Abstract
Mucosal surfaces provide first-line defense against microbial invasion through their complex secretions. The antimicrobial activities of proteins in these secretions have been well delineated, but the contributions of lipids to mucosal defense have not been defined. We found that normal human nasal fluid contains all major lipid classes (μg/ml), as well as lipoproteins and apolipoprotein AI. The predominant less polar lipids were myristic, palmitic, palmitoleic, stearic, oleic, and linoleic acid, cholesterol, and cholesteryl palmitate, -linoleate, and -arachidonate. Normal human bronchioepithelial cell secretions exhibited a similar lipid composition. Removal of less-polar lipids significantly decreased the inherent antibacterial activity of nasal fluid against Pseudomonas aeruginosa, which was in part restored after replenishing the lipids. Furthermore, lipids extracted from nasal fluid exerted direct antibacterial activity in synergism with the antimicrobial peptide HNP-2 and liposomal formulations of cholesteryl linoleate and cholesteryl arachidonate were active against P. aeruginosa at physiological concentrations as found in nasal fluid and exerted inhibitory activity against other gram negative and gram positive bacteria. These data suggest that host-derived lipids contribute to mucosal defense. The emerging concept of host-derived antimicrobial lipids unveils novel roads to a better understanding of the immunology of infectious diseases.
Keywords: Lipid mediators, mucosa, lung, bacterial infections
Introduction
The respiratory tract is constantly challenged with air-borne and aspirated microbes and its integrity depends on continuous removal of inhaled microbes by mucociliary clearance. Airway mucosal secretions have been intensely studied and important defense functions have been attributed to antimicrobial (poly)peptides such as lysozyme, defensins, or LL37 (1). Other antimicrobial components include surfactant proteins (2) and mucins (3). Antimicrobial (poly)peptides are cationic and hydrophobic, and their functions in innate host defense are to kill microbes through membrane permeabilization and disruption of membrane bound multienzyme complexes (4,5) and to modulate the immune response through chemotactic activity and sequestration of proinflammatory microbial products (6).
In addition, bodily secretions also contain lipids. Lipids form a heterogeneous group of hydrophobic substances ranging from simple fatty acids, linear chains of carbons carrying a carboxyl group, to more complex ring-structured molecules like cholesterol and cholesteryl esters, which can be further modified by the attachment of various side groups including alcohol, phosphate, and amino-functions. Major lipid classes comprise fatty acids (fatty acyls), glycerolipids, glycerophospholipids, sphingolipids, and sterols, including cholesterol and cholesteryl esters. Among these, glycerophospholipids and sphingolipids are the most polar and hydrophilic, and triglycerides and cholesteryl esters are the least polar and most hydrophobic (7). Although some fatty acids appear as free molecules, most of the lipids in bodily fluids are bound to carrier proteins, including albumin and apolipoproteins, the latter forming lipoproteins upon lipid-binding (8). Lipoproteins are classified based on their apolipoprotein and lipid composition, which affect their density. The five major lipoproteins described in plasma are chylomicrons, very-low-density lipoproteins (VLDL), intermediate-density lipoproteins (IDL), low-density lipoprotein (LDL), and high-density lipoproteins (HDL), with increasing protein and decreasing lipid contents, respectively (9). ApoA-I is one of the nine major apolipoproteins. It is an integral component of HDL and is also found in chylomicrons and VLDL. This 29-kDa protein is the main cholesterol acceptor and activates lecithin-cholesteryl ester transferase (LCAT), which is involved in esterification of cholesterol and production of cholesteryl esters. Recent studies suggest that apolipoproteins and their lipoprotein particles function also as immune modulators, regulating cytokine production and activity of immune, endothelial, and epithelial cells (10-12).
The functions of lipids are diverse. Lipids are essential components of all biological membranes, but the membrane lipid composition differs greatly between eukaryotes and prokaryotes (13, 14). Lipids are also precursors for hormones, are used for energy storage, and they have a prominent role as messengers and regulators of inflammation (15). Furthermore, lipids in lung surfactant, predominantly phospholipids, are essential for reducing the surface tension in the alveoli, thus preventing their collapse during exhalation (16).
Previous studies implicate that lipids also have antimicrobial functions. For example, the antimicrobial activity of fatty acids has been known for a long time and is utilized in food preservation (17), and certain chronic infectious diseases have been associated with an altered lipid profile (18-22). In cystic fibrosis, a hereditary disease with abnormal mucus production and chronic lung infection with Pseudomonas aeruginosa, reduced levels of docosahexaenoic and linoleic acid in saliva and serum have been documented and corroborated by a mouse model (23). A role in neonatal protection against infection has been also attributed to lipids in milk (24) and lipids in vernix caseosa, the newborn coating (25).
However, despite these reports on the antimicrobial actions of lipids and speculations on their use as therapeutics (26) the concept of a lipid-mediated component of innate host defense is new. We propose that host-derived lipids constitute the arsenal of antimicrobial agents in innate mucosal defense. We will show here that: 1) selective removal of lipids from nasal fluid leads to diminished antibacterial activity, 2) this is partially restored by re-supplementing the fluid with lipids, and 3) selected cholesteryl esters identified in nasal fluid exert antibacterial activity in vitro.
Materials and Methods
Nasal fluid collection
Under full institutional review board approval, nasal fluid was collected from healthy adult donors (D1 – D11) without history of allergies or sinusitis into 50 ml polypropylene centrifuge tubes by vacuum suction. Since nasal fluid may be contaminated with the mucosal microbiota, samples were incubated for 15 min at 60°C to heat-inactivate the normal flora. Trypticase soy broth (2 ml) inoculated with 20 μl heat inactivated nasal fluid remained sterile after 72 hours of incubation in contrast to non-heated nasal fluid. After heat-inactivation, nasal fluid was homogenized with a tip sonicator (Fisher Dismembrator, Fisher Scientific International, Hampton, NH), three times at level 2 for 20 s), centrifuged for 5 min at 500 × g, and the resulting cell- and debris free supernatant was further centrifuged for 30 min at 11,000 × g and 4°C). Epithelial cells in nasal fluid were sparse and under these conditions the contaminating cells were not disrupted and sedimented (data not shown). Cole et al. (27) reported a significant reduction of the cationic antibacterial protein fraction in nasal fluid upon boiling. To verify that heat inactivation at 60 °C for 15 min does not significantly alter the biochemical and antibacterial properties of nasal fluid, we compared the overall protein profile, peptidoglycan hydrolyzing activity, antibacterial activity, and lipid profile of nasal fluid before and after heat inactivation. There was no major difference in the protein and lipid profile of nasal fluid demonstrable and there was no statistical significant difference in respect to the peptidoglycan hydrolyzing and antibacterial activity (data not shown).
Clarified nasal fluid was stored under nitrogen or argon at -20°C until further use. Nasal fluids were pooled for multiple analyses and functional assays to increase volume and to minimize interdonor variations in sample composition.
Epithelial cell culture
Normal human-derived bronchial epithelial cells grown at an air-liquid interface in six-well plates (AIR-606) were purchased from MatTek Corporation (Ashland, MA). Upon receipt, cells had been grown for 6 days (in a serum-free DMEM-based medium with proprietary growth factors) and apical secretions had been allowed to accumulate. After overnight incubation (37°C, 5% CO2), apical secretions from six wells were collected and pooled. Potentially contaminating cells and debris were removed by centrifugation (500 × g, 4°C, 10 min), and the sample was subjected to lipid extraction, followed by rpHPLC/ELSD, as described below.
Lipid standards
Lipid standards, selected according to the literature (28) and guided by our studies, were purchased from Matreya (Pleasant Gap, PA) or Sigma (St. Louis, MO). These were dissolved in organic solvents (chloroform or dichloromethane) in glass vials, overlaid with nitrogen or argon gas, and stored at -20°C. Free fatty acids: myristic acid (M, C14:0, number of C atoms:number of double bonds), palmitic acid (P, C16:0), palmitoleic acid (P:1, C16:1), heptadecanoic acid (H, C17:0, plant fatty acid used as internal standard), stearic acid (S, C18:0), oleic acid (O, C18:1), linoleic acid (L, C18:2), and docosahexaenoic acid (D, C22:6). Glycerolipids: dipalmitic acid (DP) and tripalmitic acid (TP). Glycerophospholipids: phosphatidyl serine (PS). Sphingolipid: sphingomyelin (SM). Sterols: cholesterol (C), cholesteryl stearate (CS), cholesteryl palmitate (CP), cholesteryl oleate (CO), cholesteryl linoleate (CL), and cholesteryl arachidonate (CA).
Lipid extraction
Lipid extractions were based on the Bligh & Dyer method (29). Briefly, per 0.4 ml sample, 1.5 ml of methanol: chloroform (2:1, v/v) were added, the sample flushed with N2 gas, and vigorously vortexed for 1 min. Then, 0.5 ml of chloroform were added, the sample flushed with N2, and vortexed for 1 min. The procedure was repeated using 0.5 ml of dH2O. Samples were then incubated on a rotary shaker for 10 min and centrifuged (500 × g, 25°C, 10 min). The lower phase was collected and an equal volume of chloroform was added to the upper phase to repeat the lipid extraction. The chloroform layers from both steps were pooled and the organic solvent was removed under a gentle stream of N2 gas at 37°C. The extracted lipids were re-suspended in a small volume of appropriate organic solvent(s) for further studies and analysis. For quantitative experiments, to control for lipid extraction efficiency, heptadecanoic acid was added (20 μg unless stated otherwise) to all samples prior to lipid extraction. Extraction efficiencies were in average 40 - 80%, and test extractions showed that the various lipid classes were recovered with similar relative efficiencies (data not shown).
Thin-layer chromatography
The TLC system employed two separation systems run serially (30). Glass-backed silica gel TLC plates (250-μm layer thickness, 60 Å, 20 × 20 cm; EMD Chemicals, Inc., Gibbstown, NJ; pre-washed in methanol and heat-activated for 10 min at 130°C in an oven) were spotted with concentrated lipid extracts and standards and first developed in chloroform:methanol:water (65:25:4 v/v/v) to 13 cm from the origin, then in hexanes:diethyl ether (36:9 v/v) to near the top. Plates were dried and lipid components visualized in iodine vapor. The darkness of the resulting spots is influenced by the quantity of each lipid and the number of unsaturated bonds, limiting the suitability of TLC for comparative lipid quantification of samples with different lipid composition.
Reverse-phase HPLC
Separation and quantification of lipids was performed with a low-pressure quarternary gradient system (Summit HPLC System, Dionex Corporation, Sunyvale, CA with Dionex PCS1Chromeleon software) on a reverse phase column (Clipeus C18, 5 μm, 250 × 3.0 mm; Higgins Analytical, Inc., Mountain View, CA, pre-equilibrated in acetonitrile (solvent A)/dichloromethane (solvent B), 87.5/12.5) with evaporative light scattering detection (ELSD 800, Alltech Associates, Inc., Deerfield, IL, operated at 40 °C, 1 or 2 bar N2). Five μl samples were injected and eluted (0.6 ml/min) with an increasing concentration of solvent B (time/%B; 4/12.5, 7/47.5, 28/50.0, 32/12.5, 35/12.5). Response curves were established for authentic standards (data not shown) and used to quantify lipids in test samples, whereby the amount detected in the HPLC chromatogram was corrected for the actual extraction efficiency evident in the recovery of the internal standard heptadecanoic acid. In this system linoleic acid (C18:2) co-elutes with myristic acid (C14:0) and palmitoleic acid (C16:1), oleic acid (C18:1) with palmitic acid (C16:0), and cholesteryl palmitate with cholesteryl oleate (data not shown). In some cases, peak fractions were collected between 2 and 10 min or 11.5 and 13 min, and further analyzed by GC/MS for free fatty acid analysis, or a commercial enzymatic cholesterol kit according to the manufacturer’s instruction (BioVision Inc., Mountain View, CA), respectively.
Free fatty acid analysis
Lipid extracts from nasal fluid (800 μl) were subjected to reverse-phase HPLC and fractions collected between 3.5 and 10 min of the gradient were dried under a stream of nitrogen, treated with 250 μl of boron trifluoride in methanol (Alltech associates, Inc.; 14% w/v; (31) for 20 min at 60 °C, extracted after addition of 2 ml water twice with 2 ml CH2Cl2, dried under nitrogen, dissolved in chloroform, and then injected (1 μl, splitless mode, Agilent 7638B autoinjector, 280°C) onto a bonded-phase non-polar fused silica capillary column (HP1, 50 m × 0.2 mm, 0.11-μm film thickness) housed within an Agilent 6890N gas chromatograph. The column was eluted (constant flow, 1 ml/min) with ultra-high purity helium, the oven was held at 50°C for 1 minute following injection, then ramped (10°C/min) to a plateau at 320°C. The end of the column (GC/MS transfer line at 280°C) was directly inserted into the electron-impact (EI, 230 °C, 70 eV) source of a quadrupole mass spectrometer (Agilent 5975 MSD) scanning from m/z 40-1050 (0.67 sec/scan). Data was analyzed with instrument-supplied software, and compound identification was achieved with the NIDST MS Search algorithm (v. 2.0). Identification of fatty acyl methyl ester derivatives in nasal fluid was based on elution times and by comparing the peak mass spectra with library spectra and the spectra from authentic standards.
Cholesteryl ester analysis
Identification of cholesteryl esters was performed according to Duffin et al. (32). Total lipid extracts from 100 μl of individual or pooled nasal fluid were re-suspended in 100-μl chloroform:methanol (4:1, v/v) containing 10 mM ammonium formate. Standards (cholesteryl palmitate, -arachidonate, -linoleate and -oleate) were dissolved similarly at 1 pmol/μl. Mass spectra were recorded by direct injection (20 μl) of samples into a stream of the same solvent (20 μl/min), entering an Ionspray™ source attached to a triple-quadrupole mass spectrometer (Sciex API III+, Thornhill, Canada). Mass (m/z 100-1000, 0.3 Da step size, 6.16 sec/scan, orifice 65 volts) and fragment and parent ion MS/MS spectra (m/z 350-1000, orifice 50 volts,0.3-Da step size, 6.67 sec/scan, instrumental setting for collision gas (argon) thickness of 100) were recorded using hyrocarbon- and particulate-depleted air (so-called zero air) for spray nebulization, and the vapors of liquid nitrogen for the curtain gas, both at a flow rate of 0.6 L/min. Data were collected (Tune version 2.5 (FPU)) and displayed (MacSpec™, version 3.3) with instrument manufacturer-supplied software. The prominent cholesteryl esters in nasal fluid were identified after comparison of the spectra from samples and standards, and with reference to spectra reported in the literature (32).
Lipoprotein analysis
To assess the presence of lipid-binding proteins, Titan Gel electrophoresis was performed with 20-μl aliquots of individual nasal fluid, lyophilized and dissolved in 2 μl of electrophoresis buffer, according to the manufacturer’s instructions (Lipoprotein Kit, Helena Laboratories). Immunoblotting was employed to specifically identify apolipoprotein A-I (apoA-I). Aliquots of nasal fluid were subjected to 12% SDS-PAGE, followed by immunoblotting without antigen fixation (32). The primary antibodies were mouse monoclonal antibodies against apoA-I, B100 and E (Chemicon International, Temecula, CA). Specific bands were visualized with alkaline phosphatase-conjugated polyclonal goat-anti mouse antibodies (Pierce Biotechnology, Inc., Rockfort, IL) and BCIP/NBT substrate (Research Products International Corp., Mount Prospect, IL), and quantified using Versadoc imaging documentation and QuantityOne software (BioRad, Hercule, CA).
Protein analysis
Protein composition and concentrations were assessed by Coomassie- and silver-stained 12% AU-PAGE (33), analyzed with a gel imaging system (Versadoc, BioRad) and BCA assay using bovine serum albumin standard protein according to the manufacturer’s instructions (Pierce), respectively.
Lipid depletion of nasal fluid by solid phase extraction
To deplete nasal fluid of non-polar lipids, SPE was performed on pooled nasal fluids using tC18 1cc Sep-Pak cartridges (Waters Corporation, Milford, MA) with a 12-port vacuum manifold and a bench-top vacuum station (Alltech). The method had been first developed using mock nasal fluid (data not shown). Cartridges were sequentially conditioned and washed with 2 ml of acetonitrile and 2 ml of dH2O, respectively. Fifty μl of pooled nasal fluid diluted 1:2 in dH2O (NF) were loaded per cartridge and the flow-through was collected. The cartridges were washed with 2 × 1 ml of dH2O and then eluted with 2 ml of 45 % acetonitrile containing 0.05 % TFA to recover hydrophobic proteins, including antimicrobial peptides like human neutrophil peptides that may have bound to the tC18 matrix. The latter two fractions were separately collected, lyophilized, dissolved in a small amount of 0.01% acetic acid, and pooled, with the flow through yielding lipid-depleted nasal fluid (LD). The protein concentrations of NF and LD were determined and NF was further diluted in dH2O to match the protein concentration of LD (typically 1:2.5). Samples of NF and LD, matched for total protein concentrations, were then compared to assess the extent of lipid depletion and their similarities in protein composition. Between 100 μl – 300 μl aliquots of NF and LD were subjected to lipid extraction and rpHPLC/ELSD. To calculate the overall lipid depletion, NF and LD were first normalized to the internal standard hepatdecanoic acid [H] and the NF and LD peak areas were calculated for peaks eluting after 3.75 min (to exclude interfering peaks associated with non-retained material). Aliquots of total protein-matched NF and LD were subjected to Coomassie-stained AU-PAGE to verify similar protein composition (2 μl); and to immunoblotting (20 μl, (33) for human neutrophil peptide HNP1-3, to ascertain that antimicrobial peptides had not been removed through our SPE procedure. In addition, 6 μl of each sample were subjected to immunoblotting to quantify apoA-I.
Antimicrobial peptides and polypeptides
Lysozyme was purified in the Porter laboratory, purified HNP-2 and lactoferrin had been kindly provided by Dr. Tomas Ganz, UCLA.
Antibacterial assays
For colony forming unit assays, mid-logarithmic cultures of Pseudomonas aeruginosa (PA, a cystic fibrosis strain originating from the Welsh laboratory, University of Iowa) were adjusted to McFarland 0.5 (~ 1 × 108 CFU/ml) in saline and nutrient-supplemented phosphate buffer (assay buffer; 100 mM NaCl, 10 mM NaPi at pH 7.4, 4 % trypticase soy broth). Siliconized microfuge tubes were used and assays were performed in duplicates. Six μl of bacterial stock suspension were added to 54 μl of complete nasal fluid (NF) or lipid-depleted nasal fluid (LD). After incubation at 37°C and 150 rpm for 30 and 60 min samples were serially diluted in assay buffer and plated in duplicates or triplicates on trypticase soy agar plates. Colonies were counted after 24 h incubation at 37°C and the original bacterial concentrations (CFU/ml) calculated. As control, bacteria were added to assay buffer and plated immediately (T0) and after incubation as above.
For experiments in which LD was re-supplemented with lipids, total lipid extract (without hepatdecanoic acid) from the corresponding NF was subjected to rpHPLC. Peak fractions were pooled, dried, and dissolved in chloroform to yield a 9-fold concentrated stock (relative to the nasal fluid volume used for lipid extraction). Of this, 6 μl were added to the respective microfuge tubes and the chloroform was allowed to evaporate. Control received 6 μl of chloroform only. Fifty-four μl of NF, LD, or assay buffer (for controls) were added and lipids were allowed to solubilize at 37°C for 30 min before bacteria were added.
To determine the antibacterial activity of nasal fluid lipids in the presence and absence of HNP-2, native liposomes were prepared freshly for each experiment from crude lipid extracts according to Sadzuka and colleagues (34). Lipid extracts or chloroform as solvent control were dried in round bottom glass tubes that had been pre-washed with 5 % acetic acid, incubated in assay buffer (1.2 fold concentrated compared to original nasal fluid volume) at 65 °C for 10 min, and subsequently sonicated at 75 °C for 30 min. After cooling to room temperature liposomes were placed on ice. Per sample tube the following was pipetted: 2.5 μl of 10-fold concentrated HNP-2 dissolved in 0.01% acetic acid or solvent only, 42.5 μl of native liposomes, solvent control or assay buffer (the latter for T0 only), and 5 μl of mid logarithmic growth phase bacteria (PA and Staphylococcus aureus ATCC 29213, cultured in TSB, adjusted to McF 0.5 in assay buffer, and further diluted in assay buffer 1:10). At T0 and after 3 h of incubation, 200 μl of assay buffer were added to each tube and the numbers of colony forming units were determined after serial dilution and plating on TSA plates. Samples were prepared in duplicates and plated twice.
To assess their antibacterial activity, cholesterol and cholesteryl esters (cholesteryl arachidonate, - linoleate, - oleate, and - palmitate; Sigma) were incorporated in liposomes to ensure consistent and homogenous suspension in an aqueous medium. Briefly, thin lipid films consisting of phosphatidylcholines, phosphatidylglycerols and cholesterol with and without test lipid (test liposomes and vehicle liposomes, respectively) were prepared by pipetting aliquots of lipid stock solutions (in methanol/chloroform) into glass tubes and evaporating the solvent at 50°C under a stream of N2 gas. The films were then placed under vacuum for at least 8 h to remove residual organic solvent. To prepare the liposomes, films were hydrated with 9% sucrose, pH 5.0, heated at 45°C - 65°C for 5-10 min and probe sonicated for 5 min. The liposomal formulations were sterile filtered using a 0.2 μm size membrane (Millex-GV, Millipore).
Initial screening was performed in a microtiter format (2006 Clinical and Laboratory Standards Institute Guidelines, M7-A7) with fluorescence read out using Syto 9 (LiveDead BacLight, Invitrogen), a non-fluorescent probe that diffuses freely into bacterial cells and exhibits green fluorescence upon binding to bacterial DNA whereby the resulting fluorescence correlates with the number of bacteria present . P. aeruginosa was grown for 24 h in trypticase soy broth, adjusted to McF 0.5 in saline and further diluted 1 : 100 in 1.1-fold concentrated cation-adjusted Mueller-Hinton broth to ~ 1 × 105 CFU/ml. Of this 90 μl were aliquotted into wells containing 10 μl of 640 μg/ml test liposomes or vehicle liposomes. Plates were incubated for 16 h at 37°C. Syto 9 was added (100 μl of a 1.25 μM solution in dH2O) and fluorescence was determined after 15 min incubation at RT (Excitation at 485 nm, Emission at 530 nm). The resulting relative fluorescence units (RFU) were blanked against Mueller-Hinton broth and liposomal formulations without bacteria. Liposomal formulations that demonstrated at least 80 % growth inhibition at 64 μg/ml were further diluted to determine their minimal inhibitory concentrations, subjected to CFU assay as described above, and tested at 64 μg/ml against a broader range of gram-positive and gram negative bacteria employing the microtiter assay . The bacterial strains were Staphylococcus aureus (SA, ATCC 29213), Enterococcus faecalis (EF, ATCC 700802), and Enterobacter cloacae (EC, ATCC 49141) cultured prior to testing as recommended by ATCC.
Data analysis
Unless stated otherwise, means, SEM, and SD were calculated and data graphed with SigmaPlot 9.0 and statistical significance was determined with SigmaStat 2.0.
Results
Human nasal fluid is rich in lipids
By thin-layer chromatography (Figure 1a) we detected all the major lipid classes in human nasal fluid: free fatty acids, glycerolipids, glycerophospholipids, sphingolipids, and sterols including cholesterol, and cholesteryl esters. Retention times during reverse-phase HPLC with evaporative light scattering detection (rpHPLC/ELSD) were consistent with the presence of myristic and/or linoleic acid, palmitic and/or oleic acid, stearic acids, cholesterol, and a variety of cholesteryl esters and triglycerides (Figure 1b). Additional peaks eluting shortly before cholesterol were tentatively assigned as diglycerides. Myristic (C14:0), palmitoleic (C16:1), palmitic (C16:0), linoleic (C18:2), oleic (C18:1) and stearic acid (C18:0) were identified by gas chromatography/mass spectrometry (GC/MS) based on retention times (Figure 1c) and by comparing the peak mass spectra with library spectra and the spectra from authentic standards (data not shown). The presence of cholesterol was confirmed by subjecting the corresponding rpHPLC fraction to a commercial enzymatic test kit (data not shown). To identify the predominant cholesteryl esters, electrospray ionization (ESI) mass spectra (Figure 1d) were acquired from lipid extracts. The mass spectra revealed signals for the (M+ NH4)+ adducts consistent with the presence of cholesteryl-linoleate, -arachidonate, and –palmitate, and these assignments were unequivocally verified by the MS/MS spectra, which in all cases revealed a strong signal for the cholesterol fragment (m/z 369.3). This behavior was indistinguishable from that of the authentic standards. Compared to rpHPLC/ELSD peak analysis of standard lipids, the individual lipid concentrations were in the μg/ml range (see Table I), free fatty acids were found at 8.1 ± 1.9 μg/ml, cholesterol at 36.9 ± 14.4 μg/ml reaching up to 90 μg/ml and individual cholesteryl esters at 24.1 ± 6.3 μg/ml (means ± S.E.M., n = 36, 6, 18, samples, respectively).
Figure 1.
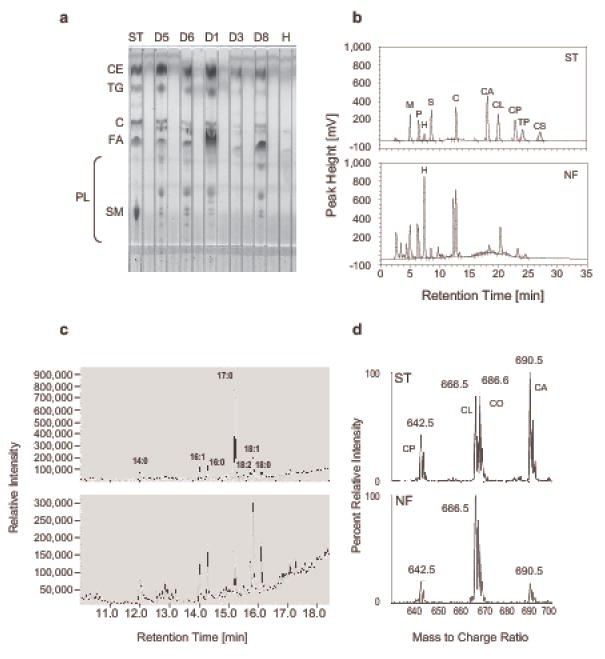
Nasal fluid is rich in lipids and contains all major lipid groups. Lipid extracts from individual or pooled nasal fluid samples (NF) were subjected to TLC (a); reverse phase HPLC/ELSD (b); gas chromatography followed by mass spectral analysis (c); and tandem mass spectrometry (d). (a) ST: standard lipids (ST, 25 μg each), cholesteryl ester (CE, cholesteryl palmitate), triglyceride (TG, tripalmitin), cholesterol (C), fatty acid (FA, palmitic acid, C16:0) and sphingomyelin (SM); PL: indicates where polar lipids migrate; D1 – D8: lipid extracts from individual nasal fluid samples (350 μl) collected from the respective donors (D), all samples had been spiked with heptadecanoic acid prior to extraction; H: 25 μg of heptadecanoic acid. (b) ST: standard lipids (5 μg each), NF: lipid extract from 150 μl pooled nasal fluid collected from donors D1, D2, D3 and spiked with 50 μg of heptadecanoic acid (H, C17:0). M: myristic acid (C14:0); P: palmitic acid (C16:0); S: stearic acid (C18:0); C: cholesterol; CA, CL, CP, CS: cholesteryl-arachidonate, -linoleate, -palmitate, and –stearate; TP: tripalmitin. (c) Total ion current profiles of methylated, rpHPLC-purified fatty acids from 400 μl NF (top panel, pooled from donors D1, D2, and D3) and 800 μl NF (lower panel, donor D6); peaks represent the methyl esters of myristate (14:0), palmitoleate (16:1), palmitate (16:0), heptadecanoate (17:0), linoleate (18:2), oleate (18:1), and stearate (18:0). (d) Representative electrospray ionization parent ion mass spectra (m/z 369.4, cholesterol) of standard cholesteryl esters (ST, top panel, each 60 nmol) and the lipid extract of 100 μl of nasal fluid (NF, lower panel, pooled from D1, D2, and D3). CP, CL, CO, and CA: cholesteryl- palmitate, linoleate, -oleate, and arachidonate; calculated molecular weights for the NH4 + adducts are 642.62, 666.62, 668.63, and 690.62 Da, respectively.
Table I. Free fatty acids, cholesterol and cholesteryl esters are found in normal human nasal fluid in μg/ml concentrations.
Nasal fluid collected from individual donors (D) and pooled nasal fluid were supplemented with an internal standard (heptadecanoic acid, a plant fatty acid absent in humans) and lipid extracts were subjected to rpHPLC/ELSD. Native lipids were identified and quantified according to established reference chromatograms and standard curves and recovery of the internal standard. Only lipids which have been confirmed by a second technique are included here, concentrations are in μg/ml.
| Fatty acids1 | Cholesterol2 | Cholesteryl esters3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Individual Nasal Fluids | Myristic4 (14:0) | Palmitic5 (16:0) | Palmitoleic4 (16:1) | Stearic (18:0) | Oleic5 (18:1) | Linoleic4 (18:2) | -palmitate | -linoleate | -arachidonate | |
| D1 | 4.8 | 10.1 | 5.8 | 8.0 | 0.3 | 0.2 | 27.8 | 11.4 | 17.9 | 8.6 |
| D2 | 19.0 | 56.3 | 27.2 | 0.0 | 6.9 | 12.0 | 71.4 | 79.8 | 86.0 | 69.9 |
| D3 | 1.3 | 3.4 | 1.8 | 0.0 | 0.0 | 0.1 | 6.4 | 4.9 | 4.7 | 4.2 |
| D5 | 6.9 | 21.2 | 9.4 | 0.0 | 0.1 | 0.1 | 90.3 | 25.2 | 41.6 | 21.1 |
| D6 | 2.7 | 6.0 | 3.5 | 6.6 | 0.1 | 0.1 | 11.2 | 7.2 | 15.6 | 7.1 |
| D8 | 23.7 | 7.6 | 26.0 | 7.6 | 0.0 | 13.9 | 14.23 | 7.36 | 13.15 | 8.04 |
|
| ||||||||||
| Mean | 9.7 | 17.4 | 12.3 | 3.7 | 1.2 | 4.4 | 36.9 | 22.7 | 29.8 | 19.8 |
| SEM | 3.8 | 8.2 | 4.7 | 1.7 | 1.1 | 2.7 | 14.4 | 11.8 | 12.3 | 10.3 |
|
| ||||||||||
| Pooled Nasal Fluid | ||||||||||
| D1,D2,D3,D7 | 10.9 | 13.1 | 12.8 | 13.7 | 0.2 | 0.2 | 17.8 | 15.7 | 28.7 | 13.3 |
Confirmed by GC/MS;
Confirmed by enzymatic assay;
Confirmed by MS/MS;
4 and 5 Co-elute in rpHPLC at 4.6 – 5 min and at 5.8 – 6.0 min, respectively. Based on GC/MS intensity distribution in nasal fluid from donors D1, D2, D3, and D6 (see Figure 1 c) myristic acid, palmitoleic and linoleic acid are assigned 10%, 45% and 45% of the peak area, respectively, and palmitic and oleic acid are each assigned 50% of the peak area.
Lipid compositions of human respiratory epithelial cell secretions and nasal fluid are similar
To further substantiate that lipids detected in nasal fluid represent respiratory epithelial cell secretions, we harvested apical secretions from commercial normal human bronchioepithelial cells grown at air-liquid interface and determined their lipid profile using TLC (Figure 2a) and rpHPLC/ELSD (Figure 2b). As in nasal fluid, cholesterol was the predominant non-polar lipid in the apical secretions, reaching concentrations of approximately 250 – 300 μg/ml in both batches tested. A pre-cholesterol peak in rpHPLC/ELSD (see figure 1b) as well as lipids with chromatographic behavior characteristic of cholesteryl esters were also detected at the low μg/ml range. In contrast, naïve cell culture medium contained fatty acids and lipids that behaved like phospholipids in TLC (Figure 2a) at much reduced concentrations, and, importantly, cholesterol and cholesteryl esters were not detectable (Figure 2a and b). This was despite analysis of a 10-fold larger volume. Hence, it appears that lipids in nasal fluid originate at least in part from local production by epithelia in the upper airways.
Figure 2.
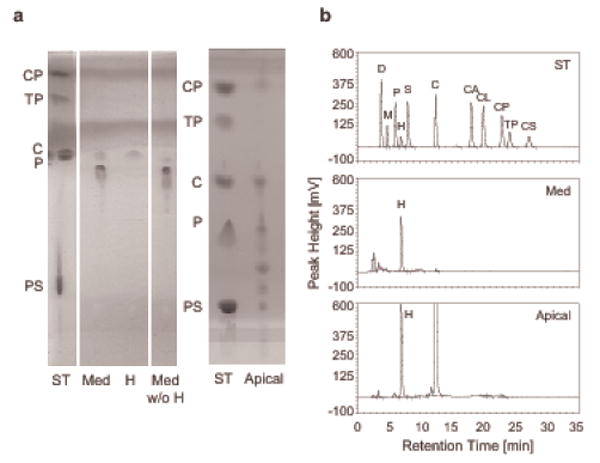
Apical secretions of human normal bronchial epithelial cells contain lipids similar to nasal fluid. Apical secretions of human bronchial epithelial cells (EpiAir-606) accumulated over 7 days in air-liquid interface culture and 2 ml of serum free naïve culture medium (Med), included as negative control, were mixed with the internal standard heptadecanoic acid (H, 20 μg), lipid extracted, and subjected to TLC (a) and rpHPLC/ELSD (b) analysis. (a) ST: standard lipids (25 μg each), CP: cholesteryl palmitate, TP: tripalmitin, C: cholesterol, P: palmitic acid (C16:0), PS: phosphatidyl serine; Med w/o H: for comparison, 2 ml of medium were extracted without prior addition of H; Apical: lipid extracts from 200 μl of apical secretions representing a growth area of 16.8 cm2. On the left TLC plate the solvent fronts appear as homogenously stained lines. (b) rpHPLC/ELSD chromatogram. ST: lipid standards, 5 μg each, D: docosahexaenoic acid, for others see Figure 1b legend; Apical: lipid extract from 300 μl of apical fluid originating from a 16.8 cm2 growth area.
Lipoproteins are also found in nasal fluid
In bodily fluids, an aqueous medium, the vast majority of lipids is bound to lipoproteins. Therefore, we surmised that lipoproteins should also be present in nasal fluid, and we subjected nasal fluid to Fat-Red-7B-stained gel electrophoretic lipoprotein analysis (Figure 3a). All samples tested contained one stronger band, which was located where chylomicrons in serum typically appear, according to the manufacturer, and also showed diffuse staining across the gel towards the anode. Subsequent immunoblotting using monoclonal antibodies against the apolipoproteins apoA-I, apoB-100 and apoE demonstrated the presence of apoA-I (Figure 3b), but not apoB-100 and E (data not shown). In addition to two major bands appearing between 20 and 30 kDa (nominal molecular weight of apoA-I is 28 kDa) minor bands of smaller molecular weight were detected possibly reflecting fragmentation of apoA-I.
Figure 3.
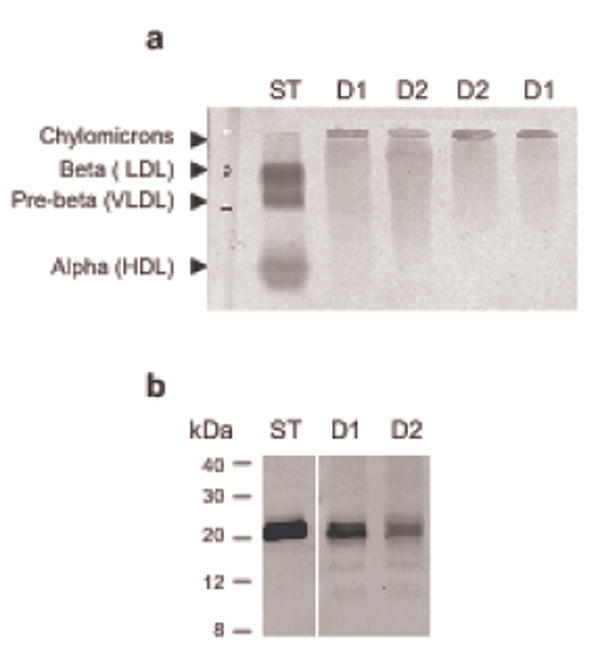
Nasal fluid contains apolipotproteins including ApoAI. Individual nasal fluid samples were subjected to agarose gel electrophoresis with Fat Red 7B staining (a), and 12% Tricine SDS-PAGE followed by Western immunoblotting (b) using mouse monoclonal antibodies against apoAI and an alkaline phosphatase/BCIP/NBT detection system. ST: serum standard, 2 μl; D1 and D2: nasal fluid, 20 μl each, collected from donors 1 and 2, respectively.
Partial removal of lipids from nasal fluid results in decreased antimicrobial activity, which can be restored by lipid re-supplementation
To ascertain whether lipids contribute to the inherent antimicrobial activity of nasal fluid, we selectively removed lipids from nasal fluid (Figure 4) and compared the antimicrobial activity of complete and lipid-depleted nasal fluid (NF and LD, respectively, Figure 5). We focused on the removal of less polar lipids (fatty acids, cholesterol, di- and triglycerides, and cholesteryl esters) because pilot studies suggested an association between higher concentrations of cholesteryl esters in nasal fluid and stronger inherent antibacterial activity (data not shown). For this we developed a solid-phase extraction (SPE) protocol using a C18 matrix that achieved a reduction of fatty acids, cholesterol/di-glycerides, cholesteryl esters/triglycerides by 54.6 ± 11.1 %, 59.6 ± 19.2 %, and 71.9 ± 15.3%, respectively (means ± S.E.M., n = 4). When comparing the rpHPLC/ELSD chromatograms of nasal fluid before (NF) and after SPE (LD), 61.57% ± 11.24% of the less polar lipids (means ± S.E.M., n = 4) were removed with this method (Figure 4a shows representative chromatograms reflecting an 89% reduction), while the overall protein profile appeared unaltered (Figure 4b). Importantly, this procedure did not reduce the contents of antimicrobial (poly)peptides as indicated by silver-stained AU-PAGE and by immunoquantification of human neutrophil peptides HNP1-3 (NF: 6.4 ± 1.1 μg/ml; LD: 6.2 ± 1.3 μg/ml, means ± S.E.M., n = 4; Figure 4c). However, apoA-I appeared to be somewhat reduced in lipid-depleted nasal fluid (Figure 4d, 32% reduction of apoA-I in LD).
Figure 4.
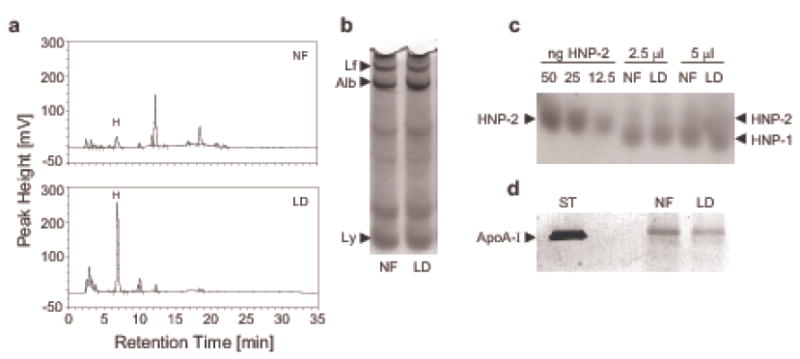
Non-polar lipids can be selectively removed from nasal fluid by solid-phase extraction. Nasal fluid was pooled from three donors (D1, D2, and D3) and subjected to solid phase extraction using a tC18 Sep-Pak cartridge to selectively remove less polar lipids. Aliquots from nasal fluid (NF) and lipid-depleted nasal fluid (LD) were adjusted to the same protein concentration and analyzed by rpHPLC/ELSD (a); silverstained AU-PAGE (b); Western immunoblotting probing for human neutrophil peptides HNP1-3 (c) and apolipoprotein apoA-I (d). (a) rpHPLC/ELSD Chromatograms of lipid extracts from 200 μl of NF and LD each; H: heptadecanoic acid (C17:0). In this example an overall lipid reduction of 89.5 % was achieved. (b) 2 μl of NF and LD were loaded, Lf: lactoferrin, Alb: albumin, Ly: lysozyme. (c) Samples were probed with a polyclonal rabbit antiserum against HNP1-3 and HNP-specific bands were visualized with an alkaline phosphatase/BCIP/NBT detection system; (d) 2 μl of serum standard (ST) and 25 μl of NF and LD each were probed for apo-AI.
Figure 5.
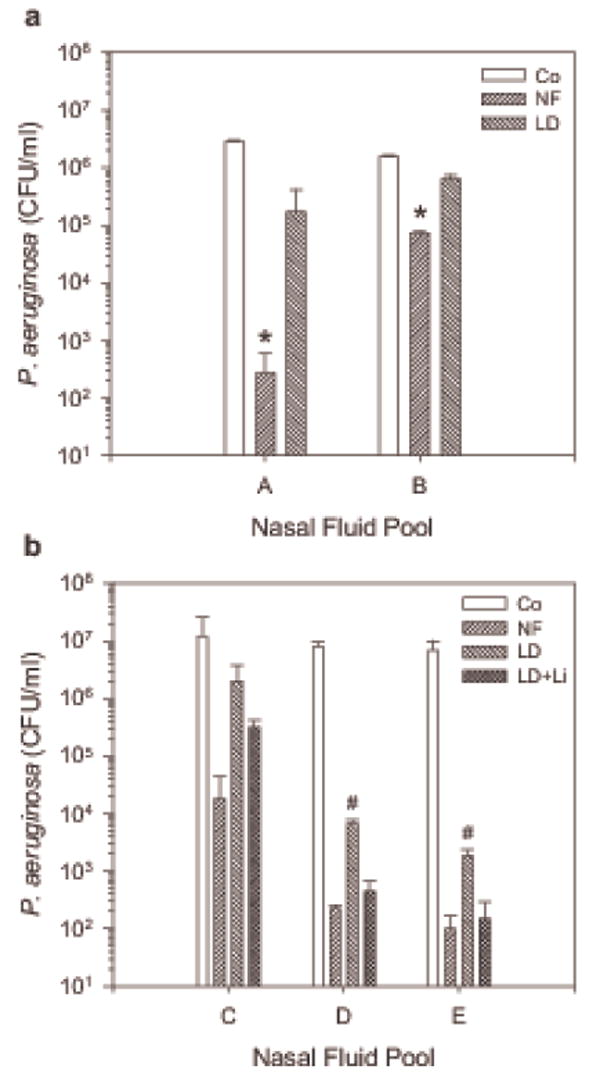
Lipids contribute to the inherent antimicrobial activity of nasal fluid. Nasal fluid (NF), lipid-depleted nasal fluid (LD), and LD supplemented with lipids extracted from the corresponding NF (LD + Li) were mixed with P. aeruginosa and CFU/ml were determined after 30 min incubation at 37°C. Co: bacteria incubated in buffer control. (a) A: NF pooled from donors D1, D2, and D3; B: NF pooled from donors D1, D2, and D5; * NF significantly reduced CFU/ml compared to the control (in paired t-test p = 0.022 for A, 0.023 for B), in contrast to LD. (b) C: NF pooled from donors D1 and D3; D: NF pooled from donors D1, D6, D9, and D10; E: NF pooled from donors D1, D5, D6, D9, and D11. Shown are the means + S.D. of two independent experiments conducted in duplicates for each nasal fluid pool. #: CFU/ml in LD are significantly increased compared to NF and LD + Li (in t-test p = 0.016 and p = 0.018, respectively, for C; p = 0.038 and p = 0.043, respectively, for D) while NF and LD + Li are statistically not different.
We tested then the antibacterial activity of NF and LD and found that the antibacterial activity of LD against P. aeruginosa was diminished (Figure 5a), but could be partially restored after replenishing LD with lipid extracts prepared from homologous NF (Figure 5b). These data suggest that less polar lipids contribute to the inherent antibacterial activity of nasal fluid.
Lipids extracted from nasal fluid exert direct antibacterial activity against P. aeruginosa in synergism with HNP-2
To further substantiate the antibacterial role of lipids in nasal fluid we subjected native liposomes prepared from lipid extracts from three different nasal fluid pools to colony forming unit assays with P. aeruginosa. Considering the presence of HNP1-3 in nasal fluid (see Figure 4c) and previous reports on synergism between fatty acids and antimicrobial peptides (25) we also included testing the effect of HNP-2 on the antibacterial activity of lipids. We found that native liposome preparations from two nasal fluid pools reduced the number of CFU/ml by over 1 log and that all three native liposome preparations acted in synergism with HNP-2 leading to a significant reduction of CFU/ml of up to 4 log in the presence of 25 μg/ml HNP-2 (Figure 6a).
Figure 6.
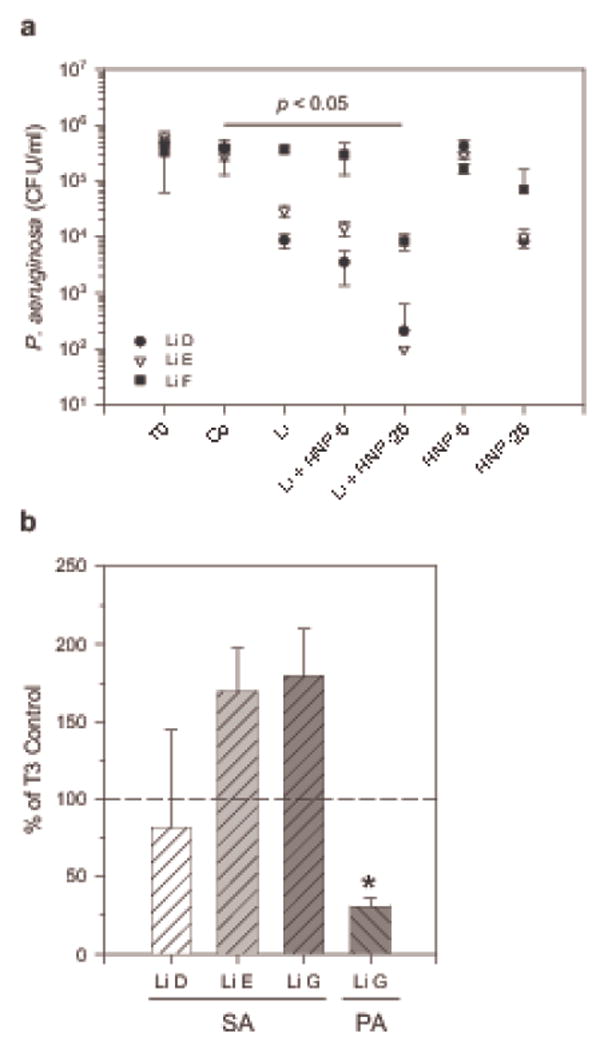
Lipids extracted from nasal fluid exert antibacterial activity against P. aeruginosa but not S. aureus. Lipids (Li) were extracted from nasal fluid pooled from donors D1, D6, D9, and D10 (Li D); from donors D1, D5, D6, D9, and D11 (Li E); from donors D1 and D6 (Li F); and from donors D1, D5, and D6 (Li G). Li C and Li D originate from nasal fluid pool NF C and NF D described in Figure 5. Extracted lipids or solvent control (Co) were heated and sonicated in assay buffer to yield liposomal preparations, and mixed with bacteria. CFU/ml were determined immediately (T0) or after 3 h (T3) incubation at 37 °C. (a) Activity of Li against PA in the presence or absence of 5 and 25 μg/ml HNP-2 (HNP-5 and HNP-25, respectively). * p < 0.05 in Kruskal-Wallis One Way ANOVA on Ranks for control versus treatments. (b) Activity of Li against SA and, for comparison, against PA. Shown are means + S.D. of two independent experiments conducted in duplicates. * p = 0.040 in t-test for Li G versus Co when tested against PA.
We also tested the activity of initially two native liposome preparations from nasal fluid against S. aureus (Figure 6b), a microorganism that is often found in the nostrils unlike P. aeruginosa. No significant activity against S. aureus was observed. To rule out a loss of lipid activity due to prolonged storage of lipid extracts prior to liposome preparation, we made native liposomes from lipids extracted from an additional nasal fluid pool (LiG) and tested the antibacterial activity against S. aureus and P. aeruginosa in parallel. As before, the number of S. aureus CFU was not significantly reduced in contrast to the number of P. aeruginosa CFU, thus suggesting a differential antibacterial activity of host derived lipids. The reduction of antibacterial activity of lipid depleted nasal fluid appeared to be mainly linked to the removal of cholesterol and cholesteryl esters. Therefore, in order to begin characterizing the lipids that carry the antibacterial function in nasal fluid, we investigated the antibacterial activity of cholesterol and cholesteryl esters packaged in liposomes in vitro.
Cholesteryl arachidonate and Cholesteryl linoleate have antibacterial activity
Initial screening of numerous liposomal formulations of cholesterol, and various cholesteryl esters using an MIC based assay with P. aeruginosa and a fluorescence read out (Figure 7a) yielded two lipids, cholesteryl arachidonate (CA) and cholesteryl linoleate (CL), that effected fluorescence inhibition of at least 80 %, which translates to a 1.5 log difference in CFU/ml between treated and untreated bacteria. This activity was dose dependent and growth inhibition of P. aeruginosa was demonstrable at concentrations as low as 4 μg/ml (Figure 7b), which is within the lower range of what we have measured in nasal fluid ex vivo. When CA and CL were subjected to a 3 hour colony forming unit assay in a low nutrient phosphate buffer with a higher bacterial inoculum (Figure 7c), CA appeared to slow bacterial growth whereas CL was bactericidal, significantly reducing the number of CFU/ml to 20 ± 16.99 % of the inoculum (p = 0.002 in One Way ANOVA for buffer vs CL liposome. When testing CA and CL against a broader range of bacterial strains both cholesteryl esters inhibited growth of S. epidermidis and furthermore, CL inhibited also E. faecalis and E. cloacae (Figure 7d). The vehicle liposomes, which are composed mainly of phospholipids, demonstrated some inhibitory activity as well.
Figure 7.
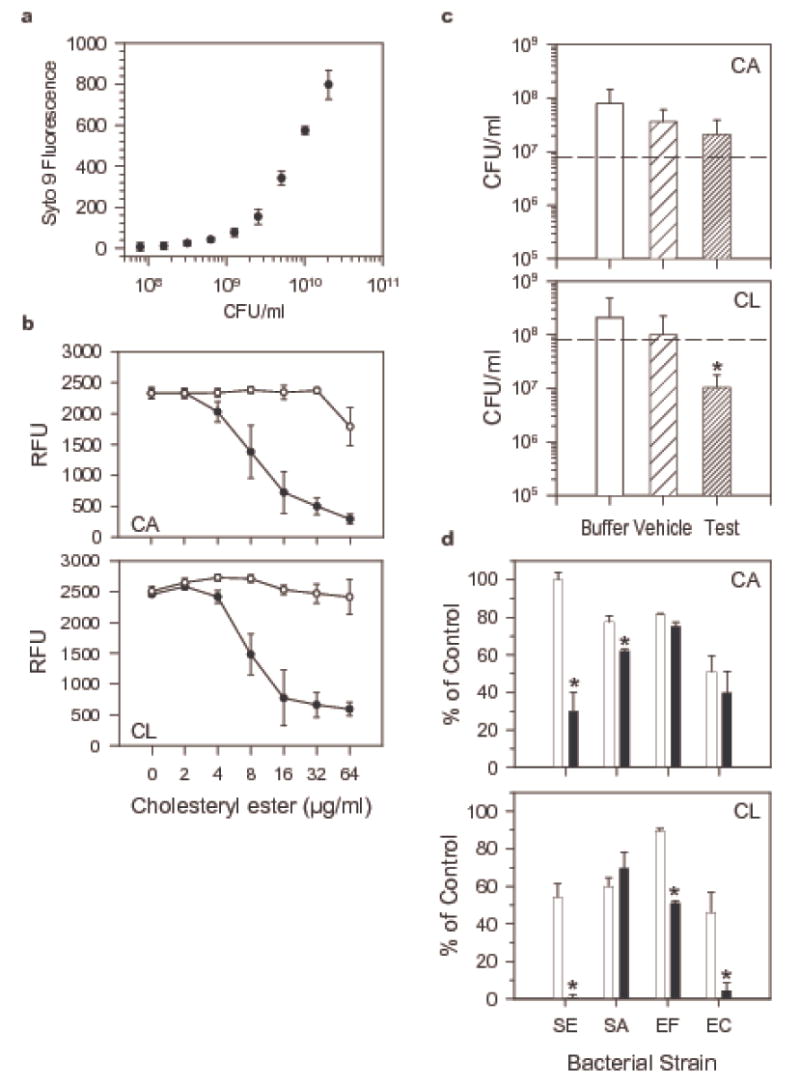
Cholesteryl arachidonate and cholesteryl linoleate exhibit antibacterial activity against gram negative and gram positive bacteria in vitro. (a) Staining with the DNA probe Syto 9 as measurement for bacterial growth. P. aeruginosa were grown in trypticase soy broth, serially diluted, and Syto 9 fluorescence (RFU) and the number of colony forming units/ml were determined in parallel. Shown are means ± S.D., n = 4. In an MIC assay uninhibited bacteria reach concentrations of 1011 CFU/ml. A reduction of RFU of 50% and 80% relates to a decrease in the number of colony forming units by 1 log and 1.5 log, respectively. (b) Dose dependent inhibitory activity of cholesteryl arachidonate (CA) and cholesteryl linoleate (CL) against P. aeruginosa. Bacteria were incubated in cation-adjusted Mueller-Hinton broth for 16 hours with and without liposomes containing cholesteryl esters (●) and vehicle liposomes (○) and relative fluorescence units were measured after addition of the DNA probe Syto 9. Shown are means + S.D., n = 3. (c) CFU assay with P. aeruginosa and the cholesteryl esters CA and CL. Bacteria were incubated for 3 hours in saline supplemented, low nutrient phosphate buffer in the presence or absence of vehicle and test liposomes and the number of CFU/ml was determined. Shown are means + S.D., n = 4 for CA and n = 3 for CL. *CFU/ml were significantly reduced to 20% + 16.99 of the inoculum, p = 0.002 in One Way ANOVA for Buffer vs CL. (d) Extended antibacterial activity testing of CA and CL in microtiter assay with Syto 9 read out. Cholesteryl esters were tested at 64 μg/ml; SE: Staphylococcus epidermidis, SA: Staphylococcus aureus, EF: Enterococcus faecalis, EC: Enterobacter cloacae. Data are expressed as % of RFU values for untreated bacteria. Shown are means + S.D., n = 3. *p < 0.001 in t-test for test liposomes compared to vehicle liposomes.
Taken together, we have demonstrated that cholesteryl arachidonate and cholesteryl linoleate exhibit direct antibacterial activity in low and high nutrient medium and that cholesteryl linoleate appears to be the more potent antibacterial lipid with a broader spectrum.
Discussion
This study provides evidence that lipids are secreted to mucosal surfaces and contribute to the inherent antimicrobial activity of mucosal secretions. To examine the potential role of lipids in innate mucosal host defense, we used nasal mucosal secretions. Nasal mucosa is a primary microbial exposure site; nasal mucosa is not exposed to alimentary lipids, its secretions are easily accessible and its antibacterial activity has been previously established in respect to antimicrobial polypeptides (27).
We found all major lipid classes in nasal fluid collected from healthy adults and, to our knowledge, this study is the first quantification of lipids in human nasal fluid. Glycerophospholipids and cholesterol, as well as to a lesser extent triglycerides and free fatty acids, have already been described in bronchioalveolar and nasal lavages, whereby the lipids were mainly thought to originate from lung surfactant reaching the upper airways through mucociliary propulsion (35-37). However, the presence of cholesteryl esters in native nasal fluid and their identification in the apical secretions of human bronchial epithelial cells suggest that the epithelial cells of the upper airways contribute to the lipids found in nasal fluid. This is also supported by the recent discovery of surfactant lamellar bodies in normal sinus mucosa (38). Nonetheless, we can not exclude transudation of plasma lipids through endothelial cells of the upper respiratory tract (39,40).
In aqueous environments, at submicellelar concentrations, lipids require carrier molecules such as albumin or lipoproteins. Our results demonstrate the presence of lipoproteins in nasal fluid. Specifically, by employing immunoblotting, apolipoprotein A-I (apoA-I) was identified, which has also been detected by Ghafouri et al. (41) in nasal fluid lavage. We found apoA-I in individual samples in multiple forms possibly reflecting proteolysis of apoA-I by enzymes in nasal fluid (42). ApoA-I is a mainly found in HDL and one of the major functions of apoA-I is cholesterol binding and reverse cholesterol transport from tissue to bile (43). This is consistent with our findings showing the presence of both cholesterol and apoA-I. However, we were not able to detect distinct lipoprotein bands co-migrating with HDL in the lipoprotein gels, but, in contrast, observed diffuse staining. Such diffuse mobility in agarose gels has been also shown for protease-modified HDL particles (42). Furthermore, our results could also reflect the presence of other lipid binding proteins. For example, lipocalin, which is produced by nasal mucosa (44,45) has been previously detected in nasal fluid (27,46). Alternatively, nasal fluid may contain unique lipoprotein particles synthesized by epithelial cells in the upper airways. The specific lipoproteins in nasal fluid and their origins remain to be defined.
To achieve reasonably selective lipid depletion from nasal fluid while minimally altering its natural, highly complex composition, we developed an SPE procedure. Though there are several widely accepted SPE protocols for the purification of lipids from biological fluids (reviewed in (47)), these do not aim to preserve the depleted fractions for further testing. Initial pilot studies suggested an association between inherent killing capacity of nasal fluid with cholesteryl ester contents and we focused on removing primarily non-polar lipids while allowing the presence of phospholipids. We found that the inherent antibacterial activity of nasal fluid was significantly diminished when non-polar lipid concentrations were reduced. Even though the SPE procedure may have altered other constituents of the nasal fluid, such as electrolytes and mucins, the observation that the re-addition of lipids restored in part the antibacterial activity, and the direct antibacterial activity of lipid extracts and commercial cholesteryl esters strongly suggest that host-derived antimicrobial lipids contribute to the inherent antimicrobial activity of nasal fluid, which was previously speculated by Widdicombe (48). Antimicrobial functions of cholesteryl esters are supported by a study by Georgel et al. (49), in which mice with a mutation in the stearoyl coenzyme A desaturase 1 gene develop spontaneous chronic dermatitis and also exhibit a decrease in cholesteryl ester content in the affected skin.
The contributions of apoA-I and possibly other lipoproteins to the observed antimicrobial activity of lipids is not clear yet. ApoA-I was partially co-depleted during SPE, in contrast to other proteins, and lipid supplement alone did not fully restore the antibacterial activity of LD. Hence, apoA-I may exert antimicrobial activity alone (50,51) or by enabling host-derived lipids to function similarly to the reported antibacterial activity of lipoproteins in conjunction with antimicrobial peptides (52).
Host-derived antimicrobial lipids may exert their activity in conjunction with antimicrobial (poly) peptides, which have a well documented membrane-perturbing activity (53). It is conceivable that antimicrobial lipids disrupt microbial membranes by embedding their hydrophobic acyl chains or side chains, and this activity could be facilitated by lesions initially created by antimicrobial (poly) peptides. Here, we have shown that lipids extracted from nasal fluid act synergistically with the antimicrobial peptide HNP-2. Tollin et al. (25) reported in vitro synergistic activity between selected fatty acids and the antimicrobial peptide LL37, and we are currently addressing in our laboratory the mode of synergistic action between fatty acids and lysozyme (manuscript submitted). Similarly, lipopeptaibols, naturally occurring lipopeptides with antimicrobial activity (54) and in vitro modified antimicrobial peptides that contain acyl chains and exhibit enhanced antimicrobial activity have been described (55). The antimicrobial properties of host-derived lipids may also extend to intracellular targets (56) and direct or indirect toxicity due to oxidation of lipids. Polyunsaturated fatty acids, such as linoleic acid and arachidonic acid, which have been found in our study either as free fatty acid or esterified in cholesteryl linoleate and cholesteryl arachidonate, are prone to peroxidation in the presence of oxygen and can form cytotoxic by-products (57-59).
Cole et al. (27) reported previously that the cationic protein extract of nasal fluid retained most of the bactericidal activity of nasal fluid. However, the lipid contents has not been analyzed in this study and it cannot be ruled out that some antibacterial lipids associated with highly hydrophobic antimicrobial proteins such as lysozyme may have been co-extracted. Even though lysozyme exerted rapid bactericidal activity and was able to fully restore the bactericidal activity of cationic protein- depleted nasal fluid when tested at high concentrations in vitro suggesting a predominant role in mucosal defense, its relative contribution to the inherent antibacterial activity of nasal fluid (and other bodily fluids) may be more complex in vivo. Cholesteryl esters exhibited prolonged activity in a cation adjusted, nutrient rich medium, unlike most antimicrobial peptides (60), and it is conceivable that antimicrobial peptides and antimicrobial lipids cover different aspects of mucosal defense, rapid initial mass destruction and retardation of the proliferation of survivors, respectively. Similarly, considering the varying spectrum of activity among both, antimicrobial (poly)peptides and cholesteryl esters, mucosal defense may relay on both components to cover the entire spectrum of potential invaders. A careful analysis of antimicrobial (poly)peptides and antimicrobial lipids in mucosal secretions and their in vitro activities will reveal their respective relative contributions to mucosal defense.
Lipid secretion may contribute to shaping the resident microbiota on mucosal surfaces like antimicrobial peptides (61). We did not observe a significant activity of nasal fluid lipids against S. aureus, which often colonizes the nostrils (62), but a pronounced activity against PA which is not routinely present in normal subjects. Furthermore, secretion of antimicrobial lipids may be also important for keeping the total number of the resident microbiota low. Mucosal surfaces populated by squamous epithelial cells with less secretory functions such as in the oral and vaginal cavity are typically heavily colonized. Hence, antimicrobial lipid secretion may be a feature of columnar epithelial cells in the airways and possibly other body sites with little colonization.
In conclusion, we have shown that lipids are present in mucosal secretions of the upper airways in significant quantities, and have provided evidence that lipids contribute to the inherent antibacterial activity of nasal fluid alone and in synergism with antimicrobial peptides. This suggests a role of host-derived lipids as direct antimicrobial effector molecules in innate mucosal immunity. The concept of antimicrobial lipids may unveil new mechanisms of host resistance to infections and microbial pathogenesis, as well as new avenues for prophylactic and therapeutic strategies in infectious diseases.
Acknowledgments
We thank Drs. Charles L. Bevins and Tomas Ganz for critical review of the manuscript, Dr. Don Puppione for helpful discussion, Sandra Alvarez for lysozyme purification, and Yessenia Velazco for technical assistance. Parts of this work had been presented at the 18th Annual CSUPERB symposium in San Jose, January 17th, 2006, and at the 106th General ASM Meeting in Orlando, Florida, May 23rd, 2006.
Footnotes
This work was supported by NIAID AI55675, NIH 1P20 MD001824, NSF HRD-0331537 (LSAMP-BD), NIH MBRS RISE R25 GM61331, and CSULA and CSUPERB Grants.
Abbreviations used in this paper: AU-PAGE, acid-urea poly acrylamide gel electrophoresis; NF, nasal fluid; LD, lipid-depleted nasal fluid; rpHPLC/ELSD, reverse phase HPLC with evaporative light scattering detection.
Disclosures The authors have no financial conflict of interests.
Publisher's Disclaimer: This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.
References
- 1.Laube DM, Yim S, Ryan LK, Kisich KO, Diamond G. Antimicrobial peptides in the airway. Curr Top Microbiol Immunol. 2006;306:153–182. doi: 10.1007/3-540-29916-5_6. [DOI] [PubMed] [Google Scholar]
- 2.Kishore U, Greenhough TJ, Waters P, Shrive AK, Ghai R, Kamran MF, Bernal AL, Reid KB, Madan T, Chakraborty T. Surfactant proteins SP-A and SP-D: structure, function and receptors. Mol Immunol. 2006;43:1293–1315. doi: 10.1016/j.molimm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Rubin BK. Physiology of airway mucus clearance. Respir Care. 2002;47:761–768. [PubMed] [Google Scholar]
- 4.Ganz T. Antimicrobial polypeptides. J Leukoc Biol. 2004;75:34–38. doi: 10.1189/jlb.0403150. [DOI] [PubMed] [Google Scholar]
- 5.Pag U, Oedenkoven M, Sass V, Shai Y, Shamova O, Antcheva N, Tossi A, Sahl HG. Analysis of in vitro activities and modes of action of synthetic antimicrobial peptides derived from an alpha-helical ‘sequence template’. J Antimicrob Chemother. 2008;61:341–352. doi: 10.1093/jac/dkm479. [DOI] [PubMed] [Google Scholar]
- 6.Mookherjee N, Hancock RE. Cationic host defence peptides: innate immune regulatory peptides as a novel approach for treating infections. Cell Mol Life Sci. 2007;64:922–933. doi: 10.1007/s00018-007-6475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van MG, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Olson RE. Discovery of the lipoproteins, their role in fat transport and their significance as risk factors. J Nutr. 1998;128:439S–443S. doi: 10.1093/jn/128.2.439S. [DOI] [PubMed] [Google Scholar]
- 9.Tulenko TN, Sumner AE. The physiology of lipoproteins. J Nucl Cardiol. 2002;9:638–649. doi: 10.1067/mnc.2002.128959. [DOI] [PubMed] [Google Scholar]
- 10.Navab M, Anantharamaiah GM, Fogelman AM. The role of high-density lipoprotein in inflammation. Trends Cardiovasc Med. 2005;15:158–161. doi: 10.1016/j.tcm.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Kim KD, Lim HY, Lee HG, Yoon DY, Choe YK, Choi I, Paik SG, Kim YS, Yang Y, Lim JS. Apolipoprotein A-I induces IL-10 and PGE2 production in human monocytes and inhibits dendritic cell differentiation and maturation. Biochem Biophys Res Commun. 2005;338:1126–1136. doi: 10.1016/j.bbrc.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 12.Gern JE, Brockman-Schneider R, Bhattacharya S, Malter JS, Busse WW. Serum and low-density lipoprotein enhance interleukin-8 secretion by airway epithelial cells. Am J Respir Cell Mol Biol. 2003;29:483–489. doi: 10.1165/rcmb.2002-0306OC. [DOI] [PubMed] [Google Scholar]
- 13.Mouritsen OG, Zuckermann MJ. What’s so special about cholesterol? Lipids. 2004;39:1101–1113. doi: 10.1007/s11745-004-1336-x. [DOI] [PubMed] [Google Scholar]
- 14.Sohlenkamp C, Lopez-Lara IM, Geiger O. Biosynthesis of phosphatidylcholine in bacteria. Prog Lipid Res. 2003;42:115–162. doi: 10.1016/s0163-7827(02)00050-4. [DOI] [PubMed] [Google Scholar]
- 15.Watson AD. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006;47:2101–2111. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Veldhuizen R, Nag K, Orgeig S, Possmayer F. The role of lipids in pulmonary surfactant. Biochim Biophys Acta. 1998;1408:90–108. doi: 10.1016/s0925-4439(98)00061-1. [DOI] [PubMed] [Google Scholar]
- 17.Ross RP, Morgan S, Hill C. Preservation and fermentation: past, present and future. Int J Food Microbiol. 2002;79:3–16. doi: 10.1016/s0168-1605(02)00174-5. [DOI] [PubMed] [Google Scholar]
- 18.Arikawa J, Ishibashi M, Kawashima M, Takagi Y, Ichikawa Y, Imokawa G. Decreased Levels of Sphingosine, a Natural Antimicrobial Agent, may be Associated with Vulnerability of the Stratum Corneum from Patients with Atopic Dermatitis to Colonization by Staphylococcus aureus. J Invest Dermatol. 2002;119:433–439. doi: 10.1046/j.1523-1747.2002.01846.x. [DOI] [PubMed] [Google Scholar]
- 19.Freedman SD, Blanco PG, Zaman MM, Shea JC, Ollero M, Hopper IK, Weed DA, Gelrud A, Regan MM, Laposata M, Alvarez JG, O’Sullivan BP. Association of cystic fibrosis with abnormalities in fatty acid metabolism. N Engl J Med. 2004;350:560–569. doi: 10.1056/NEJMoa021218. [DOI] [PubMed] [Google Scholar]
- 20.Strandvik B, Gronowitz E, Enlund F, Martinsson T, Wahlstrom J. Essential fatty acid deficiency in relation to genotype in patients with cystic fibrosis. J Pediatr. 2001;139:650–655. doi: 10.1067/mpd.2001.118890. [DOI] [PubMed] [Google Scholar]
- 21.Keen C, Olin AC, Edentoft A, Gronowitz E, Strandvik B. Airway nitric oxide in patients with cystic fibrosis is associated with pancreatic function, Pseudomonas infection, and polyunsaturated fatty acids. Chest. 2007;131:1857–1864. doi: 10.1378/chest.06-2635. [DOI] [PubMed] [Google Scholar]
- 22.Man MQ, Hatano Y, Lee SH, Man M, Chang S, Feingold KR, Leung DY, Holleran W, Uchida Y, Elias PM. Characterization of a Hapten-Induced, Murine Model with Multiple Features of Atopic Dermatitis: Structural, Immunologic, and Biochemical Changes following Single Versus Multiple Oxazolone Challenges. J Invest Dermatol. 2007 doi: 10.1038/sj.jid.5701011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freedman SD, Katz MH, Parker EM, Laposata M, Urman MY, Alvarez JG. A membrane lipid imbalance plays a role in the phenotypic expression of cystic fibrosis in cftr(-/-) mice. Proc Natl Acad Sci U S A. 1999;96:13995–14000. doi: 10.1073/pnas.96.24.13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isaacs CE. Human milk inactivates pathogens individually, additively, and synergistically. J Nutr. 2005;135:1286–1288. doi: 10.1093/jn/135.5.1286. [DOI] [PubMed] [Google Scholar]
- 25.Tollin M, Bergsson G, Kai-Larsen Y, Lengqvist J, Sjovall J, Griffiths W, Skuladottir GV, Haraldsson A, Jornvall H, Gudmundsson GH, Agerberth B. Vernix caseosa as a multi-component defence system based on polypeptides, lipids and their interactions. Cell Mol Life Sci. 2005;62:2390–2399. doi: 10.1007/s00018-005-5260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thormar H, Hilmarsson H. The role of microbicidal lipids in host defense against pathogens and their potential as therapeutic agents. Chem Phys Lipids. 2007 doi: 10.1016/j.chemphyslip.2007.06.220. [DOI] [PubMed] [Google Scholar]
- 27.Cole AM, Liao HI, Stuchlik O, Tilan J, Pohl J, Ganz T. Cationic polypeptides are required for antibacterial activity of human airway fluid. J Immunol. 2002;169:6985–6991. doi: 10.4049/jimmunol.169.12.6985. [DOI] [PubMed] [Google Scholar]
- 28.Gilljam H, Andersson O, Ellin A, Robertson B, Strandvik B. Composition and surface properties of the bronchial lipids in adult patients with cystic fibrosis. Clin Chim Acta. 1988;176:29–37. doi: 10.1016/0009-8981(88)90171-4. [DOI] [PubMed] [Google Scholar]
- 29.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–918. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 30.Fried B, Sherma J. Thin-layer chromatography, techniques and applications. Silver Spring; Maryland: 1986. [Google Scholar]
- 31.Morrison WR, Smith LM. Preparation of fatty acid methyl esters and dimethylacetals from lipids with boron fluoride-methanol. J Lipid Res. 1964;5:600–608. [PubMed] [Google Scholar]
- 32.Duffin K, Obukowicz M, Raz A, Shieh JJ. Electrospray/tandem mass spectrometry for quantitative analysis of lipid remodeling in essential fatty acid deficient mice. Anal Biochem. 2000;279:179–188. doi: 10.1006/abio.1999.4452. [DOI] [PubMed] [Google Scholar]
- 33.Porter E, Yang H, Yavagal S, Preza GC, Murillo O, Lima H, Greene S, Mahoozi L, Klein-Patel M, Diamond G, Gulati S, Ganz T, Rice PA, Quayle AJ. Distinct defensin profiles in Neisseria gonorrhoeae and Chlamydia trachomatis urethritis reveal novel epithelial cell-neutrophil interactions. Infect Immun. 2005;73:4823–4833. doi: 10.1128/IAI.73.8.4823-4833.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadzuka Y, Takabe H, Sonobe T. Liposomalization of SN-38 as active metabolite of CPT-11. J Control Release. 2005;108:453–459. doi: 10.1016/j.jconrel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt R, Markart P, Ruppert C, Temmesfeld B, Nass R, Lohmeyer J, Seeger W, Gunther A. Pulmonary surfactant in patients with Pneumocystis pneumonia and acquired immunodeficiency syndrome. Crit Care Med. 2006;34:2370–2376. doi: 10.1097/01.CCM.0000234036.19145.52. [DOI] [PubMed] [Google Scholar]
- 36.Batenburg JJ, Haagsman HP. The lipids of pulmonary surfactant: dynamics and interactions with proteins. Prog Lipid Res. 1998;37:235–276. doi: 10.1016/s0163-7827(98)00011-3. [DOI] [PubMed] [Google Scholar]
- 37.Sommerer D, Suss R, Hammerschmidt S, Wirtz H, Arnold K, Schiller J. Analysis of the phospholipid composition of bronchoalveolar lavage (BAL) fluid from man and minipig by MALDI-TOF mass spectrometry in combination with TLC. J Pharm Biomed Anal. 2004;35:199–206. doi: 10.1016/j.jpba.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 38.Woodworth BA, Smythe N, Spicer SS, Schulte BA, Schlosser RJ. Presence of surfactant lamellar bodies in normal and diseased sinus mucosa. ORL J Otorhinolaryngol Relat Spec. 2005;67:199–202. doi: 10.1159/000087093. [DOI] [PubMed] [Google Scholar]
- 39.Serikov VB, Jang YJ, Widdicombe JH. Estimate of the subepithelial hydrostatic pressure that drives inflammatory transudate into the airway lumen. J Appl Physiol. 2002;92:1702–1708. doi: 10.1152/japplphysiol.00645.2001. [DOI] [PubMed] [Google Scholar]
- 40.Moore TM, Chetham PM, Kelly JJ, Stevens T. Signal transduction and regulation of lung endothelial cell permeability. Interaction between calcium and cAMP. Am J Physiol. 1998;275:L203–L222. doi: 10.1152/ajplung.1998.275.2.L203. [DOI] [PubMed] [Google Scholar]
- 41.Ghafouri B, Stahlbom B, Tagesson C, Lindahl M. Newly identified proteins in human nasal lavage fluid from non-smokers and smokers using two-dimensional gel electrophoresis and peptide mass fingerprinting. Proteomics. 2002;2:112–120. [PubMed] [Google Scholar]
- 42.Lee M, Uboldi P, Giudice D, Catapano AL, Kovanen PT. Identification of domains in apoA-I susceptible to proteolysis by mast cell chymase. Implications for HDL function. J Lipid Res. 2000;41:975–984. [PubMed] [Google Scholar]
- 43.Lewis GF. Determinants of plasma HDL concentrations and reverse cholesterol transport. Curr Opin Cardiol. 2006;21:345–352. doi: 10.1097/01.hco.0000231405.76930.a0. [DOI] [PubMed] [Google Scholar]
- 44.Redl B. Human tear lipocalin. Biochim Biophys Acta. 2000;1482:241–248. doi: 10.1016/s0167-4838(00)00142-4. [DOI] [PubMed] [Google Scholar]
- 45.Fattori B, Castagna M, Megna G, Casani A, Pelosi P. Immunohistochemical localisation of tear lipocalin in human nasal mucosa. Rhinology. 1998;36:101–103. [PubMed] [Google Scholar]
- 46.Lindahl M, Stahlbom B, Tagesson C. Identification of a new potential airway irritation marker, palate lung nasal epithelial clone protein, in human nasal lavage fluid with two-dimensional electrophoresis and matrix-assisted laser desorption/ionization-time of flight. Electrophoresis. 2001;22:1795–1800. doi: 10.1002/1522-2683(200105)22:9<1795::AID-ELPS1795>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz-Gutierrez V, Perez-Camino MC. Update on solid-phase extraction for the analysis of lipid classes and related compounds. J Chromatogr A. 2000;885:321–341. doi: 10.1016/s0021-9673(00)00181-3. [DOI] [PubMed] [Google Scholar]
- 48.Widdicombe JG. Role of lipids in airway function. Eur J Respir Dis Suppl. 1987;153:197–204. [PubMed] [Google Scholar]
- 49.Georgel P, Crozat K, Lauth X, Makrantonaki E, Seltmann H, Sovath S, Hoebe K, Du X, Rutschmann S, Jiang Z, Bigby T, Nizet V, Zouboulis CC, Beutler B. A toll-like receptor 2-responsive lipid effector pathway protects mammals against skin infections with gram-positive bacteria. Infect Immun. 2005;73:4512–4521. doi: 10.1128/IAI.73.8.4512-4521.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shiflett AM, Bishop JR, Pahwa A, Hajduk SL. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J Biol Chem. 2005;280:32578–32585. doi: 10.1074/jbc.M503510200. [DOI] [PubMed] [Google Scholar]
- 51.Tada N, Sakamoto T, Kagami A, Mochizuki K, Kurosaka K. Antimicrobial activity of lipoprotein particles containing apolipoprotein Al. Mol Cell Biochem. 1993;119:171–178. doi: 10.1007/BF00926868. [DOI] [PubMed] [Google Scholar]
- 52.Sorensen O, Bratt T, Johnsen AH, Madsen MT, Borregaard N. The human antibacterial cathelicidin, hCAP-18, is bound to lipoproteins in plasma. J Biol Chem. 1999;274:22445–22451. doi: 10.1074/jbc.274.32.22445. [DOI] [PubMed] [Google Scholar]
- 53.Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 54.Peggion C, Formaggio F, Crisma M, Epand RF, Epand RM, Toniolo C. Trichogin: a paradigm for lipopeptaibols. J Pept Sci. 2003;9:679–689. doi: 10.1002/psc.500. [DOI] [PubMed] [Google Scholar]
- 55.Avrahami D, Shai Y. A new group of antifungal and antibacterial lipopeptides derived from non-membrane active peptides conjugated to palmitic acid. J Biol Chem. 2004;279:12277–12285. doi: 10.1074/jbc.M312260200. [DOI] [PubMed] [Google Scholar]
- 56.Xiong YQ, Mukhopadhyay K, Yeaman MR, Adler-Moore J, Bayer AS. Functional interrelationships between cell membrane and cell wall in antimicrobial peptide-mediated killing of Staphylococcus aureus. Antimicrob Agents Chemother. 2005;49:3114–3121. doi: 10.1128/AAC.49.8.3114-3121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Porter NA, Caldwell SE, Mills KA. Mechanisms of free radical oxidation of unsaturated lipids. Lipids. 1995;30:277–290. doi: 10.1007/BF02536034. [DOI] [PubMed] [Google Scholar]
- 58.Do TQ, Schultz JR, Clarke CF. Enhanced sensitivity of ubiquinone-deficient mutants of Saccharomyces cerevisiae to products of autoxidized polyunsaturated fatty acids. Proc Natl Acad Sci U S A. 1996;93:7534–7539. doi: 10.1073/pnas.93.15.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giamarellos-Bourboulis EJ, Plachouras D, Skiathitis S, Raftogiannis M, onyssiou-Asteriou A, Dontas I, Karayannacos PE, Giamarellou H. Ex vivo synergy of arachidonate-enriched serum with ceftazidime and amikacin on multidrug-resistant Pseudomonas aeruginosa. J Antimicrob Chemother. 2003;51:423–426. doi: 10.1093/jac/dkg026. [DOI] [PubMed] [Google Scholar]
- 60.Starner TD, Agerberth B, Gudmundsson GH, McCray PB., Jr Expression and activity of beta-defensins and LL-37 in the developing human lung. J Immunol. 2005;174:1608–1615. doi: 10.4049/jimmunol.174.3.1608. [DOI] [PubMed] [Google Scholar]
- 61.Salzman NH, Underwood MA, Bevins CL. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin Immunol. 2007;19:70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 62.Kuehnert MJ, Kruszon-Moran D, Hill HA, McQuillan G, McAllister SK, Fosheim G, McDougal LK, Chaitram J, Jensen B, Fridkin SK, Killgore G, Tenover FC. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. J Infect Dis. 2006;193:172–179. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]


