Abstract
Human leukocyte antigen DQ2 is a class II major histocompatibility complex protein that plays a critical role in the pathogenesis of Celiac Sprue by binding to epitopes derived from dietary gluten and triggering the inflammatory response of disease-specific T cells. Inhibition of DQ2 mediated antigen presentation in the small intestinal mucosa of Celiac Sprue patients therefore represents a potentially attractive mode of therapy for this widespread but unmet medical need. Starting from a pro-inflammatory, proteolytically resistant, 33-residue peptide, LQLQPFPQPELPYPQPELPYPQPELPYPQPQPF, we embarked upon a systematic effort to dissect the relationships between peptide structure and DQ2 affinity, and to translate these insights into prototypical DQ2 blocking agents. Three structural determinants within the first 20 residues of this 33-mer peptide, including a PQPELPYPQ epitope, its N-terminal flanking sequence and a downstream Glu residue, were found to be important for DQ2 binding. Guided by the X-ray crystal structure of DQ2, the L11 and L18 residues in the truncated 20-mer analogue were replaced with sterically bulky groups so as to retain high DQ2 affinity but abrogate T cell recognition. A dimeric ligand, synthesized by regiospecific coupling of the 20-mer peptide with a bifunctional linker, was identified as an especially potent DQ2 binding agent. Two such ligands were able to attenuate the proliferation of disease-specific T cell lines in response to gluten antigens, and therefore represent prototypical examples of pharmacologically suitable DQ2 blocking agents for the potential treatment of Celiac Sprue.
Keywords: Celiac Sprue, HLA-DQ2, gluten, gliadin, 33-mer, antigen presentation, inhibition
Introduction
Class II major histocompatibility complex (MHC) proteins are glycoproteins that are expressed on the surface of antigen presenting cells. They bind exogenous peptides (typically after sequential endocytosis, lysosomal assembly and exocytosis) and present them to CD4+ T- helper cells, a critical process in the mammalian adaptive immune response (1). Susceptibility to autoimmune disease in humans has strong association with specific class II MHC alleles (2, 3). For example, >90% of Celiac Sprue patients possess the DQ2 (DQA1*05/DQB1*02) allele, with DQ8 (DQA1*03/DQB1*0302) being the most common allele in the remaining patients (4, 5).
Class II MHC proteins are αβ heterodimers, which form an extended antigen binding cleft at the subunit interface to accommodate peptides that are longer than 9 amino acids. Peptides that bind tightly to HLA-DQ2 have anchor residues in the relative positions P1, P4, P6, P7, and P9 with a high preference for negatively charged residues in P4, P6, and P7 (5).
Inhibition of antigen presentation by blocking a disease-specific MHC on antigen presenting cells with peptide (and occasionally non-peptide) ligands has been previously explored as a therapeutic strategy for autoimmune diseases such as multiple sclerosis, rheumatoid arthritis, type I diabetes, and experimental autoimmune encephalomyelitis (6–9). Notwithstanding initial promise in animal models of these diseases, however, such therapeutic agents have failed to show clinical benefit (10, 11). Their lack of success is partly due to difficulties in achieving effective drug delivery, since a combination of high molecular weight and proteolytic instability of these inhibitors of protein-protein interaction limits their ability to reach the disease-affected organs, especially via an oral route.
For several reasons Celiac Sprue may be an appropriate venue for demonstrating the clinical potential of this novel mode of immunotherapy. First, to date no non-dietary treatment has been developed for this widespread, lifelong disease; as such, there is an acute unmet need (4, 5). Second, in contrast to the organs affected by most other autoimmune diseases, the small intestine is readily accessible via oral administration of a therapeutic candidate. Finally, and perhaps most importantly, among HLA mediated diseases, Celiac Sprue is unique in that an environmental trigger (dietary gluten) has been identified and extensively dissected at an immunological level (4, 5). In turn, these studies have led to the identification of proteolytically resistant gluten peptides that are generated by physiological processes and are efficiently presented to disease associated T cells in a DQ2 restricted fashion (12 –15). Thus, if these naturally occurring T cell stimulatory agents can be transformed into inhibitors of DQ2 mediated antigen presentation, they can be considered as appropriate medicinal leads for Celiac Sprue.
A Pro- and Gln-rich 33-mer peptide from α2-gliadin, LQLQPFPQPELPYPQPELPYPQPELPYPQPQPF (transglutaminase-catalyzed Gln→Glu changes underlined), is a particularly interesting lead peptide for this purpose (15–17). Its extreme resistance to breakdown by luminal proteases and intestinal brush-border enzymes allows it to persist for a considerable duration in the upper small intestine, the primary affected region of the gastrointestinal tract in a Celiac Sprue patient (15). Not only does this peptide have a high affinity for HLA-DQ2, it is displayed on the surface of antigen presenting cells with unusual robustness (16, 17). Not surprisingly, it is a potent proliferative trigger of gluten-responsive T cells from small intestinal biopsy samples of all DQ2 Celiac Sprue patients tested thus far. Although this peptide is multivalent (it has 6 overlapping copies of 3 epitopes) (12), it binds to HLA-DQ2 with a 1:1 stoichiometry (17). It has a considerably higher affinity for DQ2 than any of its constituent epitopes PFPQPELPY, PQPELPYPQ and PYPQPELPY (17). Together, these observations led us to hypothesize that the 33-mer peptide harbors secondary interactions with DQ2 outside the core antigen binding pocket. Understanding the precise nature of these interactions would therefore be a critical prerequisite for exploiting its potential as a medicinal lead in the design of DQ2 blocking agents.
In this report we have dissected the structural determinants of the high-affinity interaction between the 33-mer peptide and HLA-DQ2. Based on these findings, we designed and synthesized simple analogues of the 33-mer peptide that retain its strong affinity for DQ2 but are not recognized by 33-mer responsive T cells from Celiac biopsies. The ability of these putative blocking agents to inhibit T cell proliferation in response to gluten antigens was also demonstrated. These peptides represent the first prototypical examples of pharmacologically relevant DQ2 blocking agents for potential treatment of Celiac Sprue.
Experimental Section
DQ2 expression and purification
Soluble DQ2 molecules were expressed and purified as previously described (17). Briefly, the soluble extracellular domains of the DQ2 α and β chains were co-expressed in High Five insect cells using a pAcAB3 baculovirus expression system, and were affinity-purified using the anti-DQ2 mAb 2.12.E11 (18). The sequence QLQPFPQPELPY was fused to the N-terminus of the DQ2 β-chain by a 15-residue linker (GAGSLVPRGSGGGGS), which includes a thrombin site (19). A complementary Fos/Jun leucine zipper pair was engineered at the C-terminal ends of α and β chains, respectively, with intervening factor Xa proteolysis sites, to increase the heterodimer stability during protein expression, analogous to earlier designs by Teyton and coworkers (20) and Wucherpfennig and coworkers (21).
The concentration of HLA-DQ2 was determined by UV spectrophotometry at 280 nm using the absorption coefficient factor 75,700 cm−1M−1 as calculated from the contents of tyrosine, tryptophan and cystine in the DQ2 sequence (22). Prior to use in binding experiments with exogenous ligands, the DQ2-ligand fusion protein was first treated with ~2% w/w thrombin in pH 7.3 PBS at 0°C for 2 h.
Peptide synthesis, labeling and purification
The majority of peptides used in this study were synthesized using Boc/HBTU chemistry starting from N-α-t-Boc-L-aminoacyl-phenylacetamidomethyl (PAM) resin. Peptides were labeled at their N-termini while still attached to the resin with 5- (and 6-) carboxyfluorescein, 1-(3-Dimethylaminopropyl)-3-ethyl-carbodiimide hydrochloride (EDC·HCl), and 1-Hydroxy-7-azabenzotriazole (HOAt) in 1:1:1 ratio in dimethylformamide as the solvent (23). Following cleavage of the peptidyl resin in trifluoroacetic acid/trifluoromethanesulfonic acid/thioanisole (TFA/TFMSA/thioanisole 10:1:1, v/v/v) for 4 h, the crude peptides were precipitated in cold ether and dissolved in 1:1 v/v acetonitrile/water. The peptides were purified by reverse-phase HPLC on a semi-preparative C18 column using a water-acetonitrile gradient in 0.1% (v/v) TFA. The identity and purity of the peptides were confirmed by electrospray mass spectrometry and analytical reverse-phase HPLC. The peptides were lyophilized and stored at −20°C. Prior to use, peptide stock solutions were prepared in 10 mM PBS with 0.02% sodium azide, and their concentrations were determined by UV-Vis spectrophotometry at 495 nm using the absorption coefficient factor 80,200 cm−1M−1 for the fluorescein-labeled peptide. The integrity of the peptide stocks was monitored by analytical HPLC every few months.
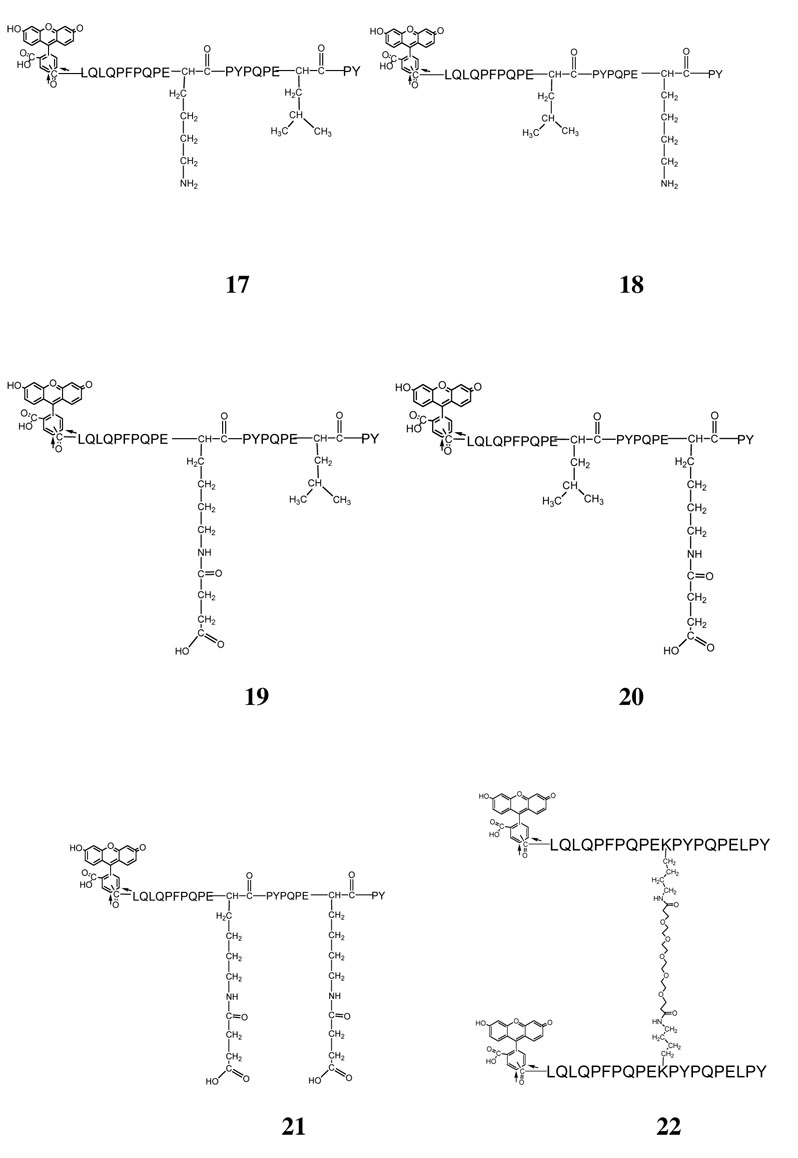
Peptide derivatives 17–22 were synthesized with N-α-t-Boc-N-ε-Fmoc-L-lysine. The Fmoc-protected side chain of lysine was deprotected after synthesis of the full-length peptide by washing the resin twice in 20% piperidine in dimethylformamide for 15 min. Then 1 g of succinimide anhydride dissolved in 2 ml dimethylformamide was added to the resin for 30 min. The extent of amide formation was monitored by the ninhydrin test.
The dimeric peptide 22 was synthesized from pure monomeric fluorescein labeled peptide 17. Fluorescein-conjugated 17 was mixed with bis-d PEG 6 NHS ester (Quanta Biodesign) in 2:1 ratio in either dimethylformamide with 10% v/v diisopropylethylamine as base, or in pH 9 phosphate solution. The reaction was monitored by analytical reverse phase HPLC using a C18 column. The product peak eluted 2 min after the monomeric starting material, and was purified by preparative reverse phase HPLC. Mass spectrometric analysis revealed that it had the expected molecular weight of 5858.8 (22+Na+, exp. MW 5858). The concentration of this fluorescent dimeric peptide was quantified by using the absorption coefficient factor 160,400 cm−1M−1 at 495 nm.
Peptide exchange assay
For peptide exchange experiments, the DQ2 heterodimer purified as described above was incubated with fluorescein-conjugated ligands in a 25:1 ratio (i.e. 4.7 µM DQ2 with 0.185 µM fluorescent peptide). Incubations were performed at 37°C in a 1:1 mixture of PBS buffer (10 mM Pi, 150 mM NaCl, pH 7.3, supplemented with 0.02% NaN3) and McIlvaine’s citrate-phosphate buffer (pH 5 or pH 7) such that the final pH was either 5.5 or 7.3, respectively. Peptide binding was measured by high performance size exclusion chromatography (HPSEC) (17). One µl of reaction mixture was diluted into 14 µl PBS; and 12.5 µl of the diluted material was injected onto a BioSep 3000 size exclusion column (Phenomenex), and eluted with PBS buffer at 1 ml/min. The DQ2-peptide complex eluted at ~8.5 min, with free peptides emerging ~2 min later. The fluorescence signal was recorded using an in-line Shimadzu RA35 fluorescent detector with excitation wavelength set at 495 nm and emission detection set at 520 nm. Peak areas corresponding to the DQ2-peptide complex and the free peptide were used to calculate the fractional yield of the DQ2-fluoresceinated peptide complex.
Peptide dissociation assay
For dissociation experiments, DQ2-fluoresceinated peptide complexes were prepared by incubating thrombin treated DQ2 (3–5 µM) with 20-fold excess fluorescein-conjugated peptides at pH 5.5 for 25 hours. The buffer composition was a 1:1 mixture of 10 mM PBS buffer and pH 5.1 McIlvaine’s citrate-phosphate buffer (24), such that the final pH was 5.5. Excess free peptide was separated from the complex on a chilled spin column (Bio-Rad) packed with Sephadex G50 superfine medium and blocked with 1% BSA solution to minimize the binding of DQ2 to the column. Spin columns were pre-washed with pH 7.3 PBS buffer, and the fluorescein-conjugated peptide + DQ2 mixture was applied to the column. The DQ2-fluoresceinated peptide complex was eluted in a volume of ~230 µl in pH 7.3 PBS buffer. Typically, this DQ2-peptide fraction contained <10% of free peptide. The solution was immediately adjusted to pH 5.5 or pH 7.3, and 20 µM of a tight DQ2 binding peptide (AAIAAVKEEAF) (17) was added to prevent the re-binding of dissociated fluorescent peptide to DQ2. Kinetic measurements of ligand dissociation were performed at 37°C, and a time course was obtained by injecting 20 µl aliquots into HPSEC column.
T cell reagents
Gluten reactive T cell lines (TCL) and T cell clones (TCC) established from small intestinal biopsies of Celiac Sprue patients were used. This includes the T cell line TCL P28 33mer (which is reactive to DQ2-αI, DQ2-αII, and DQ2-αIII epitopes of the 33-mer peptide), and the DQ2 restricted T cell clones TCC P26c αII (specific for the DQ2-αII epitope) and TCC 437.1.3.17 (specific for the DQ2-γII epitope). In addition a DR3-restricted T cell clone, TCC RN.46 (specific for the 3–13 epitope of Mycobacterium tuberculosis heat shock protein 65, KTIAYDEEARR), was used.
T cell proliferation assays
As antigen presenting cells we used HLA-DR3/DQ2 homozygous Epstein-Barr virus transformed B-lymphoblastoid cell lines (VAVY or CD114). The cells were either γ-irradiated or fixed with 1% paraformaldehyde for 10 min or with 0.05% glutaraldehyde for 90 s using 0.2M glycine to quench the reaction. The antigen presenting cells were incubated with the appropriate peptides overnight in 60 µl media containing 10% fetal bovine serum/2% human serum or 15% human serum, penicillin and streptomycin at a cell density of 2×106 cells/ml in 96-well plates. The next day, the volume was doubled to yield a cell density of 1×106 cells/ml, 50 µl of which was placed into a U-bottom 96-well plate. An equal volume of T cells (50 µl of 1×106 cells/ml) was added to each well, and cells were incubated at 37°C and 5% CO2 for 48 h, at which time 0.5 µCi/well of [methyl-3H] thymidine (Amersham, TRK120) or 1 µCi/well of [methyl-3H]thymidine (Hartmann Analytic) was added. Cells were incubated for an additional 12–14 h and then frozen. After thawing, incorporated thymidine was collected on a filter mat (Wallac) using a Tomtec cell harvester, and counted using a Wallac 1205 Betaplate or Wallac 1450 MicroBeta TriLux liquid scintillation counter. The following equation was used to fit the data to give EC50 values (N is the Hill Slope):
Results
SAR analysis of the binding of the 33-mer peptide to HLA-DQ2
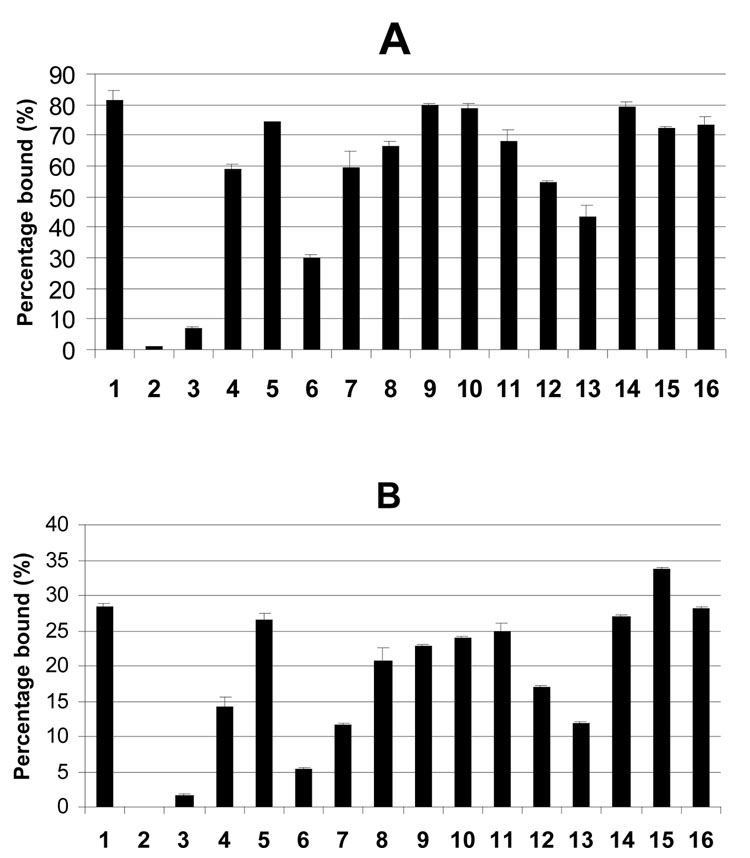
Earlier studies have demonstrated that the highly immunogenic 33-mer peptide from α2-gliadin, LQLQPFPQPELPYPQPELPYPQPELPYPQPQPF, potently displaces pre-bound ligands from the DQ2 binding site and also has a long dissociation half-life (17). These features, together with its natural proteolytic resistance, make this 33-mer peptide an attractive target for engineering an HLA-DQ2 blocking agent. To dissect its structure-activity relationships (SAR), several analogues were synthesized and initially evaluated in peptide exchange assays over a period of 45 h (Figure 1). Although the percentage of bound peptide is only reported at the 45 h end-point (i.e. at equilibrium), the half-maximum binding times for several of these peptides were obtained, and found to be comparable to each other. Therefore, the equilibrium occupancy is representative of DQ2 avidity.
Figure 1.
Equilibrium occupancy of individual peptides listed in Table 1 in the DQ2 binding site, as measured by peptide exchange assays. Measurements were made at (A) pH 5.5 and (B) pH 7.3. DQ2 (4.7 µM) was mixed with fluorescein-conjugated peptide (0.185 µM) at 37°C for 45 h, and the abundance of DQ2 bound peptide was calculated as the percentage (×100%) of total peptide.
Several noteworthy observations emerge from the data summarized in Figure 1. First, consistent with earlier data (17), it is evident that the DQ2-αII epitope (PQPELPYPQ) is the primary binding site in the 33-mer peptide 1. For example, the minimal DQ2-αII epitope bearing peptide 3 has a binding maximum of 7% at pH 5.5, whereas the minimal DQ2-αI epitope bearing peptide 2 saturates at less than 1% at pH 5.5 and is undetectable at pH 7.3. Second, comparison of the DQ2 binding properties of the minimal DQ2-αII epitope 3 with longer analogues (e.g. 6 and 7) reveals that binding efficiency increases as the C-terminal side of the epitope is extended. Similarly, comparison of the binding properties of the minimal peptide 3 with the N-terminally elongated peptide 4 suggests that the N-terminus of the 33-mer contributes significantly to its DQ2 binding potency. Together, these findings argue that structural determinants on both sides of the core DQ2-αII epitope sequence of the 33-mer influence its immunological characteristics. The importance of flanking sequences is also reinforced when one compares 21-mer peptide 7 with analogues that are incrementally extended on both sides (8, 9, 10 and 1). Interestingly, the difference between peptides 7 and 8 is more pronounced at pH 7.3 than at pH 5.5.
The 33-mer peptide 1 contains three Glu residues corresponding to three transglutaminase-catalyzed deamidation sites (16). To investigate the role of multiple deamidation sites, we synthesized and tested all possible deamidation analogues of the 33-mer (peptides 11–16). Q10E (11) has higher affinity for DQ2 than either Q17E (12) or Q24E (13) at both pH 5.5 and pH 7.3, arguing that DQ2 binding of the 33-mer is primarily centered at the N-terminal DQ2-αII epitope, although the other DQ2-αII epitopes can also bind effectively in the MHC binding site. Notably however, introduction of a second deamidation site (e.g. Q10EQ17E (14) or Q10EQ24E (15)) significantly enhances affinity relative to Q10E (11), suggesting that a second acidic residue C-terminal to the core DQ2-αII epitope is important for the optimal immunological properties of the 33-mer. This is also supported by the observation that the doubly deamidated analogue Q17EQ24E (16) approximates the 33-mer binding characteristics. Overall these results were corroborated in a competitive binding assay using detergent solubilized HLA-DQ2 molecules and a 125I-labeled DQ2 reference peptide, KPLLIIAEDVEGEY as indicator peptide (data not shown).
Taken together, the above data suggested that a peptide comprising: (i) the first DQ2-αII epitope of the 33-mer, (ii) the N-terminal sequence, and (iii) adequate C-terminal sequence to include at least one additional deamidation site, would be the shortest 33-mer analogue that could capture the essential DQ2 binding characteristics of the natural product. To test this hypothesis, we synthesized the 20-mer peptide 5, and quantitatively compared its ability to bind DQ2 in peptide exchange and dissociation assays. The results, summarized in Figure 1 and Table 2, show that peptide 5 is comparable to the 33-mer peptide 1 in all in vitro assays. The immunological relevance of these biochemical findings was verified in T cell proliferation assays using a polyclonal T-cell line that responds to all three DQ2-α epitopes equally well, and a T cell clone that responds to the DQ2-αII epitope exclusively. (Siegel et al, manuscript in preparation). As shown in Figure 2A, the T cell stimulatory characteristics of peptides 1 and 5 are very similar, both of which are substantially more potent T cell antigens than the minimal DQ2-αII epitope peptide 3. To verify that the enhanced T cell antigenicity of peptides 1 and 5 is due to superior DQ2 binding affinity rather than the ability of these peptides to stimulate a larger population of responsive T cells, the experiment was repeated using a T cell clone (TCC P26c alpha II) specific for the DQ2-αII epitope. Figure 2B shows that peptides 1 and 5 again have a comparable T cell stimulatory capacity that exceeds that of peptide 3. Thus, in view of its considerably shorter length, peptide 5 serves as an attractive lead for the design of HLA-DQ2 blocking ligands for potential treatment of Celiac Sprue.
Table 2.
Dissociation kinetic parameters of 33-mer peptide 1 as well as analogues 4 and 5 at pH 5.5 and pH 7.3 and 37 °C in the presence of 20 µM competing DQ2 strong binding peptide AAIAAVKEEAF.
| Peptide | pH 5.5 | pH 7.3 | |||||
|---|---|---|---|---|---|---|---|
| T50% (h)a | Af (%)b | kf (h−1) | As (%)c | ks (h−1) | koff (h−1) | T1/2 (h) | |
| 1 | 15±3 | 54±8 | 7±1 | 46±8 | 0.022±0.002 | 0.019±0.003 | 36.5 |
| 5 | 25±5 | 30±1 | 3.3±0.3 | 70±1 | 0.011±0.004 | 0.012±0.002 | 57.8 |
| 4 | 0.8±0.1 | 69±3 | 2.1±0.4 | 31±3 | 0.10±0.06 | 0.020±0.002 | 34.6 |
The dissociation kinetics at pH 5.5 were fit to an exponential double decay function: Y=Af×exp(−kfx)+As×exp(−ksx) using Kaleidagraph 3.5.
T50% is defined as the apparent half life when 50% of overall fluorescent DQ2 signal has disappeared, i.e. x=T50% | Y=0.5×(Af+As).
Af(%) is a measure of the fraction of ligand that dissociates in the fast phase of the dissociation curve. Af(%)=Af/(Af+As) ×100
As(%) is defined as the fraction of ligand dissociating in the slow phase of the dissociation curve. As(%)=As/(Af+As) ×100.
The dissociation kinetics at pH 7.3 follows a monophasic exponential decay function Y=A×exp(−koffx). Half life T1/2 was calculated as ln(2)/koff.
Figure 2.
T cell proliferation induced by three peptides, the 33-mer 1 (△), peptide 3 ( ), and the 20-mer 5 (◊). Paraformaldehyde-fixed DQ2 cells were used as antigen presenting cells. (A) Proliferation of a polyclonal T cell line (TCL P28 33mer) that is responsive to the DQ2-αI, DQ2-αII, and DQ2-αIII epitopes. (B) Proliferation of a T cell clone (TCC P26c αII) that recognizes the DQ2-αII epitope (peptide 3).
Design and in vitro evaluation of DQ2 blocking peptides
A major goal of this research is to design and characterize medicinally appropriate ligands that form tight, long-lived complexes with HLA-DQ2 on the surface of antigen presenting cells in the Celiac small intestine, but are not recognized by disease specific T cells. Toward this end we synthesized analogues of the 20-mer peptide 5, and evaluated their DQ2 binding properties. Our initial design targeted L11 and L18 residues of peptide 5 as modification sites. L11 was chosen based on the crystal structure of the αI-DQ2 complex, which suggested that the residue at the corresponding position (i.e. the P5 Pro residue) points away from the DQ2 protein surface (25). Consistent with this observation, epitope scanning experiments have also shown that the antigen binding site of DQ2 can accommodate a spectrum of amino acids at the P5 position (5). Therefore, the L11K analogue (17) shown in Figure 3 was synthesized. In addition, since peptide 5 can bind to HLA-DQ2 in the DQ2-αI (PFPQPELPY) and DQ2-αII (PQPELPYPQ) epitope registers, we also wished to introduce modifications in the downstream DQ2-αIII epitope. For this reason L18K (18) was also synthesized. (The P7 residue is partly exposed to the solvent.)
Figure 3.
Structures of candidate DQ2 blocking agents 17–22.
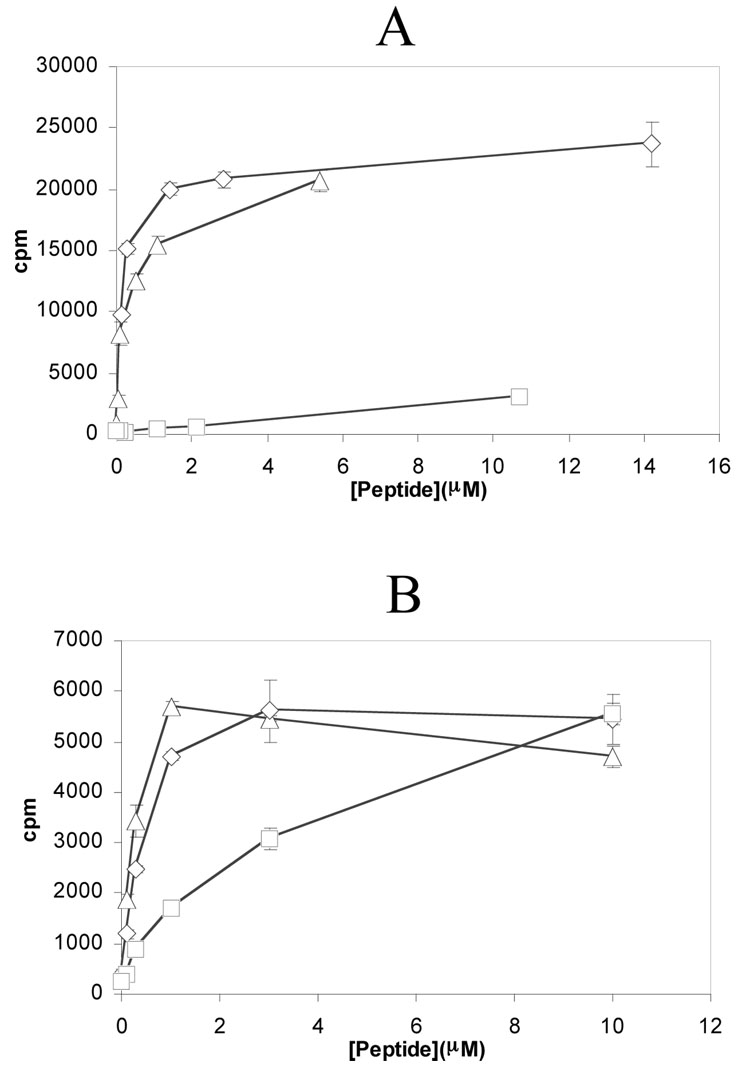
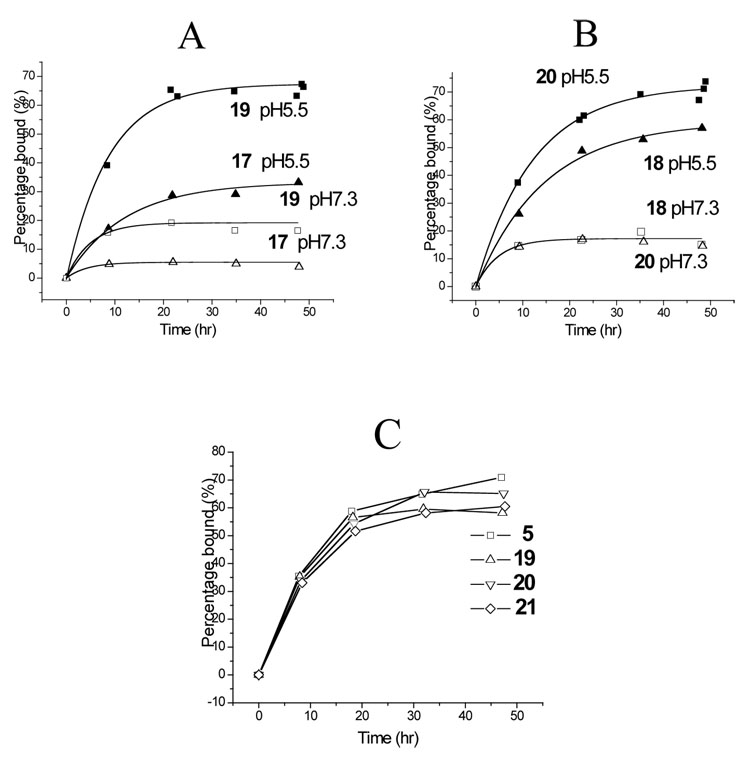
Surprisingly, both 17 and 18 bound poorly to DQ2 at either pH 5.5 or pH 7.3 (Figure 4A and 4B). We speculated that this poor affinity may be due to formation of a salt bridge between the Lys residue at positions 11 or 18 and the critical Glu residue at the preceding position. To test this hypothesis, the Lys side chains in both compounds were extended with a succinyl group, yielding peptides 19 and 20 (Figure 3) with a negative charge at residues 11 and 18, respectively. As shown in Figures 4A and 4B, peptides 19 and 20 bound to DQ2 with considerably higher affinity. We therefore also synthesized peptide 21, which possesses the above changes at both positions 11 and 18. Peptide 21 bound comparably well with DQ2 as peptide 5, both in exchange experiments (Figure 4C) and dissociation experiments (data not shown).
Figure 4.
Kinetic analysis of exchange of compounds 17–21 in the DQ2 binding site. (A) Exchange of compounds 17 ( ) and 19 ( ) onto DQ2 at pH 5.5 (filled symbols) and pH 7.3 (open symbols). (B) Exchange of compounds 18 ( ) and 20 ( ) onto DQ2 at pH 5.5 (filled symbols) and pH 7.3 (open symbols). (C) Comparative kinetics of DQ2 binding of compounds 5 ( ), 19 ( ), 20 (▽), and 21 (◊) at pH 5.5 and 37°C.
A number of independent investigations have highlighted the feasibility of enhancing ligand avidity to a biological target by the engineering of multivalent ligands (27–29). Therefore, in an attempt to further improve DQ2 affinity, a dimeric ligand (22) was synthesized by crosslinking monomeric L11K via a hexa-ethylene glycol (6-PEG) bifunctional linker through the Lys side chains (Figure 3). Not only did this ligand show improved binding affinity for DQ2 (>90% was bound after 45 hours), but remarkably, peptide 22 binds to DQ2 much faster than all gluten peptides evaluated thus far, including the 33-mer peptide 1 (Figure 5). However, no significant amount of dimeric DQ2-peptide complexes was observed via size exclusion chromatography (data not shown). In a competitive binding assay using detergent solubilized HLA-DQ2 molecules, peptide 22 bound with higher affinity than the reference DQ2 ligand KPLLIIAEDVEGEY which was considered to be one of the strongest binding peptide to DQ2, and this was particularly evident at pH 7.2 compared to pH 5.2 (data not shown).
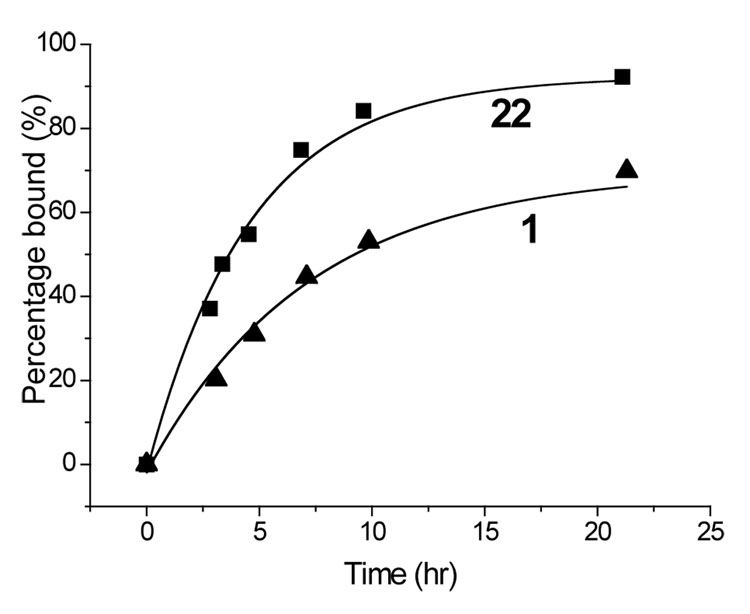
Figure 5.
Kinetic analysis of exchange of compound 22 (■) and 33-mer 1 (▲) at pH 5.5. Data at pH 7.3 was similar (not shown); at this pH peptide 1 reaches a maximum occupancy of 28% (45 h), whereas peptide 22 reaches a maximum occupancy of 40% (20 h).
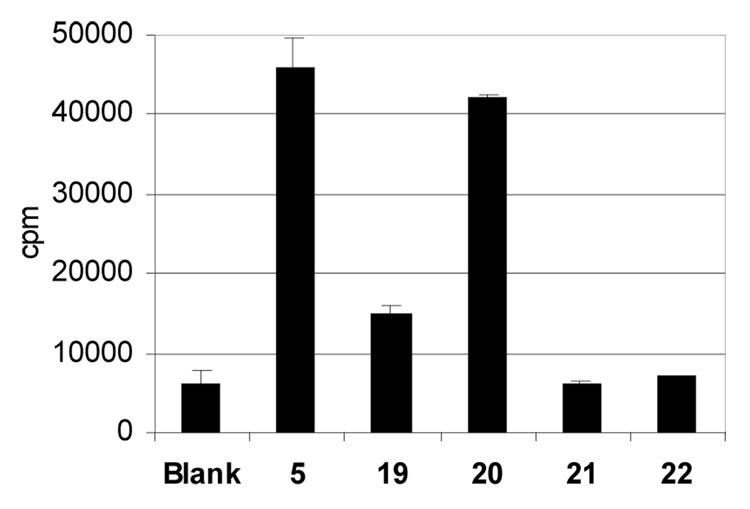
Since the modified side-chains of ligands 19–22 are oriented toward the T cell face of the DQ2-peptide complex, we anticipated that these modifications were likely to alter the T cell recognition properties of these peptides. As an initial test of this hypothesis, we wished to investigate the extent to which compounds 19–22 were able to elicit a proliferative response from gluten responsive polyclonal T cell lines derived from small intestinal biopsies of Celiac Sprue patients. For this experiment, a T cell line (TCL P28 33mer) that is strongly responsive to the 33-mer peptide 1 (or alternately peptide 5) was used. As shown in Figure 6, peptide 20 elicited the strongest T cell response, presumably because it contained an unmodified DQ2-αII (PQPELPYPQ) epitope. Peptide 19 also retained ability to stimulate the T cell line, whereas neither the doubly modified peptide 21 nor the dimeric peptide 22 elicited a T cell response (Figure 6). Together with the binding data, these results suggest that compounds 21 and 22 can be presented on the surface of DQ2 antigen presenting cells, but are relatively unrecognized by gluten specific T cells found in the small intestines of Celiac Sprue patients. Given their intrinsic proteolytic stability, these peptides were therefore evaluated as prototypical DQ2 blocking agents.
Figure 6.
Comparison of T cell proliferation in the presence of modified peptides 19, 20, 21, and 22. All peptides were tested at a concentration of 3 µM, except compound 22 was tested at 5 µM. The antigen presenting cells (VAVY) were γ-irradiated before the incubation with peptides.
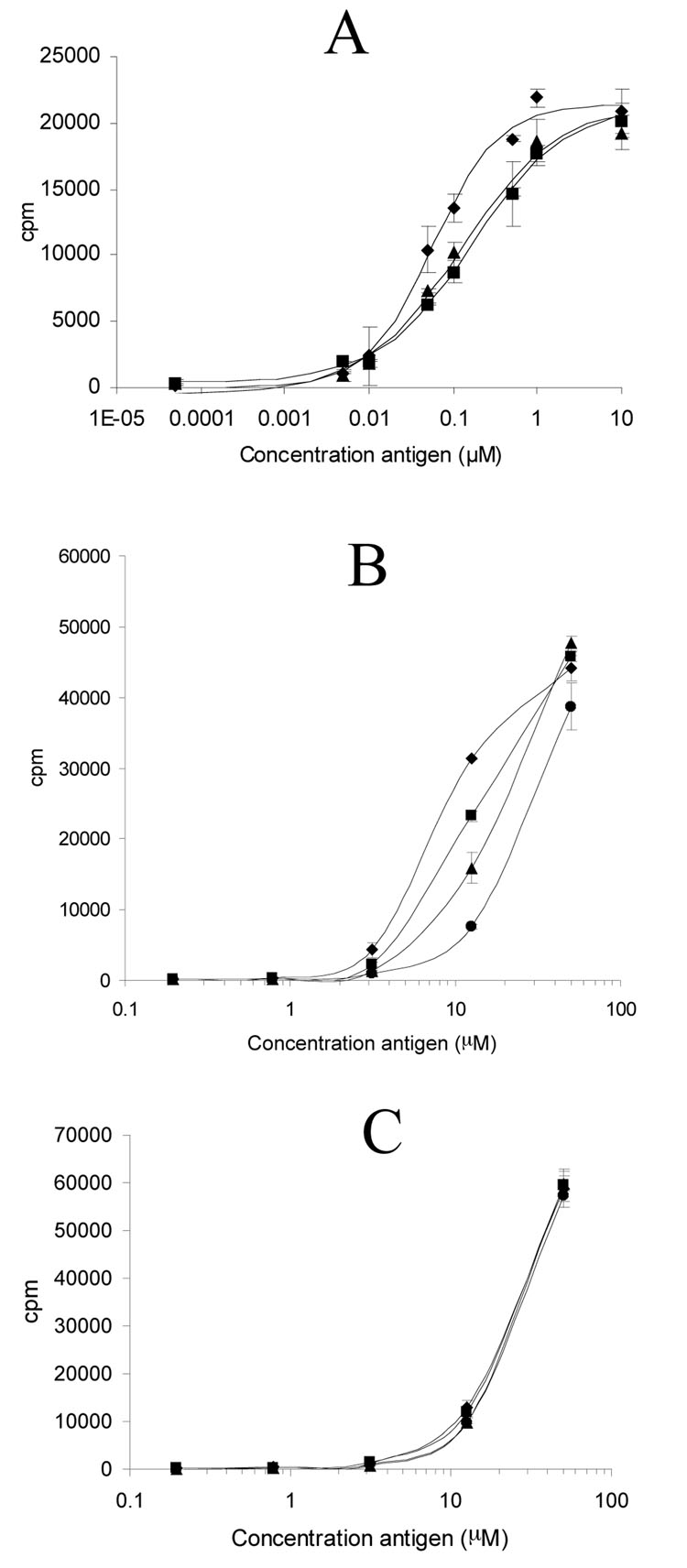
To assess the DQ2 blocking properties of the most promising compounds, 21 and 22, fixed antigen presenting cells were incubated for 12 hours with varying concentrations of antigenic peptide 5 derived from α-gliadin in the presence or absence of 5 µM blocker peptides, and the resulting antigen presenting cells were then mixed with gluten responsive T cells under appropriate culture conditions (Figure 7A). The T cell proliferation data was used to calculate EC50 values of peptide 5 in each experimental series. In the absence of any blocking agent, the EC50 value of 5 was found to be 0.057 µM. A significant increase in EC50 was observed in the presence of compounds 21 and 22 (0.122 and 0.171 µM, respectively), indicating that these synthetic peptides were indeed capable of competitively blocking T cell proliferation by potent congenetic α gluten antigens such as peptide 5 (Figure 7A).
Figure 7.
Proliferative T cell responses of an α-gliadin responsive T cell line, a DQ2-γII specific T cell clone, and a DR3-restricted T cell clone to antigens in the presence or absence of selected blocker peptides, using fixed HLA-DR3/DQ2 homozygous B-lymphoblastoid cell lines as antigen presenting cells. (A) Response of the T cell line (TCL P28 33mer) to varying concentrations of antigenic peptide 5 in the absence of any blocker peptide (◆) or in the presence of peptide 21 (▲) or peptide 22 (■) (5 µM each). (B) Response of the DQ2-γII specific T cell clone (TCC 437.1.3.17) to varying concentrations of antigenic peptide (GIIQPEQPAQL) without any blocker peptide (◆), or in the presence of the reference DQ2-binding peptide, KPLLIIAEDVEGEY (■), peptide 21 (▲), or peptide 22 (●) (5 µM each). (C) Response of the DR3-resticted T cell clone (TCC RN.46) to varying concentrations of antigenic peptide KTIAYDEEARR without any blocker peptide (◆), or in the presence of the reference DQ2-binding peptide, KPLLIIAEDVEGEY (■), peptide 21 (▲), or peptide 22 (●) (5 µM each).
The potency of peptide blockers 21 and 22 is even more promising in assays for DQ2-mediated proliferation of disease-specific T cells that respond to antigens derived from γ-gliadin. For example, 5 µM peptide 22 reduces the response of TCC 437.1.3.17, a DQ2-γII gliadin specific T cell clone, to 12.5 µM GIIQPEQPAQL by 80% (Figure 7B). Peptide 21 showed similar but less significant effect on blocking T cell response to DQ2-γII gliadin peptide. The reference DQ2 ligand KPLLIIAEDVEGEY (Mycobacterium bovis 65 kDa Hsp 243–255Y), known to be a very high affinity DQ2 ligand at pH 4.9 (31), is a considerably weaker inhibitor of antigen presentation to the same T cell clone (Figure 7B; EC50 = 9.8 µM in the absence of any blocker, EC50 = 13.6, 16.7, and 28.2 µM in the presence of 5 µM KPLLIIAEDVEGEY, peptide 21 and peptide 22, respectively).
To verify that the observed blocking effects of peptides 21 and 22 are the result of their DQ2 binding abilities, the response of a DR3-restricted T cell clone (TCC RN.46) was measured in the presence or absence of 5 µM peptide 21, peptide 22, and the reference DQ2 ligand (KPLLIIAEDVEGEY) using the same DR3/DQ2 homozygous B cell line as antigen presentation cells. None of the blockers showed an effect on the DR3-mediated antigen presentation pathway (Figure 7C), indicating the blockers 21 and 22 are specific for DQ2-mediated antigen presentation. Taken together, the above results show that peptides 21 and 22 are bona fide DQ2 blocking agents in that they not only inhibit DQ2 mediated antigen presentation by their parent DQ2-α gliadin epitope, but also by other epitopes from gluten.
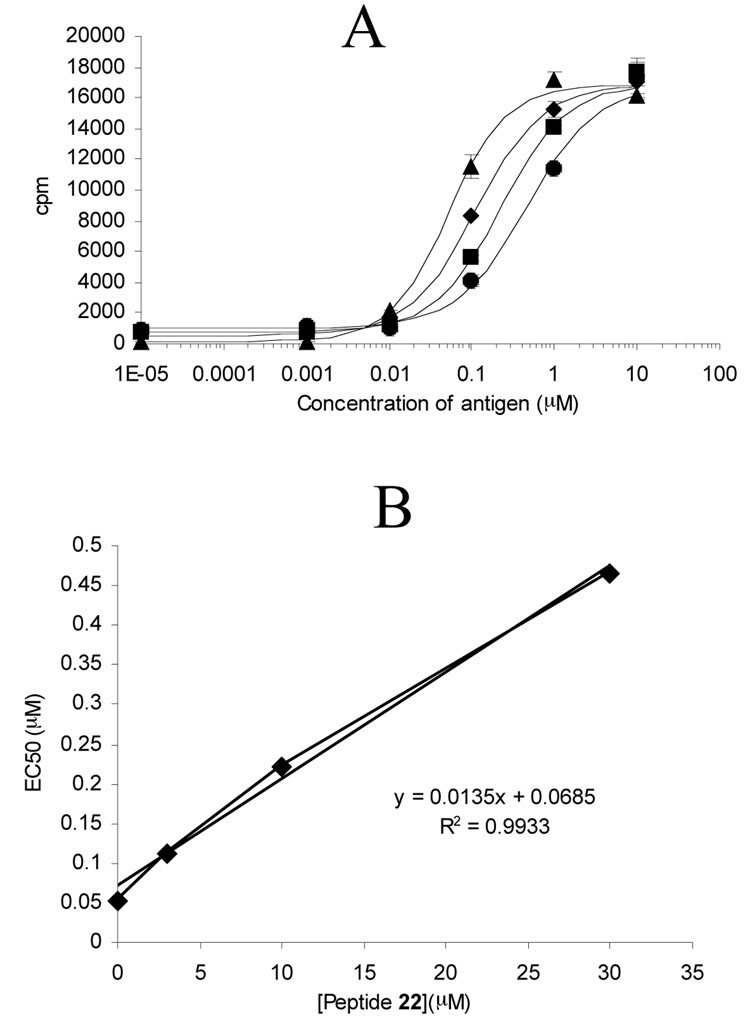
To further quantify the efficacy of peptide blocker 22, the EC50 of peptide 5 against polyclonal T cell line TCL P28 33mer was measured at varying concentrations of this blocking peptide. Within the range of 0 to 30 µM of peptide 22, the EC50 increased linearly with increasing concentrations of blocker (slope of 0.014 µM EC50/µM blocker; Figure 8A and 8B).
Figure 8.
(A) Response of a T cell line (TCL P28 33mer) to varying concentrations of antigen peptide 5 in the presence of blocker peptide 22 at 0 µM (▲), 3 µM ( ), 10 µM (■), or 30 µM (●). (B) EC50 values obtained from (A) plotted against the corresponding concentration of blocker peptide 22.
Discussion
Several recent reports have highlighted the exceptional pathogenic characteristics of a 33-mer peptide from α2-gliadin, LQLQPFPQPELPYPQPELPYPQPELPYPQPQPF, using disease specific T cells derived from small intestinal biopsies of Celiac Sprue patients (15–17). The goal of this research is to evaluate the feasibility of transforming this potent pro-inflammatory natural product into a fundamentally new type of anti-inflammatory agent for potential use in Celiac Sprue patients. To this end we have dissected the structure-function relationships of the 33-mer peptide, and have harnessed these insights, together with our recent X-ray crystal structure of HLA-DQ2, to design prototypical DQ2 blocking ligands.
The 33-mer peptide is a multivalent antigen possessing at least six overlapping copies of three distinct epitopes (designated earlier as DQ2-αI, DQ2-αII and DQ2-αIII epitopes, corresponding to the sequences PFPQPELPY, PQPELPYPQ and PYPQPELPY, respectively) (12, 15–17). However, its predominant mode of DQ2 interaction involves formation of stable monomeric complexes (1:1 stoichiometry) (17). Detailed equilibrium binding and kinetic analysis of the 33-mer peptide to HLA-DQ2 led us to hypothesize that this peptide derives its DQ2 avidity through a combination of interactions between one or more core epitopes as well as flanking sequences (17). Therefore, a first objective of this study was to identify the preferred binding epitope(s), the structural determinants in the flanking sequences, and the precise contributions of the three Glu residues, each of which is generated by post-translational deamidation of a naturally occurring Gln residue by the human transglutaminase 2 enzyme (26).
We synthesized truncated analogues (peptides 2, 3, 6–10) as well as site-directed variants (peptides 11–16) of the 33-mer peptide 1 (Table 1), and tested their ability to displace a pre-existing ligand in the binding site of a purified, soluble form of HLA-DQ2 (Figure 1). Our findings suggest that the DQ2-αII epitope centered at E10 is the most preferred register for 33-mer binding to DQ2. This is presumably due to the higher avidity of the DQ2-αII epitope for DQ2 as compared to DQ2-αI and DQ2-αIII epitopes, as well as the fact that the first DQ2-αII epitope in the 33-mer peptide can uniquely leverage secondary interactions between the N-terminal LQLQPF sequence and a yet to be determined binding site on HLA-DQ2. Additionally, a second deamidation site located in the C-terminal sequence also facilitates DQ2 binding of this DQ2-αII epitope. Thus, the 20-mer peptide 5 binds to DQ2 equally well as the 33-mer peptide 1, whereas peptide 6 (which lacks the N-terminal flank) has a considerably lower affinity for DQ2 than peptide 5 (which has this flanking sequence). The good correlation between DQ2 binding and T cell proliferative capacity was also verified by showing that peptide 5 has comparable T cell antigenicity as 1 (Figure 2).
Table 1.
Peptides derived from 33-mer 1 and their occupancy on DQ2. Gln -> Glu deamidation sites are underlined.
| No. | Peptide sequence | %pH 5.5 | %pH 7.3 |
|---|---|---|---|
| 1 | LQLQPFPQPELPYPQPELPYPQPELPYPQPQPF | 81.2 | 28.4 |
| 2 | PFPQPELPY | 1 | 0.1 |
| 3 | PQPELPYPQ | 7.2 | 1.7 |
| 4 | LQLQPFPQPELPYPQ | 59.1 | 14.2 |
| 5 | LQLQPFPQPELPYPQPELPY | 74.2 | 26.4 |
| 6 | PQPELPYPQPELPY | 30 | 5.4 |
| 7 | PQPELPYPQPELPYPQPELPY | 59.4 | 11.6 |
| 8 | PFPQPELPYPQPELPYPQPELPYPQPQP | 66.5 | 20.8 |
| 9 | LQPFPQPELPYPQPELPYPQPELPYPQPQP | 79.9 | 22.7 |
| 10 | QLQPFPQPELPYPQPELPYPQPELPYPQPQP | 78.6 | 23.9 |
| 11 | LQLQPFPQPELPYPQPQLPYPQPQLPYPQPQPF | 68 | 25 |
| 12 | LQLQPFPQPQLPYPQPELPYPQPQLPYPQPQPF | 54.9 | 17 |
| 13 | LQLQPFPQPQLPYPQPQLPYPQPELPYPQPQPF | 43.5 | 11.9 |
| 14 | LQLQPFPQPELPYPQPELPYPQPQLPYPQPQPF | 79.1 | 27 |
| 15 | LQLQPFPQPELPYPQPQLPYPQPELPYPQPQPF | 72.5 | 33.8 |
| 16 | LQLQPFPQPQLPYPQPELPYPQPELPYPQPQPF | 73.4 | 28.2 |
%pH 5.5 and %pH 7.3 are the equilibrium occupancies of the corresponding peptides in the DQ2 binding site as measured by peptide exchange assay at 37°C after 45 h at pH 5.5 and pH 7.3, respectively. The abundance of DQ2 bound peptide was calculated, from the data in Fig. 1 as the percentage (×100%) of total peptide.
The X-ray crystal structure of HLA-DQ2 bound to the DQ2-αI gliadin epitope (25) had revealed that some amino acid side chains from the epitope are deeply buried in the DQ2 binding site, whereas others face outward, presumably in the direction of the T cell receptor. L11 and L18 in peptide 5 are examples of the latter category of residues, regardless of whether they are recognized by DQ2 as part of an DQ2-αII epitope (where they are located in the P5 position) or the DQ2-αI or DQ2-αIII epitopes (where they are located in the P7 position). Leu → Lys substitutions at these positions therefore provided functional handles for further modification. Several analogues (17–21) of peptide 5 were evaluated; of these, compounds 19, 20, and 21 had similar affinity for HLA-DQ2 as unsubstituted peptide 5 (Figure 4). In T cell assays using a T cell line responsive to the DQ2-αI, DQ2-αII, and DQ2-αIII epitopes, peptide 19 and peptide 20 retained ability to stimulate the T cell line, whereas the doubly modified peptide 21 gave no or barely detectable responses (Figure 6). In the presence of 5 µM of peptide 21, the EC50 of antigenic peptide 5 against gluten specific T cells increased by >2-fold (Figure 7A).
An alternative design of potential DQ2 blocking agents involved dimerization of peptide 17 through the Lys-11 side chain. This strategy has borne fruit in the study of protein-protein interactions in other biological contexts (27–29). Our prototypical dimeric peptide 22 with a hexa-ethylene glycol bifunctional linker had a substantially enhanced affinity for HLA-DQ2 than both peptides 1 and 5 at pH 5.5 or pH 7.3. Moreover, compound 22 showed remarkably enhanced binding kinetics, reaching half maximal DQ2 occupancy at 2.5 hr instead of ~ 8 hr for the 33-mer peptide 1 (Figure 5). This rapid binding capacity could be a pharmacologically useful property in the context of inhibiting antigen presentation. In support of this anti-inflammatory potential, addition of 5 µM compound 22 also resulted in a 3-fold elevation of the EC50 of antigenic peptide 5 (Figure 7A).
Since peptides 21 and 22 are derivatives of DQ2-α gliadin epitopes, it is possible that they inhibit proliferation of T cells specific for these epitopes by mechanisms other than simple DQ2 occupancy. For example, altered peptide ligands can act as T cell antagonists and abrogate T cell proliferation by inducing a qualitatively different pattern of signal transduction (30). To rule out this possibility, we investigated the ability of peptides 21 and 22 to block DQ2-mediated antigen presentation of a DQ2-γ gliadin derived epitope. Using a DQ2-γII specific T cell clone, comparable T cell blocking activity was observed in these experiments for peptides 21 and 22 (Figure 7B). The biological specificity of peptides 21 and 22 was also verified by assays using a DR3-restricted T cell clone for read-out (Figure 7C). Perhaps most encouragingly, peptide 22 increases the inflammatory potential of a representative gluten antigen in a dose-dependent fashion (Figure 8). A 10-fold increase in half-maximal effective concentration of gluten antigen was observed within a concentration range of 22 that is readily achievable in the gut (0-30 µM).
It is interesting that both peptides 21 and 22 are superior blockers than the reference DQ2 ligand KPLLIIAEDVEGEY (Figure 7B), even though the latter peptide is one of the strongest known ligands of HLA-DQ2 (31). This could be due to the relative pH insensitivity of DQ2 binding by the gliadin analogues (16, 17). Alternatively, it may also suggest that high affinity for DQ2 (as judged by an in vitro DQ2 binding assay) is necessary but not sufficient for designing an efficient blocker, and that properties such as efficient ligand exchange are also important. Similar arguments have also been proposed by other investigators who have unsuccessfully attempted to engineer HLA-blocking agents (10).
The relative efficacy of peptide 22 as compared to peptide 21 is also noteworthy. Although 22 was designed as a ligand capable of inducing dimerization of HLA-DQ2, no such dimeric complexes were observed in assays measuring the binding affinity of soluble DQ2 and 22. While the possibility that multivalent interactions are fostered between cell surface DQ2 and this ligand cannot be excluded, we hypothesize that the efficacy of 22 stems from its superior ligand exchange properties, as illustrated in Figure 5.
In summary, our studies reported here have led to the development of an improved insight into the structure-activity relationships of the highly immunogenic 33-mer peptide from α2-gliadin, and in turn, to the design of close analogues of this peptide that show some promise in inhibiting DQ2-mediated antigen presentation to Celiac Sprue associated T cells. The design of future generations of such DQ2 blocking agents will require in-depth understanding of monomeric and oligomeric analogues of this remarkable gluten antigen to facilitate MHC-TCR interactions.
Abbreviations
- MHC
major histocompatibility complex
- HLA
human leukocyte antigen
- HPSEC
high-performance size exclusion chromatography
- APC
antigen presenting cells
- Boc
t-butyloxycarbonyl
- HBTU
2-(1H-benzotriazole-1-yl)-1,1,3,3,-tetramethyluronium hexafluorophosphate
- PAM
phenylacetamidomethyl
- PBS
phosphate buffered saline
Footnotes
This work was supported by a grant from the NIH (R21 DK65965 to C.K.) and by grants from the Research Council of Norway, Rikshospitalet University Hospital and the Fulbright Fellowship Program to L.M.S. M. Siegel is a recipient of a predoctoral fellowship from the Stanford-NIH Biotechnology Training Grant, and E. Bergseng is recipient of a fellowship from the Norwegian Foundation for Health and Rehabilitation (EXTRA fund).
References
- 1.Tiwari J, Terasaki P. HLA and disease association. New York: Springer-Verlag; 1985. [Google Scholar]
- 2.Nepom BS. Clin. Immunol. Immunopathol. 1993;67:S50. doi: 10.1006/clin.1993.1084. [DOI] [PubMed] [Google Scholar]
- 3.Todd JA, Acha-Orbea H, Bell JI, Chao N, Fronek Z, Jacob CO, McDermott M, Sinha AA, Timmerman L, Sternman L, McDevitt HO. Science. 1988;240:1003. doi: 10.1126/science.3368786. [DOI] [PubMed] [Google Scholar]
- 4.Sollid LM, Thorsby E. Gastroenterology. 1993;105:910. doi: 10.1016/0016-5085(93)90912-v. [DOI] [PubMed] [Google Scholar]
- 5.Sollid LM. Annu. Rev. Immunol. 2000;18:53. doi: 10.1146/annurev.immunol.18.1.53. [DOI] [PubMed] [Google Scholar]
- 6.Falcioni F, et al. Nature Biotechnol. 1999;17:562–567. doi: 10.1038/9865. [DOI] [PubMed] [Google Scholar]
- 7.Bolin DR, et al. J. Med. Chem. 2000;43:2135. doi: 10.1021/jm000034h. [DOI] [PubMed] [Google Scholar]
- 8.de Haan EC, Moret EE, Wagenaar-Hilbers JPA, Liskamp RMJ, Wauben MHM. Mol. Immunol. 2005;42:365. doi: 10.1016/j.molimm.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Lamont AG, Powell MF, Colon SM, Miles C, Grey HM, Sette A. J. Immunol. 1990;144:2493. [PubMed] [Google Scholar]
- 10.Ishioka GY, Adorini L, Guery JC, Gaeta FCA, LaFond R, Alexander J, Powell MF, Sette A, Grey HM. J. Immunol. 1994;152:4310. [PubMed] [Google Scholar]
- 11.(a) Jones AB, Acton JJ, Adams AD, Yuen W, Nichols EA, Schwartz CD, Wicker LS, Hermes JD. Bioorg. Med. Chem. Lett. 1999;9:2115. [PubMed] [Google Scholar]; (b) Jones AB, Acton JJ, Rivetna MN, Cummings RT, Cubbon RM, Nichols EA, Schwartz CD, Wicker LS, Hermes JD. Bioorg. Med. Chem. Lett. 1999;9:2109. doi: 10.1016/s0960-894x(99)00333-9. [DOI] [PubMed] [Google Scholar]
- 12.Arentz-Hansen H, McAdam SN, Molberg Ø, Fleckenstein B, Lundin KE, Jorgensen TJ, Jung G, Roepstorff P, Sollid LM. Gastroenterology. 2002;123:803. doi: 10.1053/gast.2002.35381. [DOI] [PubMed] [Google Scholar]
- 13.Vader W, Kooy Y, van Veelen P, de Ru A, Harris D, Benckhuijsen W, Pena S, Mearin L, Drijfhout JW, Koning F. Gastroenterology. 2002;122:1729. doi: 10.1053/gast.2002.33606. [DOI] [PubMed] [Google Scholar]
- 14.Vader W, Stepniak DT, Bunnik EM, Kooy YMC, de Haan W, Drijfhout JW, Van Veelen PA, Koning F. Gastroenterology. 2003;125:1105. doi: 10.1016/s0016-5085(03)01204-6. [DOI] [PubMed] [Google Scholar]
- 15.Shan L, Molberg Ø, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C. Science. 2002;297:2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 16.Qiao SW, Bergseng E, Molberg Ø, Xia J, Fleckenstein B, Khosla C, Sollid LM. J. Immunol. 2004;173:1757. doi: 10.4049/jimmunol.173.3.1757. [DOI] [PubMed] [Google Scholar]
- 17.Xia J, Sollid LM, Khosla C. Biochemistry. 2005;44:4442. doi: 10.1021/bi047747c. [DOI] [PubMed] [Google Scholar]
- 18.Quarsten H, McAdam SN, Jensen T, Arentz-Hansen H, Molberg Ø, Lundin KEA, Sollid LM. J. Immunol. 2001;167:4861. doi: 10.4049/jimmunol.167.9.4861. [DOI] [PubMed] [Google Scholar]
- 19.Kozono H, White J, Clements J, Marrack P, Kappler J. Nature. 1996;369:151. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- 20.Scott CA, Garcia KC, Carbone FR, Wilson IA, Teyton L. J. Exp. Med. 1996;183:2087. doi: 10.1084/jem.183.5.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalandadze A, Galleno M, Foncerrada L, Strominger JL, Wucherpfennig KW. J. Biol. Chem. 1996;271:20156. doi: 10.1074/jbc.271.33.20156. [DOI] [PubMed] [Google Scholar]
- 22.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. Protein. Sci. 1995;4:2411. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber PJ, Bader JE, Folkers G, Beck-Sickinger AG. Bioorg. Med. Chem. Lett. 1998;8:597. doi: 10.1016/s0960-894x(98)00084-5. [DOI] [PubMed] [Google Scholar]
- 24.Dawson RMC, Elliott DC, Elliott WH, Jones KM. Data for biochemical research. 3rd ed. Oxford, United Kingdom: Clarendon Press; 1986. [Google Scholar]
- 25.Kim CY, Quarsten H, Bergseng E, Khosla C, Sollid LM. Proc. Natl. Acad. Sci. USA. 2004;101:4175. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arentz-Hansen H, Körner R, Molberg Ø, Quarsten H, Vader W, Kooy YM, Lundin KEA, Koning F, Roepstorff P, Sollid LM, McAdam SN. J. Exp. Med. 2000;191:603. doi: 10.1084/jem.191.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiessling LL, Gestwicki JE, Strong LE. Curr. Opin. Chem. Biol. 2000;4:696. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 28.Kitov PI, Sadowska JM, Mulvey G, Armstrong GD, Ling H, Pannu NS, Read RJ, Bundle DR. Nature. 2000;403:669. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- 29.Mammen M, Choi SK, Whitesides GM. Angew. Chem. Int. Ed. 1998;37:2755. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Sloan-Lancaster J, Allen PM. Ann. Rev. Immunol. 1996;14:1. doi: 10.1146/annurev.immunol.14.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Johansen BH, Buus S, Vartdal F, Viken H, Eriksen JA, Thorsby E, Sollid LM. Int. Immunol. 1994;6:453. doi: 10.1093/intimm/6.3.453. [DOI] [PubMed] [Google Scholar]