Abstract
HEI10 (CCNB1IP1) was first described as a RING-finger family ubiquitin ligase that regulates cell cycle by interacting with cyclin B and promoting its degradation. Subsequently, other studies suggested specific up-regulation of HEI10 in metastatic melanoma, and demonstrated direct interaction between HEI10 and the tumor suppressor Merlin, encoded by the neurofibromatosis 2 (NF2) gene. These and other results led us to hypothesize that HEI10 also influences the processes of cell migration and metastasis. We here show that cells with depleted HEI10 both migrate more rapidly and invade more effectively than control cells,. HEI10 depletion post-transcriptionally increases the expression of a group of pro-motility regulatory proteins including p130Cas, paxillin, Cdk1 and cyclin B2, but excluding Merlin. Among these, only inhibition of Cdk1/cyclin B activity specifically reversed the motility and invasion of HEI10-depleted cells. Finally, HEI10 is abundantly transcribed in many human tissues, and particularly abundant in some tumor cell lines, suggesting it may be commonly involved in coordinating cell cycle with cell migration and invasion.
Introduction
Development of cancer requires multiple changes in the regulation of cell physiology. As defined by Hanahan and Weinberg, to form an aggressive tumor, a cell must autonomously provide growth signals, become insensitive to growth-inhibitory signals, inactivate pro-apoptotic pathways, acquire a limitless replicative potential, promote angiogenesis, and become able to invade tissues and metastasize to distant sites (Hanahan & Weinberg, 2000). Some tumor suppressors and oncogenes function specifically in one of these defined areas: for example, mutations in Bcl2 predominantly affect apoptosis (Fesik, 2005). In other cases, the action of tumor-associated proteins is more complex. For example, mutation of Ras affects not only cell proliferation, but also apoptosis and metastasis, because of the central place Ras occupies in essential cell signaling pathways. Indeed, proteomics and systems biology analyses are now finding that many important signaling proteins interact with a diverse set of partners in different functional spheres (Rual et al., 2005; Schwikowski et al., 2000), complicating simple characterization of their function.
HEI10 (Human Enhancer of Invasion, clone 10) was first identified by our group in a functional genomic screen for novel human genes that influenced cell cycle progression and/or polarization (Toby et al., 2003). Overexpression of HEI10 causes yeast to extend the G2 phase of cell cycle, and to become hyperpolarized. Our subsequent characterization of overexpressed HEI10 revealed that this protein interacts with cyclin B, and that high levels of HEI10 promote the degradation of cyclin B in vertebrate cells, and of the cyclin B ortholog Clb2p in yeast. These activities of HEI10 depended on the integrity of the HEI10 RING domain. A RING domain is a common feature of ubiquitin ligases (Jackson et al., 2000), and HEI10 was found to both interact with an ubiquitin conjugating enzyme, and to induce its own auto-ubiquitination in a purified in vitro system. Based on these data, a first model for HEI10 was as regulator of the rate of cyclin B accumulation during G2.
Other studies have suggested a more complicated function for HEI10. Mine et al. identified HEI10 as a chimeric protein fused to HMG1C, in a translocation found in uterine leiomyoma, raising the possibility that aberrant HEI10 action may contribute to cancer development (Mine et al., 2001). A study of mRNA transcripts elevated in melanoma suggested that HEI10 is highly upregulated specifically in melanoma metastases (Smith et al., 2004). Gronholm and co-workers have recently identified HEI10 as a physical interactor with the Merlin tumor suppressor protein (Gronholm et al., 2006). This last identification is particularly intriguing, as Merlin, encoded by the neurofibromatosis 2 (NF2) gene, regulates both cell proliferation and migration (Evans et al., 2000; Xiao et al., 2005). Further, the expression of HEI10 and Merlin are interdependent, and the proteins to colocalize in some phases of cell cycle, with a sub-population of HEI10 located at cortical actin at the cell periphery (Gronholm et al., 2006). Together, these studies caused us to hypothesize that HEI10 not only regulates cell cycle, but also influences cell migration and invasion.
In this report, we demonstrate that HEI10 is required for cell cycle progression. In addition, HEI10 regulates both cell migration and invasion, and governs the steady state level of several proteins known to actively promote these processes. However, contrary to expectation, HEI10 negatively regulates cell motility, in a mechanism involving post-transcriptional downregulation of p130Cas, cyclin B, and Cdk1. Coupled with an analysis of HEI10 mRNA expression in primary cells, cell lines, and tumors, our data in sum indicate HEI10 can both promote and inhibit cancerous cell growth, and serves as a novel node connecting cell cycle and cell migration.
Results
HEI10 is required for cell proliferation
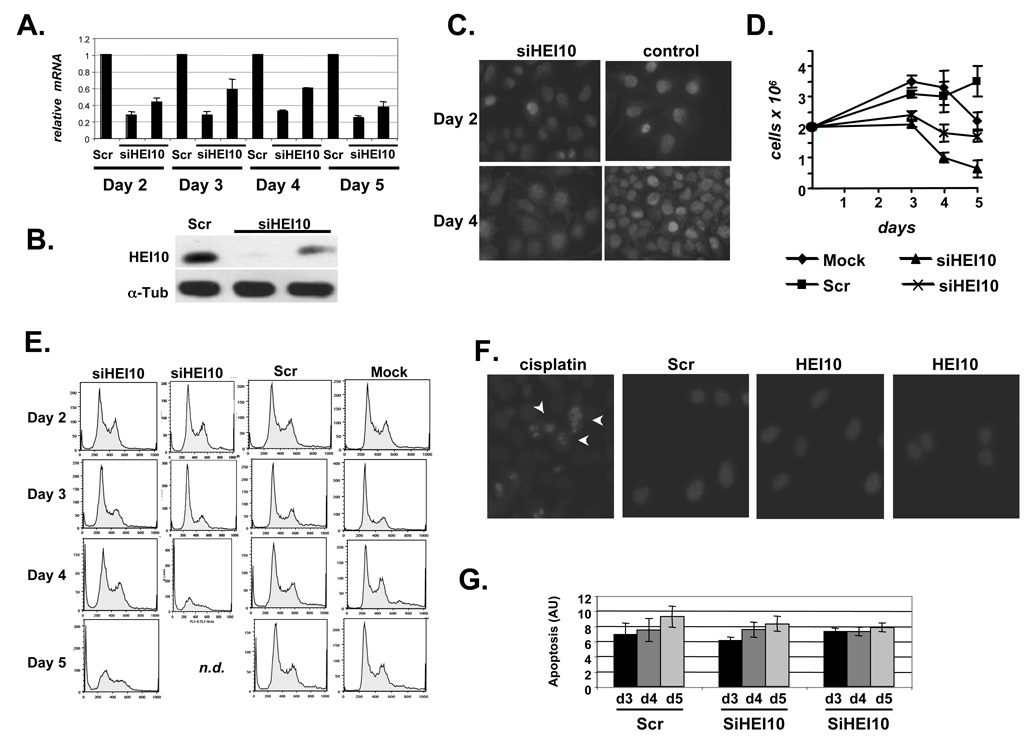
To analyze HEI10 function, we used two separate HEI10-targeted siRNAs for all experiments. Following siRNA transfection of U20S or MCF7 cells, HEI10 levels were typically reduced by ~75% for up to 5 days (Figures 1A, B, C). As HEI10 binds and promotes degradation of cyclin B (Toby et al., 2003), we first assessed whether HEI10-depletion induced proliferation defects by measuring cell doubling. For the first two days after transfection, both HEI10-depleted and control cells appeared similar (not shown). By 3 days following siRNA transfection, numbers of HEI10-depleted cells had not significantly increased in reference to seeding densities, although cells appeared morphologically normal (Figure 1D). By 4–5 days post-transfection, the number of HEI10 siRNA-transfected cells was consistently reduced below the number of cells seeded for assay (Figure 1D). FACS analysis (Figure 1E) indicated that although the cell cycle profile of HEI10-depleted cells remained similar to that of controls for 3 days, by 4–5 days after transfection the G2 cell cycle compartment was proportionally reduced, and the population contained many floating cells.
Figure 1. HEI10 depletion inhibits cell proliferation.
A. HEI10 depetion by siRNA was analyzed by RT q-PCR in all experiments described in this study. Results shown are normalized to levels of RNApolR2F. B. Western blot showing HEI10 depleted by two siRNAs, in reference to a-tubulin loading control. C. Immunofluorescence with antibody to HEI10 cells treated with siRNAs to HEI10 and control. Results were consistent between two different siRNAs to HEI10 (second not shown). Note increased number of cells in control panel at day 4 (compare to C). D. Proliferation of cell lines mock-transfected, or transfected with control siRNAs (Scr), or two siRNAs targeting HEI10. E. FACS analysis of cells treated with two HEI10-targeted siRNAs, control siRNAs, or mock-transfected, at 2, 3, 4, and 5 days after treatment. n.d., not determined because of reduced cell numbers. F. Hoechst 33342 staining of nuclei of cells treated with Scr or HEI0-directed siRNAS for 4 days, or with cisplatin (arrows indicate pycnotic nuclei). G. Relative apoptotic index produced by APOPercentage assay at days 3, 4, and 5 after HEI10 depletion. Differences between HEI10- and Scr-depleted cells are not statistically significant.
To establish whether the effect of HEI10 depletion was in restriction of cell cycle, we investigated cell death of HEI10-depleted versus control cells by measuring the relative rates of the appearance of pycnotic nuclei (Figure 1F), the cell death-associated mobilization of phospholipids (Figure 1G), and caspase-dependent cleavage of PARP or gelsolin (not shown). Based on all these measures, rate of apoptosis was not selectively enhanced by HEI10 depletion. These studies demonstrated that HEI10 is predominant required for cell proliferation. The earlier 2–3 day time points were consistently used for subsequent studies of HEI10.
Depletion of HEI10 promotes cell motility, invasion through Matrigel, and cell spreading
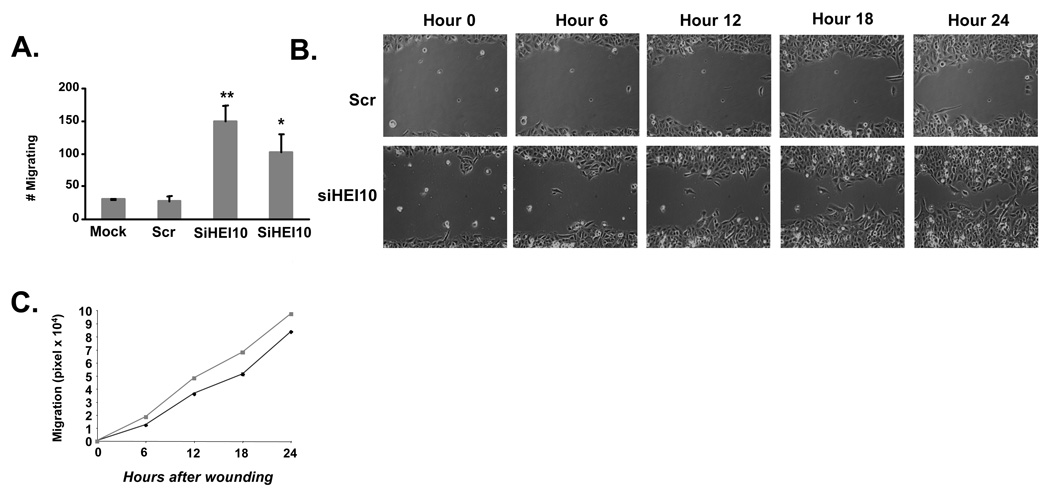
To assess whether HEI10 has a role in cell migration, HEI10- or control-depleted cells were seeded into the top of a Boyden chamber, and the number of cells migrating through the pores over 24 hours was scored (Figure 2A). HEI10 depletion increased cell migration 2.6–4.4-fold versus control-depleted cells. In a second approach, siRNA-transfected cells were allowed to form a monolayer over 48 hours. The monolayer was scratched, and live cell imaging used to track the closure of the scratch over the next 24 hours (Figures 2B, 2C). This assay also indicated faster migration of HEI10-depleted cells, with the greatest motility increase occurring at the earlier times of observation, while many cells with depleted HEI10 began to detach at later time points.
Figure 2. HEI10 depletion increases cell migration.
A. Migration of U2OS cells treated with two different HEI10-directed (siHEI10) or control (Scr) siRNAs through wells of a Boyden chamber. This experiment was performed 3 times in triplicate; numbers shown reflect average values. Similar results were obtained with siRNA treated MCF7 cells (not shown). Statistical analysis of Scr versus the two HEI10-targeted siRNAs indicates **P < 0.001 and *P< 0.01 respectively. B. Comparison of “wound” closure over 24 hours following scratching of a monolayer of cells treated 48 hours previously with two different HEI10-directed (siHEI10) or control (Scr) siRNAs. C. Quantitation of results in B. HEI10-depleted cells (squares, gray line) migrate more rapidly than Scr-depleted cells (diamonds, black line), P < 0.005 or better at all time points.
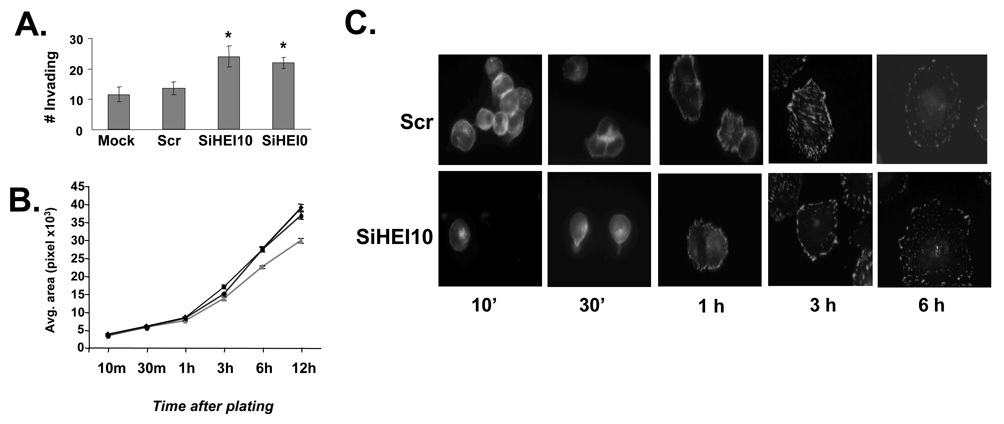
To assess the role of HEI10 in invasion, a Boyden chamber approach was again used to compare the relative movement of HEI10- versus control-depleted cells through Matrigel-coated versus uncoated filters (Figure 3A). 57–71% more HEI10-depleted versus control-depleted U2OS cells invaded through Matrigel. To assess the role of HEI10 in cell spreading, HEI10- or control-depleted cells were trypsinized 48 hours after transfection, and the extent of reattachment to cell culture plastic or poly-L-lysine was observed for the following 24 hours (Figure 3B, 3C, and not shown). Although HEI10-depleted cells consistently spread to a greater extent than controls at later time points on either support, the degree of increase was only marginally statistically significant. Moreover, the rate of appearance of indicators of cell attachment, such as accumulation of proteins such as paxillin at focal adhesions (Figure 3C), was similar in HEI10-depleted and control-depleted cells. Together, these results suggested HEI10 had a limited role in the initial spreading process, but a significant role in cell migration and invasion.
Figure 3. HEI10 depletion increases cell invasion and reattachment.
A Migration of U2OS cells treated with two different HEI10-directed (siHEI10) or control (Scr) siRNAs through Boyden chamber wells coated with Matrigel, versus uncoated wells. Experiment was performed 3 times in triplicate; numbers shown reflect average values. Statistical analysis assigns difference between Scr and HEI10-directed siRNAs as *P < 0.01 in each case. B. U2OS cells treated with two HEI10-directed (siHEI10) or control (Scr) siRNAs for 48 hours were trypsinized, then replated on tissue culture plastic, and fixed at time points up to 24 hours. Outlines of cells (stained with antibody to paxillin) were traced using the Metamorph software program, with 100 cells counted for each time point in each of 3 experiments. Statistical confidence for difference HEI10- versus Scr-depleted is P <0.02 at 6 hrs, and P <0.01 at 12 hours. Black lines with circle or diamond, HEI10 depletion; gray line with triangle, Scr depletion. C. Representative cells analyzed for the data graphed in 3B.
HEI10 depletion modulates the expression and activity of pro-motility regulatory proteins
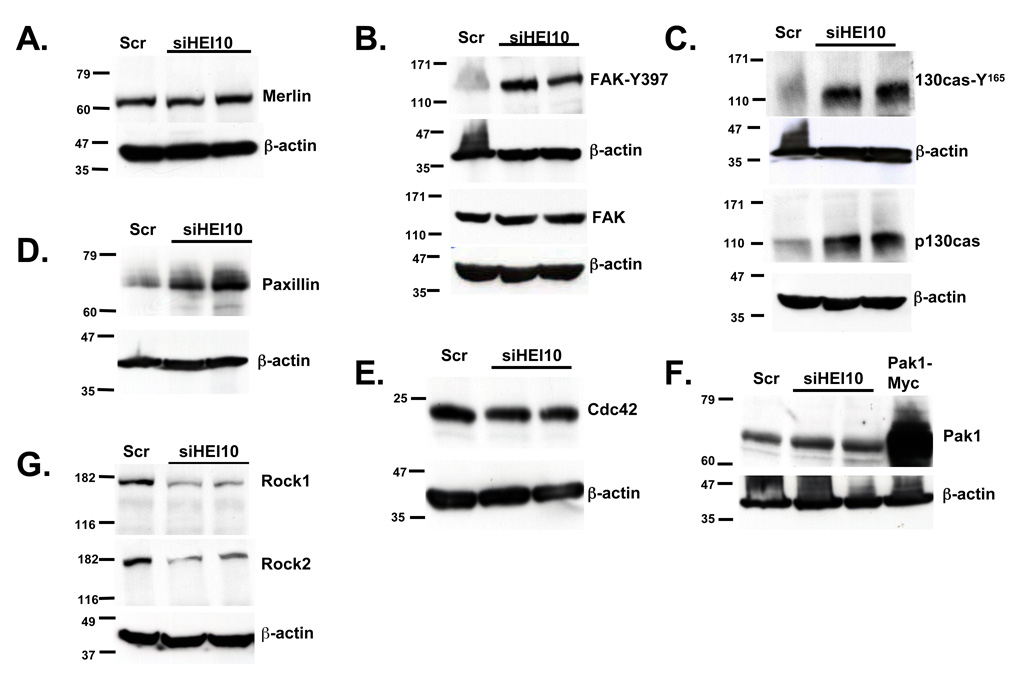
HEI10 is a ubiquitin ligase. We first hypothesized that HEI10 depletion might cause the up-regulation of pro-motility proteins such as Merlin and other cytoskeletally-localized proteins with which Merlin can associate, such as focal adhesion kinase (FAK)(James et al., 2004). However, HEI10 depletion did not affect steady state levels of Merlin or FAK (Figures 4A, 4B). This finding was consistent with the prediction of Gronholm et al., which placed HEI10 downstream of Merlin in cell signaling cascades (Gronholm et al., 2006). In contrast, the phosphorylation of FAK on Y397, associated with kinase activation (Ruest et al., 2000) and increased cellular migration, was consistently elevated by 4–6 fold following HEI10 depletion (Figure 4B). FAK is not only regulated by Merlin (Poulikakos et al., 2006), but also by association with focal-adhesion localized signaling proteins such as p130Cas (Brabek et al., 2005) and paxillin (Wade & Vande Pol, 2006). Total levels of paxillin and p130Cas were consistently elevated 5–6 fold in HEI10- versus control-depleted cells (Figures 4C, D). The levels of Y165-phosphorylated (activated) p130Cas (Bouton & Burnham, 1997) increased proportionally to the levels of total p130Cas (Figure 4B, bottom).
Figure 4. HEI10 negatively regulates p130Cas, paxillin, and FAK.
A U2OS cells were treated with two different HEI10-directed (siHEI10) or control (Scr) siRNAs for 48 hours, lysed, and total cell lysates analyzed by Western blot with antibody to Merlin. Each blot was stripped and reprobed with antibody to β-actin as a loading control. All experiments were done at least 3 times independently. B. Experiment as in A, but with blots initially probed with antibodies either to phosphorylated activated FAK (FAK-Y397), or total FAK. C. Experiment as in A.; blots initially probed with antibody to phosphorylated activated p130Cas (p130Cas Y165), or with antibody to total p130Cas, as indicated. D. Experiment as in A; blot initially probed with antibody to paxillin. E. Experiment as in A.; blot initially probed with antibody to Cdc42. F. Experiment as in A.; blot initially probed with antibody to Pak1. Pak1-Myc represents overexpressed epitope-tagged Pak1 transformed into U2OS cells as an additional size standard. G. Experiment as in A.; blot initially probed with antibody to Rock1, then stripped and reprobed with antibody to Rock2, then finally stripped and re-probed with antibody to β-actin.
We assessed additional motility regulators including, Cdc42, Pak1, ROCK1, and ROCK2 (Hirokawa et al., 2004; Kaempchen et al., 2003; Klemke et al., 1998; Worthylake & Burridge, 2003; Xiao et al., 2005; Zhao et al., 2000) (Figures 4E–G). HEI10 depletion had no effect on the steady state levels of Cdc42 or Pak1, and significantly decreased the levels of ROCK1 and ROCK2. In sum, these data indicated that HEI10 depletion did not promiscuously up-regulate expression of migration regulatory proteins. Given their upregulation, p130Cas and paxillin were the best candidates for direct targets of HEI10-dependent proteolysis. However, to date, efforts to co-immunoprecipitate HEI10 with these proteins have been unsuccessful (results not shown). Hence, HEI10 regulation of p130Cas, FAK, and other proteins may be indirect, reflecting propagation of pro-migratory signals from a separate source.
Post-transcriptional upregulation of B-type cyclins and Cdk1 by HEI10 depletion promotes migration
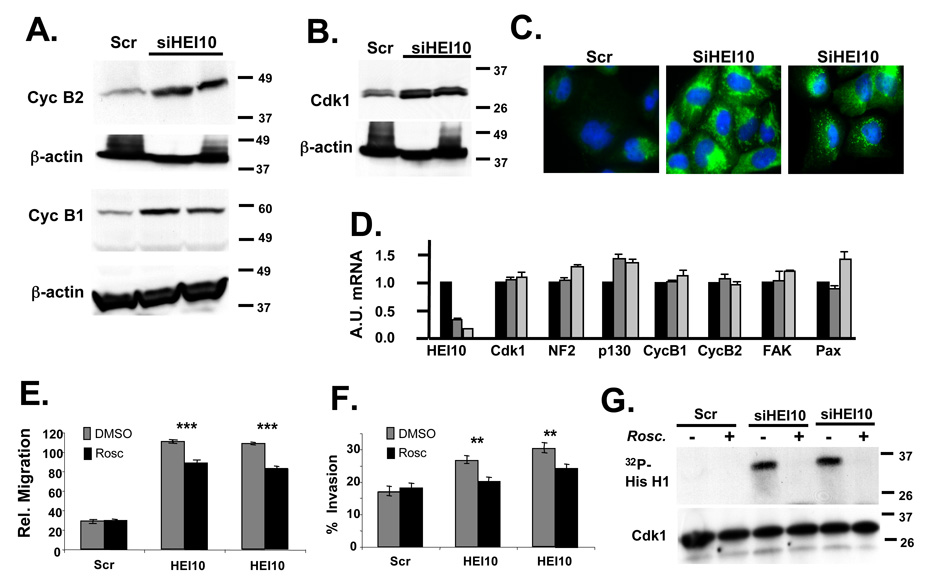
We previously demonstrated that HEI10 interacts directly with B-type cyclins (Toby et al., 2003). The recent description of a Cdk1/cyclin B2-dependent pro-motility pathway (Juliano, 2003; Manes et al., 2003) suggested an alternative direct means by which HEI10 depletion may influence cell migration. Indeed, steady state levels of cdk1, cyclinB1 and cyclin B2 were elevated 3-fold, 4–7 fold and 4-fold, respectively, in HEI10-depleted cells as compared to control cells (Figures 5A, B). Elevated levels of each of these proteins were also clearly visible by immunofluorescence of HEI10-depleted or control cells (Figure 5C, and results not shown).
Figure 5. HEI10 depletion induces Cdk1 and B-type cyclins to increase migration and invasion.
A Experiment as in 4A. Antibodies used for initial probe were to cyclin B1 and cyclin B2, as indicated; all blots were stripped and reprobed with antibody to β-actin. B. Experiment as in 4A. Antibodies used for initial probe were to Cdk1. C. Immunofluorescence of U2OS treated with Scr siRNA or two different HEI10-directed siRNAs, visualized with antibodies to cyclin B2 (green). DNA is stained with DAPI (blue). D. RT-qPCR analysis. Total RNA was prepared in parallel to the protein lysates analyzed in the experiments in Figure 4 and Figure 5. A.U., Arbitrary Units used to describe relative abundance of each mRNA listed. E, F. U2OS cells were treated with Scr- or two different HEI10-directed siRNAs in the presence of DMSO vehicle (negative control) or the Cdk1 inhibitor roscovitine. Migration (E) and invasion (F) were assayed as described in Figure 2 and Figure 3. Degree of significance of roscovitine reversal of phenotypes is P ***<0.0001 for migration (E) and **P < 0.005 for invasion (F). G. Cdk1 was immunoprecipitated with antibody to Cdk1 from cells treated as in E, and Cdk1-kinase activity assayed. Top panel shows 32P-labeled histone H1 substrate phoshorylated by Cdk1/cycB; bottom panel, total level of immunoprecipitated Cdk1.
To further probe the mechanism of HEI10 control of the expression of pro-migratory factors, cells were depleted for 48 hours with siRNA directed to HEI10 or scrambled control, then total RNA harvested and analyzed by quantitative RT-PCR. As shown in Figure 5D, the transcript levels of p130Cas, B-type cyclins, and Cdk1 were essentially unaffected by HEI10 depletion, indicating that HEI10 regulation of these proteins was post-transcriptional. At least for cyclin B/Cdk1, this most likely reflects loss of direct HEI10-dependent ubiquitination and proteolysis.
Inhibition of Cdk1 specifically limits HEI10-dependent migration and invasion
To establish which signaling pathways were particularly important for HEI10-dependent motility, we used small molecule inhibitors and/or siRNAs to inhibit specific proteins or pathways in HEI10- or control-depleted cells. We have found that roscovitine, a specific inhibitor of Cdk1 activity, selectively and strongly inhibits the migration and invasion of HEI10-depleted cells, while having no effect on migration of control cells (Figures 5E, F). Indeed, roscovitine essentially eliminates the increased invasion associated with HEI10-depletion. Supporting these results, Cdk1 from HEI10-depleted cells was substantially more active than that found in control cells (Figure 5G); roscovitine completely inhibited the enhanced Cdk1 activity. These results are congruent with and may be explained by the observation that B-cyclins are much more abundant following HEI10 depletion, driving Cdk1 activity (Figures 5A, B).
We have used a similar approach to probe the p130Cas/FAK/Src signaling pathway, using the Src kinase inhibitor PP2, and a strategy of siRNA co-depletion (e.g., of HEI10 and p130Cas). While these manipulations have influenced cell migration overall, no experiment using this strategy has inhibited migration in a manner that is specific for HEI10-depleted cells (results not shown).
In vivo expression of HEI10
Ideally, these functional analyses would be balanced with reciprocal studies analyzing the consequences of HEI10 overexpression. However, as we have previously reported (Toby et al., 2003), even moderate overexpression of HEI10 in mammalian cells leads to accumulation of most of the protein in apparently aggregated structures associated with the Golgi compartment, making interpretation of such an experiment problematic. In lieu of being able to readily overexpress HEI10, we have studied the distribution of HEI10 in different tissues and cell types, to determine if HEI10 expression levels suggest a characteristic profile of migratory or invasive behavior (Suppl. Figure 1). Particularly as elevated HEI10 mRNA expression has been suggested by one group to correlate with aggressive and melanomas (Smith et al., 2004), this seemed an important test. However, based on our current data, it is not possible to significantly correlate HEI10 mRNA expression with a specific cell lineage (Suppl. Figure 1A), or degree of cancer aggressiveness (Suppl. Figures 1B and 1C). Rather, HEI10 mRNA is generally abundant, suggesting the protein may play an important role in coordinating cell cycle and cell movement in many tissues.
Discussion
These data indicate that HEI10 negatively regulates cellular spreading, motility and invasion, but is required for cellular proliferation. Loss of HEI10 expression post-transcriptionally increased the total levels or activation of a number of proteins previously described as promoting cell motility, including p130Cas, paxillin, FAK, B-type cyclins, and Cdk1. However, only inhibition of Cdk1/cycB activity selectively reduced HEI10-associated enhancement of migration and invasion, as opposed to generally influencing these processes. We have previously demonstrated direct interaction between HEI10 and B-type cyclins. The simplest explanation for these results is that HEI10 depletion specifically stabilizes Cdk1/cyclin B complexes from HEI10-dependent proteolysis. This triggers pro-migratory signaling changes that propagate through the cell motility protein interaction network, ultimately stabilizing p130cas and paxillin (and potentially other proteins), and thereby activating FAK. Further supporting this interpretation, the study which first identified the role of CycB/Cdk1 in cell migration (Manes et al., 2003) described a phenotype for overexpression of CycB/Cdk1 very similar to that reported here for HEI10 depletion.
HEI10 directly interacts with Merlin (NF2) (Gronholm et al., 2006). In cells induced to express Merlin, HEI10 moved from the nucleus to the cytoplasm: in NF2−/− cells, HEI10 concentrated in the nucleus. Merlin acts to inhibit cell growth and proliferation; cells lacking Merlin are hyper-proliferative and migrate more actively. A model incorporating these observations with the new data presented here would postulate that in cells lacking Merlin, the concentration of HEI10 in the nucleus reduces the ability of HEI10 to interact with and downregulate extranuclear proteins affecting migration: in interphase, these proteins would include the B-type cyclins (Draviam et al., 2001). This may in part mediate the increased motility of NF2-deficient cells (Evans et al., 2000). In contrast, neither the Gronholm study nor our work here suggests that HEI10 influences the expression of Merlin, assigning HEI10 a role as a potential effector of this tumor suppressor.
We do not observe a striking variation in HEI10 mRNA expression between melanomas of different stages, or between melanomas and normal melanocyte cell lines (Suppl. Figure 1). Interesting, examining tumor cell lines derived from different tissues of origin, although central nervous system and ovarian cell lines similarly show relatively homogenous levels of HEI10 (<2-fold variation), breast and lung cancer cell lines show a much greater range of HEI10 expression. There appeared to be two distinct groups of breast cell lines, separated by ~6–8 fold difference in HEI10 expression: however, highly metastatic cell lines were found in both low and high expressor groups (e.g., MDA-MB-231, and HS-578T). A recent paper has identified amplification of sequences localized to 14q11.2 as characteristic of specific subtypes of lung cancer (Nymark et al., 2006); HEI10 localizes to this region of chromosome 14, and it will be of interest to see if HEI10 is specifically targeted by such genetic changes. Given the complex role of HEI10 in regulating cell growth, it is likely to be difficult to identify a straightforward functional relationship between expression levels and tumor aggressiveness. Nevertheless, based on the evolving delineation of interactions between HEI10, cyclin B/Cdk1, and Merlin, HEI10 clearly is positioned to influence both cell cycle and migratory properties of NF2-mutant cells, and HEI10 status may modulate the severity of disease in families with hereditary neurofibromatosis, and influence other cancers.
Materials and Methods
Cells and cell lines
U2OS (human osteosarcoma) cells and MCF7 (human breast adenocarcinoma) cell lines (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics.
siRNAs and inhibitors
Two HEI10-directed RNA oligonucleotide duplexes and a scrambled siRNA control were used (from Dharmacon, Lafayette, CO; sequences on request). For each experiment, after transfection of siRNAs (using Oligofectamine, Invitrogen) the degree of depletion of target mRNA was determined by reverse transcription real-time (quantitative) polymerase chain reaction (RT-qPCR) or by Western. Depletion was typically between 70–90%; experimental runs in which depletion <65% was observed were discarded. To inhibit Cdk1 kinase, cells were treated with 50 µM roscovotine (CALBIOCHEM®, EMD Biosciences San Diego, CA), which was added to the medium at the time of seeding cells into Boyden chambers.
Western blots, immunoprecipitation, kinase and apoptosis assays
Standard protocols were used for preparation of cell lysates for Western blotting and immunoprecipitations. Primary antibodies used in this study for Western blotting and immunofluorescence included mouse monoclonal antibodies to paxillin, cdc42, p130cas, FAK, cyclin B1, PARP, gelsolin (BD Biosciences), β-actin (Sigma), and Cdk1 (Oncogene Research Products); and rabbit polyclonal antibodies to phospho-p130casY165, PAK1 (both from Cell Signaling), cyclin B2, Rock1, Rock2 (Santa Cruz Biotechnology), HEI10 (Toby et al., 2003) and FAKY397(Covance). Secondary anti-mouse and anti-rabbit HPR conjugated antibodies were from Amersham. To quantify Western blot data densitometric analysis of ECL-exposed blots was done using NIH ImageJ (version 1.24o) software. To assess Cdk1 activity in HEI10-depleted and/or roscovitine-treated cells, protein lysates from siRNA-treated cells were immunoprecipitated with Cdk1-directed antibody, followed by in vitro kinase assay using a Cdk1 Kinase Assay Kit (Upstate), as per manufacturer’s instructions. The APOPercentage apoptosis assay kit was from Biocolor, Ltd. 24 hours pre-treatment with 5 µM cisplatin was used as a positive control to score pycnotic nuclei.
Microscopy
For immunofluorescence, cells growing on coverslips were fixed with 4% paraformaldehyde, permeabilized, blocked and incubated with antibodies using standard protocols. Secondary antibodies (including anti-mouse Alexa-488, anti-rabbit Alexa488, anti-mouse Alexa-568, anti-rabbit Alexa568) and DAPI were from Invitrogen, and Hoechst 33342 from Molecular Probes. Fluorescence microscopy was performed using a Nikon Eclipse TE 2000 microscope.
To measure cell spreading, cells were first transfected with scrambled or HEI10-targeted siRNAs. After 48 hours, cells were detached by incubation in PBS + 5mM EDTA for 15 minutes at 37°C, then re-plated onto either uncoated glass coverslips or poly-L-lysine coated cover slips (Fisher) in DMEM+10%FBS. Cells were fixed with 4% paraformaldehyde at indicated time points, and stained with anti-paxillin to visualize cell boundaries. In each group, cell area measurements were determined for ~100 cells using Metamorph imaging software. For Boyden chambers, migration analysis was performed according to the manufacturer’s (Becton Dickinson) protocol 48 hours after treatment of cells with siRNA. In parallel, as a control for changes in cell proliferation related to HEI10 depletion (see Figure 1), total cell proliferation of cells comparably treated with siRNAs was established by daily calculation of cell numbers over the course of the assay. Migration values were corrected based on proliferation index of siRNA-treated cells, i.e., made proportionate to the number of cells in the population at 3 days post-transfection. For wound healing assays, cells were transfected with siRNAs then plated at a density suitable to cause confluence after 48 hours. Wounds in the cell monolayer were made using a pipette tip, and then cells were maintained at 37°C in a stage incubator on top of a Nikon Eclipse E800 inverted microscope (Carl Zeiss, Thornwood, NY). Images were acquired for 24 hours with a Quantix cooled charge-coupled device (CCD) camera (Roper Scientific, Trenton, NJ), and analyzed using Metamorph.
All calculations of statistical significance were made using the GraphPad InStat software package (San Diego, CA) and STATA software (Stata Corps, TX). Approaches included unpaired t test (with Welch correction), the Mann-Whitney test, and multiple linear regression analysis.
RT-qPCR
Contaminating DNA from RNA preparations was removed using TURBO DNA-free™ (Ambion). RNA from cell lines was quantified using the Agilent 2100 BioAnalyzer in combination with a RNA 6000 Nano LabChip. RNA was reverse-transcribed using M-MLV reverse transcriptase (Ambion) and a mixture of anchored oligo-dT and random decamers. 5′-nuclease assays using Taqman chemistry were run on a 7900 HT sequence detection system (Applied Biosystems). The sequences of the primers and probes used, and assay details, are provided in Supplemental Table 1.
Supplementary Material
Acknowledgments
We gratefully acknowledge Margret Einarson, Cynthia Spittle, Meenhard Herlyn, and Hong Wu for help with the collection and preparation of cell lines for RNA analysis. We thank Ilya Serebriiskii for help with some experiments. We greatly appreciate the efforts of Dr. Brian Egleston in the statistical analysis of data. We thank Dr. Tony Yeung, director of the FCCC Biochemistry and Biotechnology facility for Real-Time PCR and DNA synthesis services This work and the authors were supported by NIH RO1 CA63366, and by Translational Pilot Project funding from the Tobacco Settlement from the State of Pennsylvania (to EAG); and by an Appropriation from the Commonwealth of Pennsylvania, by NIH core grant CA-06927 and support from the Pew Charitable Fund (to Fox Chase Cancer Center). DD was a recipient of the Plain and Fancy Fellowship of the Fox Chase Cancer Center Board of Associates.
References
- Bouton AH, Burnham MR. Detection of distinct pools of the adapter protein p130CAS using a panel of monoclonal antibodies. Hybridoma. 1997;16:403–411. doi: 10.1089/hyb.1997.16.403. [DOI] [PubMed] [Google Scholar]
- Brabek J, Constancio SS, Siesser PF, Shin NY, Pozzi A, Hanks SK. Crk-associated substrate tyrosine phosphorylation sites are critical for invasion and metastasis of SRC-transformed cells. Mol Cancer Res. 2005;3:307–315. doi: 10.1158/1541-7786.MCR-05-0015. [DOI] [PubMed] [Google Scholar]
- Draviam VM, Orrechia S, Lowe M, Pardi R, Pines J. The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J Cell Biol. 2001;152:945–958. doi: 10.1083/jcb.152.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Sainio M, Baser ME. Neurofibromatosis type 2. J Med Genet. 2000;37:897–904. doi: 10.1136/jmg.37.12.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5:876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- Gronholm M, Muranen T, Toby GG, Utermark T, Hanemann CO, Golemis EA, et al. A functional association between merlin and HEI10, a cell cycle regulator. Oncogene. 2006 doi: 10.1038/sj.onc.1209475. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hirokawa Y, Tikoo A, Huynh J, Utermark T, Hanemann CO, Giovannini M, et al. A clue to the therapy of neurofibromatosis type 2: NF2/merlin is a PAK1 inhibitor. Cancer J. 2004;10:20–26. doi: 10.1097/00130404-200401000-00006. [DOI] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, et al. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends in Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- James MF, Beauchamp RL, Manchanda N, Kazlauskas A, Ramesh V. A NHERF binding site links the betaPDGFR to the cytoskeleton and regulates cell spreading and migration. J Cell Sci. 2004;117:2951–2961. doi: 10.1242/jcs.01156. [DOI] [PubMed] [Google Scholar]
- Juliano R. Movin' on through with Cdc2. Nat Cell Biol. 2003;5:589–590. doi: 10.1038/ncb0703-589. [DOI] [PubMed] [Google Scholar]
- Kaempchen K, Mielke K, Utermark T, Langmesser S, Hanemann CO. Upregulation of the Rac1/JNK signaling pathway in primary human schwannoma cells. Hum Mol Genet. 2003;12:1211–1221. doi: 10.1093/hmg/ddg146. [DOI] [PubMed] [Google Scholar]
- Klemke RL, Leng J, Molander R, Brooks PC, Vuori K, Cheresh DA. CAS/Crk coupling serves as a "molecular switch" for induction of cell migration. J Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manes T, Zheng DQ, Tognin S, Woodard AS, Marchisio PC, Languino LR. Alpha(v)beta3 integrin expression up-regulates cdc2, which modulates cell migration. J Cell Biol. 2003;161:817–826. doi: 10.1083/jcb.200212172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine N, Kurose K, Konishi H, Araki T, Nagai H, Emi M. Fusion of a sequence from HEI10 (14q11) to the HMGIC gene at 12q15 in a uterine leiomyoma. Jpn. J. Cancer Res. 2001;92:135–139. doi: 10.1111/j.1349-7006.2001.tb01075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark P, Wikman H, Ruosaari S, Hollmen J, Vanhala E, Karjalainen A, et al. Identification of specific gene copy number changes in asbestos-related lung cancer. Cancer Res. 2006;66:5737–5743. doi: 10.1158/0008-5472.CAN-06-0199. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Xiao GH, Gallagher R, Jablonski S, Jhanwar SC, Testa JR. Re-expression of the tumor suppressor NF2/merlin inhibits invasiveness in mesothelioma cells and negatively regulates FAK. Oncogene. 2006 doi: 10.1038/sj.onc.1209587. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Ruest PJ, Roy S, Shi E, Mernaugh RL, Hanks SK. Phosphospecific antibodies reveal focal adhesion kinase activation loop phosphorylation in nascent and mature focal adhesions and requirement for the autophosphorylation site. Cell Growth Differ. 2000;11:41–48. [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- Smith AP, Weeraratna AT, Spears JR, Meltzer PS, Becker D. SAGE identification and fluorescence imaging analysis of genes and transcripts in melanomas and precursor lesions. Cancer Biol Ther. 2004;3:104–109. doi: 10.4161/cbt.3.1.661. [DOI] [PubMed] [Google Scholar]
- Toby GG, Gherraby W, Coleman TR, Golemis EA. A novel RING-finger protein, Human Enhancer of Invasion 10 (HEI10), alters mitotic progression through regulation of cyclin B levels. Mol. Cell. Biol. 2003;23:2109–2122. doi: 10.1128/MCB.23.6.2109-2122.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade R, Vande Pol S. Minimal features of paxillin that are required for the tyrosine phosphorylation of focal adhesion kinase. Biochem J. 2006;393:565–573. doi: 10.1042/BJ20051241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthylake RA, Burridge K. RhoA and ROCK promote migration by limiting membrane protrusions. J Biol Chem. 2003;278:13578–13584. doi: 10.1074/jbc.M211584200. [DOI] [PubMed] [Google Scholar]
- Xiao GH, Gallagher R, Shetler J, Skele K, Altomare DA, Pestell RG, et al. The NF2 tumor suppressor gene product, merlin, inhibits cell proliferation and cell cycle progression by repressing cyclin D1 expression. Mol Cell Biol. 2005;25:2384–2394. doi: 10.1128/MCB.25.6.2384-2394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZS, Manser E, Loo TH, Lim L. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol Cell Biol. 2000;20:6354–6363. doi: 10.1128/mcb.20.17.6354-6363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.