Abstract
Chondroitin sulfate (CS) is a glycosaminoglycan consisting of repeating uronic acid, N-acetylgalactosamine disaccharide units [(HexAβ/α(1–3) GalNAcβ (1–4)] n. CS chains are polydisperse with respect to chain length, sulfate content and glucuronic acid epimerization content, resulting in a distribution of glycoforms for a chain bound to any given serine residue. Usually, CS glycoforms exist, differing in sulfation position and uronic acid epimerization. This work introduces a novel LC/MS/MS platform for the quantification of mixtures of CS oligosaccharides. The CS polysaccharides were partially depolymerized and labeled with either the light (d0) or heavy (d4) form of 2-anthranilic acid (2-AA). Excess reagent was removed and mixtures of the CS standard (d0) and unknown (d4) were made. The CS mixture was subjected to size exclusion chromatography (SEC) with on-line electrospray ionization mass spectrometrometric detection in the negative ion mode. Tandem mass spectra were acquired and quantification of unknown samples within the mixture was made using relative ion abundances of specific diagnostic ions. High accuracy and precision of the glycomics platform were demonstrated using glycoform mixtures made from standard CS preparations. The CS glycoform analysis method was then applied to cartilage extract, versican, and several dermatan sulfate preparations. This work presents the first application of a glycomics platform for the quantification of CS oligosaccharide mixtures to obtain specific information on the positions of GalNAc sulfation and uronic acid epimerization.
Glycomics is applied biology and chemistry that focuses on the structure and function of carbohydrates. Carbohydrates have vital functions in the body; in particular, glycosylation serves to diversify protein functions. Despite their abundance, much less information is known about carbohydrates, as compared to genes and proteins, largely due to the absence of a simple code that determines their structures. Mass spectrometry (MS), high performance liquid chromatography and other analytical tools have advanced rapidly to support the growth of biomolecular applications in the field of proteomics. However, technologies and methods to address the most challenging problems in glycomics, particularly with respect to glycoform quantitative measurements, remain undeveloped.
Glycosaminoglycans (GAGs) are linear polysaccharides that consist of repeating disaccharide units that are attached to proteoglycan core proteins on adherent animal cell surfaces and in extracellular matrices (1–5). GAGs play significant roles in the modulation of cellular signals through interactions with proteins, including growth factors and growth factor receptors (6–8). They mediate cell-cell and cell-matrix interactions and are crucial to cell development and function (9, 10). GAG fine structure, consisting of patterns of sulfation and uronic acid epimerization, is variable and diverse. This is the result of maturation reactions involving sulfation, epimerization and glycosylation, which are dependent on cell type and not the core protein. Depending on GAG type, disaccharide units may be epimerized at the C-5 position of uronic acid residues and may be N- or O-sulfated and/or N-acetylated. The mammalian GAGs are hyaluronan, dermatan sulfate (DS), chondroitin sulfate (CS), heparan sulfate (HS), heparin, and keratan sulfate (KS).
CS is a GAG linked to serine residues in core proteins by way of a xylosyl linker; it consists of repeating disaccharide units of [(HexAβ/α(1–3)GalNAcβ(1–4)] that are polymerized in chains of size varying from 20–50 kDa depending on the core protein, tissue location, and disease contexts. The chains consist of mixtures of domains with high or low iduronic acid content with differing patterns of sulfation (11). Many types of CS exist; however, the three main classifications of CS found in higher animals include: CS type A (CSA), CS type B (CSB, otherwise known as DS, or dermatan sulfate), and CS type C (CSC). CSA is most commonly sulfated (90%) at the 4-position of GalNAc, and CSC is most commonly sulfated (90%) at the 6-position of GalNAc. CSB contains a high percentage of repeats in which the uronic acid residue is epimerized to iduronic acid; a fraction of such repeats are sulfated at the 2-position of iduronic acid. CSB is most often sulfated on the 4-position of GalNAc. Previous research has shown evidence that suggests that isomeric CS glycoforms differing in position and degree of sulfation play specific and distinct functional roles during development (12–14). The sulfation patterns of CS chains bound to the cartilage proteoglycan aggrecan have been shown to change with age. Mixtures of 4- and 6-sulfated GalNAc residues were shown to be present in juvenile cartilage, whereas that from adults contained almost entirely 6-sulfated GalNAc residues (15). For cartilage aggrecan, changes in sulfation pattern have been correlated not only with aging and development, but also with the development of osteoarthritis (16). However, due to the lack of analytical techniques, it has not been possible to directly determine sulfation and uronic acid positional isoforms in CS oligosaccharides. Thus, detailed structural information on the variability of CS glycoform expression in extended sequences during biological processes is lacking.
MS is a valuable tool for structural and quantitative analysis of biomolecules, such as GAGs, as it offers high analytical versatility, sensitivity and precision. Negative mode electrospray ionization (ESI) in the presence of ammonium salts has proven effective for the analysis of sulfated carbohydrates (17–19). Intact negative ions are formed without in-source fragmentation, thereby allowing tandem mass spectra to be acquired through the use of collision-induced dissociation (CID) (20). ESI is also advantageous because it allows for online coupling of mass spectrometry with chromatographic separation methods. Methods have already been developed for the use of size exclusion chromatography (SEC) and tandem mass spectrometry for the determination of sulfation position in CS oligosaccharides (18, 21). It has previously been shown that CS disaccharides sulfated at the 4- or 6-position of the GalNAc residue, CSA or CSC respectively, produce distinct CID product ion patterns (18, 22, 23). Oligosaccharides produced from the three glycoforms of CS (CSA, CSB or CSC) may be differentiated based on product ion abundance (24, 25).
A specific challenge arises in the tandem mass spectrometric analysis of CS oligosaccharides. Lyase enzymes may be used to generate mixtures of oligosaccharides from intact CS/DS chains. The monoisotopic peaks for the most abundant charge state of CS oligosaccharides that differ in length are observed at the same m/z value (m/z 458), resulting in the observation of overlapping isotopic clusters. In the present work, CS partial depolymerization products are derivatized by a reductive amination reaction. This step has two additional advantages: the first is that alkylated CS oligosaccharides have unique m/z values for their most abundant charge state. The second is that incorporation of the derivative offers a means for stable isotopic labeling. The use of a stable isotope allows direct comparison of two samples, one labeled with the light form and one with the heavy, present in a mixture. In this work, unknown samples are compared to standard preparations using stable isotope labeling.
The present work demonstrates an analytical LC/MS/MS platform that incorporates a stable isotopic labeling technique for the quantification of CS glycoforms starting with 1–10 microgram quantity of sample. The platform introduced herein is advantageous in that the quantification of glycoforms within CS/DS samples is calculated directly from tandem mass spectral data. Extended sequence analysis and quantification of CS/DS chains have previously been investigated through the use of differential lyase digestion with chondroitinase ACI and chondroitinase B followed by ion-pair reversed-phase HPLC (IP-RP-HPLC). Following IP-RP-HPLC, 4,5-unsaturated uronic acid oligosaccharides (termed Δ-oligosaccharides) are further digested to Δ-disaccharides with chondroitinase ABC for structural and quantitative analysis by high performance capillary electrophoresis (26–28). The present platform follows a simpler analytical workflow, without the need for consecutive digestions and separations. The method is advantageous in that data are produced on sulfation position and uronic acid epimers simultaneously. Further, it is not necessary to purify the Δ-oligosaccharides. Presented here are results demonstrating the usefulness of an LC/MS/MS platform for the quantification of isomeric glycoforms of CS/DS from standard mixtures and applications to biologically relevant connective tissue samples.
Experimental Procedures
Materials
CS type A (GlcA, GalNAc-4-sulfate), CSB (IdoA, GalNAc-4-sulfate) CSC (GlcA, GalNAc-6-sulfate), and chondroitinase ABC and AC1 were obtained from Seikagaku America/Associates of Cape Cod (Falmouth, MA). DS samples were generous gifts of Anders Malmström, Lund University, Lund, Sweden. Versican was a generous gift from Roberto Perris, University of Parma, Italy. Purified Δ-disaccharide standards were purchased from V-labs (Covington, LA). 2-aminoacridone (AMAC) and 2-anthranilic acid (d0-2AA) were purchased from Fluka Chemika (Buchs, Switzerland), cartilage extract and sodium cyanoborohydride was from Aldrich Chemicals Co. (St. Louis, MO), and 2-anthranilic-3,4,5,6-d4 acid (d4-2AA) was from C/D/N Isotopes (Quebec, Canada) and Cellulose Packing Material Micro Spin Columns were purchased from Harvard Apparatus (Holliston, MA).
GAG release from core protein
Sulfated GAGs were released from the core protein of both cartilage extract (from bovine articular cartilage) and versican (from bovine aorta) samples with a 1.0 M NaBH4, 0.05 M NaOH solution at 45°C for 16 hours. The pH was adjusted to 5.0 using glacial acetic acid. The core protein was then separated from the GAG using a Pepclean™ C-18 spin column (Pierce, Rockford, IL). The sulfated GAGs (CS) were then separated from O-linked glycans by ethanol precipitation (−20°C, overnight).
Preparation of Δ-Unsaturated Chondroitin Sulfate Oligosaccharides
CSA, CSB, or CSC (5 μL, 2 mg/mL) was mixed with water (87 μL), Tris-HCl buffer (5 μL, 1M, pH 7.4), ammonium acetate (0.5 μL, 1M) and chondroitinase ABC (2.5 μL, 4 mU/μL), and digested at 37 °C to an absorbance value (232 nm) equal to 0.341 and boiled for 1 minute. This degree of digestion was found to reproducibly depolymerize 10 μg of CSA, CSB and CSC to 30% reaction completion, as determined by absorbance at 232 nm. Partial depolymerization products were lyophilized for subsequent reductive amination.
Derivatization of Oligosaccharides with d0- and d4-2-AA
All oligosaccharides were derivatized with d0- or d4-2-AA according to the method of Bigge et al. (29). Briefly, a dried CSA sample was dissolved in 10 μL of a reaction reagent containing d0-2AA in DMSO/glacial acetic acid (7:3) and 1.0 M sodium cyanoborohydride. All other partially depolymerized CS (unknown) samples were derivatized using d4-2AA under the same conditions. The glycan solutions were then centrifuged for 3 minutes and incubated at 65°C for 3 hours. In an attempt to optimize the published reductive amination protocol, water and ammonium hydroxide were added to reactions one hour into the incubation time to create a more neutral or basic environment. Also, two consecutive derivatizations were used to give the portion of the glycan not reacting the first time a more optimal opportunity to react during a second round. The relative derivatization yields were found via mass spectrometry on a quadrupole-ion trap mass spectrometer (QITMS). Samples were sprayed in a 30% MeOH, 0.2% NH4OH solution under nano-spray conditions (24). Mass spectra revealed peaks at m/z 459.1 and 578.8 representing the unlabeled and the labeled CS disaccharides, respectively. The relative yield was found by calculating the percent total ion abundance (%TIA) of the labeled and unlabeled compounds. None of the procedure modifications improved on the published method.
Derivatization Sample Clean-Up
Excess reagents were removed from derivatized glycans via cellulose microspin columns. The column was first hydrated with five 200 μL volumes of water, rinsed with five 200 μL volumes of 30% acetic acid solution, and then with 3 200 μL volumes of acetonitrile. The 2-AA derivatized reaction mixture was applied to the column allowing 15 minutes for it to adsorb to the cellulose. Excess reagents were washed off with three 200 μL volumes of acetonitrile followed by two 200 μL volumes of 96% acetonitrile. The derivatized glycan was then eluted with 2 100 μL volumes of water and dried.
LC/MS Analysis
The mixed, derivatized glycan samples were fractionated using high performance SEC with on-line tandem mass spectrometric detection. Briefly, the column (Superdex Peptide 3.2/30, Amersham Biosciences, Piscataway, NJ) was equilibrated in 10% acetonitrile, 0.05 M ammonium formate solution at 40 μL/min and the oligosaccharide mixture (10 μL) was injected with UV detection at 310 nm.
The HPLC system was connected to a Bruker Daltonics (Billerica, MA) Esquire 3000 QITMS equipped with a standard electrospray ion source. The spray voltage was set at 3 kV; capillary temperature was set to 250°C; the nebulizer gas (He) was set to 10 psi and the drying gas (N2) was set to 5 L/min. The capillary exit was set to −50.9 V and the skimmer potential was set to −10 V, so as to prevent in-source fragmentation. The ion signal was optimized using a trap drive of 60. Sample introduction into the mass spectrometer was achieved by connecting the SEC column to the sample inlet of the QIT electrospray source with peek™ tubing. To minimize dead volume the column was clamped to a ring stand as close as possible to the MS sample inlet. The HPLC flow was split prior to the sample inlet, allowing 10 μL/min into the mass spectrometer. All scans were acquired in the negative ion mode using the automated MSn feature of the ion trap. To enable data dependent scanning, specific parent ions (representing hexasaccharides, tetrasaccharides and disaccharides) were listed into the auto MSn method. The width of the isolation and fragmentation windows was set to 12.0 u so that CID spectra of both heavy and light forms of derivatized CS species within the mixture were acquired simultaneously. The ion trap was set at an accumulation time of 150 u/s. The instrument was set at a scan speed of 13,000 m/z/sec, adequate to resolve heavy and light paired fragment ions and their isotopic distribution. The instrument was set to cycle between the MS and MS/MS modes until the entire mixture had eluted through the column. The MS/MS scans were summed for the most abundant charge state for the CS oligosaccharides; this corresponds to one negative charge per sulfate group.
Calculation of Equation Coefficients
Tandem mass spectral data was collected on 2AA-labeled CSA, CSB and CSC standards in order to establish diagnostic ions. An internal standard, d0-2AA-CSA, was used for every sample. d4-2AA-CSB and d4-2AA-CSC were run with each series of samples, in triplicate. The percent total ion abundance of the diagnostic ions in each 2-AA labeled standard was then calculated and the results represent the equation coefficients, described further in the Results section. The coefficients are therefore re-calculated for every corresponding series of samples.
Exhaustive digestion of CS/DS
To 10 μg of CS/DS was mixed 87 μL water, 5 μL 1M Tris-HCl buffer (pH 7.4), 0.5 μL 1M NH4OAc and 10 mU of chondroitinase ABC, 10 mU AC1, and 2 mU Chondroitinase B and digestion was allowed to proceed at 37°C for 3 hours. The reaction was stopped by incubation with boiling water for two minutes and was then derivatized with AMAC.
AMAC derivatization
Derivatization with AMAC was performed following the procedure described by Militsopoulou (30). Briefly, to the lyophilized CS/DS disaccharide sample were added 5 μL 0.1 M AMAC solutions in acetic acid: DMSO (3:17, v/v) and 5 μL of a freshly prepared 1M NaBH3CN solution in water. The mixture was vortexed for 3 minutes and was incubated at 45°C for 4 hrs after which 10 μL of DMSO was added. Excess reagent was removed using cellulose spin columns as described for 2-AA above.
Capillary Electrophoresis - Laser Induced Fluorescence (CE-LIF) Analysis
The AMAC derivatized CS/DS samples were analyzed using a Beckman Coulter (Fullerton, CA) P/ACE MDQ capillary electrophoresis instrument. The uncoated fused silica capillary tubing (75 μm ID, 60 cm total length) was regenerated with 0.1M HCl, 0.1M NaOH, and HPLC grade water before each run and analyses were performed using 50mM NaH2PO4 buffer, pH 3.5, at 30 kV using reverse polarity and detected using the AMAC fluorophore (λex = 485, λem = 510). Baseline separation of the CS/DS isomers and good reproducibility were also obtained when fresh anode and cathode buffer solutions were used for each run. A tri-sulfated heparin disaccharide (ΔHSIS) was used as an internal standard for each run as it has a unique migration time compared to the CS/DS disaccharides.
Results
LC/MS/MS Platform
Each CS unknown sample was divided into three equal aliquots, and each aliquot was subjected to the analytical workflow shown in Figure 1. Each aliquot was partially depolymerized using chondroitinase ABC, to produce a mixture consisting of abundant (4–5)-unsaturated (Δ-) hexasaccharides, tetrasaccharides, and disaccharides. The reference standard CS (CSA, Seikagaku) was then derivatized via a reductive amination reaction with d0-2AA, while the unknown CS aliquots were derivatized via a reductive amination reaction with d4-2AA. The excess derivatization reagent was removed using a cellulose micro-spin column. Mixtures of the standard (d0) and unknown (d4) CS samples were then made. The isotopically labeled CS mixtures were separated using SEC with on-line negative ion ESI-MS/MS detection.
Figure 1.
The protocol for the LC/MS/MS platform presented.
Distinguishing Diagnostic Ions for Isomeric CS glycoforms
It has previously been shown that native oligosaccharides yield tandem mass spectrometric fragment ions, the abundances of which are diagnostic of the CS-type (18, 22, 23). Because this prior work utilized native CS oligosaccharides, it was necessary to show that reductively aminated CS yields different fragment ion abundances according to sulfation and epimerization patterns. Therefore, 10-μg samples of CSA (GlcA-GalNAc4S)n, CSB (IdoA-GalNAc4S)n and CSC (GlcA-GalNAc6S)n standards were subjected to the analytical scheme shown in Figure 1. Tandem mass spectra for the 2-AA delta-unsaturated depolymerized tetrasaccharide (termed Δdp4) peaks are shown in Figure 2 for (a) d0-2AA-CSA, (b) d4-2AA-CSB and (c) d4-2AA-CSC. From visual inspection it can easily be determined that abundant fragment ions for 2-AA-Δdp4 oligosaccharides distinguish CS glycoforms, including: Y11− for CSA, Y32− for CSB, and [M-H-SO3]2− for CSC. The Y11− ion is present in all three species; however, it is significantly higher in abundance for the 2-AA-Δdp4 oligosaccharides derived from CSA relative to those from the CSB and CSC, and is therefore diagnostic of CSA. The same is true for the Y32− and the [M-H-SO3]2− ions for CSB and CSC, respectively. In order to use such diagnostic ions as a means for quantification it is necessary to calculate the percent total ion abundance of all three ions in the spectra of each CS species. The resulting percent total diagnostic ion abundances, for all three CS species, shown in Figure 2d, demonstrates that each form of CS Δdp4 oligosaccharide produces a unique signature; the pattern observed for an unknown is produced by a linear combination of product ion abundances reflecting the composition of the mixture with respect to Δdp4 oligosaccharides containing CSA-, CSB-, and CSC-like disaccharide repeats (25). A complete list of ion m/z values observed in the tandem mass spectra of 2-AA-Δdp4 CS standards is presented in Table I.
Figure 2.
A. Tandem mass spectrum of CSA d0-2AA-Δdp4. B. Tandem mass spectrum of CSB d4-2AA-Δdp4. C. Tandem mass spectrum of CSC d4-2AA-Δdp4. D. Bar graph representation of the contribution of each diagnostic ion in the pure CS standards. CSA (white), CSB (gray), and CSC (dark gray). E. Structure of a CS tetrasaccharide and its diagnostic fragment ions. The Uronic Acid (UroA) and N-acetylgalactosamine (GalNAc) sulfate are symbolized by the open diamond and the open square, respectively.
Table I.
CID diagnostic product ion data for the 2-AA-Δdp4 CSA, CSB and CSC standards.
| Ion | Composition | Calculated m/z | Observed m/z |
|---|---|---|---|
| [d0-2AA-Y1-SO3]1− | C15O7H22N2 | 341.13 | 341.1 |
| [d4-2AA-Y1-SO3]1− | C15O7H22N2D4 | 345.16 | 345.1 |
| [d0-2AA-Y1]1− | C15O10H22N2S | 421.08 | 421.2 |
| [d4-2AA-Y1]1− | C15O10H22N2SD4 | 425.12 | 425.2 |
| [d0-2AA-Y3]2− | C29O24H43N3S2 | 439.58 | 439.6 |
| [d4-2AA-Y3]2− | C29O24H43N3S2D4 | 441.59 | 441.6 |
| [d0-2AA-0,2X1]1− | C17O11H19N2S1 | 458.06 | 458.1 |
| [d4-2AA-0,2X1]1− | C17O11H19N2S1D4 | 462.18 | 462.4 |
| [d0-2AA-M-H-SO3]2− | C35O26H49N3S | 478.61 | 478.6 |
| [d4-2AA-M-H-SO3]2− | C35O26H49N3SD4 | 480.62 | 480.7 |
| B31− | C20H26NO19S | 616.08 | 616.1 |
Binary mixtures of CS tetrasaccharide standards
In order to validate the LC/MS/MS platform, the same CSA, CSB and CSC standards used above were subjected to the workflow shown in Figure 1. The isotopically labeled CS species were then mixed together in certain ratios, termed standard mixtures, in order to assess the validity of the method of quantification. All isotopically labeled mixtures were subjected to the entire analytical workflow in triplicate in order to determine the total experimental standard deviation. Each sample was mixed with d0-2AA oligosaccharides derived from CSA. The first mixture analyzed was a 1:1 mixture of 5 μg d0-2AA-CSA and 5 μg d4-2AA-CSC. The CS mixture was separated by SEC with on-line ESI-MS/MS detection. The resulting MS total ion-chromatogram can be seen in Figure 3a. The signal-to-noise ratio for 2-AA-Δdp6 was not high enough to yield confident results. The 2-AA-Δdp4 and 2-AA-Δdp2 spectra were of sufficient signal-to-noise ratios to be useful for glycoform mixture analysis. The Δdp4 oligosaccharides produce more information than the Δ-disaccharides due to the existence of an internal uronic acid residue; therefore, 2-AA-Δdp4 oligosaccharides will be discussed herein.
Figure 3.
A. Total Ion Chromatogram of a 10 μg binary 1:1 mixture of d0-2AA-CSA: d4-2AA-CSC. B. Tandem mass spectrum of the tetramer mixture of 1:1 d0-2AA-CSA: d4-2AA-CSC. Diagnostic ions (Y11−, Y32−, and [M-H-SO3]2−) are paired by 2 or 4 u depending on whether they are doubly or singly charged, respectively. C. Expanded mass range of the diagnostic ions. CSA labeled with the light form (d0) of 2-AA was incorporated as an internal standard. In the case of the CS standard mixture above, the CSA standard sample labeled with d0-2AA was mixed with a CSC sample (termed unknown) labeled with d4-2AA.
The tandem mass spectrum acquired for the 2-AA-Δdp4 mixture, 1:1 d0-2AA-CSA: d4-2AA-CSC, is shown in Figure 3b. From the product ion isotopic ratios in the tandem mass spectrum one can see that a mixture of glycoforms is present (see Figure 3c). The light (d0) CSA and the heavy (d4) CSC precursor ions yield fragment ions that are paired by either 4 u if singly charged ion, or 2 u if doubly charged. Therefore, one can see in Figure 3c that two peaks exist at m/z 421.2 and 425.2 that represent the light and heavy Y11− ion, respectively. A pair of peaks differing by 2 u at m/z 478.7 and m/z 480.7, are representative of the [M-H-SO3]2− ion.
In order to quantify the mixture present in the sample, the percent total ion abundances of light and heavy diagnostic ions of interest were found by listing all ions and abundances within the spectrum, and then removing all that do not contain the reducing end, therefore considering only ions paired by 4 or 2 u. These ions are then separated into heavy and light columns. The percent total ion abundances were calculated separately for heavy and light diagnostic ions. This resulted in two groups of ions, one significantly more abundant for CSA Δdp4 (light ions) and one more abundant for CSC Δdp4 (heavy ions). The 1:1 CSA: CSC mixture was subjected to the entire analytical workflow in triplicate to produce average percent ion abundances with standard deviations.
Quantification
In order to quantify glycoforms within the mixture, the percent total ion abundances of the diagnostic ions in both the pure standards and the standard mixture were used in a system of three equations and three unknowns. The diagnostic ion abundances for the three standard CS samples were used to determine the equation coefficients. The system of equations is a method originally used to quantify mixtures of oil distillates (31). The procedure was later adapted and specifically modified for the compositional analysis and quantification of glycosaminoglycans using electrospray ionization-quadrupole ion trap mass spectrometry (22, 32, 33). It results in the determination of the relative quantity of each component in a given glycosaminoglycan mixture. A system of equations based on the same principles is stated below:
In the above equations a, b, and c represent the percentage of CSA, CSB, and CSC, respectively, in a mixture. The constants Y1CSA, Y1CSB, and Y1CSC represent the abundance of the Y11− diagnostic ion in the pure standards. Y3CSA, Y3CSB, and Y3CSC represent the abundance of Y32− diagnostic ion in the pure standards, and M-H-SO3CSA, M-H-SO3CSB, and M-H-SO3CSC represent the abundance of [M-H-SO3]2− diagnostic ion in the pure standards. CY1, CY3 and CM-H-SO3 represent the abundance of the heavy and light diagnostic ions measured in the unknown CS sample.
Upon insertion of the values of the diagnostic ion abundances for both the pure standards and the 1:1 mixture of d0-2-AA-CSA: d4-2-AA-CSC, the system of equations yields:
The values of a, b and c were then calculated to equal 1.368, 0.1338, and 1.424, respectively. The percentage of CSA in the mixture was then calculated by dividing the value of a by the sum of the values of a, b and c. The percentages of CSB and CSC were then calculated in the same manner. The percentages of CSA, CSB and CSC in the first mixture were found to be 46.8%, 4.5% and 48.7%, respectively. The original CS mixture was previously separated into three aliquots and subjected in parallel to the protocol shown in Figure 1. The final isomer percentages are represented as an average of the determination made for three aliquots of each mixture subjected to the entire analytical workflow to give a standard deviation that is representative of the total experimental error. Nine different binary mixtures created from CS standards were subjected to the LC/MS/MS platform in this manner and the results are shown in Table II.
Table II.
The percent composition of CSA-like (GlcAGalNAc4S), CSB-like (IdoAGalNAc4S), CSC-like (GlcAGalNAc6S) isomeric glycoforms in ten different artificially made CS mixtures.
| CSA-like GlcAGalNAc4S | CSB-like IdoAGalNAc4S | CSC-like GlcAGalNAc6S | |
|---|---|---|---|
| 3:7 CSA:CSC | 36.9% ± 1.5 | 0.8% ± 0.8 | 62.2% ± 0.5 |
| 1:1 CSA:CSC | 48.7% ± 1.7 | 3.8% ± 1.2 | 49.0% ± 3.3 |
| 7:3 CSA:CSC | 66.0% ± 3.1 | 3.8% ± 1.3 | 30.1% ± 1.9 |
| 3:7 CSA:CSB | 34.5% ± 2.7 | 64.8% ± 2.4 | 0.7% ± 1.0 |
| 1:1 CSA:CSB | 53.9% ± 1.7 | 46.0% ± 2.3 | 0.7% ± 0.5 |
| 7:3 CSA:CSB | 66.4% ± 1.8 | 32.9% ± 1.9 | 0.8% ± 0.4 |
| 3:7 CSB:CSC | 2.0% ± 1.2 | 31.8% ± 3.1 | 66.2% ± 4.3 |
| 1:1 CSB:CSC | 2.6% ± 2.5 | 48.3% ± 1.9 | 49.1% ± 2.9 |
| 7:3 CSB:CSC | 4.2% ± 2.1 | 64.9% ± 1.1 | 30.9% ± 2.4 |
| 1:1:1 CSA:CSB:CSC | 34.9% ± 3.3 | 34.7% ± 5.3 | 30.4% ± 2.3 |
Quantification of Ternary mixtures of CS tetrasaccharides standards
Following the quantification of nine binary mixtures, it was necessary to validate that the LC/MS/MS platform is also applicable to ternary mixtures. Therefore a 1:1:1 isotopically labeled CS mixture of 3.3 μg d0-2AA-CSA, 3.3 μg d4-2AA-CSB and 3.3 μg d4-2AA-CSC was generated and subjected in triplicate to the same protocol described previously. The resulting tandem mass spectrum of the mixture tetrasaccharide is shown in Figure 4a. Paired peaks representing the diagnostic ions are present at m/z 421.0 and 425.0 (Y11−), m/z 439.4 and 441.4 (Y32−), and m/z 478.5 and 480.5 ([M-H-SO3]2−). The percent total ion abundance of each heavy and light diagnostic ion was inserted into the above system of three equations and three unknowns, and solved for a, b and c. The percentages of CSA, CSB and CSC in the ternary mixture were found to be, as shown in Table II, 34.9% ± 3.3, 34.7% ± 5.3 and 30.4% ± 2.3, respectively.
Figure 4.
Expanded mass regions for diagnostic ions of different CS samples. A. Tandem mass spectrum of a 10-μg tetramer ternary mixture of 1:1:1 d0-2AA-CSA: d4-2AA-CSB: d4-2AA-CSC. B. Tandem mass spectrum of a 10-μg tetramer mixture of cartilage extract. Heavy (d4) diagnostic ions used for quantification include: Y11− at m/z 425.1, Y32− at m/z 441.5 and [M-H-SO3]2− at m/z 480.6. C. Tandem mass spectrum of a 10-μg tetramer mixture of Versican. CSA labeled with the light form (d0) of 2-AA was incorporated as an internal standard. In the case of the CS standard mixture (A), the CSA standard samples labeled with d0-2AA were mixed with CSB and/or CSC samples (termed unknown) labeled with d4-2AA in varying ratios. In the case of proteoglycan samples (B and C), 3 μg of the CSA-d0-2AA internal standard was added to each sample prior to LC/MS analysis. The CSA-d0-2AA internal standard is represented by the peak at m/z 421.1.
Applying the LC/MS/MS platform to the quantification of biologically relevant proteoglycan samples
Cartilage Extract
CS/DS chains from triplicate 10 μg samples of bovine cartilage extract were released using alkaline borohydride, and subjected to the analytical workflow described in Figure 1 using a d4-2AA label. Prior to mass spectrometric analysis each 10 μg d4-2AA derivatized sample was mixed with 3 μg of the d0-2AA derivatized CSA, termed the reference standard. The diagnostic ions representative of 2-AA-Δdp4 oligosaccharides derived from CSA, CSB and CSC (Y11−, Y32−, and [M-H-SO3]2−, respectively) are shown to be present and quantifiable in the tandem mass spectrum of the CS/DS chains released from cartilage extract, shown in Figure 4b. The percent total ion abundance of the heavy form of each diagnostic ion was calculated, and inserted into the same system of three equations and three unknowns described for the CS standards. The contributions of the diagnostic ions from the d0-2AA Δdp4 derived CS standards were again inserted into the three equations, and solved for the percent of CSA, CSB and CSC. The percentages of the isomeric glycoforms of CSA, CSB and CSC were found to be 30.2% ± 2.8, 1.8% ± 0.7, and 68.0% ± 2.6, respectively. The standard deviation is again representative of the entire experimental error within the method.
To minimize experimental variability a reference standard, d0-2AA-CSA was spiked into each d4-2AA labeled proteoglycan or GAG sample that was quantified. The reference standard produced light (d0) fragment ions in the tandem mass spectra (i.e. The Y11− diagnostic ion for CSA was produced at an m/z 421.2; whereas the Y11− diagnostic ion for the proteoglycan sample was produced at an m/z 425.2). The percent total ion abundance of the light diagnostic fragment ion was calculated in each proteoglycan CS/DS sample subjected to the LC/MS/MS platform. The abundance of the reference standard product ion was then compared with that of each sample to ascertain that, in fact, the samples produced similar ionization efficiencies and therefore these samples can be compared for quantification.
Versican
CS/DS chains from triplicate 10 μg samples of bovine aorta versican were next subjected to the LC/MS/MS platform with an internal standard, in the same manner as that of the cartilage extract samples. The diagnostic ions representative of CSA, CSB and CSC (Y11−, Y32−, and [M-H-SO3]2−, respectively) were found to be present and quantifiable in the tandem mass spectrum of the CS/DS chains released from versican, as shown in Figure 4c. The percentages of the isomeric glycoforms of CSA, CSB and CSC were found to be 86.3% ± 2.1, 9.3% ± 2.5, and 4.7% ± 2.6, respectively.
Dermatan Sulfate
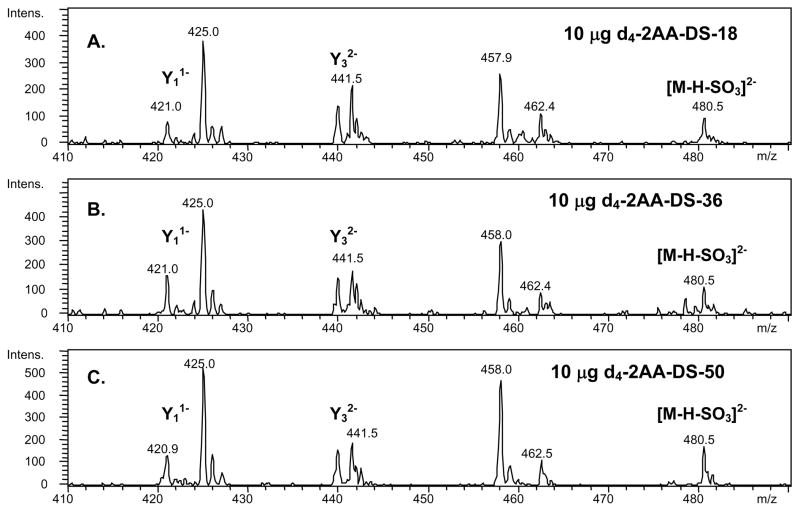
Several DS preparations from bovine skin, namely DS18, DS36 and DS50 were subjected in triplicate to the analytical workflow described previously. The DS chains were purified from skin, and the associated numeral refers to the percent ethanol at which the chains precipitated (34–36). The resulting 2-AA-Δdp4 tandem mass spectra for the DS preparations are shown in Figures 5a–c. From the signature ion patterns, DS18, DS36 and DS50 contain high percentages of the IdoA-GalNAc4S repeat. The complete results representing the quantification of the isomeric glycoforms of CSA, CSB and CSC in each DS samples are presented in Table III.
Figure 5.
A. Tandem mass spectrum of a 10-μg tetramer mixture of DS-18. Heavy (d4) diagnostic ions used for quantification include: Y11− at m/z 425.0, Y32− at m/z 441.5 and [M-H-SO3]2− at m/z 480.5. B. Tandem mass spectrum of a 10-μg tetramer mixture of DS-36. C. Tandem mass spectrum of a 10-μg tetramer mixture of DS-50. CSA labeled with the light form (d0) of 2-AA was incorporated as an internal standard. The CSA-d0-2AA internal standard (3 μg) was added to each sample prior to LC/MS analysis. The CSA-d0-2AA internal standard is represented by the peak at m/z 421.1.
Table III.
MS Ion Signature. The percent composition of CSA-like (GlcAGalNAc4S), CSB-like (IdoAGalNAc4S), CSC-like (GlcAGalNAc6S) isomers for biologically relevant tetramer proteoglycan samples known to contain chondroitin sulfate GAG chains. CSA, CSB and CSC standards were used to compare the relative characteristic of each biological sample.
| CSA-like GlcAGalNAc4S | CSB-like IdoAGalNAc4S | CSC-like GlcAGalNAc6S | |
|---|---|---|---|
| Cartilage Extract | 30.2 ± 2.8 | 1.8 ± 0.7 | 68.0 ± 2.6 |
| Versican | 86.3 ± 2.1 | 9.0 ± 2.5 | 4.7 ± 2.6 |
| DS-18 | 11.4 ± 5.2 | 85.4 ± 7.2 | 3.2 ± 2.5 |
| DS-36 | 26.1 ± 4.5 | 72.1 ± 4.5 | 1.9 ± 1.6 |
| DS-50 | 27.3 ± 2.9 | 68.5 ± 0.6 | 4.2 ± 2.4 |
Disaccharide Analysis by CE-LIF
It is useful to compare the results of the analytical platform described here with traditional disaccharide analysis of the same samples. The MS ion signature methodology entails comparison of an unknown sample relative to standard CS/DS preparations. It was necessary to place these results in the context of those obtained using classical disaccharide analysis. Disaccharide analysis of standards and samples was achieved by exhaustive depolymerization of the whole polymer chain using combined chondroitinase ABC, chondroitinase AC1 and chondroitinase B digestion, followed by fluorescent derivatization using AMAC and CE-LIF analysis of the disaccharide compositions. Migration times of Δ-disaccharides generated from standard CS preparations CSA, CSB, CSC and those from proteoglycan samples were verified against those of AMAC-derivatized commercially purified-disaccharide standards. An AMAC-derivatized tri-sulfated disaccharide derived from heparin (ΔHSIS), was used as an internal standard for each run, as it has a unique migration time compared to the CS/DS samples analyzed. Table IV lists the Δ–disaccharide compositions of the whole polymer chain of every sample analyzed within this study. The Δ-disaccharide composition percentages were calculated using the CE fluorescence peak areas, normalized to the area of the internal standard.
Table IV.
The CE-LIF percentage composition of ΔHexA-GalNAc4S, ΔHexA-GalNAc6S, ΔHexA2S-GalNAc4S and ΔHexA2S-GalNAc6S after an exhaustive digestion of the whole polymer chain of the proteoglycan. Percentage Δ-disaccharide compositions were calculated using CE fluorescence peak areas normalized to the area of the internal standard.
| ΔHexAGalNAc4S | ΔHexAGalNAc6S | ΔHexA2SGalNAc4S | ΔHexA2SGalNAc6S | |
|---|---|---|---|---|
| CSA | 96.56 ± 0.19 | 3.44 ± 0.20 | 0 | 0 |
| CSB | 93.67 ± 0.72 | 3.56 ± 0.14 | 2.75 ± 0.77 | 0 |
| CSC | 20.60 ± 0.51 | 74.06 ± 0.82 | 0 | 5.33 ± 1.10 |
| Cartilage Extract | 31.49 ± 1.02 | 68.26± 0.95 | 0.25 ± 0.08 | 0 |
| Versican | 84.82 ± 3.37 | 3.82 ± 0.83 | 11.37 ± 4.20 | 0 |
| DS18 | 95.16 ± 3.30 | 3.56 ± 3.01 | 1.28 ± 0.44 | 0 |
| DS36 | 91.76 ± 0.27 | 1.51 ± 0.40 | 6.73 ± 0.14 | 0 |
| DS50 | 86.60 ± 1.32 | 11.59 ± 1.48 | 1.79 ± 0.18 | 0 |
For the GlcA-containing CS standards, CSA is shown to contain approximately 96.6% ΔHexA- GalNAc4S and 3.5% ΔHexA-GalNAc6S while CSC is a mixture of approximately 20.6% ΔHexA-GalNAc4S, 74.1% ΔHexA-GalNAc6S and 5.3% of the 2-sulfated disaccharide, ΔHexA2SGalNAc6S (Table IV). The CSB standard, which is an IdoA-containing compound, shows a high abundance of the ΔHexA-GalNAc4S. Formation of Δ-disaccharides destroys information on the uronic acid epimerization; thus the distinction between iduronic acid (IdoAGalNAc4S) and glucuronic acid (GlcAGalNAc4S) repeats is lost.
Discussion
A novel feature of the LC/MS/MS platform introduced herein is the incorporation of a stable isotope for the quantification of CS glycoforms. It is necessary for CS partial depolymerization products to undergo derivatization for two primary reasons: to eliminate redundancy of m/z values for oligosaccharides of different lengths, and to incorporate a means for stable isotopic labeling. The incorporation of a stable isotopic label allows for the platform to be used with internal standard CS oligosaccharides so as to compare glycoform variations, such as those associated with healthy and diseased states. All samples are run using the same internal standard CS oligosaccharides, thus compensating for variations in instrumental performance.
Tandem mass spectrometry is used to produce structural and quantitative information about sulfated GAGs, by virtue of the abundances of specific diagnostic product ions. Thus, it is possible to distinguish among the three isomeric forms of CS/DS based on 2-AA-Δdp4 product ion abundances. In previous work, diagnostic ions were generated from native CS/DS oligosaccharides (25). The present work demonstrates that derivatization with 2-AA increases the abundances of diagnostic fragment ions (Y11−, Y32−, and [M-H-SO3]2−) and thereby the certainty of glycoform quantification. In addition, the differences in diagnostic ion abundances between CS/DS Δdp4 glycoforms are greater in magnitude for the 2-AA derivatized samples relative to the native oligosaccharides.
The LC/MS methodology used herein provides the advantage that oligosaccharide sulfation and epimerization patterns are measured simultaneously using an on-line platform. The availability of CS standards lends specificity to the analysis given that the CSA, CSB, and CSC glycoforms are positional variants. The validity of our initial assumption that differences in ionization efficiency are negligible was demonstrated on a series of glycoform mixtures generated from CS standards.
The LC/MS/MS platform was first tested on CS standards in order to validate the use of the technique. The highest purity commercially available CS standards are stated to be not less than 90% pure (37). Therefore, a potential 10% gap in purity allows for the detection of small percentages of other isomers not originally assumed to be in the mixture. Disaccharide analysis was performed as described in the methods on each CSA, CSB and CSC standard sample. CSA was found to be 96.56 ± 0.19% pure Δdi4S and CSC to be 74.06 ± 0.82% pure Δdi6S (complete set of results shown in Table IV). Therefore, the LC/MS/MS data measures the extent to which a CS glycoform mixture resembles each of the three CS standards. This approach provides the advantage that uronic acid epimerization and GalNAc sulfation state are determined in a single direct measurement.
Disaccharide analysis by CE-LIF was accomplished not only to determine the purity of the CS standard samples used for quantification, but also as a way to validate the quantification of the proteoglycan samples proposed by our novel LC/MS/MS platform. The LC/MS/MS approach measures uronic acid epimers using CS-2AA-Δdp4 oligosaccharides. Epimerization information is destroyed when the CS chains are converted completely to disaccharides. Therefore the ΔHexAGalNAc4S measured by CE-LIF is produced from both the GlcAGalNAc4S and IdoAGalNAc4S repeats. Disaccharide analysis of samples high in iduronic acid content shows a high percentage of the ΔDi4S disaccharide (Table IV). LC/MS/MS analysis of such samples shows much lower percentages of CSA-like and high percentages of CSB-like repeats (Table III). Thus, the LC/MS/MS approach provides significantly more information than does disaccharide analysis. Both methods were used to analyze bovine aorta versican, bovine articular cartilage extract and a series of bovine DS samples. These samples were chosen to correlate the 2-AA-Δdp4 LC/MS/MS results using CS/DS chains expressed in tissue with different glucuronic acid C5-epimerase activities.
The most abundant CS chains in cartilage are those on the large aggregating proteoglycan, aggrecan. In adult articular cartilage CS 6-sulfation is known to dominate over non-sulfated and 4-sulfated GalNAc residues (38). The level of IdoA sulfation is also known to be low (39). Previous research has shown that the GAG composition of aggrecan in normal human adult articular cartilage was predominantly Δdi6S while Δdi4S was low in all normal samples (40). Disaccharides, ΔdiSB and ΔdiSD (GlcA2SGalNAc4S and GlcA2SGalNAc6S, respectively) were detected in normal samples but only in minor quantities of approximately 1%. Following chondroitin lyase digestion, cartilage GAG products were shown to contain ~70% ± 2% Δdi6S and ~30% Δdi4S (40). The present LC/MS/MS results show articular cartilage as 68.0% ± 2.6 CSC-like, 30.2% ± 2.8 CSA-like and 1.8% ± 0.7 CSB-like. Such numbers establish clearly that in cartilage, Δdi4S derives from CSA-like GlcAGalNAc4S repeats; given the low abundance of those derived from CSB. It can be concluded that the LC/MS/MS methodology used herein provides another dimension of traditional analysis by determining epimerization positions directly. This capability will be applicable to studies of cartilage development and disease processes in which sulfation and epimerization levels change.
Versican is expressed in the extracellular matrix of a variety of tissues and organs (41), and has previously been reported to contain ~15% L-iduronic acid content in follicular fluid (42). The present LC/MS/MS results show versican to contain a high percentage of CSA-like GlcAGalNAc4S (86.3% ± 2.1), and a low percentage of CSC-like GlcAGalNAc6S (4.7% ± 2.6) repeats. The CSB-like signature was found to be 9.0% ± 2.5, slightly less than the 15% published previously. The ratio of the C5-epimerase activity to the 4-O-sulfotransferase activity has been shown to be 15 fold higher in fibroblasts, which produce versican, than in articular cartilage (43). The results are therefore consistent with the higher C5-epimerase activity resulting in versican containing a higher percentage of CSB-like signature ions than cartilage.
DS has previously been indicated to contain high levels of IdoA-GalNAc4S repeats, with lower levels of IdoA2S-GalNAc4S and GlcA (β1–3) GalNAc6S (β1–4) repeats (35, 36, 44–46). Previous tandem mass spectrometric analysis on the DS preparations investigated in this work showed that the abundances of CSB-like repeats was found to be 90.0%, 75.0%, and 50.0% for DS18, DS36, and DS50 respectively (47). These results were obtained using underivatized chondroitinase ABC-generated oligosaccharides. The results obtained in the present work using the LC/MS/MS platform for the dermal DS samples show 85.4, 72.1 and 68.5% respectively, have CSB-like IdoAGalNAc4S character (Table III, column 2) and that CSC-like GlcAGalNAc6S repeats are detected in very low abundances. In the present work, the reductive amination step was observed to increase the abundances of product ions that are diagnostic for different CS glycoforms. Further, the difference in the abundances of these product ions was observed to be greater than for native oligosaccharides. Based on these observations, the present method employing reductive amination and an on-line LC/MS/MS platform is thus shown to produce more accurate glycoform quantification. The corresponding disaccharide analysis of the present DS samples indicates that ΔDi4S comprises 95.2, 91.8, and 86.6%, respectively, of the disaccharides formed by chondroitinase ABC digestion, and a small percentage of ΔDi6S is also detected (Table IV).
For the CE-LIF disaccharide analysis, the samples were subjected to exhaustive digestion using chondroitinases ABC, B, and ACI. For the MS-based analysis, the samples were subjected to limited digestion using chondroitinase ABC. The results for the CE-LIF measurement of ΔDi6S and the MS ion signature measurement of CSC-like isomers (Tables III and IV) are consistent for cartilage extract, versican, DS18 and DS36. For DS50, however, the measurement of ΔDi6S (11.6%) is significantly higher than that obtained for CSC-like isomer content (4.2%). Given the consistency of the MS ion signature method for standard mixtures (Table II) and among the other GAG samples, it is likely that the discrepancy reflects structural differences between DS50 and the other samples. CS disaccharides other than those reported in Table IV were not detected, and this is consistent with the tetrasaccharides analyzed in Table III composed entirely of HexAGalNAc4S and HexAGalNAc6S repeats. In theory, ΔDi6S may be produced from either a GlcAGalNAc6S or an IdoAGalNAc6S repeat. A DS repeat consisting of IdoA2S-GalNAc6S has been identified in invertebrates and antibodies against this repeat found to indicate its present in mouse brain (48, 49). A DS repeat of IdoA-GalNAc4S,6S is found in hagfish (50). Although it is conceivable that an IdoA-GalNAc6S repeat is present in the porcine skin from which the DS samples were extracted, such a repeat has never been described in mammalian systems. If present, it is unknown whether such a repeat would produce a unique ion signature or one very similar to that of CSC-like GlcAGalNAc6S repeats. Efforts are underway to obtain invertebrate DS from which a tetramer containing an IdoAGalNAc6S repeat may be prepared so as to determine its tandem mass spectrometric diagnostic ion abundances.
The methodology introduced herein advances previous techniques for CS/DS analysis by producing information on both sulfation position and uronic acid epimerization in a single measurement. The stable isotopic labeling allows for internal standard CS oligosaccharides to be used in all quantification steps. The technique allows for a high throughput analysis of CS GAGs on a 10-μg scale using high performance SEC. The next step in this research is to further diminish the sample requirement by using a more sensitive chromatography method. Improved sensitivity will also allow analysis of oligosaccharides greater than Δdp4.
The present work also demonstrates concepts that are likely to be useful as general analytical principles in glycomics. Specifically, it is valuable to measure changes in the expression of glycoforms through increases or decreases in the abundances of specific tandem mass spectrometric product ions. Thus, changes in glycoform expression are measured using reductive amination labels that include stable isotope labels to compare product ion abundances in a given sample against those of a reference glycan mixture. Standard preparations of CS were used as the references in this work to provide detailed information on the types of disaccharide repeats present in a given sample. For other glycan classes, the level of information that will result from the glycoform analysis will depend on how much structural detail is known about the reference glycan mixture. Even for glycan classes for which detailed information on the significance of the product ion patterns is lacking, an approach based on stable isotope labeling and tandem MS will be useful to produce ions that indicate changes in glycan expression that accompany biological perturbation. Thus, an LC/MS/MS approach similar to that described here will likely provide a general means of correlating the expression of glycan expression with biological change.
Abbreviations
- AMAC
2-aminoacridone
- CE
capillary electrophoresis
- CID
collision-induced dissociation
- CS
chondroitin sulfate
- CSA
chondroitin sulfate type A
- CSB
chondroitin sulfate type B
- CSC
chondroitin sulfate type C
- dp
degree of polymerization
- DS
dermatan sulfate
- GAG
glycosaminoglycan
- LC/MS/MS
liquid chromatography-tandem mass spectrometry
- LIF
laser-induced fluorescence
- SEC
size-exclusion chromatography
- 2-AA
2-anthranilic acid
Footnotes
Funding from NIH grants P41 RR10888 and R01 HL74197 is gratefully acknowledged. Bruker Daltonics donated the Esquire 3000 ion trap mass spectrometer used in this work.
References
- 1.Kraemer PM. Heparan Sulfates of Cultured Cells. I. Membrane-Associated and Cell-Sap Species in Chinese Hamster Cells. Biochemistry. 1971;10:1437–1445. doi: 10.1021/bi00784a026. [DOI] [PubMed] [Google Scholar]
- 2.Kraemer PM. Heparan Sulfates of Cultured Cells. II. Acid-Soluble and - Precipitable Species of Different Cell Lines. Biochemistry. 1971;10:1445–1451. doi: 10.1021/bi00784a027. [DOI] [PubMed] [Google Scholar]
- 3.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of Cell Surface Heparan Sulfate Proteoglycans. Annu Rev Biochem. 1999;68:729–77. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 4.Perrimon N, Bernfield M. Specificities of Heparan Sulphate Proteoglycans in Developmental Processes. Nature. 2000;404:725–8. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 5.Iozzo RV. Proteoglycans: Structure, Biology and Molecular Interactions. Marcel Dekker; New York: 2000. [Google Scholar]
- 6.Lander AD. Proteoglycans: Master Regulators of Molecular Encounter? Matrix Biol. 1998;17:465–72. doi: 10.1016/s0945-053x(98)90093-2. [DOI] [PubMed] [Google Scholar]
- 7.Kawashima H, Hirose M, Hirose J, Nagakubo D, Plaas AH, Miyasaka M. Binding of a Large Chondroitin Sulfate/Dermatan Sulfate Proteoglycan, Versican, to L-Selectin, P-Selectin, and Cd44. J Biol Chem. 2000;275:35448–35456. doi: 10.1074/jbc.M003387200. [DOI] [PubMed] [Google Scholar]
- 8.Trowbridge JM, Rudisill JA, Ron D, Gallo RL. Dermatan Sulfate Binds and Potentiates Activity of Keratinocyte Growth Factor (Fgf-7) J Biol Chem. 2002;277:42815–42820. doi: 10.1074/jbc.M204959200. [DOI] [PubMed] [Google Scholar]
- 9.Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. Proteoglycans and Glycosaminoglycans. [Google Scholar]
- 10.Conrad HE. Heparin Binding Proteins. Academic Press; New York: 1998. [Google Scholar]
- 11.Malmström A, Fransson LA. Biosynthesis of Dermatan Sulfate. I. Formation of L-Iduronic Acid Residues. J Biol Chem. 1975;250:3419–3425. [PubMed] [Google Scholar]
- 12.Mark MP, Butler WT, Ruch JV. Transient expression of a chondroitin sulfate-related epitope during cartilage histomorphogenesis in the axial skeleton of fetal rats. Dev Biol. 1989;133:475–488. doi: 10.1016/0012-1606(89)90051-1. [DOI] [PubMed] [Google Scholar]
- 13.Tufvesson E, Malmström A. Biglycan Isoforms with Differences in Polysaccharide Substitution and Core Protein in Human Lung Fibroblasts. Eur J Biochem. 2002;269:3688–3696. doi: 10.1046/j.1432-1033.2002.03058.x. [DOI] [PubMed] [Google Scholar]
- 14.Kitagawa HK, Tsutsumi, et al. Developmental Regulation of the Sulfation Profile of Chondroitin Sulfate Chains in the Chicken Embryo Brain. J Biol Chem. 1997;272:31377–31381. doi: 10.1074/jbc.272.50.31377. [DOI] [PubMed] [Google Scholar]
- 15.Belcher C, Yaqub R, Fawthorp F, Bayliss M, Doherty M. Synovial Fluid Chondroitin and Keratan Sulphate Epitopes, Glycosaminoglycans, and Hyaluronan in Arthritic and Normal Knees. Ann Rheum Dis. 1997;56:299–307. doi: 10.1136/ard.56.5.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinmei M, Miyauchi S, Machida A, Miyazaki K. Quantitation of chondroitin 4-sulfate and chondroitin 6-sulfate in pathologic joint fluid. Arthritis Rheum. 1992;35:1304–8. doi: 10.1002/art.1780351110. [DOI] [PubMed] [Google Scholar]
- 17.Chai W, Luo J, Lim CK, Lawson AM. Characterization of Heparin Oligosaccharide Mixtures as Ammonium Salts using Electrospray Mass Spectrometry. Anal Chem. 1998;70:2060–6. doi: 10.1021/ac9712761. [DOI] [PubMed] [Google Scholar]
- 18.Zaia J, Costello CE. Compositional Analysis of Glycosaminoglycans by Electrospray Mass Spectrometry. Anal Chem. 2001;73:233–239. doi: 10.1021/ac000777a. [DOI] [PubMed] [Google Scholar]
- 19.Kim YS, Ahn MY, Wu SJ, Kim DH, Toida T, Teesch LM, Park Y, Yu G, Lin J, Linhardt RJ. Determination of the structure of oligosaccharides prepared from acharan sulfate. Glycobiology. 1998;8:869–77. doi: 10.1093/glycob/8.9.869. [DOI] [PubMed] [Google Scholar]
- 20.Zamfir A, Seidler DG, et al. Structural Investigation of Chondroitin/Dermatan Sulfate Oligosaccharides from Human Skin Fibroblast Decorin. Glycobiology. 2003;13:733–742. doi: 10.1093/glycob/cwg086. [DOI] [PubMed] [Google Scholar]
- 21.Zaia J, McClellan JE, Costello CE. Tandem Mass Spectrometric Determination of the 4S/6S Sulfation Sequence in Chondroitin Sulfate Oligosaccharides. Anal Chem. 2001;73:6030–6039. doi: 10.1021/ac015577t. [DOI] [PubMed] [Google Scholar]
- 22.Desaire H, Leary J. Detection and Quantification of the Sulfated Disaccharides in Chondroitin Sulfate by Electrospray Tandem Mass Spectrometry. J Am Soc Mass Spectrom. 2000;11:916–20. doi: 10.1016/S1044-0305(00)00168-9. [DOI] [PubMed] [Google Scholar]
- 23.Lamb DJ, Wang HM, Mallis LM, Linhardt RJ. Negative Ion Fast-Atom Bombardment Tandem Mass Spectrometry to Determine Sulfate and Linkage Position in Glycosaminoglycan derived Disaccharides. J Am Soc Mass Spectrom. 1992;3:797–803. doi: 10.1016/1044-0305(92)80002-3. [DOI] [PubMed] [Google Scholar]
- 24.Zaia J, Li XQ, Chan SY, Costello CE. Tandem Mass Spectrometric Strategies for Determination of Sulfation Positions and Uronic Acid Epimerization in Chondroitin Sulfate Oligosaccharides. J Am Soc Mass Spectrom. 2003;14:1270–1281. doi: 10.1016/S1044-0305(03)00541-5. [DOI] [PubMed] [Google Scholar]
- 25.Miller MJC, Costello CE, Malmström A, Zaia J. A Tandem Mass Spectrometric Approach to Sequence Determination of Chondroitin/Dermatan Sulfate oligosaccharide Glycoforms. 2005 doi: 10.1093/glycob/cwj093. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karamanos NK, Vanky P, Syrokou A, Hjerpe A. Identity of Dermatan and Chondroitin Sequences in Dermatan Sulfate Chains Determined by Using Fragmentation with Chondroitinases and Ion-Pair High-Performance Liquid Chromatography. Anal Biochem. 1995;225:220–30. doi: 10.1006/abio.1995.1147. [DOI] [PubMed] [Google Scholar]
- 27.Lamari FN, Militsopoulou M, Mitropoulou TN, Hjerpe A, Karamanos NK. Analysis of glycosaminoglycan-derived disaccharides in biologic: samples by capillary electrophoresis and protocol for sequencing glycosaminoglycans. Biomed Chromatogr. 2002;16:95–102. doi: 10.1002/bmc.144. [DOI] [PubMed] [Google Scholar]
- 28.Karamanos NK, Axelsson S, Vanky P, Tzanakakis GN, Hjerpe A. Determination of Hyaluronan and Galactosaminoglycan Disaccharides by High-Performance Capillary Electrophoresis at the Attomole Level. Applications to Analyses of Tissue and Cell Culture Proteoglycans. J Chromatogr A. 1995;696:295–305. doi: 10.1016/0021-9673(94)01294-o. [DOI] [PubMed] [Google Scholar]
- 29.Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and Efficient Fluorescent Labeling of Glycans Using 2-Amino Benzamide and Anthranilic Acid. Anal Biochem. 1995;230:229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- 30.Militsopoulou M, Lamari FN, Hjerpe A, Karamanos NK. Determination of twelve Heparin- and Heparan Sulfate-Derived Disaccharides as 2-Aminoacridone Derivatives by Capillary Zone Electrophoresis using Ultraviolet and Laser-Induced Fluorescence Detection. Electrophoresis. 2002;23:1104–1109. doi: 10.1002/1522-2683(200204)23:7/8<1104::AID-ELPS1104>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 31.Roboz J. Introduction to Mass Spectrometry: Instrumentation and Techniques. InterScience; New York: 1968. [Google Scholar]
- 32.Desaire H, Leary J. Utilization of MS3 Spectra for the Multicomponent Quantification of Diasteromeric N-Acetylhexosamines. J Am Soc Mass Spectrom. 2000;11:1086–1094. doi: 10.1016/S1044-0305(00)00179-3. [DOI] [PubMed] [Google Scholar]
- 33.Leary J, Saad OM. Compositional Analysis and Quantification of Heparin and Heparan Sulfate by Electrospray Ionization Ion Trap Mass Spectrometry. Anal Chem. 2003;75:2985–2995. doi: 10.1021/ac0340455. [DOI] [PubMed] [Google Scholar]
- 34.Davidson E, Hoffman P, et al. The Acid Mucopolysaccharides of Connective Tissue. Biochim Biophys Acta. 1956;21:506–518. doi: 10.1016/0006-3002(56)90188-3. [DOI] [PubMed] [Google Scholar]
- 35.Fransson LA, Roden L. Structure of Dermatan Sulfate. I. Degradation by Testicular Hyaluronidase. J Biol Chem. 1967;242:4161–4169. [PubMed] [Google Scholar]
- 36.Fransson LA, Roden L. Structure of Dermatan Sulfate. II. Characterization of Products Obtained by Hyaluronidase Digestion of Dermatan Sulfate. J Biol Chem. 1967;242:4170–4175. [PubMed] [Google Scholar]
- 37.Schiller S, Slover GA, Dorfman A. A Method for the Separation of Acid Mucopolysaccharides: Its Application to the Isolation of Heparin from the Skin of Rats. J Biol Chem. 1961;236:983–987. [PubMed] [Google Scholar]
- 38.Bayliss MT, Osborne D, Woodhouse S, Davidson C. Sulfation of Chondroitin Sulfate in Human Articular Cartilage. The Effect of Age, Topographical Position and Zone of Cartilage on Tissue Composition. J Biol Chem. 1999;274:15892–15900. doi: 10.1074/jbc.274.22.15892. [DOI] [PubMed] [Google Scholar]
- 39.Lauder RM, Huckerby TN, Neiduszynski IA. A Fingerprinting Method for Chondroitin/Dermatan Sulfate and Hyaluronan Oligosaccharides. Glycobiology. 2000;10:393–401. doi: 10.1093/glycob/10.4.393. [DOI] [PubMed] [Google Scholar]
- 40.Plass AHK, West LA, Wong-Palms S, Nelson FRT. Glycosaminoglycan Sulfation in Human Osteoarthritis. Disease-Related Alterations at the Non-Reducing Termini of Chondroitin and Dermatan Sulfate. J Biol Chem. 1998;273:12642–12649. doi: 10.1074/jbc.273.20.12642. [DOI] [PubMed] [Google Scholar]
- 41.Wu YJ, La Pierre DP, Wu JW, Yee AJ, Yang BB. The Interaction of Versican with its Binding Partners. Cell Res. 2005;15:483–494. doi: 10.1038/sj.cr.7290318. [DOI] [PubMed] [Google Scholar]
- 42.Eriksen G, Carlstedt I, Mörgelin M, Uldbjerg N, Malmström A. Isolation and Characterization of Proteoglycans from Human Follicular Fluid. Biochem J. 1999;340:613–620. [PMC free article] [PubMed] [Google Scholar]
- 43.Tiedemann K, Larsson T, Heinegard D, Malmström A. The Glucuronyl C5-Epimerase Activity is the Limiting Factor in the Dermatan Sulfate Biosynthesis. Arch Biochem Biophys. 2001;391:65–71. doi: 10.1006/abbi.2001.2376. [DOI] [PubMed] [Google Scholar]
- 44.Hoffman P, Linker A, et al. The Acid Mucopolysaccharides of Connective Tissues. II. Further Experiments on Chondroitin Sulfate B. Arch Biochem Biophys. 1957;69:435–440. doi: 10.1016/0003-9861(57)90508-8. [DOI] [PubMed] [Google Scholar]
- 45.Coster L, Malmström A, et al. The Co-Polymeric Structure of Pig Skin Dermatan Sulphate. Distribution of L-iduronic Acid Sulphate Residues in Co-Polymeric Chains. Biochem J. 1975;145:379–389. doi: 10.1042/bj1450379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fransson LA. Structure of Dermatan Sulfate. 3. The Hybrid Structure of Dermatan Sulfate from Umbilical Cord. J Biol Chem. 1968;243:1504–1510. [PubMed] [Google Scholar]
- 47.Thorsson-Westergren G, Onnervik PO, Fransson LA, Malmström A. Proliferation of Cultured Fibroblasts is Inhibited by L-Iduronate-Containing Glycosaminoglycan. J Cell Phys. 1991;147:523–530. doi: 10.1002/jcp.1041470319. [DOI] [PubMed] [Google Scholar]
- 48.Mourão PA, Pavão MS, Mulloy B, Wait R. Chondroitin ABC Lyase Digestion of an Ascidian Dermatan Sulfate. Occurrence of Unusual 6-O-sulfo-2-acetamido-2-deoxy-3-O-(2-O-sulfo-alpha-L-idopyranosyluronic acid)-beta-D-galactose Units. Carbohydr Res. 1997;300:315–21. doi: 10.1016/s0008-6215(97)00061-x. [DOI] [PubMed] [Google Scholar]
- 49.Bao X, Pavão MS, Dos Santos JC, Sugahara K. A Functional Dermatan Sulfate Epitope Containing Iduronate(2-O-sulfate)α1–3GalNAc(6-O-sulfate) Disaccharide in the Mouse Brain: Demonstration Using a Novel Monoclonal Antibody Raised Against Dermatan Sulfate of Ascidian Ascidia Nigra. J Bio Chem. 2005;280:23184–93. doi: 10.1074/jbc.M503036200. [DOI] [PubMed] [Google Scholar]
- 50.Ueoka C, Nadanaka S, Seno N, Khoo KH, Sugahara K. Structural Determination of Novel Tetra- and Hexasaccharide Sequences Isolated from Chondroitin Sulfate H (Oversulfated Dermatan Sulfate) of Hagfish Notochord. Glycoconj J. 1999;16:291–305. doi: 10.1023/a:1007022229813. [DOI] [PubMed] [Google Scholar]