations and monitor the degree of convergence in the data, two independent trajectories were obtained for WT sequence and each mutant. The trajectories were run between 15 and 20ns, depending on the system. The first two ns were considered as the equilibration period and not included in further analysis. Trajectories were first analyzed independently to test if they converged to the same most populated structure. For all systems reported here, they did. Second, trajectories were concatenated to yield better statistics. To monitor convergence trends, population of the most populated cluster C1 was computed as a running average for the two independent trajectories as well as for the combined trajectory. Figure 3 shows these averages for the WT sequence. It is seen that in trajectory 1, the population of C1 rapidly reaches 40% after about 3ns of simulation time and it stays at this level through the remainder of the simulation. In trajectory 2, the population rapidly increases to 30% at 3ns of simulation time, stays at this level until about 10ns and then gradually increases to about 40% in the last 5ns of the simulation. For the combined trajectory, population of cluster C1 remains almost constant over more than 30ns of simulation time, indicating that sufficient convergence in conformational ensembles has occurred. To rule out the possibility that averaging over non-equilibrium ensembles is performed in the initial parts of both trajectories, block analysis was performed. The fact that populations tend to reach plateaus after about 3ns of simulation time indicates that the relaxation time τ of our system under each given condition is roughly of the same order of magnitude. Averaging over τ periods should therefore produce qualitatively similar results as averaging over the entire trajectory. To test if this is the case for the studied systems, the combined (concatenated) trajectory was split into 10 non-overlapping periods, each approximately 3ns long. The most populated conformations were determined for each period and analyzed. These conformations were seen to belong to the C1 cluster with the population averaging to 44%. Splitting of the trajectories allows us to estimate the error in the C1 population through the standard deviation, assuming that the consecutive parts are statistically independent. This yielded errors of 1.59% for the WT, 0.95% for E22K, 1.26% for E22G, 2.83% for E22Q and 2.72% for D23N.
Fig. 3.
Plot of the running average C1 population for Aβ21-30WT. Data for two independent replica-exchange trajectories are presented. Additionally, the average resulting from combining the two trajectories is also shown. The population levels stay approximately constant over 30 ns averaging time.
Effects of force fields and solvent models
In addition to satisfactory convergence, another critical component of a simulation study is the accuracy of the model employed. The wild type Aβ(21-30) peptide and the OPLS/AA force field used in this study, in combination with the TIP3P water model, yield good agreement with structures derived experimentally.10 However, no experimental structures are available for the mutant systems investigated. We therefore evaluate the performance of the OPLS/TIP3P combination compared to other force-field/solvent models. To estimate how the main results reported in this study are affected by different force fields/solvent models, we consider the Aβ(21-30) peptide with charged termini, investigated by our group previously.10 Two more force field combinations are tested: GROMOS96 G43a1 united-atom force field (G)43 combined with the SPC water model44 and AMBER99SB (A)45,46 force-field in combination with the TIP3P water model.36 Both force fields are used widely and their strengths and weaknesses have been discussed previously.47
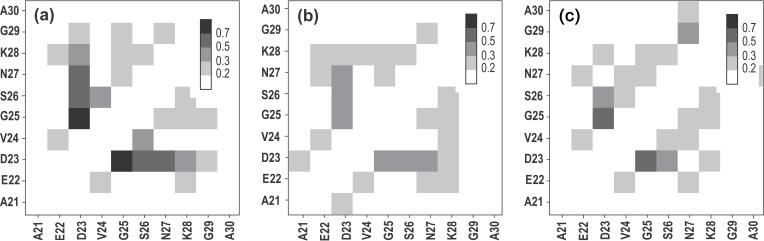
The setup of simulations A and G is similar to what was used in our previous study.10 40 replicas were spaced at temperatures between 300K and 600K. A total of 36ns of equilibrium sampling time was collected for the G set and 58ns for the A set. The block clustering technique described above was used to analyze conformations recorded in these simulations. Both Gromos and Amber force fields generate conformations with the highest degree of structuring observed in their central 7 residues. The two most structured segments EDVGS and DVGSN are seen to be populated to a lesser extent than when the OPLS force-field is used (14% in the G setup and 15% and 17%, respectively, in the A simulation setup). In all force fields, the EDVSGN segment adopts bend and turn configurations, but their mutual RMSD is ∼2Å. This is to be expected because bends and turns are not as rigid as other secondary structures (e.g., helices) as they are stabilized by a smaller number of hydrogen bonds. The contribution of these bonds to the overall system energetics differs among force fields, thus variations in the final structures should be observed. However, the interactions that drive formation of structure in all three force fields are similar. Interactions occurring among side chains in Aβ(21-30) as seen in our simulations using OPLS, Gromos, and Amber force fields are illustrated in Fig. 4, which shows side-chain contact maps. The maps were computed for heavy atoms only, to be consistent with the Gromos force field, using a 6Å cut-off for a contact. All three maps confirm our conclusion that structuring in Aβ(21-30) occurs primarily in the central EDVGSN segment but that the extent of the structuring is strongest for the OPLS force field. There are significant variations in the contact intensities among three force fields, especially in the contacts formed by terminal residues. All three maps, however, have one element in common, the side chain of D23 interacts with the side chain of S26 (and with backbone atoms preceding this residue) and drives formation of the EDVGSN bend. The intensity of this contact varies among the force fields. It is strongest in OPLS simulations and extends further along the sequence to include N27 and K28 side chains. This contact is weakest in the Amber simulations. A strong D23-S26 contact was also reported in a recent NMR study on Aβ(21-30).48 Based on the data presented in Fig. 4, it is clear that the OPLS/TIP3P force-field/solvent model combination produces results qualitatively similar with those obtained using other popular force fields. Our conclusion therefore is that the results presented in this work for Aβ(21-30) peptide and its mutants are not very sensitive to the particular choice of the force-field.
Fig. 4.
Probability map for side chain contact formation obtained for Aβ(21-30) using (a) OPLS/AA35/TIP3P36 combination, (b) GROMOS96 G43a43/SPC44 combination and AMBER99SB45/TIP3P combination. Here, the contact maps are calculated using all the structures from each simulation. Only heavy atoms of the side chains were considered in computing the probabilities. A contact was assumed formed when any two atoms of a pair of side chains were separated by 6Å or less. Differences are seen among the maps, but all three are dominated by the contacts of D23 with S26 and neighboring residues, which drive the formation of the EDVGSN bend.
ACKNOWLEDGMENTS
Simulations were performed using the TACC Lonestar Cluster (NSF Teragrid MCA05S027). Support from the NSF (No. MCB 0642086 To JES), the NIH (No. AG027818 to DBT and MB and No. GM083600 to AB), the David and Lucile Packard foundation (to JES) and the University of North Carolina Charlotte and North Carolina Biotechnology Center (to AB) are gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hardy J, Selkoe DJ. The Amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Murakami K, Irie H, Morimoto A, Ohigashi H, Shindo M, Nagao M, Shimizu T, Shirasawa T. Neurotoxicity and physicochemical properties of Aβ mutant peptides from cerebral amyloid angiopathy. J. Biol. Chem. 2003;278:46179–46187. doi: 10.1074/jbc.M301874200. [DOI] [PubMed] [Google Scholar]
- 3.Hou LM, Shao HY, Zhang YB, Li H, Menon NK, Neuhaus EB, Brewer JM, Byeon IJL, Ray DG, Vitek MP, Iwashita T, Makula RA, Przybyla AB, Zagorski MG. Solution NMR studies of the Aβ(1-40) and Aβ(1-42) peptides establish that the Met35 oxidation state affects the mechanism of amyloid formation. J. Am. Chem. Soc. 2004;126:1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 4.Riek R, Guntert P, Dobeli H, Wipf B, Wuthrich K. NMR studies in aqueous solution fail to identify significant conformational differences between the monomeric forms of two Alzheimer peptides with widely different plaque-competence, Aβ(1-40)ox and Aβ(1-42)ox. Eur. J. Biochem. 2001;268:5930–5936. doi: 10.1046/j.0014-2956.2001.02537.x. [DOI] [PubMed] [Google Scholar]
- 5.Sgourakis NG, Yan Y, McCallum SA, Wang C, Garcia AE. The Alzheimer’s peptides Aβ40 and 42 adopt distinct conformation in water: A combined MD/NMR study. J. Mol. Biol. 2007;368:1448–1457. doi: 10.1016/j.jmb.2007.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard SJ. The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta. 1998;1382:191–206. doi: 10.1016/s0167-4838(97)00175-1. [DOI] [PubMed] [Google Scholar]
- 7.Fontana A, Laureto P. P. d., Filippis VD, Scaramella E, Zambonin M. Probing the partly folded states of proteins by limited proteolysis. Fold. Des. 1997;2:R17–R26. doi: 10.1016/S1359-0278(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 8.Teplow DB, Nakayama C, Leung PC, Harshey RM. Structure-function relationships in the transposition protein B of bacteriophage Mu. J. Biol. Chem. 1988;263:10851–10857. [PubMed] [Google Scholar]
- 9.Lazo ND, Grant MA, Condron MC, Rigby AC, Teplow DB. On the nucleation of amyloid β-protein monomer folding. Protein Sci. 2005;14:1581–1596. doi: 10.1110/ps.041292205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumketner A, Bernstein SL, Wyttenbach T, Lazo ND, Teplow DB, Bowers MT, Shea J-E. Structure of the 21-30 fragment of amyloid β-protein. Protein Sci. 2006;15:1239–1247. doi: 10.1110/ps.062076806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borreguero JM, Urbanc B, Lazo ND, Buldyrev SV, Teplow DB, Stanley HE. Folding events in the 21-30 region of amyloid β-protein (Aβ) studied in silico. Proc. Natl. Acad. Sci. USA. 2005;102:6015–6020. doi: 10.1073/pnas.0502006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz L, Urbanc B, Borreguero JM, Lazo ND, Teplow DB, Stanley HE. Solvent and mutation effects on the nucleation of amyloid β-protein folding. Proc. Natl. Acad. Sci. USA. 2005;102:18258–18263. doi: 10.1073/pnas.0509276102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Mousseau N, Derreumaux P. The conformations of the amyloid-β (21-30) fragment can be described by three families in solution. J. Chem. Phys. 2006;125:084911. doi: 10.1063/1.2337628. [DOI] [PubMed] [Google Scholar]
- 14.Baumketner A, Shea J-E. Folding landscapes of the Alzheimer amyloid-β(12-28) peptide. J. Mol. Biol. 2006;362:567–579. doi: 10.1016/j.jmb.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 15.Baumketner A, Krone MG, Shea J-E. Role of the familial Dutch mutation E22Q in the folding and aggregation of the 15-28 fragment of the Alzheimer amyloid-β protein. Proc. Natl. Acad. Sci. USA. 2008;105:6027–6032. doi: 10.1073/pnas.0708193105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumketner A, Shea J-E. The structure of the Alzheimer amyloid-β(10-35) peptide probed through replica-exchange molecular dynamics simulations in explicit solvent. J. Mol. Biol. 2007;366:275–285. doi: 10.1016/j.jmb.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Massi F, Peng JW, Lee JP, Straub JE. Simulation study of the structure and dynamics of the Alzheimer’s amyloid peptide congener in solution. Biophys. J. 2001;80:31–44. doi: 10.1016/S0006-3495(01)75993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarus B, Straub JE, Thirumalai D. Dynamics of Asp23-Lys28 salt-bridge formation in Aβ10-35 monomers. J. Am. Chem. Soc. 2006;128:16159–16168. doi: 10.1021/ja064872y. [DOI] [PubMed] [Google Scholar]
- 19.Han W, Wu YD. A strand-loop-strand structure is a possible intermediate in fibril elongation: Long time simulations of amyloid-β peptide (10-35) J. Am. Chem. Soc. 2005;127:15408–15416. doi: 10.1021/ja051699h. [DOI] [PubMed] [Google Scholar]
- 20.Grant MA, Lazo ND, Lomakin A, Condron MM, Arai H, Yamin G, Rigby AC, Teplow DB. Familial Alzheimer’s disease mutations alter the stability of the amyloid β-protein monomer folding nucleus. Proc. Natl. Acad. Sci. USA. 2007;104:16522–16527. doi: 10.1073/pnas.0705197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 22.Nymeyer H, Gnanakaran S, Garcia AE. Atomic simulations of protein folding, using the replica exchange algorithm. Methods Enzymol. 2004;383:119–149. doi: 10.1016/S0076-6879(04)83006-4. [DOI] [PubMed] [Google Scholar]
- 23.Garcia A, Onuchic J. Folding a protein in a computer: An atomic description of the folding/unfolding of protein A. Proc. Natl. Acad. Sci. USA. 2003;100:13898–13903. doi: 10.1073/pnas.2335541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitera JW, Swope W. Understanding folding and design: Replica-exchange simulations of “Trp-cage” fly miniproteins. Proc. Natl. Acad. Sci. USA. 2003;100:7587–7592. doi: 10.1073/pnas.1330954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zagorski MG, Barrow CJ. NMR studies of amyloid β-peptides: Proton assignments, secondary structure, and mechanism of an α-helix-β-sheet conversion for a homologous 28-residue, N-terminal fragment. Biochemistry. 1992;31:5621–5631. doi: 10.1021/bi00139a028. [DOI] [PubMed] [Google Scholar]
- 26.Zheng J, Jang H, Ma B, Tsai C-J, Nussinov R. Modeling of the Alzheimer Aβ17-42 fibril architecture: Tight intermolecular sheet-sheet association and intramolecular hydrated cavities. Biophys. J. 2007;93:3046–3057. doi: 10.1529/biophysj.107.110700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model of Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchete N-V, Tycko O, Hummer G. Molecular dynamics simulations of the Alzheimer’s β-amyloid protofilaments. J. Mol. Biol. 2005;353:804–821. doi: 10.1016/j.jmb.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 29.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobelii H. 3D structure of Alzheimer’s amyloid-β(1-42) fibrils. Proc. Natl. Acad. Sci. USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern-recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 31.Zhang SS, Casey N, Lee JP. Residual structure in the Alzheimer’s disease peptide: Probing the origin of a central hydrophobic cluster. Folding Des. 1998;3:413–422. doi: 10.1016/S1359-0278(98)00054-6. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S, Iwata K, Lachenmann MJ, Peng JW, Li S, Stimson ER, Lu Y.-a., Felix AM, Maggio JE, Lee JP. The Alzheimer’s peptide Aβ adopts a collapsed coil structure in water. J. Struct. Biol. 2000;130:130–141. doi: 10.1006/jsbi.2000.4288. [DOI] [PubMed] [Google Scholar]
- 33.Massi F, Klimov D, Thirumalai D, Straub JE. Charge states rather than propensity for β-structure determine enhanced fibrillogenesis in wild-type Alzheimer’s β-amyloid peptide compared to E22Q Dutch mutant. Protein Sci. 2002;11:1639–1647. doi: 10.1110/ps.3150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sciarretta KL, Gordon DJ, Petkova AT, Tycko R, Meredith SC. Aβ40-lactam (D23/K28) models a conformation highly favorable for nucleation of amyloid. Biochemistry. 2005;44:6003–6014. doi: 10.1021/bi0474867. [DOI] [PubMed] [Google Scholar]
- 35.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105:6474–6487. [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of single potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 37.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 38.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 39.Miyamoto S, Kollman PA. SETTLE: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 40.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 41.Nose S, Klein ML. Constant pressure molecular-dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 42.Daura X, Gademann K, Jaun B, Seebach D, Gunsteren W. F. v., Mark AE. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Edit. 1999;38:236–240. [Google Scholar]
- 43.Scott WRP, Hunenberger PH, Tironi IG, Mark AE, Billeter SR, Fennen J, Torda AE, Huber T, Kruger P, van Gunsteren WF. The GROMOS biomolecular simulation program package. J. Phys. Chem. A. 1999;103:3596–3607. [Google Scholar]
- 44.Berendsen HJC, Postma JPM, Gunstern W. F. v., Hermans J. In: Pullman B, editor. Intermolecular Forces: Proceedings of the Fourteenth Jerusalem Symposium on Quantum Chemistry and Biochemistry held in Jerusalem, Israel, April 13-16, 1981; D. Reidel, Dordrecht, Holland. 1981. [Google Scholar]
- 45.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorin EJ, Pande VS. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys. J. 2005;88:2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rueda M, Ferrer-Costa C, Meyer T, Perez A, Camps J, Hospital A, Gelpi JL, Orozco M. A consensus view of protein dynamics. Proc. Natl. Acad. Sci. USA. 2007;104:796–801. doi: 10.1073/pnas.0605534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fawzi NL, Phillips AH, Ruscio JZ, Doucleff M, Wemmer DE, Head-Gordon T. Structure and dynamics of the Aβ21-30 peptide from the interplay of NMR experiments and molecular simulations. J. Am. Chem. Soc. 2008;130:6145–6158. doi: 10.1021/ja710366c. [DOI] [PMC free article] [PubMed] [Google Scholar]