Abstract
Purine biosynthesis requires ten enzymatic steps in higher organisms while prokaryotes require an additional enzyme for step six. In most organisms steps nine and ten are catalyzed by the purH gene product, a bifunctional enzyme with both 5-formaminoimidazole-4-carboxamide-5′-monophosphate ribonucleotide (FAICAR) synthase and inosine monophosphate (IMP) cyclohydrolase activity. Recently it was discovered that Archaea utilize different enzymes to catalyze steps nine and ten. An ATP dependent FAICAR synthetase is encoded by the purP gene and IMP cyclohydrolase is encoded by the purO gene. We have determined the X-ray crystal structures of FAICAR synthetase from Methanocaldococcus jannaschii complexed with various ligands, including the tertiary substrate complex and product complex. The enzyme belongs to the ATP grasp superfamily and is predicted to use a formylphosphate intermediate formed by an ATP-dependent phosphorylation. In addition, we have determined the structures of a PurP ortholog from Pyrococcus furiosus, which is functionally unclassified, in three crystal forms. With approximately 50% sequence identity, P. furiosus PurP is structurally homologous to M. jannaschii PurP. A phylogenetic analysis was performed to explore the possible role of this functionally unclassified PurP.
The purine biosynthetic pathway generates inosine monophosphate, which is subsequently converted to either adenosine monophosphate or guanosine monophosphate. Buchanan worked out the details of the vertebrate pathway in the 1950's identifying ten enzymatic conversions (1). Later, Stubbe and coworkers showed that in prokaryotes the conversion of aminoamidazole ribonucleotide to carboxyaminoamidazole ribonucleotide catalyzed by PurE in step 6 requires an additional enzyme (PurK) (2–4), resulting in a total of eleven enzymatic conversions. In Escherichia coli each step of the pathway is catalyzed by a monofunctional enzyme, with the exception of the last two steps, while in vertebrates steps 2 (PurD), 3 (PurN) and 5 (PurM) comprise a trifunctional enzyme (5), steps 6 (PurE) and 7 (PurC) comprise a bifunctional enzyme (6, 7) and steps 9 and 10 are catalyzed by the bifunctional enzyme PurH (8–11). Additional species dependent gene fusions have been observed.
Other deviations from the vertebrate purine biosynthetic pathway have also been observed. In vertebrates, formyltransferase reactions occur in steps 3 (PurN) and 9 (PurH), with N10-formyltetrahydrofolate as the donor. However, some organisms lacking tetrahydofolate as a cofactor utilize an ATP-dependent formate ligation: PurT for step 3 (12–14) and PurP for step 9 (15). Both the tetrahydofolate dependent and ATP-dependent formyltransferase reactions are found in E. coli (12). In vertebrates, PurH catalyzes step 10, while in methanogenic bacteria PurO, which is structurally dissimilar to PurH (9, 11), catalyzes step 10 (16, 17). Examination of the available genomes suggests that other variations in the purine biosynthetic pathway remain to be discovered.
Enzyme structures for all of the known purine biosynthetic activities have been determined with the exception of PurP, which converts aminocarboxyimidazole ribonucleotide (AICAR) to formylaminocarboxyimidazole ribonucleotide (FAICAR) in step 9. PurP from Methanocaldococcus jannaschii has been biochemically characterized (15) and sequence comparisons indicate the presence of PurP orthologs in a number of related organisms. Sequence comparisons reveal that PurP is a member of the ATP grasp superfamily (18–20). The purine biosynthetic enzymes PurD, PurT, PurK and PurC are also members of the ATP grasp superfamily (14, 21–24).
Here we report the structure of PurP from M. jannaschii (MjPurP) complexed with substrates, products and analogues. Among the ATP grasp members MjPurP shows a novel hexameric arrangement in which loops from threefold related monomers fold over onto adjacent monomers. The ATP binding site of PurP is similar to those for other members of the ATP grasp superfamily. The AICAR/FAICAR binding site is comprised of the conserved residues His27, Arg264, Ser266, and Arg314. We also report structures from three crystal forms of a PF1517 (PfPurP), one of two PurP orthologs found in Pyrococcus furiosus. Although the MjPurP and PfPurP active sites are highly conserved, and PfPurP binds both ATP and AICAR, PfPurP does not catalyze the FAICAR synthetase reaction and its function remains unknown. A phylogenetic analysis of PurP orthologs is also presented.
MATERIALS AND METHODS
Overexpression and Purification of MjPurP
Methanocaldococcus jannaschii purP gene Mj0136 was cloned into the expression vector pET19b and overexpressed in Escherichia coli B834(DE3), a methionine auxotrophic strain (15). For overexpression of native protein, cells were grown in LB medium supplemented with 100 µg/mL ampicillin. For overexpression of selenomethionine (SeMet) substituted protein, cells were grown in M9 minimal salts supplemented with 4% (w/v) glucose, 2 mM MgSO4, 0.1 mM CaCl2, 1% BME vitamin solution (GibcoBRL), 25 µg/mL FeSO4·7H2O, 40 µg/mL of each of the L-amino acids (L-selenomethionine substitutes for L-methionine). The cells were induced with 0.1 mM isopropyl β-D-thiogalactoside (IPTG) for 6 h at 25°C once the absorbance of the cell culture reached an OD600 of 0.8. The recombinant protein was purified by metal-chelate affinity chromatography using a cobalt column (Clontech). Polyhistidine tagged MjPurP was eluted from the column with buffer A (50 mM sodium phosphate, pH 7.0, 300 mM NaCl, 1 mM β-mercaptoethanol, and 300 mM imidazole). The fractions containing MjPurP were combined and exchanged into 10 mM Tris-HCl, pH 7.6, 1 mM MgCl2, and 1 mM dithiothreitol. For the native protein, 1 L of cell culture produced ~ 20 mg of homogeneous MjPurP, while for the SeMet protein, only 2 mg of MjPurP were obtained per liter of cell culture. The purified protein was then concentrated to 10 mg/mL using 10 kDa cutoff microcon concentrators (Amicon).
Overexpression and Purification of PfPurP
Pyrococcus furiosus purP gene at loci Pf1517 was amplified by PCR from genomic DNA and subcloned into pT7-7 vector. The gene containing plasmid was overexpressed in E. coli BL21(DE3). The cells were grown in LB medium in the presence of 100 µg/mL ampicillin at 37°C until the absorbance of the cell culture reached an OD600 of 0.8, at which point the cells were induced with 0.2 mM IPTG for an additional 6 h at 25°C. The cells were harvested by centrifugation and sonicated on ice in buffer B (20 mM Tris-HCl, pH 7.5). The crude cell extract was pre-purified by heating in a water bath at 70°C for 30 min, followed by centrifugation at 15000g for 20 min to remove the insoluble material. Heat-stable cell extract was applied to a MonoQ HR anion exchange column (1 × 10 cm). After washing the column with 15 column volumes of buffer B, the bound protein was eluted with a 15 column volume linear gradient from 0 to 1 M NaCl in buffer B at a flow rate of 0.4 mL/min. The fraction containing homogeneous PfPurP was buffer exchanged into 10 mM Tris-HCl, pH 7.5, and 2 mM MgCl2, and concentrated to 15 mg/mL.
Crystallization of MjPurP
Crystallization was performed using the hanging drop vapor diffusion method at 18°C with drops containing 1.5 µL of protein solution and 1.5 µL of reservoir solution. MjPurP was subjected to a series of sparse matrix screens (Hampton Research, Emerald Biostructures) in order to determine initial crystallization conditions. Both SeMet and native protein crystallized from 1.2 – 1.4 M (NH4)2SO4, 0.2 M NaCl, and 0.1 M sodium acetate at pH 4.1 – 4.3. To obtain the ligand complexed structures, ATP, AMPPCP, ADP, AMP, AICAR, and FAICAR were used for co-crystallization with MjPurP at 1 mM concentration for FAICAR and 5 mM for the others. The FAICAR was prepared by the enzymatic formylation of AICAR with ATP and formate catalyzed by MjPurP followed by purification of the FAICAR on a MonoQ column, while all other compounds were purchased from Sigma-Aldrich Co. Crystals, in the shape of rhombohedral prisms, usually appear in a week and reach a maximum size of 300 µm × 300 µm × 80 µm in two weeks. These crystals belong to the space group R32 with unit cell dimensions of a = 109.2 Å and c = 255.7 Å on average. Each asymmetric unit (a.u.) contains one protomer, corresponding to a solvent content of 65% and Matthews coefficient (25) of 3.5 Å3/Da.
The soaking experiments were performed at 18°C for 1 h. The stabilizing solution contains 1.5 M (NH4)2SO4, 0.2 M NaCl, 20 mM MgCl2, and 0.1 M sodium acetate at pH 4.1. The ligand concentration in the soaking solutions was 50 mM for ATP, AMPPCP, ADP and AMP, 20 mM for AICAR, and 6 mM for FAICAR. For the AMPPCP-AICAR complex, 20 mM ammonium formate was also present in the solution.
Crystallization of PfPurP
The crystallization experiment for PfPurP was similar to those for MjPurP. PfPurP crystallized in three crystal forms. The first crystal form grew from 30 – 32% 2-methyl-2,4-pentanediol (MPD), 200 mM NaCl, 100 mM Tris-HCl, pH 7.0. Crystals usually appear in two days and reach a maximum size of 200 µm × 200 µm × 80 µm in one week. The crystals are in the shape of rhombohedral prisms, and belong to the space group R32. The unit cell dimensions are a = 123.4 Å and c = 375.5 Å. Each a.u. contains two protomers, corresponding to a solvent content of 65% and Matthews coefficient of 3.5 Å3/Da.
A second crystal form was obtained from 30 – 35% MPD and 100 mM Na+/K+ phosphate, pH 6.2, in presence of 10 mM ATP. This crystal form belongs to the space group R32 with unit cell dimensions of a = 122.5 Å and c = 560.9 Å. In this crystal form, there are also two protomers in the a.u., corresponding to a solvent content of 76% and Matthews coefficient of 5.2 Å3/Da.
The third PfPurP crystal form grew from 10% (v/v) 2-propanol, 200 mM Li2SO4, and 100 mM sodium phosphate-citrate at pH 4.2, in the presence of 5 mM ADP. The crystals are in the space group P21 with unit cell dimensions of a = 75.7 Å, b = 126.8 Å, c = 121.4 Å, and β = 102.9°. There are six protomers in the a.u., corresponding to a solvent content of 51% and Matthews coefficient of 2.5 Å3/Da.
X-ray Intensity Measurements
For cryoprotection, the MjPurP crystals were briefly transferred into a buffer containing 12% glycerol, 12% ethylene glycol, 1.6 M (NH4)2SO4, 0.2 M NaCl, and 0.1 M sodium acetate at pH 4.1. The crystals were then flash frozen by plunging them into liquid nitrogen. The PfPurP crystals in both R32 forms were directly frozen without cryoprotection, while those in monoclinic form were cryoprotected by 15% glycerol added to the mother liquor. Data sets were collected either at the Advanced Photon Source (APS) beamline 24-ID-C using a ADSC Quantum 315 detector, or at the Cornell High Energy Synchrotron Source (CHESS) beamline F1 using a ADSC Quantum 270 detector. For the single wavelength SeMet data set of MjPurP, the energy was selected to maximize Δf" of the incorporated selenium, and a total of 360° of data were collected. A total of 90 – 150° of data were collected for each of the other data sets. The oscillation per image ranged from 0.5 to 1°, depending on the mosaicity of the crystal. The crystals of MjPurP diffracted to around 2.0 Å; however, the diffraction pattern was usually anisotropic, resulting in low completeness in the high resolution shells. The crystals of PfPurP diffracted to 1.7 – 1.9 Å for the crystal form of R32 in the small unit cell, 2.5 Å for R32 in the large unit cell, and 2.3 Å for the P21 crystal form. The HKL2000 suite of programs was used for integration and scaling (26). Data processing statistics are summarized in Table 1.
Table 1.
Data collection statistics.
| Protein Complex | MjPurP AICAR-AMPPCP (SeMet) | MjPurP AICAR - AMPPCP | MjPurP AICAR - ATP | MjPurP FAICAR-ADP | MjPurP AMP-AMP | PfPurP AICAR-AMP | PfPurP AMP-AMP | PfPurP Pi-ATP | PfPurP Pi-ADP |
|---|---|---|---|---|---|---|---|---|---|
| Wavelength (Å) | 0.97922 | 0.97922 | 0.97922 | 0.97922 | 0.97922 | 0.97918 | 0.97918 | 0.97918 | 0.91770 |
| Resolution (Å) | 2.5 | 2.1 | 2.1 | 2.4 | 2.3 | 1.9 | 1.7 | 2.5 | 2.3 |
| Space group | R32 | R32 | R32 | R32 | R32 | R32 | R32 | R32 | P21 |
| Unit cell | |||||||||
| a (Å) | 109.5 | 109.4 | 109.6 | 108.0 | 109.4 | 123.7 | 123.2 | 122.5 | 75.7 |
| b (Å) | 126.8 | ||||||||
| c (Å) | 256.2 | 256.0 | 255.8 | 254.3 | 256.3 | 375.4 | 376.3 | 560.9 | 121.4 |
| β (°) | 102.9 | ||||||||
| Total reflections | 401282 | 557779 | 161067 | 103716 | 140027 | 602085 | 834589 | 420106 | 288596 |
| Unique reflections | 20490 | 34334 | 31509 | 20897 | 26365 | 85987 | 119607 | 55984 | 95364 |
| completeness (%) | 98.0 (82.3) | 98.5 (90.7) | 90.3 (61.6) | 92.0 (57.4) | 99.6 (100) | 99.3 (94.4) | 99.0 (97.0) | 97.9 (81.9) | 96.7 (99.0) |
| Rsym * (%) | 9.7 (25.0) | 7.9 (28.0) | 5.4 (14.6) | 7.9 (23.4) | 6.8 (36.4) | 7.4 (36.5) | 5.2 (28.4) | 5.7 (28.3) | 7.6 (24.3) |
| I/σ | 42.2 (6.8) | 37.6 (7.8) | 26.1 (5.6) | 21.0 (3.7) | 28.0 (4.4) | 25.0 (3.0) | 36.6 (3.9) | 32.0 (3.0) | 15.5 (4.5) |
| Redundancy | 19.6 (12.6) | 16.2 (11.4) | 5.1 (4.6) | 5.0 (3.9) | 5.3 (4.7) | 7.0 (5.9) | 7.0 (6.0) | 7.5 (6.1) | 3.0 (2.9) |
Values for the highest resolution shell are given in parentheses.
Rsym = ∑∑i | Ii−<I> | /∑<I>, where <I> is the mean intensity of the N reflections with intensities Ii and common indices h,k,l.
Structure Determination and Refinement
To determine the structure of MjPurP, single wavelength anomalous dispersion (SAD) phasing, density modification and automatic model building were performed at 2.5 Å using the program autoSHARP (27). Approximately 300 out of a total of 361 residues were built with correct sidechains in the initial model, which was manually adjusted and further completed using the interactive graphics program Coot (28). The model refinement was performed through alternating cycles of manually rebuilding using Coot, and restrained refinement using CNS (29) and Refmac5 (30). The native data set of MjPurP complexed with AMPPCP-AICAR at 2.1 Å was used to extend the phases, and the resulting model was used to refine against the other data sets. Ligands were directly constructed into the corresponding difference electron density in each structure. Water molecules were included after the ligand was added.
The structure of PfPurP in the small unit cell of space group R32 was determined by molecular replacement using the program MOLREP (31). A protomer of MjPurP was used as the search model. The initial rigid body refinement and restrained refinement with the molecular replacement solution resulted in an R-factor of 35%. The structures were further refined using the same procedure as described above for MjPurP, and the refined model was used to determine the structures of PfPurP in the other two crystal forms by molecular replacement. Model refinement statistics for the MjPurP and PfPurP structures are summarized in Table 2. The graphic figures of the structures were prepared using PyMOL (32).
Table 2.
Refinement statistics.
| Protein Complex | MjPurP AICAR - AMPPCP | MjPurP AICAR - ATP | MjPurP FAICAR-ADP | MjPurP AMP-AMP | PfPurP AICAR-AMP | PfPurP AMP-AMP | PfPurP Pi-ATP | PfPurP Pi-ADP |
|---|---|---|---|---|---|---|---|---|
| PDB code | 2R7K | 2R7L | 2R7N | 2R7M | 2R84 | 2R85 | 2R86 | 2R87 |
| Space group | R32 | R32 | R32 | R32 | R32 | R32 | R32 | P21 |
| Resolution (Å) | 2.1 | 2.1 | 2.4 | 2.3 | 1.9 | 1.7 | 2.5 | 2.3 |
| No. of total atoms | 3035 | 3015 | 2942 | 2976 | 6308 | 6576 | 5535 | 17028 |
| No. of water atoms | 108 | 128 | 58 | 94 | 624 | 656 | 133 | 541 |
| R factora (%) | 20.8 | 20.9 | 21.2 | 20.4 | 17.5 | 16.6 | 21.9 | 20.3 |
| Rfree b (%) | 24.8 | 26.0 | 28.0 | 25.7 | 19.5 | 18.6 | 23.9 | 24.4 |
| r.m.s.d. from ideal geometry | ||||||||
| bonds (Å) | 0.006 | 0.006 | 0.007 | 0.007 | 0.007 | 0.008 | 0.007 | 0.007 |
| angles (°) | 1.018 | 1.004 | 1.125 | 1.093 | 1.072 | 1.140 | 1.024 | 1.067 |
| Ramachandran plot (%) | ||||||||
| most favored regions | 91.0 | 92.0 | 89.1 | 91.3 | 91.1 | 90.6 | 90.7 | 89.1 |
| additional allowed regions | 8.7 | 7.7 | 10.3 | 8.4 | 8.6 | 9.1 | 8.7 | 10.3 |
| generously allowed regions | 0 | 0 | 0.3 | 0 | 0 | 0 | 0.3 | 0.3 |
| disallowed regionsc | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 |
| Average B factors (Å2) | 48.1 | 44.1 | 51.3 | 46.2 | 21.8 | 17.5 | 47.9 | 22.3 |
R factor = ∑hkl || Fobs | − k | Fcal || / ∑hkl | Fobs |, where Fobs and Fcal are observed and calculated structure factors respectively.
For Rfree the sum is extended over a subset of reflections (5%) excluded from all stages of refinement.
The residue in the disallowed region of the Ramachandran plot corresponds to His27 for MjPurP and His11 for PfPurP.
RESULTS
Overview of MjPurP and PfPurP Structures
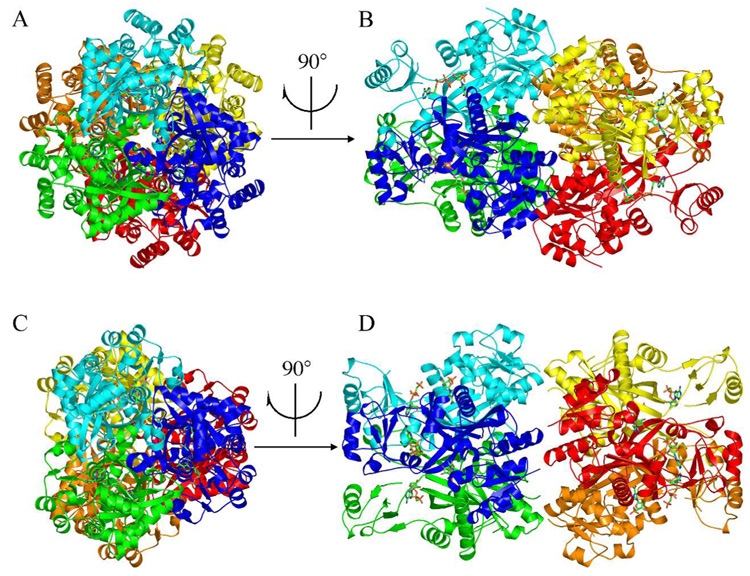
The crystal structure of MjPurP is a homohexamer (Figure 1A & B), consistent with the molecular weight analysis using size exclusion gel filtration. PfPurP appeared to be predominantly trimeric in solution; however, PfPurP in all three crystal forms showed a common hexameric arrangement (Figure 1C & D). While the trimeric substructures of MjPurP and PfPurP are nearly identical, the hexamers are slightly different. MjPurP has ~ 2.5 times more buried surface area between the two trimers than PfPurP (Table 3), resulting an overall more compact structure. Based upon the trimer superposition of the two structures, the same compact hexameric structure can be constructed for PfPurP without causing significant close contacts, by rotating one trimer approximately 30° and translating it towards the opposing trimer.
Figure 1.
The structures of MjPurP and PfPurP. The hexameric crystal structures of MjPurP (A and B) and PfPurP (C and D) are shown in ribbon diagrams and colored by the protomers. Compared to MjPurP, the hexameric arrangement of PfPurP has significantly less interactions at the trimer interface, and is possibly a crystallization artifact rather than biologically relevant.
Table 3.
Buried surface area (Å2).
| Interface | MjPurP | PfPurP |
|---|---|---|
| 3-fold (subunit A – B) | 4100 | 4100 |
| 2-fold (trimer – trimer) | 4600 | 1800 |
PurP protomeric fold
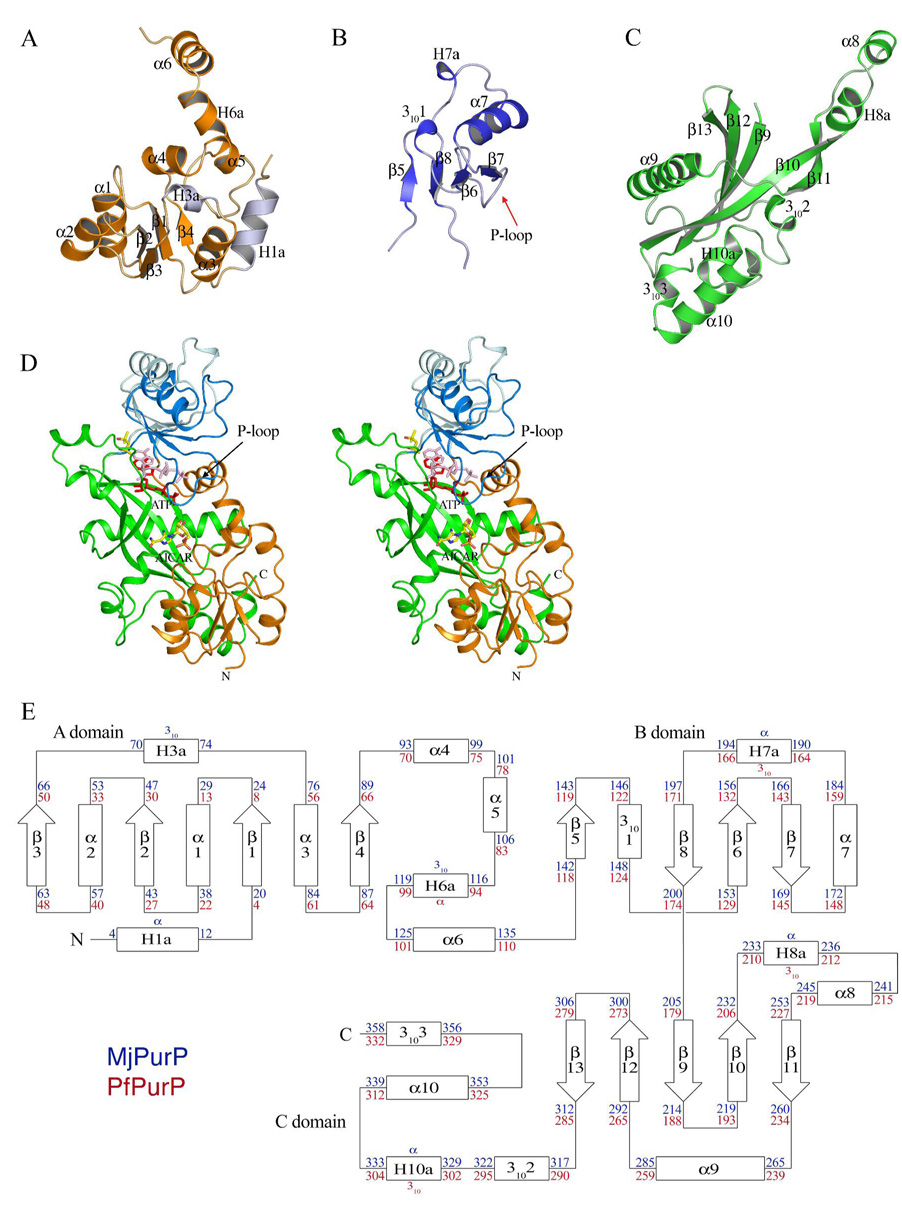
MjPurP and PfPurP belong to the ATP grasp superfamily. Similar to other superfamily members, the molecular architecture of MjPurP and PfPurP consists of three motifs: the A, B, and C domains (Figure 2). Domain A is formed by residues 1 – 137/1 – 113 (the numbers correspond to MjPurP/PfPurP throughout). The core of domain A adopts a truncated Rossman fold, with a central four-stranded parallel β-sheet (3↑2↑1↑4↑) flanked by three α-helices. The structure of MjPurP has two additional flanking helices (N-terminal α-helix H1a and 310-helix H3a; helices present in only one of the PurP structures are designated H). In addition to the core, the last 40 amino acids of domain A contain four more helices: α4, α5, H6a (α-helix in MjPurP and 310-helix in PfPurP), and α6. Helices α4 and α5 pack against the core motif, while H6a and α6 wrap around domain C and serve as a linker between domain A and domain B. Domain B is the smallest domain of the three, consisting of residues 138 – 204/114 – 180. The structure of domain B is an αβ two-layered sandwich, comprised of a four-stranded antiparallel β-sheet (5↑8↓6↑7↓) and three flanking helices on the solvent exposed face: 3101, α7, and H7a (α-helix in MjPurP and 310-helix in PfPurP). Domain C is composed of residues 205 – 361/179 – 334. The core of domain C is a twisted β-sheet consisting of five antiparallel strands (13↓12↑9↓10↑11↓), and flanked by a long helix α9 on one side and four helices on the other side, 3102, H10a (α-helix in MjPurP and 310-helix in PfPurP), α10, and 3103. Two helices, H8a (α-helix in MjPurP and 310-helix in PfPurP) and α8, extend away from the core motif. These two helices and strands β10 and β11 contribute significantly to the trimer interface by interacting with the same region from the two adjacent protomers related by the three-fold crystallographic symmetry.
Figure 2.
The conserved protomer architecture of MjPurP and PfPurP. (A) A domain. The two insertion helices of MjPurP are colored in light blue. (B) B domain. (C) C domain. (D) The stereodiagram of the protomer colored by domains. The B domain in closed and open conformations are in dark blue and light blue, respectively, and the corresponding ADP/ATP in red and pink, respectively. (E) Topology diagram. The conserved secondary structural elements between MjPurP and PfPurP are numbered consecutively. The residue numbers are indicated in blue for MjPurP and red for PfPurP.
Active Site Cleft
The assembly of the A, B, and C domains forms an active site cleft of approximately 25 Å × 15 Å × 10 Å, with the long β-sheet from the C domain as the bottom. The AICAR/FAICAR binding pocket is formed between domains A and C, and a small part of domain C from an adjacent protomer. The secondary structural elements involved in the AICAR/FAICAR binding site include α4, β11, and α9*, and loops connecting β1 and α1, β2 and α2, β11* and α9* (the asterisk indicates a symmetry related protomer throughout). The ATP binding site is sandwiched between the β-sheets from the B domain (β6, β7, and β8) and C domain (β10, β12, and β13). Helix H8a* and the loop region following it also contribute to the ATP binding site.
Domain Movement Observed for Domain B
Two main conformations were observed in the structures of MjPurP and PfPurP: a closed conformation for all MjPurP structures and the Pi-ADP complex of PfPurP, and an open conformation for the rest of PfPurP structures. In the closed conformation, the ATP binding site and the AICAR/FAICAR binding site are close together, while in the open conformation the two sites are farther apart (Figure 2D). Loop 136 – 141/111 – 117 and loop 201 – 204/175 – 178 appear to serve as hinges allowing the B domain to move and consequently to open and close over the substrate binding site.
The P-loop
The structures of MjPurP complexed with AICAR-ATP and with AICAR-AMPPCP (the nomenclature follows ligands bound at the AICAR/FAICAR site and the ATP site, separated by a hyphen) have clear density for the nucleotide triphosphate molecule, indicating that the ligand is relatively well ordered and fully occupied in the active site. In contrast the predicted P-loop for ATP binding is partially disordered. Among all the MjPurP structures, the AICAR-AMPPCP complexed structure shows the most complete P-loop with only three residues missing (161 RGG 163). For PfPurP, the complete polypeptide chain was built for the Pi-ADP complex (space group P21) and subunit A of the Pi-ATP complex (space group R32, large unit cell). However, the P-loop (135 GAKGG 139) and the β- and γ-phosphates exhibit relatively high temperature factors and weaker electron densities, suggesting significant flexibility. The structures of PfPurP crystallized in the small unit cell of space group R32 were determined at higher resolution (1.7 – 1.9 Å); however, the electron density is only interpretable for the AMP moiety even though ATP or AMPPCP was present in the crystallization condition. Consistent with the disordered phosphate groups, residues 134 – 140 from the P-loop also lack interpretable electron density and were omitted from the final model in this crystal form of PfPurP.
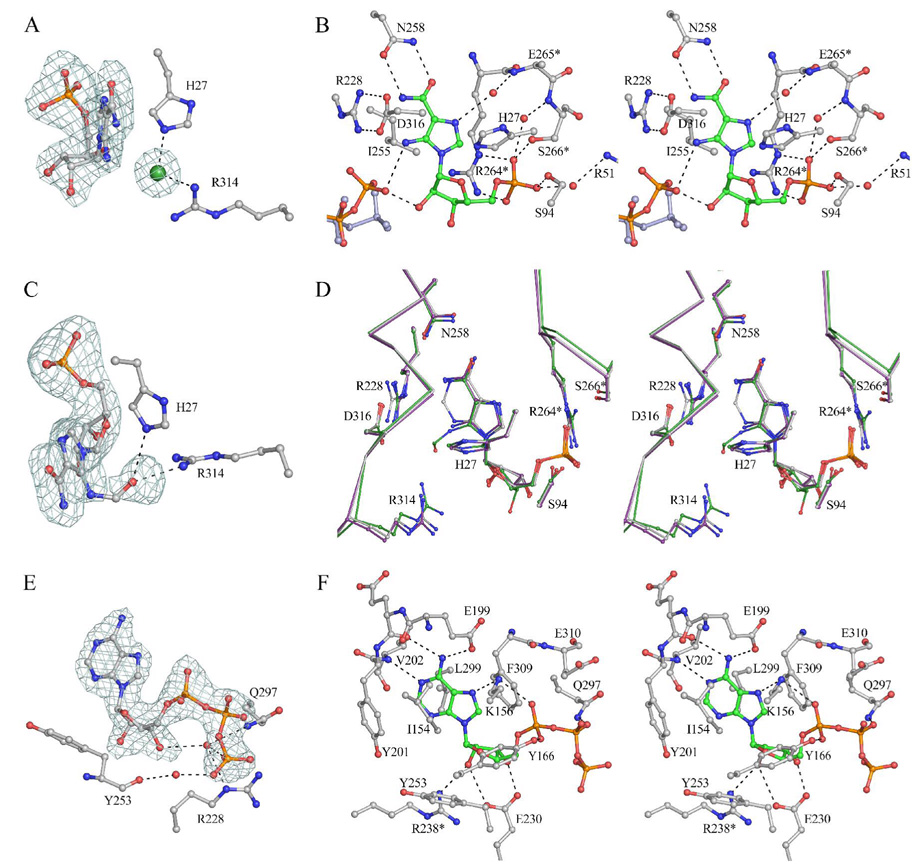
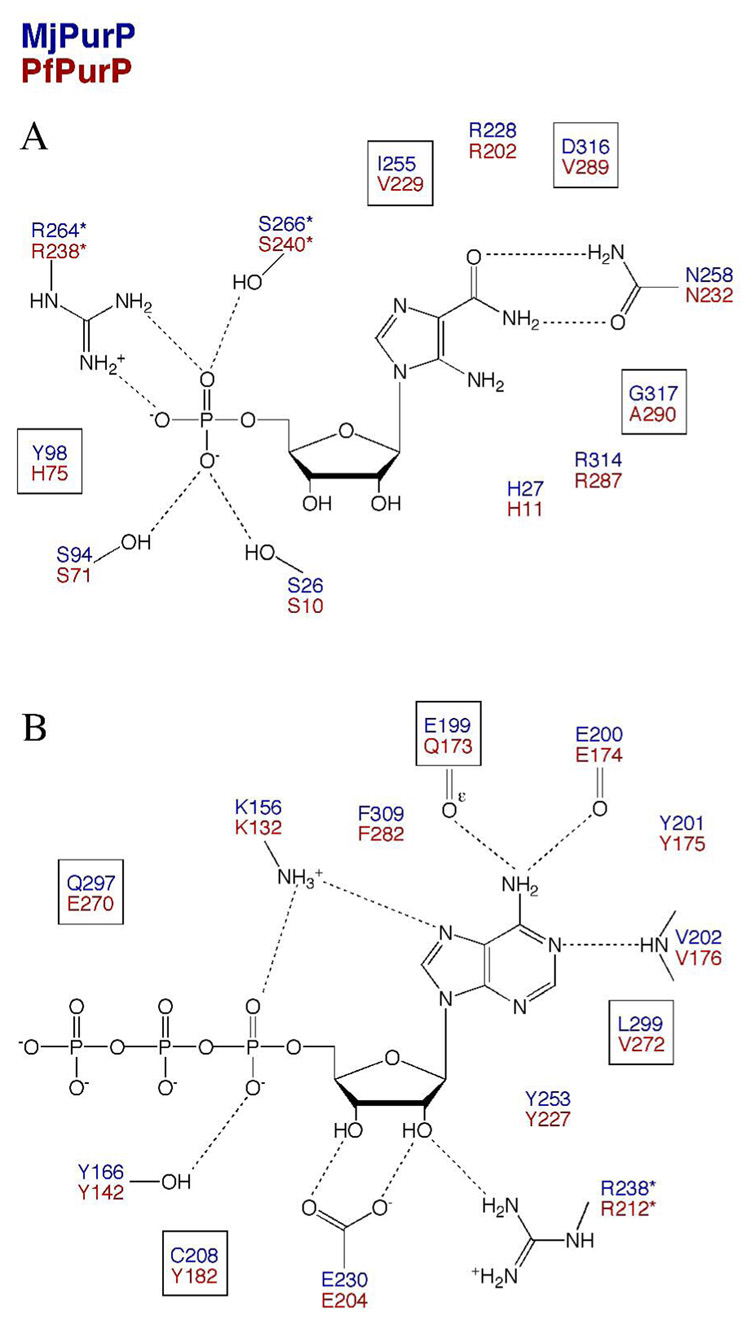
The AICAR/FAICAR Binding Site of MjPurP
We have determined the structures of MjPurP in four different ligand complexes: AICAR-ATP, AICAR-AMPPCP, FAICAR-ADP, and AMP-AMP. The substrate AICAR binds essentially the same in the AICAR-ATP and the AICAR-AMPPCP complex structures. The ribose moiety of the AICAR molecule is in a slightly twisted 3′-endo conformation (Figure 3A). Surprisingly, the ribose hydroxyl groups are solvent exposed and lack hydrogen bond interaction with any active site residues (Figure 3B). The aminoimidazole carboxamide moiety is mostly anchored through a pair of hydrogen bonds between the carboxamide and the Asn258 side chain. His27, Ile255, Asp316, and Gly317 also help to orient the imidazole ring through van der Waals interactions. In addition, a water molecule that hydrogen bonds to the carbonyl oxygen of His27 and the backbone amide of Glu265* also hydrogen bonds to the imidazole. A chloride anion, presumably an artifact due to the high concentration of NaCl in the crystallization condition, was modeled near the AICAR imidazole ring based on the intensity of electron density and the positively charged binding environment: His27 and Arg314 interact with the chloride anion through electrostatic interactions/hydrogen bonds (Figure 3A). The 5′-monophosphate is positioned by a total of six hydrogen bond interactions. Ser94 and Ser266* each provide a hydrogen bond to the phosphate with the sidechain hydroxyl group; the guanidinium group of Arg264* donates two hydrogen bonds; and the last two hydrogen bonds are provided by water molecules, which in turn hydrogen bond to the Arg51 sidechain and the backbone amide of Ser266*.
Figure 3.
The active site of MjPurP. (A) Fo – Fc density contoured at 3 σ around AICAR and the chloride anion (green) of the AICAR-ATP structure. (B) The stereodiagram of the AICAR binding site for the AICAR-ATP structure. The γ-phosphate of AMPPCP from the AICAR-AMPPCP structure is superimposed and colored in light blue. Hydrogen bonds are indicated by dashed lines. (C) Fo – Fc density contoured at 3 σ around FAICAR of the FAICAR-ADP structure. (D) The superposition of AICAR (purple), FAICAR (green), and AMP (grey) at the active site. (E) Fo – Fc density contoured at 3.5 σ around ATP of the AICAR-ATP structure. (F) The stereodiagram of the ATP binding site.
In the structure of MjPurP complexed with the products FAICAR and ADP, the binding geometry of FAICAR is similar to that of AICAR, except that the ribose moiety of FAICAR is in a slightly twisted 4′-endo conformation (Figure 3C & 3D). The substrate binding site residues superimpose well between the two structures. The most significant differences are for Arg314: the Cα position moves approximately 0.5 Å closer to the ligand upon FAICAR binding. The guanidinium group of Arg314 is in the vicinity of the formyl group of FAICAR and donates a hydrogen bond with a distance of 2.9 Å. However, Arg314 has weak density for the guanidinium group in the FAICAR-ADP complex structure, which suggests high thermal motion. This thermal motion is probably related to the FAICAR binding, since Arg314 has clear electron density and relatively low temperature factor in the AICAR-ATP structure. The relatively weak density for the formyl group carbon atom suggests that the formyl group also undergoes some thermal motion. Besides Arg314, the His27 sidechain and the backbone amide of Gly317 are also potential hydrogen bond donors to the formyl group.
In the structure of the MjPurP complex with AMP, one AMP molecule is bound in the AICAR/FAICAR binding site and the second AMP is bound in the ATP binding site. The binding geometry of AMP at the AICAR/FAICAR binding site is essentially the same as that of AICAR, except that the Asn258 sidechain is flipped to form hydrogen bonds with both the N1 atom and the N6 amine of the adenine base.
The ATP Binding Site of MjPurP
While the AICAR molecule is somewhat solvent exposed, the ATP molecule is mostly buried within the active site (Figure 3E & 3F). The ATP binding environment can be divided into three components: the base binding site, the ribose binding site, and the triphosphate binding site. The adenine base is oriented through a mixture of hydrophobic interactions and hydrogen bonds. Ile154 and Tyr201 pack against the adenine base from one side, while Leu299 and Phe309 pack on the other side. There are total of four hydrogen bonds between the base and the enzyme. The N1 atom forms a hydrogen bond to the backbone amide of Val202. The N6 amino group donates two hydrogen bonds to the Glu199 sidechain carboxylate and the carbonyl group of Glu200. The N7 atom accepts a hydrogen bond from Lys156. The ribose moiety is in the 3′-endo conformation and the hydroxyl groups of the ribose form a total of three hydrogen bonds to Glu230 and Arg238*. In addition, Tyr253 and Phe309 stack against the hydrophobic faces of the ribose. The α-phosphate accepts two hydrogen bonds from Lys156 and Tyr166 and the β-phosphate forms one hydrogen bond with the carboxamide of Gln297. The γ-phosphate does not form any direct hydrogen bond with the protein; however, the γ-phosphate forms two hydrogen bonds with the AICAR molecule (Figure 3B). In addition, water molecules are found to bridge between the γ-phosphate and active site residues, including Arg228, Tyr253, and Gln297 (Figure 3E).
The nonhydrolyzable ATP analog AMPPCP binds at the active site in a similar geometry as for ATP, with the only difference coming from the positioning of the γ-phosphate. The γ-phosphate of AMPPCP binds more towards the solvent and makes only one hydrogen bond to the 2′-hydroxyl group of AICAR (Figure 3B). Although Mg+2 ions were present in the crystallization and soaking solutions, there is no evidence of Mg+2 binding in any of the MjPurP structures.
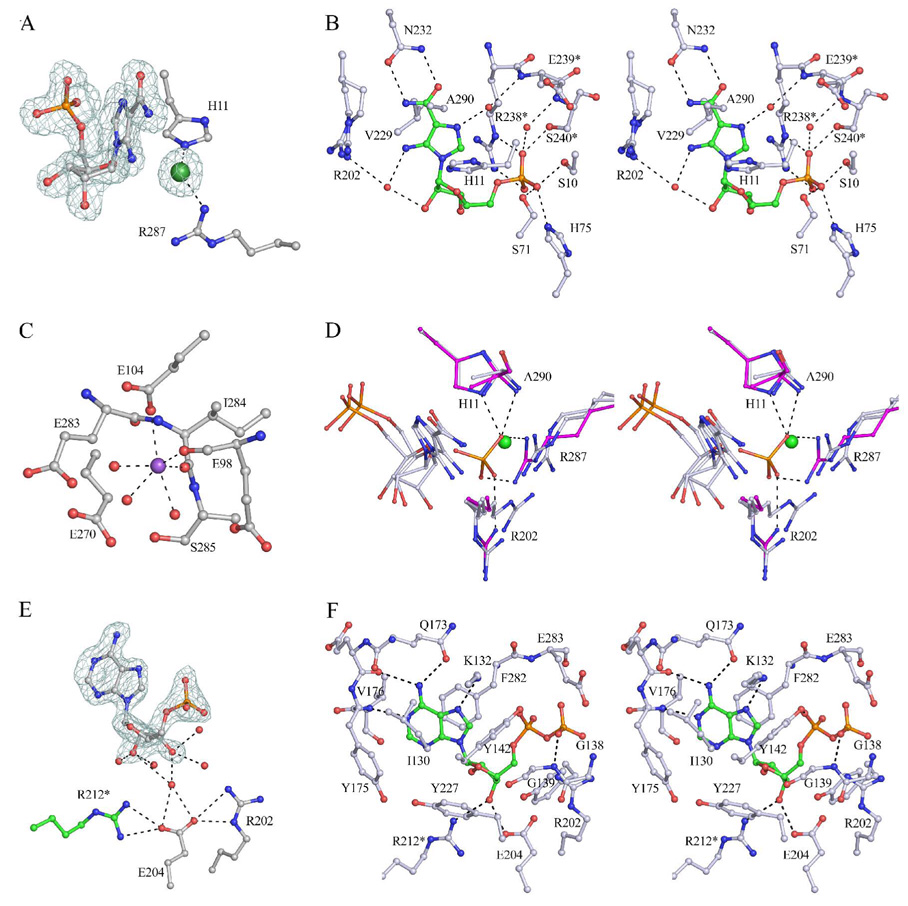
The AICAR Binding Site of PfPurP
The AICAR molecule was modeled in two conformations that differ mostly in the conformation of the ribose moiety: the 3′-endo and 2′-endo sugar conformation represent approximately 70% and 30% of the occupancy, respectively. Despite the conformational difference, the two conformers bind to the enzyme through similar interactions. Given only four residues different (Tyr98/His75, Ile255/Val229, Asp316/Val289, and Gly317/Ala290, corresponding to MjPurP/PfPurP throughout), the AICAR binding scheme is remarkably similar to that of MjPurP, with most interactions through the aminoimidazole carboxamide moiety and the 5′-monophosphate (Figure 4A, 4B, & 5A). His75 and Ser10, whose structural equivalent Tyr98 and Ser26 in MjPurP are positioned too far for hydrogen bonding interactions, provide two additional hydrogen bonds to the 5′-monophosphate. As is the case for the structure of MjPurP, a chloride anion is also found near the AICAR molecule, forming salt bridges or hydrogen bonds with His11 and Arg287.
Figure 4.
The active site of PfPurP. (A) Fo – Fc density contoured at 3.5 σ around AICAR and the chloride anion. For clarity only the 3′-endo conformer of AICAR is shown. (B) The stereodiagram of the AICAR binding site. Arg202 is built in alternate conformations. (C) The putative sodium ion (purple) binding site. The binding of the sodium ion is presumably a crystallization artifact. (D) The AICAR binding sites of AICAR-AMP structure (silver blue) and Pi-ATP structure (magenta) superimposed. The phosphate oxygen from the Pi-ATP structure overlays with the chloride atom from the AICAR-AMP structure. (E) AMP bound at the ATP binding site in the open conformation, with Fo – Fc density contoured at 3.5 σ. (F) The stereodiagram of the ATP binding site of the Pi-ADP structure in the closed conformation.
Figure 5.
The active site comparison of MjPurP and PfPurP. The active site residues are indicated in blue for MjPurP and red for PfPurP. In the AICAR binding site (A), only four residues are different between the two structures, and are highlighted by boxes: Tyr98/His75, Ile255/Val229, Asp316/Val289, and Gly317/Ala290 (corresponding to MjPurP/PfPurP throughout). In the ATP binding site (B), also only four residues are different and highlighted: Glu199/Gln173, Cys208/Tyr182, Gln297/Glu270, and Leu299/Val272.
In the Pi-ATP and Pi-ADP structures of PfPurP, inorganic phosphates, presumably coming from the crystallization conditions, are bound at both the AICAR binding site and the chloride binding site. The overlay of the structures showed that the second phosphate group has an oxygen atom superimposed with the chloride and forms a total of five hydrogen bonds with His11, Arg202, Arg287, and the mainchain amide of Ala290 (Figure 4D). In the AMP-AMP structure of PfPurP, the monophosphate nucleotide is bound at the AICAR binding site as well as the ATP binding site; however, in the AICAR binding site the orientation of the AMP is flipped relative to the substrate. Besides the interactions with the 5′-monophosphate, an additional hydrogen bond is formed between the N6 amino group and the carbonyl group of Ala74. The binding of the AMP molecule at the AICAR binding site is only observed at high concentration of the nucleotide (50 mM) and is probably not biochemically relevant.
The ATP Binding Site of PfPurP
The Pi-ADP complex structure in space group P21 is the only PfPurP structure in the closed conformation. In this conformation the ADP molecule forms two hydrogen bonds through the 3′-hydroxyl group to Glu204 and Arg212*. The 2′-hydroxyl group forms a hydrogen bond to a water molecule that in turn hydrogen bonds to the carbonyl group of Gly271. The P-loop encircles the β-phosphate and forms a hydrogen bond through Gly139. The rest of the ATP binding interactions resemble that of MjPurP (Figure 4F & 5B), with only four residue substitutions (Glu199/Gln173, Cys208/Tyr182, Gln297/Glu270, and Leu299/Val272).
In the remaining PfPurP structures, which are all in the open conformation, the ribose moiety of ATP does not make any direct hydrogen bond interactions to the enzyme; however, a water molecule was found to bridge between Glu204 and the hydroxyl group of the ribose (Figure 4E).
A metal ion was observed between the ATP binding site and the AICAR binding site for the structures of PfPurP in the open conformation, coordinated by the carboxylate group of Glu104, the carbonyl groups of Glu98 and Ile284, and three water molecules in a octahedral geometry (Figure 4C). The coordination bond distances range from 2.3 to 2.6 Å. Based on the binding geometry and the size of the electron density, a sodium atom that presumably came from the crystallization buffer was modeled at the metal binding site; however, the density might also represent a partially occupied magnesium ion.
DISCUSSION
Structural Comparison of PurP with Other ATP Grasp Superfamily Members
Many ATP grasp superfamily members have been structurally characterized, including glycinamide ribonucleotide synthetase (PurD) (21), phosphoribosylglycinamide transformylase (PurT) (14), N5-carboxyaminoimidazole ribonucleotide synthetase (PurK) (22), phosphoribosylaminoimidazolesuccinocarboxamide synthetase (PurC) (23, 24), biotin carboxylase (20), carbamoyl phosphate synthetase (33–35), D-Ala-D-Ala ligase (36), and glutathione synthetase (37, 38). Interestingly, PurD, PurT, PurK, PurC, and PurP all belong to the purine biosynthetic pathway. Using the structure of MjPurP as the reference, a structural homology search and comparison was performed with DALI (39) and the results are summarized in Table 4. PurD, PurT, PurK, and PurP are more structurally similar to each other, while PurC only shares a similar topology with the C domain, although PurD, PurT, and PurK have an additional C-terminal domain of approximately 70 residues compared to PurP.
Table 4.
Dali search and structure comparison (39) using the structure of M. jannaschii PurP (2R7K) as the reference. Structures of hypothetical proteins or Z score below 10.0 are not included.
| Structure | PDB ID | Za | r.m.s.db | Aligned residues | % Identity |
|---|---|---|---|---|---|
| PfPurP (closed) | 2R87 | 42.3 | 1.4 | 322 | 50 |
| PfPurP (open) | 2R84 | 40.0 | 2.2 | 319 | 50 |
| TkPurP (unknown function) | 2PBZ | 34.9 | 1.7 | 287 | 36 |
| PurD | 1GSO | 19.8 | 3.5 | 282 | 11 |
| PurT | 1EZ1 | 19.5 | 3.7 | 269 | 14 |
| Carbamoyl phosphate synthetase | 1KEE | 18.4 | 4.1 | 270 | 16 |
| Biotin carboxylase | 1BNC | 17.9 | 5.2 | 270 | 15 |
| PurK | 1B6R | 17.1 | 3.1 | 244 | 11 |
| D-Ala-D-Ala ligase | 1IOW | 14.4 | 3.8 | 250 | 14 |
| Glutathione synthetase | 1GSA | 14.3 | 3.5 | 247 | 13 |
| Lysine biosynthesis enzyme Lysx | 1UC8 | 13.9 | 3.2 | 226 | 12 |
| Synapsin Ia fragment | 1AUV | 13.3 | 3.5 | 236 | 11 |
| inositol 1,3,4-triphosphate 5/6 kinase | 1Z2N | 12.5 | 3.9 | 232 | 10 |
Z, strength of structural similarity in standard deviations above expected.
r.m.s.d., root mean square deviation.
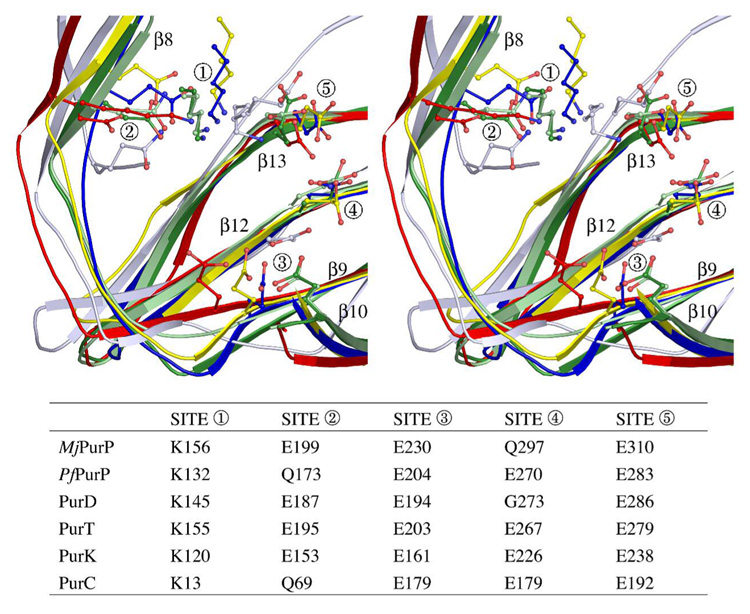
ATP or ADP complex structures have been determined for all ATP grasp members from the purine biosynthetic pathway except for PurD; however, an ordered P-loop has only been observed for PurT, PurC and PfPurP. Despite the low sequence conservation among the five enzymes, structural alignment of PurC, PurD, PurK, PurT, and PurP revealed significant similarities at the ATP binding site shown in Figure 6. Because Mg+2 was present in the crystallization solutions and is required for activity, it is surprising that no Mg+2 ions were found in the active site of PurP. This may be a crystallization artifact caused by the interference of high salt concentrations. Based upon the structural comparison with other ATP grasp members, Glu310 of MjPurP is probably involved in Mg+2 binding in solution (Figure 6).
Figure 6.
Superposition of ATP binding motif of PurD (1GSO, red), PurT (1EZ1, blue), PurK (1B6S, yellow), PurC (2GQS, silver blue), MjPurP (dark green) and PfPurP (light green). For site 1, a structurally conserved lysine residue from the P-loop is associated with α-phosphate binding. For site 2, a glutamate or glutamine residue is responsible of base binding through a hydrogen bond to the N6 amine. This conserved glutamate or glutamine resides on β8 (using the nomenclature of PurPs) except for PurC, in which case the glutamine comes from an adjacent β-strand. For site 3, a glutamate residue, however coming from different β-strands (β10 for MjPurP and PfPurP, β9 for PurD, PurT, and PurK, and β12 for PurC), forms or potentially can form hydrogen bonds to the hydroxyl groups of the ribose. Sites 4 and 5 are involved in Mg+2 binding in the structures of PurT and PurK.
PurD, PurT, PurK, PurC, and PurP all recognize 5′-monophosphate substrates specifically. Structures of the PurP, PurT and PurC complexed with a 5′-monophosphate ligand are available. While the 5′-monophosphate ligands are generally in the same part of the fold, the details of binding are different for the four structures, which presumably have evolved to accommodate their specific substrates.
Domain B and Substrate Binding Site Closure
The open and closed conformations observed in the structures of PfPurP suggest that a hinge-like movement of domain B is associated with active site closure. In both conformations an ATP or ADP molecule is bound at the active site; however, a MPD molecule identified on the domain interface of domains B and C, which is presumably a structural artifact, may help stabilize the open conformation (Figure 2D). Similar conformational changes in the B domain between the unliganded and ATP bound structures have been observed for PurK (22), carbamoyl phosphate synthetase (33–35), and glutathione synthetase (37, 38). These observations suggest that B domain closure is probably a common feature of the ATP grasp superfamily and is associated with ATP binding.
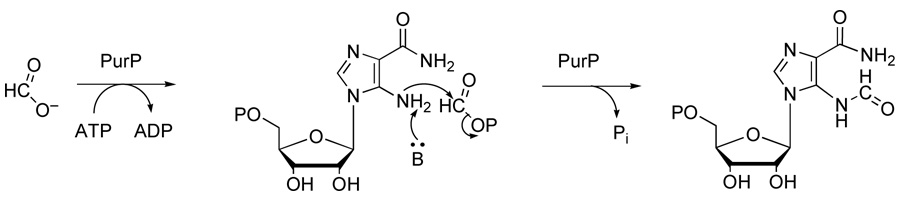
Mechanistic Implications
PurP catalyzes an ATP-dependent ligation, and is structurally and functionally unrelated to the bifunctional enzyme PurH (9). As is characteristic of ATP grasp enzymes, the substrate of PurP is activated by ATP-dependent phosphorylation. A formylphosphate intermediate is predicted, as is the case for E. coli PurT (13) (Scheme 1). The reaction would also be mechanistically analogous to the formylglycinamide ribonucleotide (FGAR) amidotransferase (PurL) reactions (40, 41) involved in the conversion of FGAR to formylglycinamidine ribonucleotide (FGAM), and the aminoimidazole ribonucelotide (AIR) synthetase (PurM) reaction converting FGAM to AIR (21, 42). Both of these enzymes are believed to utilize iminophosphate intermediates. The PurM and PurL enzymes belong to a different superfamily of enzymes than ATP grasp.
Scheme 1.
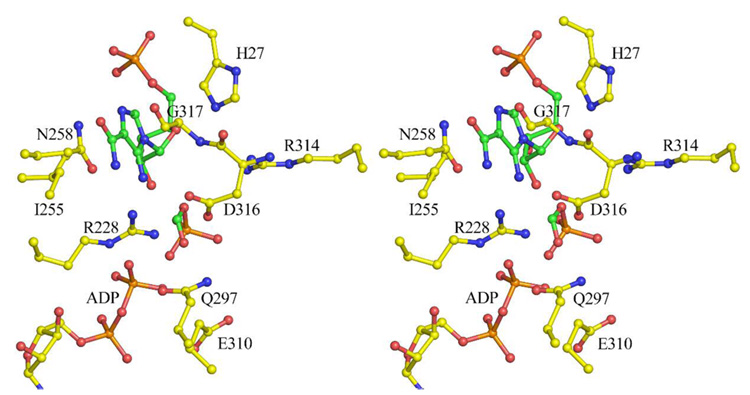
Attempts to crystallize the PurP-AICAR-formate-AMPPCP complex were unsuccessful; however, the MjPurP-AICAR-AMPPCP complex binds chloride anion, which likely occupies the formylphosphate or formate binding site and prevents formate from binding. The PfPurP-AICAR-AMP complex also showed a chloride in the same position as the chloride in MjPurP; however, in both the PfPurP-Pi-ADP and PfPurP-Pi-ATP complexes, a phosphate occupies the chloride binding site. Modeling studies based on these observations, in which a formylphosphate intermediate is positioned in the chloride binding site, show good active site geometry (Figure 7). The FAICAR synthetase reaction requires a base near the 5-amino group of AICAR. His27 of MjPurP, which is conserved throughout all PurP sequences, possibly provides this function.
Figure 7.
Modeled formylphosphate intermediate for the FAICAR synthetase reaction at the MjPurP active site. The intermediates were modeled to optimize the reaction trajectories while using the observed AICAR/FAICAR binding sites as a constraint.
Sequence Analysis of PurP-like Genes
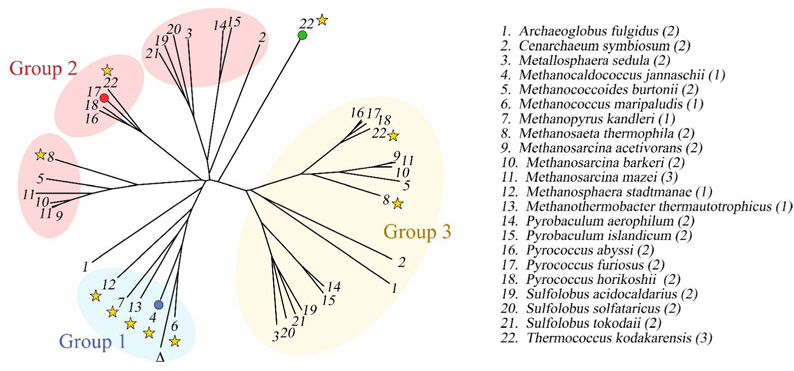
PurP is found in most Archaea with more than one copy. P. furiosus, for example, has two purP genes (Pf0421 and Pf1517). In preliminary studies, neither P. furiosus gene product showed detectable FAICAR synthetase activity. This observation raises the possibility that the duplicated genes might have alternative functions. To explore this possibility, we performed a phylogenetic analysis of PurP sequences. Using MjPurP as the search sequence, a BLAST search (43) revealed 41 putative PurP sequences after removing duplicates and partial sequences. These sequences come from a total of 22 archaeal organisms, 17 of which contain either two or three purP genes, and share approximately 30 – 90% sequence identity. A sequence alignment and phylogenetic tree (Figure 8) were generated using CLUSTALW (44). The purP genes divide into three groups with two outliers. The first group has five members: M. jannaschii, Methanococcus maripaludis, Methanopyrus kandleri, Methanosphaera stadtmanae, and Methanothermobacter thermautotrophicus. They each contain a single purP gene and we conclude that these five PurPs are FAICAR synthetases. Fifteen organisms, including P. furiosus, have two purP genes with one group 2 PurP and one group 3 PurP. PfPurP (Pf1517) belongs to group 2, which can be further divided into three subgroups; and the second P. furiosus PurP (Pf0421) belongs to group 3. No gene product from group 2 or group 3 has been functionally characterized. One of the PurP from Thermococcus kodakarensis, which represents an outlier from the alignment, has recently been structurally characterized with ATP bound at the active site (PDB 2PBZ) by the New York Structural GenomiX Research Consortium (NYSGXRC). Structure 2PBZ shares approximately 30% sequence identity to both MjPurP and PfPurP, and forms a trimer.
Figure 8.
The phylogenetic analysis of PurP orthologs. The species containing PurP like sequences are numbered in alphabetical order, with the number of PurP alternate forms listed in parentheses. MjPurP, PfPurP, and PDB structure 2PBZ are highlighted by blue, red, and green dots, respectively. The purO gene containing species are indicated by stars. The symbol Δ indicates a partial PurP sequence from Methanococcus vannielii, whose complete genome is not available at this point.
Further analysis of the sequence alignment revealed that key active site residues are mostly conserved for group 1 and 2 members but not for group 3. Asn258/232, the asparagine residue interacting with the carboxamide of AICAR, is conserved for groups 1 and 2 but replaced by a histidine residue in all the group 3 sequences. In addition, the motif KX6GR(K)G present in group 1 and 2 corresponding to the P-loop is replaced by KX8ERG(A) at the aligned position for group 3. These observations raise the questions (1) whether or not group 2 members also function as FAICAR synthetases, but perhaps utilizing a different formyl source, and (2) whether group 3 members are silent genes, catalyze a different reaction, or have other noncatalytic functions.
Additional questions remain to be answered regarding the identification of the enzymes that catalyze the last two steps of purine biosynthesis in Archaea. PurO, the known IMP cyclohydrolase for the final step is not commonly present in Archaea. With the exception of Methanosaeta thermophila and Thermococcus kodakarensis, the purO gene is found only in the five species with a single purP gene (group 1) and four halobacteria organisms where the purP gene is missing altogether. Therefore, PurO probably functions as an uncommon catalyst for the last step of purine biosynthesis. MjPurP is the only enzyme with confirmed AICAR transformylase activity in Archaea to date. As a signature gene PurPs are widely present in Archaea and, given the high sequence homology among the PurP groups, it is tempting to speculate that group 2 PurPs are responsible for FAICAR synthetase activity or might even catalyze the ATP-dependent ring closure to form IMP. However, it is also possible that enzymes yet to be identified in Archaea catalyze these two reactions. The answer to these intriguing questions awaits further biochemical and structural investigations.
ACKNOWLEDGEMENTS
We thank Leslie Kinsland for assistance in the preparation of the manuscript. We thank the NE-CAT staff at beamline 24-ID-C of the Advanced Photon Source and MacCHESS for assistance with data collection.
ABBREVIATIONS
- AICAR
5-aminoimidazole-4-carboxamide-5′-monophosphate ribonucleotide
- FAICAR
5-formaminoimidazole-4-carboxamide-5′-monophosphate ribonucleotide
- PurP
FAICAR synthetase, AICAR transformylase
- PurO
IMP cyclohydrolase
- PurD
glycinamide ribonucleotide synthetase
- PurT
phosphoribosylglycinamide transformylase
- PurK
N5-carboxyaminoimidazole ribonucleotide synthetase
- PurC
phosphoribosylaminoimidazolesuccinocarboxamide synthetase
- PurH
bifunctional AICAR transformylase and IMP cyclohydrolase
- FGAR
formylglycinamide ribonucleotide
- PurL
FGAR amidotransferase
- FGAM
formylglycinamidine ribonucleotide
- AIR
aminoimidazole ribonucelotide
- PurM
AIR synthetase
- a.u.
asymmetric unit
- r.m.s.d.
root mean square deviation
- MPD
2-methyl-2,4-pentanediol
Footnotes
This work was supported by National Institutes of Health grants RR15301 and GM073220. SEE is indebted to the W. M. Keck Foundation and the Lucille P. Markey Charitable Trust.
The Brookhaven Protein Data Bank codes are 2R7K, 2R7L, 2R7M, 2R7N, 2R84, 2R85, 2R86, and 2R87.
REFERENCES
- 1.Buchanan JM, Hartman SC. Enzymatic Reactions in the Synthesis of Purines. Advances in Enzymology. 1959;21:199–261. [Google Scholar]
- 2.Mueller EJ, Meyer E, Rudolph J, Davisson VJ, Stubbe J. N5-carboxyaminoimidazole ribonucleotide: evidence for a new intermediate and two new enzymatic activities in the de novo purine biosynthetic pathway of Escherichia coli. Biochemistry. 1994;33:2269–2278. doi: 10.1021/bi00174a038. [DOI] [PubMed] [Google Scholar]
- 3.Meyer E, Leonard NJ, Bhat B, Stubbe J, Smith JM. Purification and characterization of the purE, purK, and purC gene products: identification of a previously unrecognized energy requirement in the purine biosynthetic pathway. Biochemistry. 1992;31:5022–5032. doi: 10.1021/bi00136a016. [DOI] [PubMed] [Google Scholar]
- 4.Meyer E, Kappock TJ, Osuji C, Stubbe J. Evidence for the direct transfer of the carboxylate of N5-carboxyaminoimidazole ribonucleotide (N5-CAIR) to generate 4-carboxy-5-aminoimidazole ribonucleotide catalyzed by Escherichia coli PurE, an N5-CAIR mutase. Biochemistry. 1999;38:3012–3018. doi: 10.1021/bi9827159. [DOI] [PubMed] [Google Scholar]
- 5.Aimi J, Qiu H, Williams J, Zalkin H, Dixon JE. De novo purine nucleotide biosynthesis: cloning of human and avian cDNAs encoding the trifunctional glycinamide ribonucleotide synthetase-aminoimidazole ribonucleotide synthetase-glycinamide ribonucleotide transformylase by functional complementation in E. coli. Nucleic Acids Res. 1990;18:6665–6672. doi: 10.1093/nar/18.22.6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Firestine SM, Davisson VJ. Carboxylases in de novo purine biosynthesis. Characterization of the Gallus gallus bifunctional enzyme. Biochemistry. 1994;33:11917–11926. doi: 10.1021/bi00205a030. [DOI] [PubMed] [Google Scholar]
- 7.Chen ZD, Dixon JE, Zalkin H. Cloning of a chicken liver cDNA encoding 5-aminoimidazole ribonucleotide carboxylase and 5-aminoimidazole-4-N-succinocarboxamide ribonucleotide synthetase by functional complementation of Escherichia coli pur mutants. Proc. Natl. Acad. Sci. U. S. A. 1990;87:3097–3101. doi: 10.1073/pnas.87.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni L, Guan K, Zalkin H, Dixon JE. De novo purine nucleotide biosynthesis: cloning, sequencing and expression of a chicken PurH cDNA encoding 5-aminoimidazole-4-carboxamide-ribonucleotide transformylase-IMP cyclohydrolase. Gene. 1991;106:197–205. doi: 10.1016/0378-1119(91)90199-l. [DOI] [PubMed] [Google Scholar]
- 9.Greasley SE, Horton P, Ramcharan J, Beardsley GP, Benkovic SJ, Wilson IA. Crystal structure of a bifunctional transformylase and cyclohydrolase enzyme in purine biosynthesis. Nat. Struct. Biol. 2001;8:402–406. doi: 10.1038/87555. [DOI] [PubMed] [Google Scholar]
- 10.Rayl EA, Moroson BA, Beardsley GP. The human purH gene product, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase. Cloning, sequencing, expression, purification, kinetic analysis, and domain mapping. J. Biol. Chem. 1996;271:2225–2233. doi: 10.1074/jbc.271.4.2225. [DOI] [PubMed] [Google Scholar]
- 11.Wolan DW, Cheong CG, Greasley SE, Wilson IA. Structural insights into the human and avian IMP cyclohydrolase mechanism via crystal structures with the bound XMP inhibitor. Biochemistry. 2004;43:1171–1183. doi: 10.1021/bi030162i. [DOI] [PubMed] [Google Scholar]
- 12.Marolewski A, Smith JM, Benkovic SJ. Cloning and characterization of a new purine biosynthetic enzyme: a non-folate glycinamide ribonucleotide transformylase from E. coli. Biochemistry. 1994;33:2531–2537. doi: 10.1021/bi00175a023. [DOI] [PubMed] [Google Scholar]
- 13.Marolewski AE, Mattia KM, Warren MS, Benkovic SJ. Formyl phosphate: a proposed intermediate in the reaction catalyzed by Escherichia coli PurT GAR transformylase. Biochemistry. 1997;36:6709–6716. doi: 10.1021/bi962961p. [DOI] [PubMed] [Google Scholar]
- 14.Thoden JB, Firestine S, Nixon A, Benkovic SJ, Holden HM. Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase. Biochemistry. 2000;39:8791–8802. doi: 10.1021/bi000926j. [DOI] [PubMed] [Google Scholar]
- 15.Ownby K, Xu H, White R. A Methanocaldococcus jannaschii archeal signature gene encodes for a 5-formaminoimidazole-4-carboxamide-1-β-D-ribofuranosyl 5′-monophosphate synthetase: A new enzyme in purine biosynthesis. J. Biol. Chem. 2005;280:10881–10887. doi: 10.1074/jbc.M413937200. [DOI] [PubMed] [Google Scholar]
- 16.White RH. Purine biosynthesis in the domain Archaea without folates or modified folates. J. Bacteriol. 1997;179:3374–3377. doi: 10.1128/jb.179.10.3374-3377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang YN, Tran A, White RH, Ealick SE. A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase. Biochemistry. 2007;46:5050–5062. doi: 10.1021/bi061637j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murzin AG. Structural classification of proteins: new superfamilies. Curr. Opin. Struct. Biol. 1996;6:386–394. doi: 10.1016/s0959-440x(96)80059-5. [DOI] [PubMed] [Google Scholar]
- 19.Artymiuk PJ, Poirrette AR, Rice DW, Willett P. Biotin carboxylase comes into the fold. Nat. Struct. Biol. 1996;3:128–132. doi: 10.1038/nsb0296-128. [DOI] [PubMed] [Google Scholar]
- 20.Waldrop GL, Rayment I, Holden HM. Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochemistry. 1994;33:10249–10256. doi: 10.1021/bi00200a004. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Kappock TJ, Stubbe J, Ealick SE. X-ray crystal structure of glycinamide ribonucleotide synthetase from Escherichia coli. Biochemistry. 1998;37:15647–15662. doi: 10.1021/bi981405n. [DOI] [PubMed] [Google Scholar]
- 22.Thoden JB, Kappock TJ, Stubbe J, Holden HM. Three-dimensional structure of N5-carboxyaminoimidazole ribonucleotide synthetase: a member of the ATP grasp protein superfamily. Biochemistry. 1999;38:15480–15492. doi: 10.1021/bi991618s. [DOI] [PubMed] [Google Scholar]
- 23.Zhang R, Skarina T, Evdokimova E, Edwards A, Savchenko A, Laskowski R, Cuff ME, Joachimiak A. Structure of SAICAR synthase from Thermotoga maritima at 2.2 Å reveals an unusual covalent dimer. Acta Crystallogr. F. 2006;62:335–339. doi: 10.1107/S1744309106009651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ginder ND, Binkowski DJ, Fromm HJ, Honzatko RB. Nucleotide complexes of Escherichia coli phosphoribosylaminoimidazole succinocarboxamide synthetase. J. Biol. Chem. 2006;281:20680–20688. doi: 10.1074/jbc.M602109200. [DOI] [PubMed] [Google Scholar]
- 25.Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated Structure Solution With autoSHARP. Methods Mol. Biol. 2006;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project-Number 4. The CCP-4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 32.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 33.Thoden JB, Miran SG, Phillips JC, Howard AJ, Raushel FM, Holden HM. Carbamoyl phosphate synthetase: caught in the act of glutamine hydrolysis. Biochemistry. 1998;37:8825–8831. doi: 10.1021/bi9807761. [DOI] [PubMed] [Google Scholar]
- 34.Thoden JB, Holden HM, Wesenberg G, Raushel FM, Rayment I. Structure of carbamoyl phosphate synthetase: a journey of 96 Å from substrate to product. Biochemistry. 1997;36:6305–6316. doi: 10.1021/bi970503q. [DOI] [PubMed] [Google Scholar]
- 35.Thoden JB, Wesenberg G, Raushel FM, Holden HM. Carbamoyl phosphate synthetase: closure of the B-domain as a result of nucleotide binding. Biochemistry. 1999;38:2347–2357. doi: 10.1021/bi982517h. [DOI] [PubMed] [Google Scholar]
- 36.Fan C, Park IS, Walsh CT, Knox JR. D-alanine:D-alanine ligase: phosphonate and phosphinate intermediates with wild type and the Y216F mutant. Biochemistry. 1997;36:2531–2538. doi: 10.1021/bi962431t. [DOI] [PubMed] [Google Scholar]
- 37.Hara T, Kato H, Katsube Y, Oda J. A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 Å resolution. Biochemistry. 1996;35:11967–11974. doi: 10.1021/bi9605245. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi H, Kato H, Hata Y, Nishioka T, Kimura A, Oda J, Katsube Y. Three-dimensional structure of the glutathione synthetase from Escherichia coli B at 2.0 Å resolution. J. Mol. Biol. 1993;229:1083–1100. doi: 10.1006/jmbi.1993.1106. [DOI] [PubMed] [Google Scholar]
- 39.Holm L, Sander C. Protein structure comparison by alignment of distance matrixes. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 40.Morar M, Anand R, Hoskins AA, Stubbe J, Ealick SE. Complexed structures of formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima describe a novel ATP binding protein superfamily. Biochemistry. 2006;45:14880–14895. doi: 10.1021/bi061591u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anand R, Hoskins AA, Stubbe J, Ealick SE. Domain organization of Salmonella typhimurium formylglycinamide ribonucleotide amidotransferase revealed by X-ray crystallography. Biochemistry. 2004;43:10328–10342. doi: 10.1021/bi0491301. [DOI] [PubMed] [Google Scholar]
- 42.Li C, Kappock TJ, Stubbe J, Weaver TM, Ealick SE. X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution. Structure. 1999;7:1155–1166. doi: 10.1016/s0969-2126(99)80182-8. [DOI] [PubMed] [Google Scholar]
- 43.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]